Abstract
Methyl 2-cyano-3,11-dioxo-18β-olean-1,12-dien-30-oate (CDODA-Me) is a synthetic derivative of glycyrrhetinic acid, a triterpenoid phytochemical found in licorice extracts. CDODA-Me inhibited growth of RKO and SW480 colon cancer cells and this was accompanied by decreased expression of Sp1, Sp3 and Sp4 protein and mRNA and several Sp-dependent genes including survivin, vascular endothelial growth factor (VEGF), and VEGF receptor 1 (VEGFR1 or Flt-1). CDODA-Me also induced apoptosis, arrested RKO and SW480 cells at G2/M, and inhibited tumor growth in athymic nude mice bearing RKO cells as xenografts. CDODA-Me decreased expression of microRNA-27a (miR-27a), and this was accompanied by increased expression of two miR-27a-regulated mRNAs, namely ZBTB10 (an Sp repressor) and Myt-1 which catalyzes phosphorylation of cdc2 to inhibit progression of cells through G2/M. Both CDODA-Me and antisense miR-27a induced comparable responses in RKO and SW480 cells, suggesting that the potent anticarcinogenic activity of CDODA-Me is due to repression of oncogenic miR-27a.
Keywords: CDODA-Me, anticarcinogenicity, miR-27a, colon cancer, cell cycle
INTRODUCTION
MicroRNAs (miRNAs) are 20 to 25 bp oligonucleotides that interact with complementary binding sites in 3'-untranslated regions of target mRNAs to inhibit their expression by blocking translation or by decreasing mRNA stability 1, 2. MiRNA interactions with mRNA requires the overlap of 6 to 8 base pairs and, due to this relatively low stringency, computational studies show that miRNAs can potentially interact with several hundred mRNAs.
Despite this lack of specificity, miRNAs have a profound effect on gene expression and cellular homeostasis and, in cancer cells, expression of several critical oncogenes and tumor suppressor genes are regulated by miRNA expression 3–6. MiR-221 and miR-222 target the cyclin-dependent kinase inhibitor p27 6 and miR-21 decreases expression of several mRNAs including the tumor suppressor gene tropomyosin 1 3.
Recent studies in this laboratory showed that miR-27a targets ZBTB10 mRNA, a putative zinc finger protein that suppresses specificity protein (Sp) transcription factors and Sp-dependent gene expression 7. The Sp transcription factors Sp1, Sp3 and Sp4 are highly expressed in cancer cell lines, and results of RNA interference studies show that Sp proteins regulate expression of angiogenic genes such as vascular endothelial growth factor (VEGF), VEGF receptor 1 (VEGFR1, Flt-1), VEGFR2 (KDR), and the antiapoptotic gene survivin 8–14.
Betulinic acid and the non-steroidal anti-inflammatory drug tolfenamic acid inhibit prostate and pancreatic cell and tumor growth through activation of proteasome-dependent degradation of Sp1, Sp3 and Sp4 proteins 13, 14. In this study, we show that methyl 2-cyano-3,11-dioxo-18β-olean-1,12-dien-30-oate (CDODA-Me) is highly cytotoxic to colon cancer cells and also decreases Sp and Sp-dependent genes and proteins. However, these effects are proteasome-independent. We now show for the first time that CDODA-Me acts through downregulation of miR-27a and this is accompanied by enhanced expression of ZBTB10 and Myt-1 which arrests colon cancer cells at G2/M phase. The cell culture studies are complemented by inhibition of tumor growth and decreased miR-27a expression in athymic nude mice bearing RKO cells as xenografts and treated with CDODA-Me (15 mg/kg/d).
MATERIALS AND METHODS
Plasmids, antibodies and reagents
Sp1 and Sp3 promoter constructs were kindly provided by Drs. Carlos Cuidad and Veronique Noe (University of Barcelona, Barcelona, Spain). The pVEGF-133 construct contain VEGF promoter insert (positions −131 to +54) linked to luciferase reporter gene. The pSurvivin-269 was kindly provided by Dr. M. Zhou (Emory University, Atlanta, GA). The pCMV6-XL4-ZBTB10 expression vector and empty vector (pCMV6-XL4) were from Origene (Rockville, MD). Antibodies for Sp1, Sp3, Sp4, VEGF and VEGFR1 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). c-PARP and survivin antibodies were from Cell Signaling Technology Inc. (Danvers, MA). Monoclonal β-actin antibody was purchased from Sigma-Aldrich. Reporter lysis buffer, and luciferase reagent for luciferase studies were supplied by Promega (Madison, WI). β-Galactosidase (β-Gal) reagent was obtained from Tropix (Bedford, MA), and LipofectAMINE 2000 reagent was purchased from Invitrogen (Carlsbad, CA). Western Lightning chemiluminescence reagent was obtained from PerkinElmer Life and Analytical Sciences (Boston, MA). The PPARγ agonist 2-chloro-5-nitro-N-4-pyridinyl-benzamide (T007) was synthesized by condensation of 4-aminopyridine and 2-chloro-5-nitrobenzoyl chloride, followed by thin-layer chromatography, and the purity (98%) was confirmed by gas chromatography-mass spectrometry (GC-MS). CDODA-Me was synthesized as previously described and was > 98% pure as determined by GC-MS (15).
Cell proliferation and transfection assay and western blot analysis
The RKO and SW480 colon cancer cell lines were previously characterized at the M.D. Anderson Cancer Center (Houston, TX) and kindly provided by Dr. Stanley Hamilton. RKO and SW480 colon cancer cells (2 × 104 per well) were plated in 12-well plates and allowed to attach for 24 hr. The medium was then changed to DMEM/Ham’s F-12 medium containing 2.5% charcoal-stripped FBS, and either vehicle [dimethyl sulfoxide (DMSO)] or different concentrations of the compound were added. Fresh medium and compounds were added every 48 hr, and cells were then trypsinized and counted after 48 and 96 hr using a Coulter Z1 cell counter. Transfection experiments in RKO and SW480 cells used 0.4 µg of reporter gene constructs and 0.04 µg of β-Gal and LipofectAMINE 2000 reagent (Invitrogen). Results of cell proliferation and transfection studies are expressed as means ± S.E. for at least three replicate determinations for each treatment group. Western blots were determined with whole cell lysates essentially as described 9–13.
Northern blot analysis
For miRNA analysis, 20 µg total RNA per lane was electrophoresed on 15% TBE urea polyacrylaminde gel (Invitrogen), electrophoretically transferred in 0.5X TBE at 300 mA for 45 minutes to GeneScreen Plus membrane (PerkinElmer, Boston, MA), UV cross-linked, and hybridized in ULTRAhyb-Oligo hybridization buffer (Ambion, Austin, TX) at 42 °C with 32P end-labeled DNA oligonucleotides complementary to the miRNA under examination. Blots were washed at 42 °C in 2× SSC and 0.5% SDS for 30 min with gentle agitation.
Semiquantitative RT-PCR
RKO and SW480 colon cancer cells were transfected with either as-miRNA-27a or with pCMV6-XL4 control and pCMV6-XL4-ZBTB10 expression plasmid using Lipofectamine 2000 following manufacturer’s protocol. Total RNA was extracted using RNeasy Mini Kit (Qiagen, Inc.), and 2 µgm of RNA was used to synthesize cDNA using Reverse Transcription System (Promega). Primers were obtained from IDT and used for amplification were as follows: Sp1 (sense 5'- ATG GGG GCA ATG GTA ATG GTG G -3'; antisense 5'- TCA GAA CTT GCT GGT TCT GTA AG -3'), Sp3 (sense 5'- ATG ACT GCA GGC ATT AAT GCC G -3'; antisense 5'- TGT CTC TTC AGA AAC AGG CGA C -3'), Sp4 (sense 5'- ATG GCT ACA GAA GGA GGG AAA AC -3'; antisense 5'- TTG ACC AGG GGT GGA AGA ATT AC -3'), ZBTB10 (sense 5'- GCT GGA TAG TAG TTA TGT TGC -3'; antisense 5'- CTG AGT GGT TTG ATG GAC AGA G -3'), VEGF (sense 5'- CCA TGA ACT TTC TGC TGT CT T -3'; antisense 5'- ATC GCA TCA GGG GCA CAC AG -3'), VEGFR1 (sense 5'- ATG GAG CGT AAG AAA GAA AAA ATG -3'; antisense 5'- TCA AGT ACC TCC TTT TCT CAC AT -3'), Survivin (sense 5'- ATG GCC GAG GCT GGC TTC ATC -3'; antisense 5'- ACG GCG CAC TTT CTT CGC AGT T -3') and GAPDH (sense 5'- ACG GAT TTG GTC GTA TTG GGC G -3'; antisense 5'- CTC CTG GAA GAT GGT GAT GG -3'). PCR products were electrophoresed on 1% agarose gels containing ethidium bromide and visualized under UV transillumination.
Quantitative real-time PCR of mRNA and miRNA
cDNA was prepared from the RKO and SW480, cell lines using Reverse Transcription System (Promega). Each PCR was carried out in triplicate in a 20-µl volume using SYBR Green Mastermix (Applied Biosystems, Foster City, CA) for 15 min at 95 °C for initial denaturing, followed by 40 cycles of 95 °C for 30 s and 60 °C for 1 min in the Applied Biosystems 7900HT Fast Real-time PCR System. The ABI Dissociation Curves software was used following a brief thermal protocol (95 °C for 15 s and 60 °C for 15 s, followed by a slow ramp to 95 °C) to control for multiple species in each PCR amplification. Values for each gene were normalized to expression levels of TATA-binding protein. The primers used for real-time PCR were Myt-1 (sense 5'- CCT TCC AAG AGT AGC TCC AAT TC -3'; antisense 5'- GCC GGT AGC TCC CAT ATG G -3') and TATA-binding protein (sense 5'- TGC ACA GGA GCC AAG AGT GAA -3'; antisense 5'- CAC ATC ACA GCT CCC CAC CA -3'). miRNA was extracted using the mirVana miRNA extraction kit (Applied Biosystems). Quantification of miRNA (RNU6B and miRNA-27a) was done using the Taqman miRNA kit (Applied Biosystems) according to the manufacturer’s protocol with real-time PCR. U6 small nuclear RNA was used as a control to determine relative miRNA expression.
Transfection assay
Colon cancer cells (1.5× 105) were seeded in 12-well plates using DMEM:Ham’s F-12 media containing 2.5% charcoal stripped serum. After 24 hr, cells were transfected with different constructs (pSp1-For4, pSp1-For2, pSp1-For1, pSp3-For5, pSp3-For2, pVEGF-131-Luc, pSVV-259-Luc (0.4 µg), β-galactosidase (0.04 mg) using Lipofectamine 2000 according to manufacturer’s protocol. Five hr after transfection, cells were treated with control or XΔOΔA–Mε for 22–24 hr and luciferase activity (normalized to β-galactosidase) was determined using Lumicount luminometer (PerkinElmer Life and Analytical Sciences).
shRNA study
shRNA (shGFP and shZBTB10) were purchased from Origene (Rockville, MD). RKO colon cancer cells (1×105) were seeded and left for 24 hr for cells to attach to the plate. shRNA (4 µg ) and various amounts of DNA (pSp1-For4-Luc (0.4 µg) and β-galalactosidase (0.04 µg) were co-transfected into the cells using Lipofectamine 2000. After 24 hr, cells were then treated with either control or XΔOΔA–Mε and luciferase activity (normalized to β-galactosidase) was determined.
Fluorescence-activated cell sorting analysis (FACS)
RKO and SW480 cells were treated with either the vehicle (DMSO) or the compound for 24 hr or with asmiR27a. Cells were trypsinized, centrifuged, and resuspended in staining solution containing 50 µg/mL propidium iodide, 4 mmol/L sodium citrate and 30 units/mL RNase. After incubation at room temperature for 1 hr, cells were analysed on a FACS Vantage SE DiVa made by Becton Dickinson (BD), using BD FACSDiva Software V4.1.1. Propidium iodide (PI) fluorescence was collected through a 610SP bandpass filter, and list mode data were acquired on a minimum of 50,000 single cells defined by a dot plot of PI width vs. PI area. Data analysis was performed in BD FACSDiva Software V4.1.1 using PI width vs. PI area to exclude cell aggregates.
Xenograft studies in athymic mice
Mice were used in accordance with institutional guidelines when they were 8 – 12 wk old. To produce tumors, RKO cells were harvested from subconfluent cultures by a brief exposure to 0.25% trypsin and 0.02% EDTA and only suspensions consisting of single cells with >90% viability were used for the injections. A xenograft was established by s.c. injection of the cells (5 × 106) into the flanks of individual mice and, after 6 days, mice were randomized into two groups of 5 mice per group and dosed by oral gavage with corn oil or 15 mg/kg/d CDODA-Me (dissolved in corn oil) 5 days a week for 22 days. The mice were weighed, and tumor size was measured every third day with calipers to permit calculation of tumor volumes: V = LW2/2, where L and W were length and width, respectively. Final tumors weights were determined at the end of the dosing regimen. Tumor tissues and selected body organs (liver and kidney) were either stored in RNAlater solution (per manufacture's recommendations) for later microRNA analysis, snap frozen and stored at −80°C, or fixed in 10% formalin and embedded in paraffin.
RESULTS
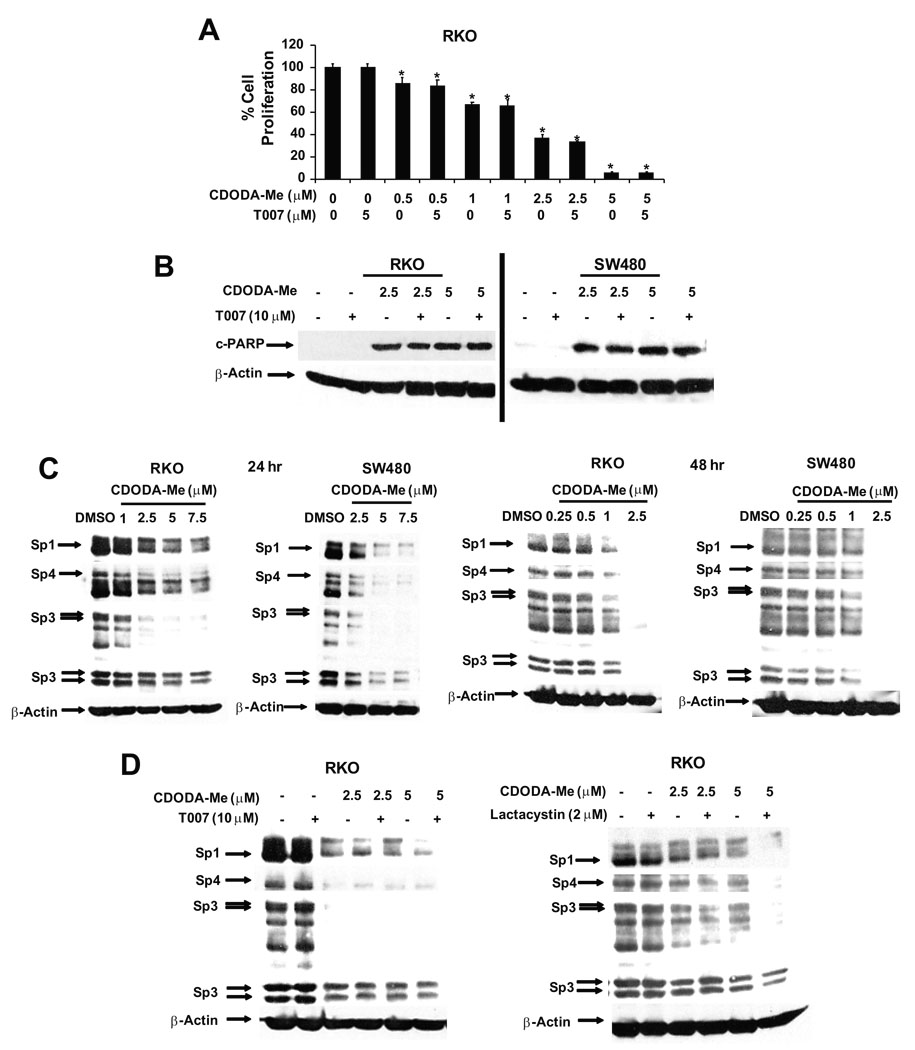
CDODA-Me is a PPARγ agonist in colon cancer cell lines 15. Although CDODA-Me decreased proliferation (Fig. 1A) and induced caspase-dependent PARP cleavage in RKO and SW480 cells (Fig. 1B), these responses were not affected after cotreatment with the PPARγ agonist T007. Receptor-independent effects have also been observed for other PPAR agonists in colon cancer cells 16. Recent studies with tolfenamic acid and the triterpenoid betulinic acid show that many of the growth inhibitory and proapoptotic responses in pancreatic and prostate cancer cells are due to decreased expression of Sp proteins 12–14. Results summarized in Figure 1C show that CDODA-Me induced a concentration- and time-dependent decrease in Sp1, Sp3 and Sp4 proteins in RKO and SW480 cells and, in RKO cells, decreases were observed with concentrations lower than 1.0 µM after treatment for 48 hr. The role of PPARγ and activation of proteasomes in mediating the effects of CDODA-Me on Sp protein expression was also investigated in RKO cells (Fig. 1D). CDODA-Me-induced downregulation of Sp1, Sp3 and Sp4 in RKO cells was not affected after cotreatment with the PPARγ antagonist T007 or the proteasome inhibitor lactacystin, and similar results were observed in SW480 cells (data not shown). The proteasome inhibitor MG132 also did not block Sp protein downregulation in RKO and SW480 cells treated with CDODA-Me (data not shown), suggesting that CDODA-Me-dependent Sp protein degradation is proteasome-independent.
Figure 1.
CDODA-Me inhibits growth, induces cleaved PARP, and degradation of Sp proteins. (A) Decreased cell proliferation in RKO cells. Cells were seeded and treated with solvent (DMSO) or different concentrations of CDODA-Me (0.5–5 µM) alone or in combination with T007 for 4 days. Cell proliferation is expressed as the percentage of CDODA-Me-treated cells remaining compared to DMSO (set at 100%). Results are expressed as means ± SE for three replicate determinations for each treatment group and significantly (p < 0.05) decreased proliferation is indicated by an asterisk. Similar results were observed for SW480 cells (data not shown). Induction of cleaved PARP (B) and decreased expression of Sp1, Sp3 and Sp4 (C, D). RKO and SW480 cells were treated with DMSO, CDODA-Me (1–5 µM), T007 (10 µM), Lactacystin (2 µM), or combinations as indicated for 24 hr or 48 hr and whole cell lysates were analyzed by Western blot analysis as described in the Materials and Methods.
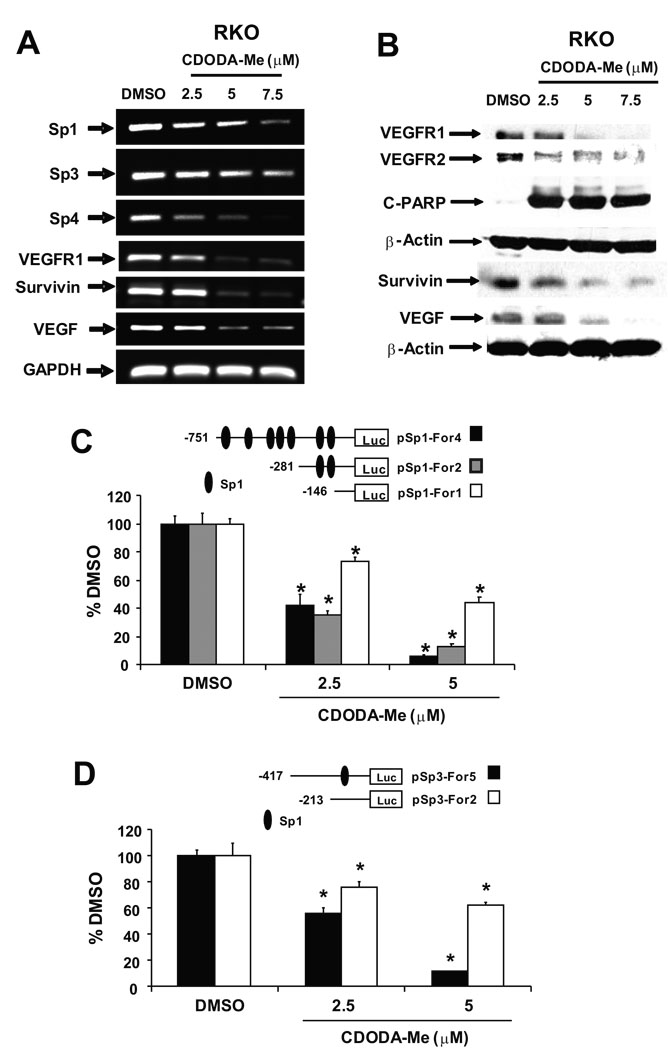
Using RKO cells as a model, treatment with CDODA-Me decreased expression of Sp1, Sp3 and Sp4 mRNA levels and expression of at least three Sp-dependent mRNAs including survivin, VEGFR1 (Flt-1), and VEGF (Fig. 2A). Figure 2B shows that CDODA-Me also decreased expression of Sp1, Sp3 and Sp4 proteins after treatment for 24 hr, and similar effects were observed for levels of the Sp-dependent genes VEGFR1, VEGF and survivin proteins (Fig. 2B). Both the Sp1 and Sp3 promoters contain multiple GC-rich sites, and Figure 2C shows that CDODA-Me decreased luciferase activity in RKO cells transfected with pSp1For4, pSp1For2 and pSp1For1 constructs which contain the −751 to −20, −281 to −20, and −146 to −20 regions (respectively) of the Sp1 gene promoter. Similarly, CDODA-Me also decreased luciferase activity in RKO cells transfected with pSp3For5 and pSp3For2 constructs which contain the −417 to −38 and −213 to −38 regions (respectively) of the Sp3 gene promoter. These results demonstrate that CDODA-Me decreases Sp1, Sp3 and Sp4 transcription.
Figure 2.
Effects of CDODA-Me on Sp and Sp-dependent expression. CDODA-Me decreases expression of Sp1, Sp3 and Sp4 and Sp-dependent angiogenic/survival gene mRNAs (A) and proteins (B). RKO cells were treated with different concentrations of CDODA-Me and after 24 hr, mRNA and protein were extracted and analyzed by semi-quantitative RT-PCR and Western blots, respectively, as described in the Materials and Methods. CDODA-Me decreases Sp1 (C) and Sp3 (D) promoter activity. RKO cells were transfected with various constructs, treated with different concentrations of CDODA-Me, and luciferase activity determined as described in the Materials and Methods. Results are expressed as means ± SE for three replicate determinations for each treatment group and significantly (p < 0.05) decreased activity is indicated by an asterisk.
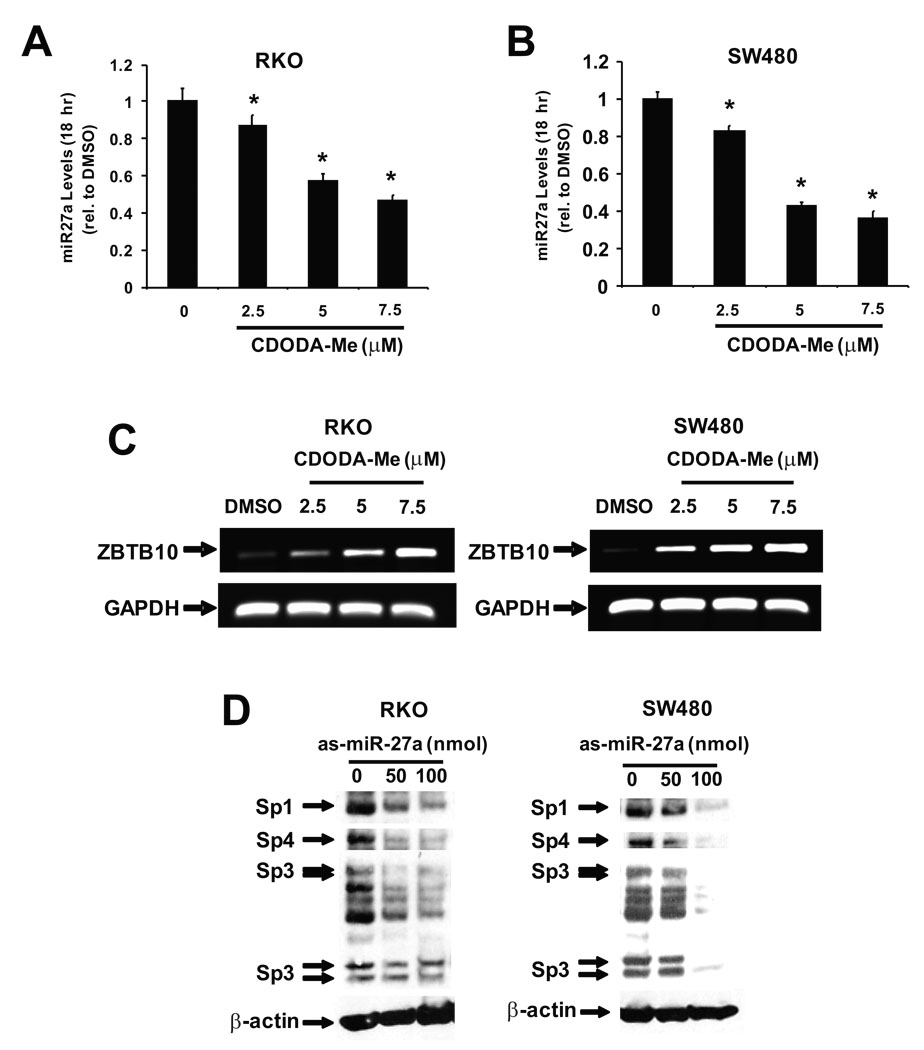
It was recently reported that microRNA-27a (miR-27a) suppresses ZBTB10 mRNA levels in breast cancer cells and treatment with antisense miR-27a (as-miR-27a) increases expression of ZBTB10 and decreases expression of Sp mRNA and proteins 7. Results illustrated in Figures 3A and 3B show that CDODA-Me decreased miR-27a in RKO and SW480 cells as determined by quantitative real time PCR and similar results were observed in Northern blot analysis (data not shown). In addition, treatment of RKO or SW480 cells with CDODA-Me also induced ZBTB10 mRNA levels (Fig. 3C). Thus, the effects of CDODA-Me on miR-27a and ZBTB10 expression in colon cancer cells are comparable to those observed in breast cancer cells transfected with as-miR-27a which also increases ZBTB10 and decreases Sp protein expression 7. Results in Figure 3D confirm that like CDODA-Me (Figs. 1C and 1D), as-miR-27 also decreased expression of Sp1, Sp3 and Sp4 proteins in RKO and SW480 colon cancer cells.
Figure 3.
Effects of CDODA-Me and as-miR27a. CDODA-Me decreases miR-27a (A and B) and increases ZBTB10 (C) expression. RKO and SW480 colon cancer cells were treated with DMSO or different concentrations of CDODA-Me and after 18 hr, total RNA was extracted and analyzed for miR27a by real time PCR and ZBTB10 by semi-quantitative RT-PCR as described in the Materials and Methods. Northern blot analysis also showed that CDODA-Me decreased miR-27a in colon cancer cells (data not shown). (D) as-miR27a decreases Sp1, Sp4 and Sp3 proteins. RKO cells were transfected with 50 and 100 ng as-miR27a and after 24 hr, whole cell lysates were analyzed by Western blot analysis for Sp1, Sp4 and Sp3 proteins as described in the Materials and Methods.
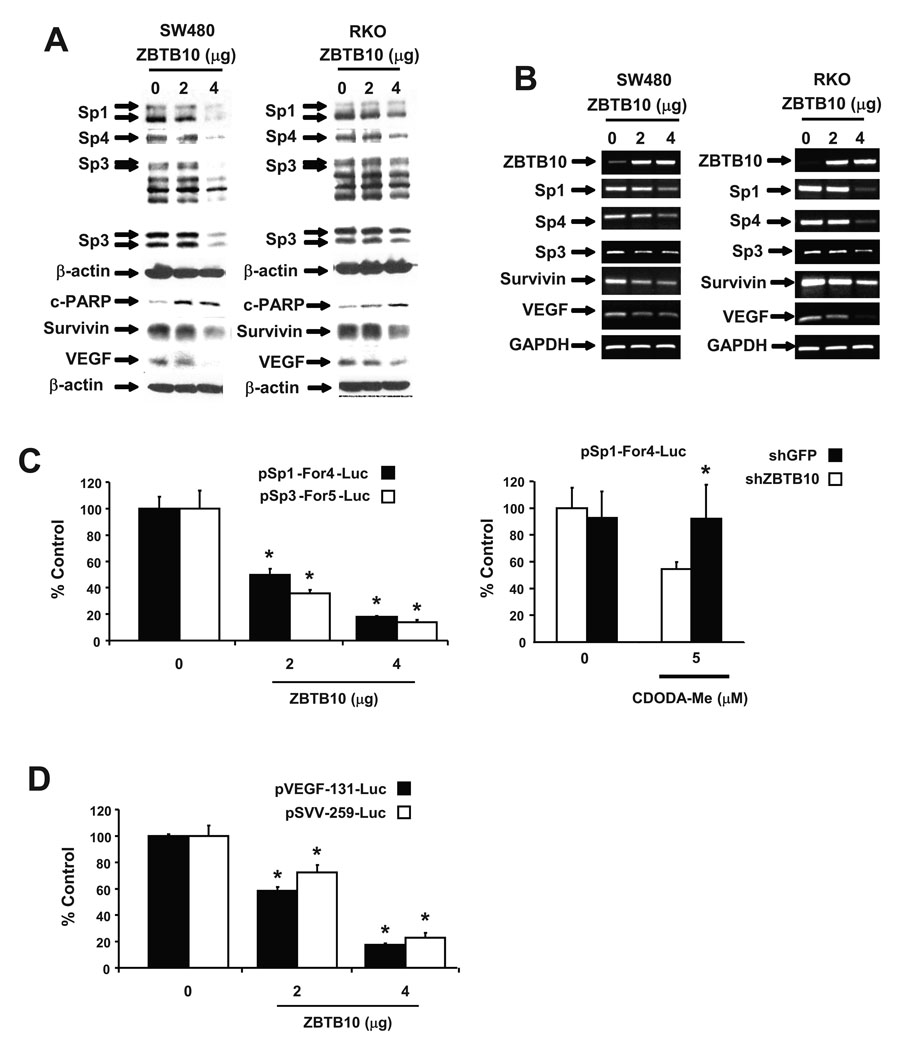
The direct effects of ZBTB10 as a "putative" Sp repressor were further investigated in colon cancer cells. Transfection of SW480 and RKO cells with 2 or 4 µg ZBTB10 expression plasmid decreased expression of Sp1, Sp3 and Sp4 proteins and also decreased the Sp-dependent VEGF and survivin proteins and induced PARP cleavage (Fig. 4A). In parallel studies, ZBTB10 overexpression also decreased Sp1, Sp3 and Sp4 survivin and VEGF mRNA levels (Fig. 4B) and the results complemented the effects of ZBTB10 overexpression on Sp and Sp-dependent proteins (Fig. 4A). ZBTB10 expression also decreased luciferase activity in RKO cells transfected with the pSp1-For4-luc and pSp3-For5-luc constructs demonstrating that ZBTB10 directly decreases promoter activity (Fig. 4C) and this complements the same effects of ZBTB10 on Sp1 and Sp3 mRNA levels (Fig. 4B). The role of ZBTB10 in mediating CDODA-Me-dependent responses was investigated in RKO cells transfected with pSp1-For4-luc and treated with CDODA-Me and cotransfected with a non-specific shGFP or shZBTB10 (Fig. 4C). The results show that CDODA-Me-induced downregulation of luciferase activity was significantly reversed after transfection with shZBTB10. CDODA-Me decreased luciferase activity in RKO cells transfected with constructs containing VEGF and survivin promoter inserts (Fig. 4D) and similar results were observed in SW480 cells (data not shown).
Figure 4.
ZBTB10 decreases expression of Sp proteins and Sp-dependent angiogenic and survival genes. ZBTB10 expression decreases expression of Sp and angiogenic/survival proteins (A) and mRNA (B). RKO and SW480 cells were transfected with ZBTB10 expression plasmid and after 24 hr, protein and mRNA were extracted and analyzed by Western blots and semi-quantitative RT-PCR, respectively, as described in the Materials and Methods. (C) Role of ZBTB10 in mediating CDODA-Me-dependent downregulation of Sp1 and Sp3 promoter activity. RKO cells were transfected with pSp1-For4-luc or pSp3-For5-luc and cotransfected with ZBTB10 (left) or cotransfected with pSp1-For4-luc and shGFP or shZBTB10 and treated with DMSO or CDODA-Me (right), and luciferase activity was determined as described in the Materials and Methods. (D) ZBTB10 expression decreases VEGF and survivin promoter activity. RKO cells were transfected with various constructs and ZBTB10 expression plasmid, and luciferase activity was determined as described in the Materials and Methods. Results in (C) and (D) are expressed as means ± SE for three replicate determinations for each treatment group and significantly (p < 0.05) decreased activity is indicated by an asterisk.
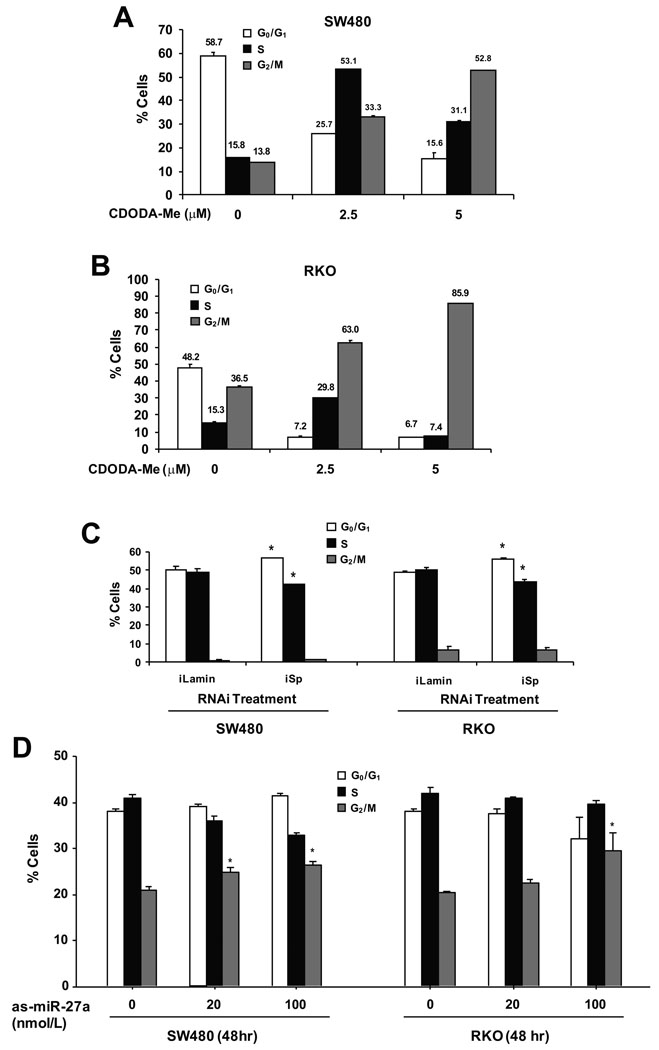
Figures 5A and 5B summarizes the effects of CDODA-Me on distribution of SW480 and RKO cells in G0/G1, S and G2/M phases of the cell cycle. Compared to treatment with DMSO, CDODA-Me induced a concentration-dependent decrease in the percentage of cells in G0/G1 and an increase of cells in G2/M. The percentage of cells in S phase increased and then decreased after treatment with 2.5 and 5.0 µM CDODA-Me, respectively; however, the dominant effects of CDODA-Me were associated with a block in progression through G2/M. The potential role of decreased Sp protein on mediating the effects of CDODA-Me on distribution of SW480 and RKO cells in different phases of the cell cycle was determined by RNA interference using a combination of small inhibitory RNAs for Sp1 (iSp1), Sp3 (iSp3) and Sp4 (iSp4) as previously described for knockdown of these proteins in other cancer cell lines 8–14. Transfection of SW480 and RKO with iSp1/iSp3/iSp4 (combined; iSp) significantly decreased expression of all three proteins (least efficiency observed for Sp4) (see Supplemental data) and, compared to the results for iLamin (non-specific RNA), Sp knockdown caused a significant G0/G1 to S phase arrest (Fig. 5C). These results are comparable to previous studies in MCF-7 breast cancer cells transfected with iSp1 17 which also exhibited G0/G1 to S phase arrest. However, the effects of Sp protein knockdown contrasted the effects of CDODA-Me which induced a G2/M arrest in both RKO and SW480 cells (Figs. 5A and 5B). Since CDODA-Me and as-miR-27a induced similar responses in colon cancer cells (Fig. 1 – Fig. 4), we also investigated the effects of as-miR-27a on distribution of RKO and SW480 cells in G0/G1, S and G2/M phases. The results (Fig. 5D) demonstrate that like CDODA-Me, as-miR-27a induced a G2/M arrest in colon cancer cells. Transfection of as-miR-27a (100 nM RKO; 200 nM SW480) increased accumulation of cells in G2/M and this was accompanied by a decrease in percentage of cells in S (SW480) and G0/G1 (RKO) phases. However, the magnitude of the G2/M arrest observed in colon cancer cells transfected with as-miR-27a was lower than observed for CDODA-Me, suggesting that the compound-induced response may also be due to other factors.
Figure 5.
Modulation of cell cycle progression by CDODA-Me, RNA interference and as-miR-27a. Effects of CDODA-Me. SW480 (A) and RKO (B) cells were treated for 24 hr with DMSO (0), 2.5 and 5.0 µM CDODA-Me, and analyzed by FACS analysis as described in the Materials and Methods. (C) iSp modulates Sp protein expression and the cell cycle in SW480 and RKO cells. Cells were transfected with iSp, a combination of small inhibitory RNAs for Sp1, Sp3 and Sp4 or a non-specific oligonucleotide (iLamin), and analyzed for Sp proteins by Western blots (to confirm Sp knockdown) (see Supplemental data) and FACS analysis as described in the Materials and Methods. (D) as-miR-27a modulates cell cycle progression. SW480 and RKO cells were transfected with different amounts of as-miR-27a and, after 48 hr, analyzed by FACS as described in the Materials and Methods. All experiments in this Figure [(A) – (D)] were repeated three times, and results are expressed as means ± SE. Significant (p < 0.05) changes compared to untreated (0) or iLamin-treated cells are indicated by asterisks.
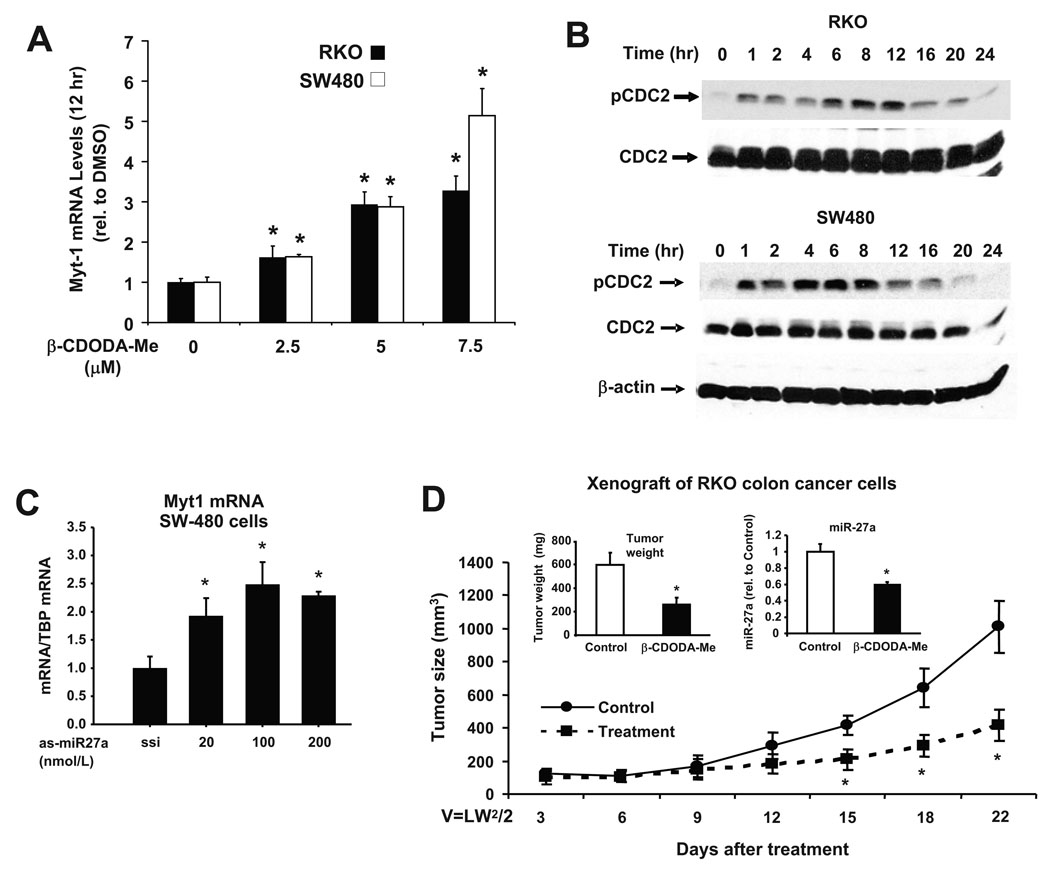
In MDA-MB-231 breast cancer cells transfected with as-miR-27a, cells were arrested at G2/M and this was due to upregulation of Myt-1 7. Myt-1 is also a target for miR-27a and catalyzes phosphorylation of cdc2 to inhibit progression through the G2/M checkpoint 18. Results in Figure 6A show that CDODA-Me induced Myt-1 mRNA expression in RKO and SW480 cells, and this was accompanied by the time-dependent induction of cdc2 phosphorylation in these cell lines (Fig. 6B). Myt-1 and Wee-1 are potential miR-27a targets that inactivate cdc2; however, CDODA-Me and as-miR-27a induced Myt-1 levels (Figs. 6A and 6C) but did not affect Wee-1 expression in SW480 or RKO cells (data not shown). We also investigated the in vivo activity of CDODA-Me as an inhibitor of tumor growth in athymic nude mice bearing RKO cells as xenografts. CDODA-Me (15 mg/kg/d) inhibited tumor growth and tumor weight and miR-27a expression (Fig. 6D), and this was not accompanied by any body or organ weight loss associated with toxic side effects (data not shown). Thus, CDODA-Me, like other compounds such as tolfenamic and betulinic acids, decrease Sp protein expression 13, 14; however, CDODA-Me acts through targeting miR-27a downregulation in colon cancer cells and tumors. This represents a novel drug-miR interaction with potential for development of new approaches for clinical treatment of colon cancer.
Figure 6.
In vitro and in vivo effects of CDODA-Me on G2/M arrest and inhibition of tumor growth. Effects of CDODA-Me (A and B) and as-miR-27a (C) on Myt-1 and cdc2 phosphorylation. Colon cancer cells were treated with different amounts of CDODA-Me or as-miR-27a for the indicated times, and Myt-1 expression and cdc2 phosphorylation were determined by real time PCR or Western blots, respectively, as described in the Materials and Methods. The effects of as-miR-27a on Myt-1 mRNA expression in RKO cells was similar to that observed in SW480 cells, and as-miR-27a also increased cdc2 expression as described (7) (data not shown). (D) CDODA-Me inhibits tumor growth (volume) and weight/miR-27a expression in a mouse xenograft model. Athymic nude mice bearing RKO cells as xenografts were treated with corn oil (solvent control) or CDODA-Me (15 mg/kg/d), and tumor volumes, tumor weights, and miR-27a expression were determined as described in the Materials and Methods. Results are expressed as means ± SE for replicate (at least three or more) determinations for each treatment group, and significantly (p < 0.05) decreased tumor volume or weight and miR-27a is indicated by an asterisk.
DISCUSSION
Sp transcription factors are critical for early embryonic development and for regulating expression of multiple genes including those important for cell proliferation and differentiation 19. The age-dependent expression of Sp proteins has not been extensively investigated; however, a recent study reported that levels of these proteins decrease during cellular senescence and aging 20. Sp1 protein is overexpressed in sub-sets of patients with several tumor types compared to non-tumor tissue, and Sp1 is a negative prognostic factor for cancer patient survival 21–26. Several studies report that Sp1 is overexpressed in tumors of some patients with gastric, breast, thyroid and colorectal cancer and overexpression of Sp1 in tumors is a predictor for a poor prognosis. Sp1 is also overexpressed in malignant human fibroblast cell lines and results of Sp1 overexpression or knockdown in fibroblasts and fibrosarcoma cells has established a causal linkage between Sp1 overexpression and malignant transformation 27.
Research in this laboratory has shown Sp1, Sp3 and Sp4 are highly expressed in cancer cell lines, and RNA interference studies clearly demonstrate that these transcription factors cooperatively regulate prosurvival, growth promoting, and angiogenic genes, suggesting that targeting Sp protein degradation may be a viable strategy for cancer chemotherapy 7–14.
Both betulinic acid and tolfenamic acid inhibit growth of prostate and pancreatic tumors and cells, and these effects are linked to induction of proteasome-dependent degradation of Sp1, Sp3 and Sp4 proteins which is accompanied by decreased expression of Sp-dependent genes such as survivin, VEGF and VEGFR1 11–14. However, ongoing studies with betulinic and tolfenamic acids in other cancer cell lines indicate that their effects on decreased Sp protein and mRNA levels are primarily proteasome-independent. CDODA-Me is a triterpenoid acid derived synthetically from glycyrrhetinic acid and this compound activates PPARγ and inhibits proliferation of colon, pancreatic and bladder cancer cells 15, 28. Results in Figure 1 show that CDODA-Me induced PARP cleavage and inhibited colon cancer cell growth and the responses were not blocked by the PPARγ antagonist T007 (Figs. 1A and 1B) or other PPARγ antagonists (data not shown). CDODA-Me also decreased Sp1, Sp3 and Sp4 protein expression in SW480 and RKO cells, and these responses were also not affected by T007 or proteasome inhibitors (Figs. 1C and 1D). CDODA-Me decreased Sp proteins and mRNA levels and also decreased protein and mRNA levels of the Sp-dependent genes VEGF, VEGFR1 and survivin (Figs. 2A and 2B). CDODA-Me decreased transactivation in colon cancer cells transfected with GC-rich constructs containing inserts from the Sp1 and Sp3 gene promoters demonstrating that Sp1 and Sp3 transcription factors regulate their own expression.
The effects of CDODA-Me on Sp proteins and Sp-dependent genes in colon cancer cells were reminiscent of the effects of antisense miR-27a (as-miR-27a) in ER-negative MDA-MB-231 breast cancer cells 7. In MDA-MB-231 cells transfected with as-miR-27a, there was a parallel increase in a zinc finger transcription factor, ZBTB10, which also binds GC-rich promoter sequences 29 and inhibits expression of Sp1, Sp3 and Sp4 and Sp-dependent genes 7. Figures 3A – 3C show that CDODA-Me decreased miR-27a and increased ZBTB10 expression in RKO and SW480 colon cancer cells. Moreover, as previously reported in MDA-MB-231 cells, as-miR-27a or ZBTB10 overexpression decreased Sp proteins and mRNA levels and Sp-dependent genes (e.g. survivin and VEGF) in colon cancer cells (Fig. 3D, Fig. 4A and 4B). Both CDODA-Me and ZBTB10 decreased luciferase activity in colon cancer cells transfected with the GC-rich pSp1-For4 and pSp3-For5 constructs (Fig. 2C, 2D and Fig. 4C). Moreover, the effects of CDODA-Me on luciferase activity in RKO cells transfected with pSp1-For4-luc were reversed in cells cotransfected with shZBTB10 (Fig. 4C), thus confirming the linkage between CDODA-Me and ZBTB10 in downregulation of Sp proteins. CDODA-Me also decreased transactivation in colon cancer cells transfected with constructs (pSp1-For1 and pSp3-For2) that do not contain GC-rich Sp binding sites (Figs. 2C and 2D). We observed similar results in breast cancer cells transfected with as-miR-27a 7, suggesting that CDODA-Me-dependent downregulation of miR-27a (Figs. 3A and 3B) may increase expression of other factors associated with downregulation of Sp1 and Sp3 and these are currently being investigated.
The effects of CDODA-Me on distribution of RKO and SW480 cells in the G0/G1, S and G2/M phases of the cell cycle (Figs. 5A and 5B) showed that the dominant effect was accumulation of cells in G2/M. Decreased Sp1 expression in MCF-7 cells by RNA interference arrests cells in G0/G1 9 and, in colon cancer cells transfected with small inhibitory RNAs for Sp1, Sp3 and Sp4 (combined), we observed a significant block in G0/G1 to S phase progression but no effects on G2/M (Fig. 5C). Similar results were observed in MDA-MB-231 breast cancer cells transfected with ZBTB10 7. However, transfection of MDA-MB-231 or colon cancer cells with as-miR-27a resulted in G2/M arrest (Fig. 5D), and this mimicked the effects of CDODA-Me (Figs. 5A and 5B). Growth arrest in colon cancer cells treated with CDODA-Me was greater than observed for as-miR-27a, suggesting that the compound may activate other pathways and these are currently being investigated.
Since miR-27a potentially targets both Myt-1 and Wee-1, two kinases that inhibit cdc2 and progression of cells through the G2/M checkpoint, we investigated the effects of CDODA-Me on cdc2 and phospho-cdc2 expression. CDODA-Me induced Myt-1 but not Wee-1 expression in both RKO and SW480 cells (Fig. 6A), and this was accompanied by phosphorylation of cdc2 in RKO and SW480 cells (Fig. 6B). As-miR-27a also induced Myt-1 and cdc2 phosphorylation in these cell lines (Fig. 6C and data not shown) as previously observed in MDA-MB-231 breast cancer cells 7. Thus, like as-miR-27a, CDODA-Me acts through decreased expression of miR-27a, resulting in enhanced expression of ZBTB10 and Myt-1 which subsequently induce downstream growth inhibitory, proapoptotic and antiangiogenic genes and pathways in colon cancer cells. These in vitro responses induced by CDODA-Me were complemented by the inhibition of tumor growth and tumor weight in athymic nude mice bearing RKO cells as xenografts (Fig. 6D), and miR-27a expression was also decreased in tumors from CDODA-Me-treated animals compared to tumors from corn oil-treated mice.
In summary, results of this study show that CDODA-Me decreases expression of Sp proteins and Sp-dependent genes and induces G2/M arrest in colon cancer cells, and these responses are due, in part, to repression of miR-27a and increased expression of ZBTB10 and Myt-1. CDODA-Me also decreased tumor growth and this was also accompanied by decreased miR-27a expression in the tumor and this represents one of the first examples of an in vivo drug-miR interaction resulting in tumor growth inhibition. Other compounds such as betulinic and tolfenamic acids also decrease Sp proteins in prostate and pancreatic cancer cells 12, 14, and there is evidence that a hydroxamic acid histone deacetylase inhibitor decreases expression of miR-27a and other miRs in SKBR3 cells 30. In addition, our recent studies also show that curcumin induces proteasome-dependent degradation of Sp proteins in bladder cancer cells and tumors 31. Current studies are focused on potential clinical applications of CDODA-Me-miR interactions for treatment of colon cancer and delineation of other pathways and other miRs that contribute to the anticancer activity of CDODA-Me and related compounds.
Supplementary Material
Acknowledgments
Funding: This research was supported by the National Institutes of Health (R01CA108718, R01CA112337 and R01ES09106,) and the Texas Agricultural Experiment Station.
Footnotes
Conflict of Interest Statement: None declared.
REFERENCES
- 1.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 2.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da PI, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 3.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 4.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 7.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein (Sp) transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 8.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Abdelrahim M, Safe S. Cyclooxygenase-2 inhibitors decrease vascular endothelial growth factor expession in colon cancer cells by enhanced degradation of Sp1 and Sp4 proteins. Mol Pharmacol. 2005;68:317–329. doi: 10.1124/mol.105.011825. [DOI] [PubMed] [Google Scholar]
- 10.Higgins KJ, Abdelrahim M, Liu S, Yoon K, Safe S. Regulation of vascular endothelial growth factor receptor-2 expression in pancreatic cancer cells by Sp proteins. Biochem Biophys Res Commun. 2006;345:292–301. doi: 10.1016/j.bbrc.2006.04.111. [DOI] [PubMed] [Google Scholar]
- 11.Abdelrahim M, Smith R, III, Burghardt R, Safe S. Role of Sp proteins in regulation of vascular endothelial growth factor expression and proliferation of pancreatic cancer cells. Cancer Res. 2004;64:6740–6749. doi: 10.1158/0008-5472.CAN-04-0713. [DOI] [PubMed] [Google Scholar]
- 12.Abdelrahim M, Baker CH, Abbruzzese JL, Sheikh-Hamad D, Liu S, Cho SD, Yoon K, Safe S. Regulation of vascular endothelial growth factor receptor-1 (VEGFR1) expression by specificity proteins 1, 3 and 4 in pancreatic cancer cells. Cancer Res. 2007;67:3286–3294. doi: 10.1158/0008-5472.CAN-06-3831. [DOI] [PubMed] [Google Scholar]
- 13.Abdelrahim M, Baker CH, Abbruzzese JL, Safe S. Tolfenamic acid and pancreatic cancer growth, angiogenesis, and Sp protein degradation. J Natl Cancer Inst. 2006;98:855–868. doi: 10.1093/jnci/djj232. [DOI] [PubMed] [Google Scholar]
- 14.Chintharlapalli S, Papineni S, Ramaiah SK, Safe S. Betulinic acid inhibits prostate cancer growth through inhibition of specificity protein transcription factors. Cancer Res. 2007;67:2816–2823. doi: 10.1158/0008-5472.CAN-06-3735. [DOI] [PubMed] [Google Scholar]
- 15.Chintharlapalli S, Papineni S, Jutooru I, McAlees A, Safe S. Structure-dependent activity of glycyrrhetinic acid derivatives as peroxisome proliferator-activated receptor γ (PPARγ) agonists in colon cancer cells. Mol Cancer Therap. 2007;6:1588–1598. doi: 10.1158/1535-7163.MCT-07-0022. [DOI] [PubMed] [Google Scholar]
- 16.Chintharlapalli S, Papineni S, Konopleva M, Andreef M, Samudio I. Safe S. 2-Cyano-3,12-dioxoolean-1,9-dien-28-oic acid and related compounds inhibit growth of colon cancer cells through peroxisome proliferator-activated receptor γ-dependent and -independent pathways. Mol Pharmacol. 2005;68:119–128. doi: 10.1124/mol.105.011437. [DOI] [PubMed] [Google Scholar]
- 17.Abdelrahim M, Samudio I, Smith R, Burghardt R, Safe S. Small inhibitory RNA duplexes for Sp1 mRNA block basal and estrogen-induced gene expression and cell cycle progression in MCF-7 breast cancer cells. J Biol Chem. 2002;277:28815–28822. doi: 10.1074/jbc.M203828200. [DOI] [PubMed] [Google Scholar]
- 18.Booher RN, Holman PS, Fattaey A. Human Myt1 is a cell cycle-regulated kinase that inhibits Cdc2 but not Cdk2 activity. J Biol Chem. 1997;272:22300–22306. doi: 10.1074/jbc.272.35.22300. [DOI] [PubMed] [Google Scholar]
- 19.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and Krüppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 20.Oh JE, Han JA, Hwang ES. Downregulation of transcription factor, Sp1, during cellular senescence. Biochem Biophys Res Commun. 2007;353:86–91. doi: 10.1016/j.bbrc.2006.11.118. [DOI] [PubMed] [Google Scholar]
- 21.Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J Clin Invest. 2002;109:525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao JC, Wang L, Wei D, Gong W, Hassan M, Wu TT, Mansfield P, Ajani J, Xie K. Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin Cancer Res. 2004;10:4109–4117. doi: 10.1158/1078-0432.CCR-03-0628. [DOI] [PubMed] [Google Scholar]
- 23.Zannetti A, Del VS, Carriero MV, Fonti R, Franco P, Botti G, D'Aiuto G, Stoppelli MP, Salvatore M. Coordinate up-regulation of Sp1 DNA-binding activity and urokinase receptor expression in breast carcinoma. Cancer Res. 2000;60:1546–1551. [PubMed] [Google Scholar]
- 24.Chiefari E, Brunetti A, Arturi F, Bidart JM, Russo D, Schlumberger M, Filetti S. Increased expression of AP2 and Sp1 transcription factors in human thyroid tumors: a role in NIS expression regulation? BMC Cancer. 2002;2:35. doi: 10.1186/1471-2407-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosoi Y, Watanabe T, Nakagawa K, Matsumoto Y, Enomoto A, Morita A, Nagawa H, Suzuki N. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int J Oncol. 2004;25:461–468. [PubMed] [Google Scholar]
- 26.Kanai M, Wei D, Li Q, Jia Z, Ajani J, Le X, Yao J, Xie K. Loss of Krüppel-like factor 4 expression contributes to Sp1 overexpression and human gastric cancer development and progression. Clin Cancer Res. 2006;12:6395–6402. doi: 10.1158/1078-0432.CCR-06-1034. [DOI] [PubMed] [Google Scholar]
- 27.Lou Z, O'Reilly S, Liang H, Maher VM, Sleight SD, Mccormick JJ. Down-regulation of overexpressed Sp1 protein in human fibrosarcoma cell lines inhibits tumor formation. Cancer Res. 2005;65:1007–1017. [PubMed] [Google Scholar]
- 28.Chadalapaka G, Jutooru I, McAlees A, Stefanac T, Safe S. Structure-dependent inhibition of bladder and pancreatic cancer cell growth by 2-substituted glycyrrhetinic and ursolic acid derivatives. Bioorg Med Chem Lett. 2008;18:2633–2639. doi: 10.1016/j.bmcl.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tillotson LG. RIN ZF, a novel zinc finger gene, encodes proteins that bind to the CACC element of the gastrin promoter. J Biol Chem. 1999;274:8123–8128. doi: 10.1074/jbc.274.12.8123. [DOI] [PubMed] [Google Scholar]
- 30.Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 31.Chadalapaka G, Jutooru I, Chintharlapalli S, Papineni S, Smith R, III, Li X, Safe S. Curcumin decreases specificity protein expression in bladder cancer cells. Cancer Res. 2008;68:5345–5354. doi: 10.1158/0008-5472.CAN-07-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.