Summary
α-synuclein is an intrinsically disordered protein that appears in aggregated forms in the brains of patients with Parkinson's Disease. The conversion from monomer to aggregate is complex and aggregation rates are sensitive to changes in amino acid sequence and environmental conditions. It has previously been observed that α-synuclein aggregates faster at low pH than at neutral pH. Here, we combine NMR spectroscopy and molecular simulations to characterize α-synuclein conformational ensembles at both neutral and low pH in order to understand how the altered charge distribution at low pH changes the structural properties of these ensembles and leads to an increase in aggregation rate. The N-terminus, which has a small positive charge at neutral pH due to a balance of positively and negatively charged amino acid residues, is very positively charged at low pH. Conversely, the acidic C-terminus is highly negatively charged at neutral pH and becomes essentially neutral and hydrophobic at low pH. Our NMR experiments and REMD simulations indicate that there is a significant structural reorganization within the low pH ensemble relative to that at neutral pH in terms of long range contacts, hydrodynamic radius, and the amount of heterogeneity within the conformational ensembles. At neutral pH there is a very heterogeneous ensemble with transient contacts between the N-terminus and the NAC, however at low pH there is a more homogeneous ensemble which exhibits strong contacts between the NAC and the C-terminus. At both pHs, transient contacts between the N- and C-termini are observed, the NAC region shows similar exposure to solvent, and the entire protein shows similar propensities to secondary structure. Based on the comparison of the neutral and low pH conformational ensembles, we propose that exposure of the NAC region to solvent and the secondary structure propensity are not factors that account for differences in propensity to aggregate in this context. Instead, the comparison of the neutral and low pH ensembles suggests that the change in long-range interactions between the low and neutral pH ensembles, the compaction of the C-terminal region at low pH and the uneven distribution of charges across the sequence are key to faster aggregation.
Keywords: Parkinson's disease, α-synuclein, NMR, replica exchange molecular dynamics
Introduction
Many human diseases are associated with proteins that convert from their normally soluble form to aggregates that accumulate in the affected organs. The final forms of the aggregates include fibrillar plaques known as amyloid. This disease class includes Alzheimer's disease, Parkinson's disease, Huntington's disease, Prion disease, and type II diabetes 1; 2; 3; 4. Parkinson's disease (PD) is the second most prevalent of the late onset neurodegenerative diseases 5. α-synuclein is the primary protein component of the Lewy body deposits that are the diagnostic hallmark of PD and is expressed in high levels in the brain 6. α-synuclein can adopt two forms, the free form found in the cytoplasm and a highly helical membrane bound form 7; 8. The membrane-bound α-synuclein may be crucial for physiological functions 9; 10; 11; 12 and has been shown to be associated with the recruitment of dopamine in the presynapse and with the synaptic signal transmission 13. The aggregation of α-synuclein has been shown to occur via a nucleation dependent mechanism during which there is an initial lag phase followed by a growth phase 14. The rate of fibrillization is highly dependent on the sequence identity of the protein as well as the solution conditions and the details of the mechanism remain unclear.
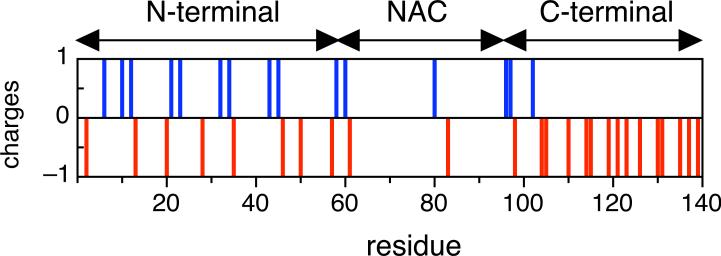
One approach to understanding the progression of fibril formation is to study human α-synuclein and variants under different conditions in order to define the distribution of conformational states in the monomeric ensemble and how these may be related to the aggregation propensities. Human α-synuclein in the free form has been shown to be an intrinsically disordered protein (IDP) 15 16 and like other IDPs is characterized by a low sequence complexity, low overall hydrophobicity and high net charge 17. It consists of three regions: an N-terminal region (residues 1−60) with a highly conserved hexamer motif KTKEGV forms α-helices in association with membranes 8; 18; 19. A central hydrophobic region, known as the “non-amyloid β component” (NAC) (residues 61−95) is proposed to be primarily responsible for aggregation 20. The C-terminal region (residues 96−140) is acidic, proline-rich 21 and also contains three highly conserved tyrosine residues. The charged residues are unevenly distributed within the sequence (Figure 1) and result in a net charge of −9 at neutral pH (Table 1). The N-terminus has a high charge density at neutral pH (18 charged residues out of 60), but the balance between positive and negative charges leads to a net charge of +4, while the NAC, with a net charge of −1, only has three charged residues. The C-terminus at neutral pH has an even higher percentage of charged residues than the N-terminus (18 out of 45 residues), and a preponderance of negatively charged Asp and Glu residues results in a net charge of −12. The uneven distribution of charges within the three regions implies that the charge profile og α-synuclein is strongly dependent of on pH. The charge distribution of the NAC, which has very few charged residues, is not greatly effected by the change in pH. However, the N-terminus region which contains the highest fraction of positively charged residues, becomes the region with the highest charge density at low pH, while the C-terminus, at low pH becomes more like the NAC, with both a low charge density and a low net charge.
Fig 1. Distribution of charged residues of α-synuclein.
The distribution of positive (blue) and negative (red) charged residues of α-synuclein at pH 7.4 is shown.
Table 1.
Distribution of charges in the sequence of α–synuclein
| pH | N | NAC | C | α–synuclein | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Net | Total | % | Net | Total | % | Net | Total | % | Net | Total | % | |
| Neutral | +4 | 18 | 30.0 | −1 | 3 | 8.6 | −12 | 18 | 40.0 | −9 | 39 | 27.8 |
| Low | +11 | 11 | 18.3 | +1 | 1 | 2.9 | +3 | 3 | 6.7 | +15 | 15 | 10.7 |
Net charge, total charge (total number of charged residues), and percentage of charged residues in the N, NAC, and C regions, and the full α-synuclein sequence at neutral pH and low pH.
Developing a better understanding of how sequence dependent effects alter the conformational propensities of monomeric α-synuclein, and correlating these effects with changes in aggregation rates, can provide insights into the mechanistic basis for abnormal folding and human disease. The conformational states of the synuclein family, consisting of human α-synuclein 15, β–synuclein 22; 23 and γ–synuclein 24 as well as the familial mutations of synuclein A53T 25; 26, A30P 25; 26 and E46K 27; 28 and mouse α-synuclein 29, which contains seven mutations relative to human α-synuclein, have been studied extensively using NMR and other spectroscopic approaches. Human α-synuclein at neutral pH has a relatively more compact conformation than is expected for a random coil conformation and transient α-helical structure has been detected in the N-terminal region of the protein 22; 23; 29; 30; 31. Transient long-range interactions between the C- and N-terminal regions and also between the C-terminal and NAC regions have been proposed based on paramagnetic relaxation enhancement (PRE) measurements 23; 29; 30; 32; 33. Observation of these long-range interactions have led to the proposal that the contacts between the NAC and the C-terminal regions shield the hydrophobic NAC region and therefore delay the aggregation of α-synuclein under these conditions 23; 30; 32. Destabilization of these long range contacts in mouse α-synuclein 29 and in A53T and A30P mutations 25 involved in early onset disease have been correlated with faster aggregation supporting the notion that protection of the NAC region may be important for delaying aggregation 25. An alternative view is that there is little difference in long-range contacts between the wild type and mutant forms of α-synuclein and that changes in aggregation rate can be attributed to local changes in the physico-chemical properties of individual residues 28.
At low pH, α-synuclein has been shown to aggregate faster than at neutral pH with fibrils taking days to form rather than several weeks 31. Fink has observed, via CD, FT-IR, and SAXS measurements, a 1.3-fold compaction in the radius of gyration of α-synuclein at low pH as well as an increase in β structure 31. He has suggested that the compaction of the protein into a partially folded conformation is responsible for the increased rate of aggregation at low pH.
NMR spectroscopy 34; 35; 36; 37 and computer simulations 38; 39; 40 have been developed in recent years to characterize conformational ensembles of IDPs. Here we use NMR and computational approaches to compare the conformational ensembles of α-synuclein at neutral and low pH. These studies help identify the role of electrostatic interactions in the pH dependent structural changes of α-synuclein and to relate these to the different aggregation propensities as a function of pH. The NMR measurements, when combined with structures generated by computer simulations, provide the first visualization of the conformational ensemble of α-synuclein at low pH. The results suggest that there is a significant structural reorganization relative to the neutral pH ensemble in terms of long-range intra-chain contacts and hydrodynamic radius. At neutral pH the highly charged C-terminal tail makes contact with the other regions of the protein in only a small population of structures. In contrast, at low pH, the C-terminus is locally collapsed. The secondary structure propensity and exposure of the NAC region to solvent is not affected by the pH change. The structural changes observed at low pH highlight the effects that the very different charge characteristics in the N-terminal, NAC, and C-terminal chain segments have on the pH dependent structural reorganization of α-synuclein.
Results
Acid induced changes in structure and dynamics probed by NMR
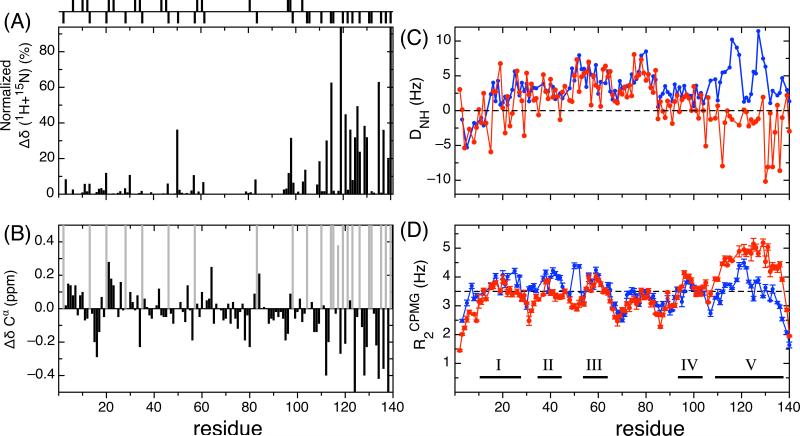
The 1H-15N HSQC spectrum at pH 2.5 and 15 °C exhibits similar spectral features to the HSQC spectrum at pH 7.4 15; the narrow 1HN chemical shift range and poor dispersion suggest that α-synuclein is intrinsically disordered at pH 2.5. Backbone assignments were carried out based on 15N-15N connections using the 3D HNN triple resonance experiments 41. 134 out of 140 residues (no Met1 and 5 Prolines) were assigned in the 1H-15N HSQC spectrum and most of the 1H and 15N chemical shift changes arise in the C-terminal region of the protein (Figure 2A) where there are 3 Asp and 12 Glu residues. The major changes in Cα shifts (ΔδCα, Figure 2B) from pH 7.4 to pH 2.5 can be directly tied to the protonation of Asp and Glu residues at low pH. Once those changes are accounted for, the remaining small ΔδCα's in the N-terminus and NAC regions alternate between positive and negative values, indicating the secondary structure propensities are essentially the same at low and neutral pH. There is however a stretch of small negative values in the C-terminus region which represents a small decrease in the β propensity in this region of the protein at low pH.
Fig 2. NMR parameters for α-synuclein at neutral and low pH.
Distribution of charged residues shown in figure 1 is displayed at the top of the figure. Comparison of chemical shifts (A, B), RDC (C) and R2CPMG (D) data of α-synuclein at acidic and neutral pH. (A) Average differences of HN and 15N chemical shifts at pH 2.5 and 7.4 with the formula: and the values are normalized. (B) Cα chemical shift differences between pH 7.4 and 2.5 (ΔδCα = δCα7.4−δCα2.5) are shown. Asp and Glu residues are colored in light gray. (C) 1H-HN residual dipolar couplings at pH 2.5 and 7.4 are shown in red and blue dot-lines, respectively. (D) R2CPMG relaxation rates at pH 2.5 and 6.1 are displayed with red and blue dot-lines, respectively. Average value (3.5 Hz) of R2CPMG at pH 2.5 is indicated. Regions of five defined segments are shown in bars and numbered.
Residual dipolar coupling (RDC) constants have been employed to probe the local structural ordering 42; 43 and also long-range contacts 30; 44 of IDPs. We have measured the DNH values of α-synuclein at pH 7.4 and pH 2.5 using an n-alkyl-poly(ethylene glycol)/n-alkyl alcohol mixture (C8E5/1-octanol) as the alignment medium. Significant differences can be seen in the RDC profiles at pH 7.4 and pH 2.5 particularly at the C-terminus and the leading portion of the N-terminus (Figure 2C). At pH 2.5, the DNH values are close to zero or negative at residues 100−127 and even more negative (∼ −8 to −10 Hz) at the end of C-terminus. The DNH values in the NAC region and the N-terminal region are similar to the DNH values measured at pH 7.4 indicating that these regions are relatively insensitive to the environmental changes.
15N backbone transverse relaxation experiments using the CPMG pulse train (R2CPMG) indicate that the low and near neutral pH forms of α-synuclein exhibit different local dynamics along the sequence (figure 2D). We have compared the R2CPMG experiments at pH 2.5 and pH 6.1 rather than pH 7.4 as we have shown (unpublished data) that the intrinsic R2CPMG values at pH 7.4 are modulated by the fast hydrogen exchange rates that exist at this pH. A more accurate view of the intrinsic R2CPMG values can be obtained at pH 6.1 where the hydrogen exchange rates are slower. The R2CPMG values can be classified into 5 segments (10−25, 32−42, 52−62, 93−105, and 109−136) using R2CPMG=3.0 Hz as cutoff. The overall features of the R2CPMG data are similar for the two pH values with the exception of the C-terminal end from residues 105 to the end of the protein. Here the R2CPMG values are uniformly greater at low pH than at neutral pH suggesting that the C-terminal end of the protein is experiencing restricted motion and is more rigid at low pH. In the NAC region, for both pHs, the R2CPMG values are the smallest indicating that this region is the most flexible.
The C-terminus is collapsed at pH 2.5
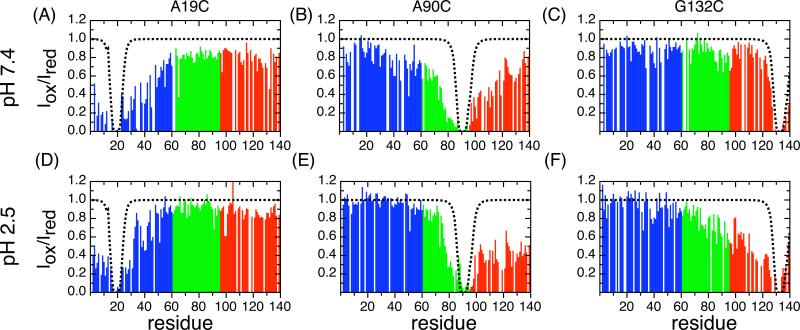
Site-directed spin labeling and paramagnetic relaxation enhancement (PRE) at three positions, A19, A90 and G132, are used to obtain information about long-range interactions within the unfolded protein. The PRE effects are compared at pH 7.4 and pH 2.5 and show differences, especially in terms of the interactions observed at the C-terminal end of the protein. At pH 7.4 (Figure 3A) the spin label at A19C results in diminished intensity throughout the entire sequence with a maximum observed value of Iox/Ired ∼0.8. At the N-terminus, there is a slow increase in Iox/Ired up to residue 60 at which point the intensity plateaus at ∼0.8. The same spin label at pH 2.5 (Figure 3D), has a progressively faster increase in PRE intensity at the N-terminus and plateaus at a higher level close to 1.0 in the NAC region and C-terminal to it. There is a small reduction in intensity (Iox/Ired ∼0.8) at the very end of the C-terminus from residues 110−140. At neutral pH there are numerous short range contacts between residue 19 and the N-terminus as well as transient contacts between residue 19 and the NAC and C-terminal regions consistent with previous experimental results 23; 30. At low pH the N-terminal region has fewer interactions with the N-terminal spin label, the NAC region is not interacting with the N-terminal spin label and is only making very local transient contact at a small region of the C terminal (residues 115−130).
Fig 3. PRE intensity ratios for spin-labeled α-synuclein.
Paramagnetic relaxation experiments of amide protons in human α-synuclein at pH 7.4 (A, B and C) and pH 2.5 (D, E and F) using MTSL spin labels for cysteine mutants A19C,A90C and G132C. Measured PRE intensity ratios (Iox / Ired) are presented in the top (pH 7.4) and bottom (pH 2.5) panels. White gaps in each plot are unassignable residues including 5 prolines. N-terminal, NAC and C-terminal regions are colored in blue, green and red respectively. Dashed lines are the theoretical PRE values of α-synuclein without any long-range contacts calculated as described 29.
The PRE results at A90C in the NAC region (Figure 3B, 3E) show distinct features at neutral and low pH. At pH 7.4, the PRE Iox/Ired pattern is similar to the published data obtained by placing the PRE label at position A85 28 or A90 30. The A90C spin label probe at pH 7.4 interacts primarily with the second half of the N-terminal region (40−60) as well as with the C-terminal region. In contrast at pH 2.5 there are essentially no interactions with the N-terminal region and increased interactions with the C-terminal (Iox/Ired 0.4−0.6) relative to the neutral pH.
The spin label at position G132C (Figure 3C, 3F) has a very different intensity profile at neutral and low pH in particular in the C-terminal region of the protein. At neutral pH the spin label does not lead to strong signal attenuation anywhere in the sequence. Three small dips in the intensity ratio (Iox/Ired = ∼0.8) are observed, one in the NAC region from residues 80−100 and two in the N-terminus, around residues 30−40 and near residue 5. This profile has similar trends to those seen by Zweckstetter 30 and Eliezer 23 who have studied the effects of spin labels at positions A140C and P120C respectively, however the decrease in the intensity ratio is somewhat smaller for the data observed here with the spin label at position 132. The PRE effects due to the G132C spin label at low pH (Figure 3F) are quite different; there is no signal attenuation in the N-terminus, but there is strong signal attenuation in the C-terminus (Iox/Ired ∼0.4) which continues, gradually weakening (Iox/Ired goes from 0.4 to 0.9), into the NAC region. The weak signal attenuation from G132C at neutral pH suggests that the protein is making infrequent transient C- to N-terminal contacts. However, the pH 2.5 the PRE profile shows a cluster of long range contacts between the C terminal spin label and the C-terminal end and NAC region.
Pulse-field gradient NMR (PFG-NMR) translational diffusion experiments were performed at low and neutral pH in order to obtain the effective hydrodynamic radius (Rh) of the proteins. The Rh value at 15 °C and pH 2.5 (Rh=30.1 Å) is smaller than that at pH 7.4 (Rh=31.7 Å) suggesting that low pH induces a conformational compaction relative to the neutral pH value. Fink has also observed a compaction at low pH relative to neutral pH 31; he saw a decrease in the radius of gyration from 40 Å at pH 7.5 to 30 Å at pH 3.0. The large gap in the radius of gyration values relative to the hydrodynamic radii is because the radius of gyration, which measures the average distance of each residue from the center of mass of the protein, is more sensitive to the extension of the chain than the hydrodynamic radius, which approximates the radius of a sphere whose diffusion constant equals that of the protein 45.
Visualization of the conformational ensemble of α-synuclein by REMD at low pH
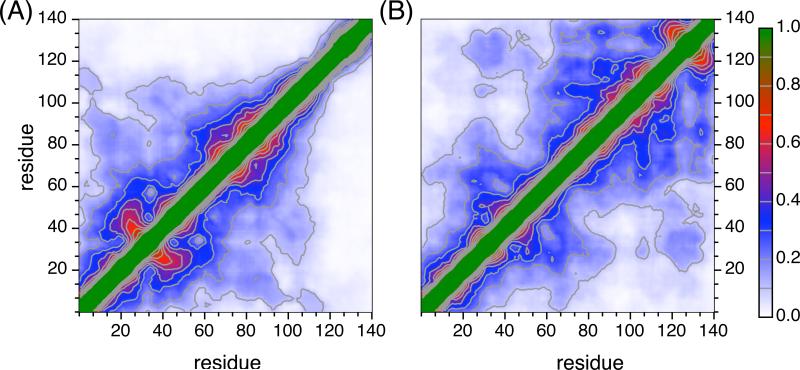
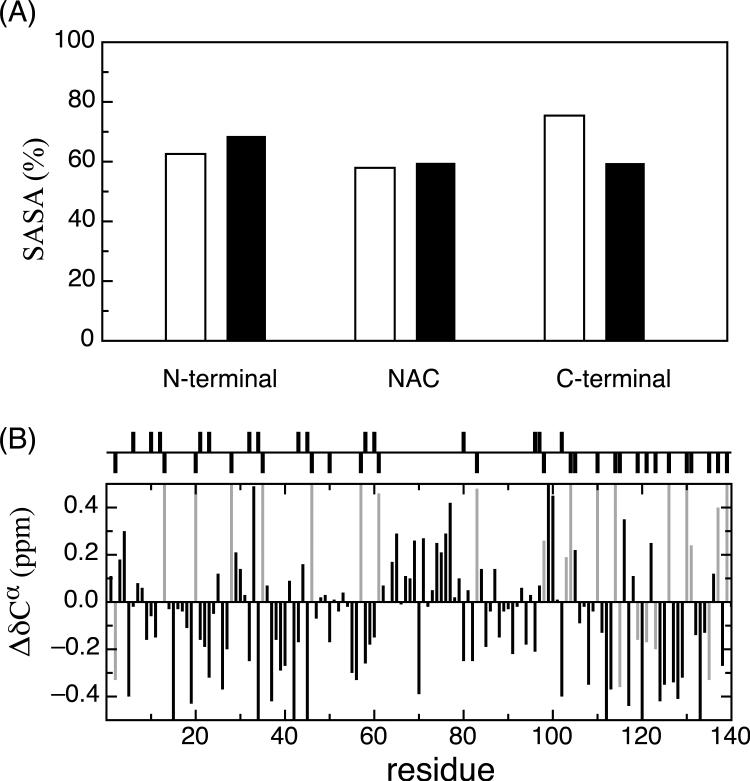
Replica exchange molecular dynamics (REMD) simulations were employed to generate conformational ensembles of α-synuclein at neutral and low pH. Long-range contacts within the neutral and low pH ensembles are consistent with the long range contacts observed experimentally through PRE measurements. Figure 4 shows the percentage of the population within the low and neutral pH ensembles with inter-residue contacts (Cα-Cα distances) within 20 Å. A cutoff of 20 Å was chosen to facilitate comparison with experimental PRE measurements which can detect contacts within 20−25 Å of the nitroxide spin label. At neutral pH (Figure 4A), there are contacts observed throughout the N-terminus and NAC regions, while the C-terminal tail (residues 115−140) is largely extended, making contact with the other regions in only ∼5% of the population. In the contact map for the low pH ensemble (Figure 4B) the region of the protein that shows many intra-molecular contacts has switched relative to the neutral pH ensemble. At low pH the C-terminus is in regular contact with itself and the NAC region, as well as a small section of the N-terminus near residue 40, but only approaches the most N-terminal part of the protein in a small fraction of the population. The section of the N-terminus near residue 40 makes transient contact not only with the C-terminus but also with the N-terminal half of the NAC. However, the leading portion of the N-terminus, from residues 1−40, forms a separate cluster of long-range contacts and has fewer contacts with the rest of the protein. The switch from an extended C-terminus at neutral pH to a somewhat less extended N-terminus at low pH is also seen in calculations of the solvent accessible surface area (Figure 5A). The C-terminus at neutral pH has the highest solvent accessibility (75.4%) of any region at either pH. However, at low pH the N-terminus is the region most exposed to the solvent (68.6%), while the NAC and C-terminus have an equal amount of solvent accessibility (59%). Interestingly the NAC solvent exposure is very similar at both neutral and low pH. It appears that the NAC region “swaps” intermolecular contacts with the N-terminal region at neutral pH for corresponding contacts with the C-terminal residues at low pH.
Fig 4. REMD simulation results exhibit good agreement with experimental data.
Contact maps of α-synuclein ensembles at neutral pH (A) and low pH (B) calculated using REMD. The color gradient from white, blue, red to green represents percentages of the conformational ensembles that have inter-residue Cα-Cα distances within 20 Å.
Fig 5. Solvent accessible surface areas at neutral and low pH.
(A) Comparison of percent solvent accessible surface areas (SASA) for the N-terminal, NAC and C-terminal regions of the neutral (opened bars) and low (filled bars) pH ensembles. A significant change in the solvent accessibility is observed in the C-terminal region which is less exposed, supporting the idea of a collapsed conformation in this region. (B) Calculated differences in average chemical shifts between the low and neutral pH ensembles (Cαneutral – Cαlow). Asp and Glue residues are colored in light gray. The chemical shifts were calculated using ShiftX 70.
Simulated PRE intensity ratios for the neutral and low pH conformational ensembles (Figure S1) portray the same trends seen in the contact maps. At neutral pH, the intensity ratio profiles show significant contacts between the N-terminus and NAC regions, while the C-terminal spin label site is only in contact with the rest of the sequence in a very small percentage of the population. Interestingly, the simulated ensemble does show a dip in the intensity ratio from G132C in the N-terminus near residue 10. At low pH, the calculated PRE profiles are consistent with fewer contacts between the N-terminus and the NAC, but show very significant drops in the intensity ratio in the C-terminus from the A90C spin label, and in the NAC from the G132C spin label. Overall, the calculated intensity ratio profiles, like the contact maps, exhibit large changes in the C-terminus when going from neutral to low pH and smaller changes in the N-terminus.
The inter-residue contact maps (Figure 4A, 4B) provide information about the long-range contacts of the ensemble. Local structural characteristics, particularly secondary structure propensities, are often related to chemical shifts that are affected by the local environment of the nucleus being monitored 24; 46. We calculated the Cα chemical shifts for the low and neutral pH ensembles. There are large changes in Cα (ΔδCα, Figure 5B) that can be attributed to the change in the charge state of the Asp and Glu residues, just as with the experimental Cα measurements. However, even accounting for the protonation of the negatively charged residues, there are more differences in the Cα shifts of the simulation ensembles than are observed experimentally. The calculated ΔδCα's show that the biggest chemical shift difference is in the C- terminal region and that it corresponds to decreased β, similar to the experimental observation, but also feature stretches of negative ΔδCα in the N-terminus and positive ΔδCα in the NAC region, indicative of decreased β propensity and decreased α propensity respectively. The overall β structure of the ensembles, as calculated by STRIDE 47, decreases as the pH is lowered, consistent with the trend predicted by the experimental ΔδCα measurement. There is no significant helical structure observed for either simulated ensemble. Though there is slightly more variation in the chemical shifts of the simulated ensembles compared to the experiments, in both cases the largest differences are found in the C-terminus and the magnitude of the chemical shift differences throughout the sequence is very small. This suggests that there is little difference in secondary structure propensity at neutral and low pH, with the biggest change a slight decrease in β propensity in the C-terminus at low pH.
Relative packing of the N-, NAC and C-terminal regions of α-synuclein were evaluated by determining compaction ratios (see Methods). The values (Table 2) are consistent with the picture provided by the contact maps. At neutral pH, the N-terminus and NAC regions have similar compaction ratios, 0.55 and 0.5, but the C-terminus is much more expanded, with a compaction ratio of 0.81. In contrast, at low pH, the C-terminus has a compaction ratio similar to that of the NAC, 0.52 and 0.56 respectively, while the N-terminus is more expanded, with a compaction ratio of 0.63. Here again, the region most affected by the change in pH is the C-terminus, whose compaction at low pH is reflected in the change the compaction ratio from 0.81 to 0.52.
Table 2.
Compaction ratios of REMD simulated ensemble of α-synuclein
| pH | N | NAC | C |
|---|---|---|---|
| Neutral | 0.55 | 0.5 | 0.81 |
| Low | 0.63 | 0.56 | 0.52 |
Compaction ratios of the N, NAC and C regions for the low and neutral pH α-synuclein simulation ensembles, calculated as the ratio of average end to end distance in the low (or neutral) pH ensemble to the same quantity for the denatured state ensemble.
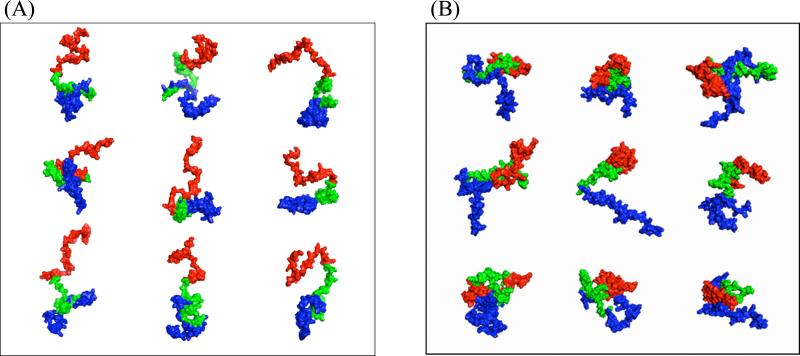
Molecular simulations allow for the visualization of individual structures within the conformational ensembles. We used a clustering algorithm 48 to select structures from the REMD simulations. The conformations from the neutral pH ensemble shown in figure 6A were chosen from the top four clusters, representing over 53% of the ensemble. The representative structures from the low pH ensemble (Figure 6B), chosen from the top three clusters and representing more than 60% of the total population, provide the first visualization of structures from an ensemble consistent with experimental observations at low pH. These figures highlight the structural changes at low pH relative to neutral pH previously observed in both the PRE measurements and the contact maps for the simulation ensembles, namely, the compaction of the C-terminal region at low pH coupled with an N-terminal region which is less extended than the C-terminus at neutral pH.
Fig 6. Selected representative conformations of α-synuclein from the neutral and low pH ensembles.
Representative structures chosen from (A) the top four clusters of the neutral pH ensemble and (B) the top three clusters from the low pH ensemble. These clusters account for more than 53% and 60% of the respective structural ensembles. The N-terminus is shown in blue, the NAC in green and the C-terminus is shown in red.
Discussion
We have used two approaches, NMR spectroscopy and computer simulation, to characterize the conformational ensembles of the IDP α-synuclein at both neutral and low pH. By NMR we have measured chemical shifts, RDCs, R2CPMG, and PREs of α-synuclein at pH 7.4 and 2.5 in order to compare the secondary structure and long-range interactions within the protein and to explore differences in dynamics. We have also used replica exchange molecular dynamics with implicit solvent to simulate α-synuclein at neutral and low pH. The simulations allow us to visualize the conformational ensembles at low pH and compare these directly to the conformations populated at neutral pH.
α-synuclein at neutral pH has been extensively studied by NMR 15; 29; 30; 32. Our experimental measurements at pH 7.4 reproduce the same general features observed in previous studies. The effect of the N-terminal spin label is seen throughout the remainder of the N-terminus and results in a small, uniform reduction in the intensity ratio in both the NAC and the C-terminus. We interpret the uniformly reduced intensity ratio throughout the NAC and C-terminus as a consequence of transient contacts between A19C and each residue from 60−140; it is a reflection of the heterogeneity of the structural ensemble since it implies that A19C is not involved in unique long range contacts with any particular residues in the C-terminal part of the protein. An alternative explanation for the plateau is that there is a scaling problem between the oxidized and reduced experiments that lowers the baseline but is not indicative of any actual contact between A19C and the residues of the NAC and C-terminal regions. However, the fact that a similar effect is seen in the PRE intensity ratios calculated from the REMD ensembles (Figure S1) lends credibility to the observed results, since there can be no question of a scaling problem in the simulations. The neutral pH conformational ensemble generated by the simulations qualitatively portrays the same long-range interactions observed by the NMR experiments. Intermittent contacts between the N-terminus and the other regions of the protein are seen both in the contact maps and the calculated PRE intensity ratios. Moreover, in both the experimental and the calculated PRE plots, the C-terminal probe at G132C induces very little reduction in intensity from the line corresponding to the intensity ratio expected for a random coil, except for a small dip in the N-terminus near residue 20. This suggests that with the exception of a small population of conformations in which the N- and C-termini are in contact, the C-terminus is largely extended into solution.
While our analysis of the neutral pH REMD ensemble suggests that it qualitatively captures the long-range interactions observed experimentally, it is quite different from the ensemble previously obtained by Griesinger et. al. 30. In their study agreement with experiments was enforced through the use of PRE-derived distance constraints. The structures they show, representatives of seven clusters accounting for more than 50% of their conformational ensemble, are all shaped like rings, with the N- and C-termini in contact with each other. That two such different ensembles can be obtained which both appear to agree with PRE data is a reflection of the fact that the determination of structural ensembles from PRE data is a very underdetermined problem 49. PRE intensities are related to the distance between each residue and the nitroxide spin label via a 1/r6 average over the conformational ensemble. As a consequence of this 1/r6 distance dependence, PRE intensities are dominated by contributions from more compact conformations. Small changes in the population of conformations containing residues a short distance from the spin label will have a big effect on the 1/r6 average; this decreases the constraints on other conformers and can allow for the inclusion of large populations of expanded conformations in an ensemble matching a given PRE intensity ratio profile. This means that the distances derived from the spin labels attached to the protein in PRE experiments can be satisfied by many different conformational ensembles. It has recently been demonstrated that if it were experimentally feasible to attach many more spin labels, covering a large percentage of the protein, then there would be enough data generated to be able to uniquely identify the true conformational ensemble 49.
NMR experiments and REMD simulations indicate that there is a significant structural reorganization within the low pH ensemble relative to that at neutral pH in terms of long-range contacts, hydrodynamic radius, and the amount of heterogeneity within the conformational ensembles. The rearrangement of long-range contacts is observed by comparing the PRE measurements and REMD contact maps at low pH with the corresponding descriptions of the neutral pH ensemble. Both the PREs and the contact maps show that the largest change takes place in the C-terminus. Whereas the C-terminal region in the neutral pH ensemble is largely extended, only making contact with the other regions transiently, at low pH there are numerous contacts between the C-terminus and the tailing portion of the NAC (figure 3E, 3F). The C-terminus at low pH is also more compact, has decreased flexibility based on 15N relaxation data, and is less exposed to the solvent than at neutral pH. The response of the N-terminal region to the change in pH is less dramatic. While the N-terminus at low pH is both more extended and more exposed to solvent than at neutral pH, neither of these changes are as large as those observed in the C-terminus. These results highlight the change in the long range interactions between the neutral and low pH ensembles; at neutral pH there is a very heterogeneous ensemble with transient contacts between the N-terminus and the NAC, however at low pH there is a more homogeneous ensemble which exhibits strong contacts between the NAC and the C-terminus. Transient contacts between the N- and C-termini are observed at both pHs.
NMR experiments and REMD simulations show that the low pH ensemble of α-synuclein has a smaller average hydrodynamic radius than the neutral pH ensemble. From the experimental data and the visualization of the REMD structures it is clear that there is no global collapse of the protein at low pH but rather the compaction is local and exists primarily in the C-terminal domain and the C-terminal region of the NAC. Fink has proposed, based on CD and FT-IR measurements, that the low pH form of α-synuclein has increased β content relative to the neutral pH form, a relatively compact conformation and can be thought of as a pre-molten globule state 31. The detailed atomic description presented here indicates that the compaction is primarily restricted to the last 60 residues of the protein and that the N-terminal region is more exposed. In addition there is no evidence at the individual residue level of an increase in β conformation in the low pH ensemble relative to the neutral pH ensemble.
The structural reorganization at low pH can be rationalized by considering the distribution of charges that exist in the low pH form relative to the neutral pH form. As seen in the charge density table (table 1), the three regions of α-synuclein are impacted differently by lowering the pH. In particular, there is a large change in the total charge density in the C-terminus that decreases from 40% charged residues at neutral pH to only 6.7% charged residues at low pH. In contrast, the effect of low pH on the N-terminus is less pronounced with a shift from 30% charged residues at neutral pH to 18.3% charged residues at low pH, and a total charge that is high at both pHs. Overall, the decrease in the total charge density of the C-terminus is considerably more than the change at the N-terminus and accounts for the fact that the reorganization of the C-terminus is the most pronounced, and that of the N-terminus less so.
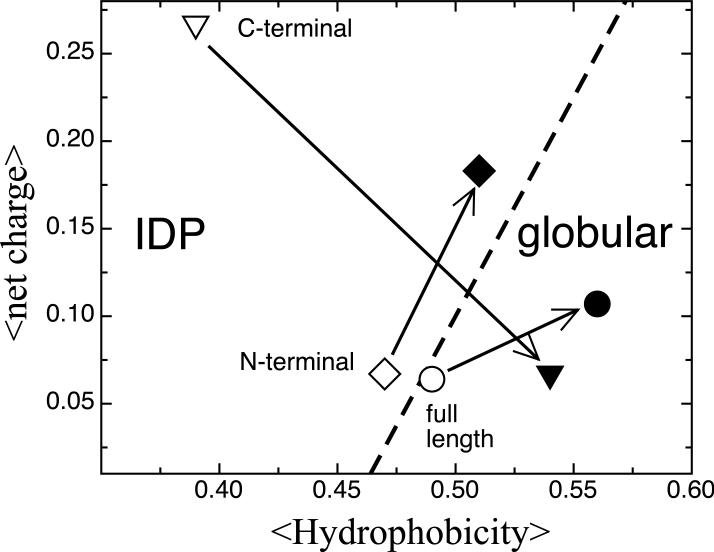
Uversky has empirically observed that IDPs and globular proteins fall into two distinct regions as a function of their mean net hydrophobicity versus mean net charge 50. He noted that in the plot of hydrophobicity versus charge α-synuclein is one of the few examples of an IDP that is just over the line into the ordered region of the plot (Figure 7). However, we consider the three functional regions of α-synuclein separately, and note that the C-terminal region at neutral pH, with a net charge of −12, resides very definitively in the IDP region of the Uversky plot. At low pH however, the C-terminus loses most of its net charge and becomes highly hydrophobic. The very dramatic movement of the C-terminus to the ordered region may be due to the high density of glutamates in the sequence that become very hydrophobic at low pH. The globular like quality of the C-terminus that is predicted in the plot is reflected in the NMR experiments and REMD calculations where the C-terminal compaction is seen both in the experimental PRE measurements and the contact map of the simulated low pH ensemble. The full α-synuclein sequence at low pH moves more fully into the ordered region of the charge v. hydrophobicity plot due to an increase in hydrophobicity. This shift is mirrored by the smaller hydrodynamic radius observed at low pH. Interestingly, because of the balance between positively and negatively charged residues in the N-terminus at neutral pH, the N-terminus has both a higher net charge and slightly higher net hydrophobicity at low pH and remains in the IDP region of the plot.
Fig 7. Charge-hydrophobicity characteristics of α-synuclein.
Charge-hydrophobicity characteristics of α-synuclein at low (filled symbols) and neutral (opened symbols) pH. The hydrophobicity values for α-synuclein were calculated using the Cowan-Whittaker hydropathy indices 72 for the low and neutral pH conditions using a window size of 5 amino acids. The hydrophobicity values for the individual amino acids were normalized to a 0 to 1 scale. The mean net hydrophobicity (net charge) was calculated as the sum of normalized hydrophobicity (charge at specific pH) divided by the total number of residues in α-synuclein. The black line represents the boundary between globular proteins and IDPs determined using a list of these proteins, an analysis similar to that performed by Uversky et al 50.
α-synuclein has been shown to aggregate more rapidly at low pH and a comparison of the structural and dynamical differences may aid in understanding which factors are critical to aggregation. Aggregation may be affected by a number of properties including propensity to form transient secondary structure 23, the distribution of long range interactions 30; 32, the hydrodynamic radius of the protein 31, the flexibility of different regions and their exposure to solvent 29; 51, and the distribution of charges along the protein 52. We find that the Cα chemical shifts of the neutral and low pH forms are quite similar, and this suggests that secondary structure propensity is not a driving force to increase the aggregation rate at low pH. It has previously been suggested that the increased rates of aggregation observed for single point mutations 25 or upon binding of divalent metal ions 53 or polycations 30; 54 are due to the release of the long range N- to C-terminal contacts and subsequent exposure of the NAC. However, the results of the REMD simulations suggest that the NAC region has comparable exposure to solvent at both neutral and low pH; NMR R2CPMG measurements suggest that the NAC region has comparable flexibility in both pH forms. Similar exposure of the NAC region to solvent in both the slower, neutral pH, and faster, low pH, aggregating species suggests that protection or exposure of the NAC region may not have a large impact on the aggregation propensity in this context.
At low pH where the charge profiles of both the N- and C-termini are significantly different from their neutral pH counterparts it is possible that initiation of aggregation may arise through a different mechanism. Previously we proposed for mouse α-synuclein at neutral pH that aggregation may be initiated through intermolecular N-terminal contacts. While the N-terminus has a balance of positively and negatively charged residues at neutral pH in both mouse and human α-synuclein, it carries a large positive charge at low pH. Without complementary negative charges anywhere in the sequence, it may be reasonable to assume that the N-terminus is unlikely to initiate intermolecular contacts at low pH. One scenario that is consistent with the structural changes and charge distribution at low pH is that the locally collapsed C-terminus, which loses its net charge and becomes highly hydrophobic under these conditions, is involved in the earliest stages of α-synuclein aggregation at low pH. These data indicate the importance of the charge distribution in directing both the mechanism and the rates of aggregation 52.
Material and methods
Sample preparation
Expression and purification of human α-synuclein in E.coli were followed human by described protocol 29. Further purification using DEAE or Q anion exchange chromatography was applied to ensure high purity of α-synuclein for low pH studies. α-synuclein was eluted at 200−300 mM NaCl gradient. Solution containing pure α-synuclein was dialyzed against with 20 volumetric times of PBS (pH 7.4) at 4 °C and repeated 3 times. Dialyzed and concentrated α-synuclein (200 μM) was transferred to 1.5 ml tubes and quickly frozen by cryogenic liquid nitrogen for −80 °C storage. For each NMR sample, the −80 °C stored proteins were unfrozen at 4 °C, polished using a 100 kDa filter (Millipore), and then dialyzed to pH 2.5 (10 mM phosphate, 140 mM NaCl) with the addition of 10% D2O.
NMR experiments for assignment
NMR experiments including triple resonance experiments and 15N backbone dynamics experiments were performed on a Varian 800 MHz spectrometer equipped with triple gradient probe. PFG-NMR diffusion, PRE and RDC measurements were acquired on a Varian 600 MHz spectrometer with a cold probe or on a Bruker 700 MHz spectrometer. Acquired spectra were processed by NMRPipe 55 and analyzed by Sparky 56.
650 μM 13C-15N labeled α-synuclein at pH 2.5 was used to perform triple resonance experiments for backbone assignment at 15 °C. The 3D spectra were acquired via the conventional strategy 57 using HNCACB, CBCA(CO)NH, HNCO, and HN(CA)CO, and a new approach using HNN 41 via 15N-15N connections. Data collection time for each 3D spectrum at 15 °C is shorter than 20 hour (with 8 or 16 transient scans) to avoid aggregation during acquisition. HSQC spectra were collected to check the stability of α-synuclein before acquiring the next 3D spectrum.
Chemical shifts were calibrated using DSS dissolved at pH 2.5 as a reference 58. Comparing the secondary structural propensity using the Cα chemical shifts deviation from the random coil shifts is not appropriate since the random coil Cα chemical shifts measured at pH 5.0 59 and 2.3 60; 61 exhibit significant differences. Directly monitoring the changes of Cα chemical shifts (ΔδCα = δCα7.4−δCα2.5) was used to inspect the secondary structure propensities at pH 7.4 and 2.5.
Translational diffusion coefficients and hydrodynamic radii
PFG-NMR experiments incorporated with longitudinal Eddy current pulse schemes and convection compensation was used to measure the translational diffusion coefficients (Dtrans) 62. Samples containing 300 μM α-synuclein and 35 300 mM 1,4-dioxane were dissolved in PBS buffers at pH 7.4 and 2.5 (100% D2O, pD uncorrected) for PFG-NMR experiments at 15 °C. 25 1D PFG-NMR spectra were acquired over a range of gradient strengths of 2 to 17 G/cm or 5 to 50 G/cm for 1,4-dioxane or α-synuclein, respectively. Integrated peak volumes of 1,4-dioxane and α-synuclein (methyl groups only) were fitted using VNMRJ 2.1 (Varian Inc. CA) and used to calculate Dtrans 62. Since α-synuclein and dioxane were dissolved in one solution, the viscosity effect on Dtrans can be ignored. Therefore, the Stokes-Einstein equation can be simplified to get the hydrodynamic radius of α-synuclein by the relationship:
where Rh(dixoane) is 2.12 Å.
15N relaxation
R2CPMG 15N relaxation experiments were acquired using 250 μM 15N labeled α-synuclein at pH 2.5 and 6.1. Complex points for each spectrum were 1024×256 in the 1H and 15N dimensions, respectively. The relaxation times for R2CPMG were 0.01, 0.03, 0.05, 0.09, 0.13, 0.17, 0.19, 0.21 and 0.25 seconds. Recycle delays for each experiment were 2 s. Data were processed by NMRPipe and analyzed by Sparky.
Site-directed spin label
Methods for MTSL (Toronto Research Chemicals, Ontario, Canada) spin labeled 15N-α-synuclein A19C, A90C, or G132C were done as described in our previous work29. In brief, 250 μM MTSL labeled α-synuclein cysteine mutants were dialyzed to pH 2.5 or 7.4 and were divided to equal volume for experiments at paramagnetic (oxidized) and diamagnetic (reduced) states. Addition of 10 mM L-ascorbate (pH 2.5) or 10 mM dithiothreitol (DTT) (pH 7.4) was applied for generating the diamagnetic samples. It is not possible to use DTT or L-ascorbate as a reducing reagent at both pHs. DTT cannot be used at low pH as the disulfide bond is not efficiently broken at pH 2.5 because the thiol groups are highly protonated. At neutral pH, we have observed significant signal enhancements and changes of 1H and 15N chemical shifts in the presence of L-ascorbate. PRE ratios (Iox/Ired) were calculated as the intensity ratios of the same residue in the absence (Iox) or in the presence (Ired) of L-ascorbate or DTT. At low pH all mutant samples were normalized against L-ascorbate and at neutral pH all samples were normalized against DTT.
RDC measurement
N-alkyl-poly(ethylene glycol)/n-alkyl alcohol mixture 63 was chosen to prepare the liquid crystalline medium for residual dipolar coupling experiments. 5% C8E5 (pentaethylene glycol monooctyl ether) and 1-octanol were mixed with PBS buffer at pH 2.5 or 7.4. The molar ratio of C8E5 and 1-octanol is 1.05. The quadrupolar deuterium splitting constants are 23.8 and 22.7 Hz at pH 7.4 and 2.5, respectively. High resolution HSQC_IPAP spectra in the absence or in the presence of an alignment medium were collected at 15 °C with complex points of 2048 (t2) × 512 (t1) and 16 transient scans.
Replica exchange molecular dynamics (REMD) simulations at low pH
Replica exchange molecular dynamics 64; 65 simulations were used to generate the conformational ensembles of α-synuclein at low and neutral pH. In this method, a number of simulations (replicas) are run in parallel over a specified temperature range. The adjacent replicas (Ti and Tj) are allowed to exchange periodically, with an acceptance criterion based on the following Metropolis transition probability.
Where βi(j) = 1/KT i(j) and E i(j) is the potential energy of the ith (jth) replica. This method provides canonical probability distributions for the ensembles over the specified temperature range 65. The REMD method has been implemented in the IMPACT simulation package 66. The simulations were performed using the AGBNP implicit solvent 67 model and the OPLS-AA force field 68.
The simulation was performed for a total of 10 ns over 20 temperatures (300, 308, 317, 325, 334, 343, 353, 362, 372, 382, 393, 403, 414, 426, 437, 449, 461, 474, 487, 500 K), corresponding to a total simulation time of 200 ns. All simulations were started with a fully extended conformation of the α-synuclein molecule. The sequence of α-synuclein, for the low pH condition, was modified to a state corresponding to that of the molecule under these conditions, by protonating the side-chain carbonyl groups of all of the acidic residues (aspartates and glutamates). 〈–synuclein was also simulated with a purely repulsive potential, in which all interactions have been turned off except for the repulsive part of the Lenard-Jones potential, to obtain the ensemble of structures corresponding to the denatured or most expanded state. The 449 K and 414 K ensembles were used for the structural characterization of the low and neutral pH ensembles. These ensembles were chosen because their hydrodynamic radii (29.1 Å and 30.5 Å respectively) most closely matched the experimental measurements at 15 °C.
A number of analysis methods were used for the structural characterization of the low pH ensembles. The average size of the ensembles was determined by calculating the hydrodynamic radii for individual structures using Hydropro 69. The secondary structural assignments for the α-synuclein ensembles were made using STRIDE 47. Cα chemical shifts were calculated using ShiftX 70. The solvent accessible surface areas (SASA) were calculated using the program Surfv 71. The clustering of the neutral and low pH ensemble structures was performed using a hierarchical clustering method 48, with the clustering based on a distance matrix of distances between the center of mass of the three regions of synuclein (N-terminus, NAC, and C-terminus). The compaction factors for the low and neutral pH conditions were calculated as the ratio of the average end-to-end distance between the low (neutral) pH and the denatured state of α-synuclein. The end-to-end distance of the denatured state was calculated from an ensemble generated using a purely repulsive potential, which gives the largest average dimensions possible for the protein.
Supplementary Material
Acknowledgments
This work has been supported in part by NIH training grants (3T90DK070135) to DSW, and (5R90DK071502) to K-PW, and by NIH GM 45302 to JB and GM 30580 to RML.
Abbreviations
- PD
Parkinson's disease
- NAC
non- amyloid ® component
- IDP
intrinsically disordered protein
- NMR
nuclear magnetic resonance
- R2CPMG
CPMG type transverse relaxation
- RDC
residual dipolar coupling
- PRE
paramagnetic relaxation enhancement
- MTSL
(1-oxy-2,2,5,5-tetra-methyl-3-pyrroline-3-methyl)-methanesulfonate
- REMD
replica exchange molecular dynamics
- SASA
solvent accessible surface areas
- C8E5
pentaethylene glycol monooctyl ether
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
While this manuscript was under revision the paper entitled “ Charge neutralization and collapse of the C-terminal tail of alpha-synuclein at low pH” by Sebastian McClendon, Carla Rospigliosi, and David Eliezer appeared in press online in Protein Science. The work was carried out independently by both laboratories
References
- 1.Lansbury PT., Jr. Evolution of amyloid: what normal protein folding may tell us about fibrillogenesis and disease. Proc Natl Acad Sci U S A. 1999;96:3342–4. doi: 10.1073/pnas.96.7.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perutz MF. Glutamine repeats and neurodegenerative diseases: molecular aspects. Trends Biochem Sci. 1999;24:58–63. doi: 10.1016/s0968-0004(98)01350-4. [DOI] [PubMed] [Google Scholar]
- 3.Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci. 1999;24:329–32. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 4.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 5.Siderowf A, Stern M. Update on Parkinson Disease. Ann Intern Med. 2003;138:651–658. doi: 10.7326/0003-4819-138-8-200304150-00013. [DOI] [PubMed] [Google Scholar]
- 6.Rochet JC, Lansbury PT., Jr. Amyloid fibrillogenesis: themes and variations. Curr Opin Struct Biol. 2000;10:60–8. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 7.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273:9443–9. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 8.Jo E, McLaurin J, Yip CM, St George-Hyslop P, Fraser PE. alpha-Synuclein membrane interactions and lipid specificity. J Biol Chem. 2000;275:34328–34. doi: 10.1074/jbc.M004345200. [DOI] [PubMed] [Google Scholar]
- 9.Abeliovich A, Schmitz Y, FariÒas I, Choi-Lundberg D, Ho W-H, Castillo PE, Shinsky N, Verdugo JMG, Armanini M, Ryan A, Hynes M, Phillips H, Sulzer D, Rosenthal A. Mice Lacking [alpha]-Synuclein Display Functional Deficits in the Nigrostriatal Dopamine System. Neuron. 2000;25:239–252. doi: 10.1016/s0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 10.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, LaBaer J, Rochet J-C, Bonini NM, Lindquist S. {alpha}-Synuclein Blocks ER-Golgi Traffic and Rab1 Rescues Neuron Loss in Parkinson's Models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenco JM, Rawlingson A, Daniels B, Morris AJ. Regulation of Phospholipase D2: Selective Inhibition of Mammalian Phospholipase D Isoenzymes by alpha- and beta-Synucleins. Biochemistry. 1998;37:4901–4909. doi: 10.1021/bi972776r. [DOI] [PubMed] [Google Scholar]
- 12.George JM, Jin H, Woods WS, Clayton DF. Characterization of a novel protein regulated during the critical period for song learning in the zebra finch. Neuron. 1995;15:361–372. doi: 10.1016/0896-6273(95)90040-3. [DOI] [PubMed] [Google Scholar]
- 13.Yavich L, Tanila H, Vepsalainen S, Jakala P. Role of alpha-synuclein in presynaptic dopamine recruitment. J Neurosci. 2004;24:11165–70. doi: 10.1523/JNEUROSCI.2559-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood SJ, Wypych J, Steavenson S, Louis JC, Citron M, Biere AL. alpha-synuclein fibrillogenesis is nucleation-dependent. Implications for the pathogenesis of Parkinson's disease. J Biol Chem. 1999;274:19509–12. doi: 10.1074/jbc.274.28.19509. [DOI] [PubMed] [Google Scholar]
- 15.Eliezer D, Kutluay E, Bussell R, Jr., Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol. 2001;307:1061–73. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 16.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr. NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–15. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 17.Uversky VN. A Protein-Chameleon: Conformational Plasticity of α -Synclein, a Disordered Protein Involved in Neurodegenerative Disorders. Journal of Biomolecular Structure & Dynamics. 2003;21:211–234. doi: 10.1080/07391102.2003.10506918. [DOI] [PubMed] [Google Scholar]
- 18.Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem. 2005;280:9595–603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 19.Bussell R, Jr., Eliezer D. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J Mol Biol. 2003;329:763–78. doi: 10.1016/s0022-2836(03)00520-5. [DOI] [PubMed] [Google Scholar]
- 20.El-Agnaf OM, Bodles AM, Guthrie DJ, Harriott P, Irvine GB. The N-terminal region of non-A beta component of Alzheimer's disease amyloid is responsible for its tendency to assume beta-sheet and aggregate to form fibrils. Eur J Biochem. 1998;258:157–63. doi: 10.1046/j.1432-1327.1998.2580157.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim TD, Paik SR, Yang CH. Structural and functional implications of C-terminal regions of alpha-synuclein. Biochemistry. 2002;41:13782–90. doi: 10.1021/bi026284c. [DOI] [PubMed] [Google Scholar]
- 22.Bertoncini CW, Rasia RM, Lamberto GR, Binolfi A, Zweckstetter M, Griesinger C, Fernandez CO. Structural Characterization of the Intrinsically Unfolded Protein beta-Synuclein, a Natural Negative Regulator of alpha-Synuclein Aggregation. J Mol Biol. 2007;372:708–722. doi: 10.1016/j.jmb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Sung YH, Eliezer D. Residual Structure, Backbone Dynamics, and Interactions within the Synuclein Family. J Mol Biol. 2007;372:689–707. doi: 10.1016/j.jmb.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertoncini CW, Fernandez CO, Griesinger C, Jovin TM, Zweckstetter M. Familial mutants of alpha-synuclein with increased neurotoxicity have a destabilized conformation. J Biol Chem. 2005;280:30649–52. doi: 10.1074/jbc.C500288200. [DOI] [PubMed] [Google Scholar]
- 26.Bussell R, Jr., Eliezer D. Residual structure and dynamics in Parkinson's disease-associated mutants of alpha-synuclein. J Biol Chem. 2001;276:45996–6003. doi: 10.1074/jbc.M106777200. [DOI] [PubMed] [Google Scholar]
- 27.Bussell R, Jr., Eliezer D. Effects of Parkinson's disease-linked mutations on the structure of lipid-associated alpha-synuclein. Biochemistry. 2004;43:4810–8. doi: 10.1021/bi036135+. [DOI] [PubMed] [Google Scholar]
- 28.Rospigliosi CC, McClendon S, Schmid AW, Ramlall TF, BarrÈ P, Lashuel HA, Eliezer D. E46K Parkinson's-Linked Mutation Enhances C-Terminal-to-N-Terminal Contacts in [alpha]-Synuclein. J Mol Biol. 2009;388:1022–1032. doi: 10.1016/j.jmb.2009.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu KP, Kim S, Fela DA, Baum J. Characterization of conformational and dynamic properties of natively unfolded human and mouse alpha-synuclein ensembles by NMR: implication for aggregation. J Mol Biol. 2008;378:1104–15. doi: 10.1016/j.jmb.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc Natl Acad Sci U S A. 2005;102:1430–5. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uversky VN, Li J, Fink AL. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J Biol Chem. 2001;276:10737–44. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 32.Dedmon MM, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson CM. Mapping long-range interactions in alpha-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J Am Chem Soc. 2005;127:476–7. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 33.Bisaglia M, Mammi S, Bubacco L. Structural insights on physiological functions and pathological effects of {alpha}-synuclein. FASEB J. 2009;23:329–340. doi: 10.1096/fj.08-119784. [DOI] [PubMed] [Google Scholar]
- 34.Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr. Opinion Struct. Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Eliezer D. Biophysical characterization of intrinsically disordered proteins. Current Opinion in Structural Biology. 2009;19:23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bracken C, Iakoucheva LM, Romero PR, Dunker AK. Combining prediction, computation and experiment for the characterization of protein disorder. Curr Opin Struct Biol. 2004;14:570–6. doi: 10.1016/j.sbi.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Wright PE, Dyson HJ. Linking folding and binding. Current Opinion in Structural Biology. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitalis A, Wang X, Pappu RV. Quantitative Characterization of Intrinsic Disorder in Polyglutamine: Insights from Analysis Based on Polymer Theories. Biophysical Journal. 2007;93:1923–1937. doi: 10.1529/biophysj.107.110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khandogin J, Brooks CL., III Linking folding with aggregation in Alzheimer's β -amyloid peptides. Proc. Natl. Acad. Sci. USA. 2007;104:16880–16885. doi: 10.1073/pnas.0703832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baumketner A, Bernstein SL, Wyttenbach T, Bitan G, Teplow DB, Bowers MT, Shea J-E. Amyloid β -protein monomer structure: A computational and experimental study. Protein Science. 2006;15:420–428. doi: 10.1110/ps.051762406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panchal SC, Bhavesh NS, Hosur RV. Improved 3D triple resonance experiments, HNN and HN(C)N, for HN and 15N sequential correlations in (13C, 15N) labeled proteins: application to unfolded proteins. J Biomol NMR. 2001;20:135–47. doi: 10.1023/a:1011239023422. [DOI] [PubMed] [Google Scholar]
- 42.Mohana-Borges R, Goto NK, Kroon GJ, Dyson HJ, Wright PE. Structural characterization of unfolded states of apomyoglobin using residual dipolar couplings. J Mol Biol. 2004;340:1131–42. doi: 10.1016/j.jmb.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Kristjansdottir S, Lindorff-Larsen K, Fieber W, Dobson CM, Vendruscolo M, Poulsen FM. Formation of native and non-native interactions in ensembles of denatured ACBP molecules from paramagnetic relaxation enhancement studies. J. Mol. Biol. 2005;347:1053–1062. doi: 10.1016/j.jmb.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Meier S, Blackledge M, Grzesiek S. Conformational distributions of unfolded polypeptides from novel NMR techniques. J Chem Phys. 2008;128:052204. doi: 10.1063/1.2838167. [DOI] [PubMed] [Google Scholar]
- 45.Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–31. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 46.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–80. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 47.Frishman D, Argos P. Knowledge based protein secondary structure assignment. PROTEINS. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 48.Johnson S. Hierarchical clustering schemes. Psychometrika. 1967;32:241–254. doi: 10.1007/BF02289588. [DOI] [PubMed] [Google Scholar]
- 49.Ganguly D, Chen J. Structural Interpretation of Paramagnetic Relaxation Enhancement-Derived Distances for Disordered Protein States. J Mol Biol. doi: 10.1016/j.jmb.2009.05.019. In Press. http://dx.doi.org/10.1016/j.jmb.2009.05.019. [DOI] [PubMed]
- 50.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–27. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 51.Hoyer W, Cherny D, Subramaniam V, Jovin TM. Impact of the acidic C-terminal region comprising amino acids 109−140 on alpha-synuclein aggregation in vitro. Biochemistry. 2004;43:16233–42. doi: 10.1021/bi048453u. [DOI] [PubMed] [Google Scholar]
- 52.Rivers RC, Kumita JR, Tartaglia GG, Dedmon MM, Pawar A, Vendruscolo M, Dobson CM, Christodoulou J. Molecular determinants of the aggregation behavior of alpha- and beta-synuclein. Protein Sci. 2008;17:887–98. doi: 10.1110/ps.073181508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Binolfi A, Rasia RM, Bertoncini CW, Ceolin M, Zweckstetter M, Griesinger C, Jovin TM, Fernandez CO. Interaction of alpha-synuclein with divalent metal ions reveals key differences: a link between structure, binding specificity and fibrillation enhancement. J Am Chem Soc. 2006;128:9893–901. doi: 10.1021/ja0618649. [DOI] [PubMed] [Google Scholar]
- 54.Fernandez CO, Hoyer W, Zweckstetter M, Jares-Erijman EA, Subramaniam V, Griesinger C, Jovin TM. NMR of alpha-synuclein-polyamine complexes elucidates the mechanism and kinetics of induced aggregation. Embo J. 2004;23:2039–46. doi: 10.1038/sj.emboj.7600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 56.Goddard TD, Kneller DG. Sparky 3. University of California; San Francisco: [Google Scholar]
- 57.Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 58.Wishart DS, Case DA, Thomas L, James VDUS. Methods in Enzymology. Vol. 338. Academic Press; 2002. Use of chemical shifts in macromolecular structure determination. pp. 3–34. [DOI] [PubMed] [Google Scholar]
- 59.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 60.Schwarzinger S, Kroon GJ, Foss TR, Chung J, Wright PE, Dyson HJ. Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc. 2001;123:2970–8. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 61.Schwarzinger S, Kroon GJ, Foss TR, Wright PE, Dyson HJ. Random coil chemical shifts in acidic 8 M urea: implementation of random coil shift data in NMRView. J Biomol NMR. 2000;18:43–8. doi: 10.1023/a:1008386816521. [DOI] [PubMed] [Google Scholar]
- 62.Li Y, Kim S, Brodsky B, Baum J. Identification of partially disordered peptide intermediates through residue-specific NMR diffusion measurements. J Am Chem Soc. 2005;127:10490–1. doi: 10.1021/ja052801d. [DOI] [PubMed] [Google Scholar]
- 63.Ruckert M, Otting G. Alignment of Biological Macromolecules in Novel Nonionic Liquid Crystalline Media for NMR Experiments. J Am Chem Soc. 2000;122:7793–7797. [Google Scholar]
- 64.Felts A, Harano Y, Gallichio E, Levy R. Free-energy surfaces of beta hairpin and alpha-helical peptides generated by Replica Exchange Molecular Dynamics with the AGBNP implicit solvent model. Proteins: Struct., Funct., Bioinf. 2004;56:310–321. doi: 10.1002/prot.20104. [DOI] [PubMed] [Google Scholar]
- 65.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chemical Physics Letters. 1999;314:141–151. [Google Scholar]
- 66.Banks JL, Beard HS, Cao Y, Cho AE, Damm W, Farid R, Felts AK, Halgren TA, Mainz DT, Maple JR, Murphy R, Philipp DM, Repasky MP, Zhang LY, Berne BJ, Friesner RA, Gallichio E, Levy RM. Integrated Modeling Program, Applied Chemical Theory (IMPACT). Journal of Computational Chemistry. 2005;26:1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gallicchio E, Levy RM. AGBNP: An analytic implicit solvent model suitable for molecular dynamics and high-resolution modeling. Journal of Computational Chemistry. 2004;25:479–499. doi: 10.1002/jcc.10400. [DOI] [PubMed] [Google Scholar]
- 68.Kaminsky G, Friesner R, Tirado-Rives J, Jorgensen W. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem B. 2001;105:6474–6487. [Google Scholar]
- 69.GarcÌa de la Torre J, Huertas ML, Carrasco B. Calculation of Hydrodynamic Properties of Globular Proteins from Their Atomic-Level Structure. 2000. pp. 719–730. [DOI] [PMC free article] [PubMed]
- 70.Neal S, Nip AM, Zhang H, Wishart DS. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J Biomol NMR. 2003;26:215–40. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 71.Sridharan S, Nicholls A, Honig B. A new vertex algorithm to calculate solvent accessible surface areas. Biophys. J. 1992;61:A194. [Google Scholar]
- 72.Cowan R, Whittaker R. Hydrophobicity indices for amino acid residues as determined by high-performance liquid chromatography. Pept. Res. 1990;3:75–80. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.