Abstract
PTB (polypyrimidine tract binding protein) participates in cellular regulatory functions in the nucleus and the cytoplasm. It binds to internal ribosome entry sites to facilitate their use in cap-independent translation. It binds to polypyrimidine tracts in pre-mRNA introns to repress inclusion of exons. It binds to the 3′ untranslated regions of mRNAs to stabilize the message. These RNAs have various structures, yet PTB binds to all of them. Here, RNAs with structured or unstructured polypyrimidine tracts are bound to the full length PTB1 protein and two protein sub-domains, each containing two RNA Recognition Motifs. Hairpin loops from c-src and GABAA γ2 pre-mRNAs and from the 3′ terminus of Hepatitis C Virus (HCV) were compared to a single-stranded polypyrimidine tract from GABAA γ2 pre-mRNA. We conclude that PTB1 RNA binding function is modular: the N-terminal RRMs preferentially bind to short (U/C) tracts displayed in loops, while RRM3/RRM4 preferentially binds to longer flexible RNA sequences. Since it can bind to short and long polypyrimidine tracts, structured or single-stranded, PTB takes on the role of a versatile adaptor protein that facilitates formation of RNA:protein regulatory complexes.
Polypyrimidine tract binding (PTB) protein has many cellular functions, and a corresponding variety of RNA targets (1,2). In its role as a translational enhancer protein, PTB binds to Internal Ribosome Entry Sites (IRES) of poliovirus, Encephalomyocarditis virus (EMCV) and some mammalian genes. IRES elements are highly structured, and the PTB binding sites are typically found in loops at the end of stems. When PTB functions as a splicing regulator, its binding sites in a pre-mRNA can be single-stranded (e.g. GABAA γ2 intron/exon) or structured (e.g. c-src intron/N1 exon/intron). When PTB binds at the 3′ untranslated region (UTR) of an mRNA in the cytoplasm, it is thought to stabilize the message; the RNA secondary structures of these regions are not known, but will certainly contain different structural elements. In each of its cellular roles, the RNA binding sites for PTB are U/C-rich, but their structural contexts are diverse.
The domain structure of PTB contributes to its RNA binding specificity. Each of the three human PTB proteins (PTB1, PTB2, and PTB4) contains four RNA Recognition Motifs (RRM) (3,4). RRMs (also known as RNA Binding Domains, or RBDs) are globular domains of about 90 amino acids, having a βαββαβ secondary structure and an α/β sandwich fold. These proteins differ only in the alternatively spliced linker sequence between RRM2 and RRM3. Human neuronal PTB (nPTB) differs in RRM sequences, although there is high identity among all PTBs (5).
Three features of PTB proteins are particularly pertinent to their RNA binding properties. First, the sequences of the RRMs are unusual, for these domains have noncanonical RNP1 and RNP2 sequences that typically define an RRM. RNP sequences include solvent-exposed aromatic amino acids that stack with bound RNA nucleobases, and these amino acids are mostly absent in the PTB RRMs. Second, the secondary and tertiary structures of RRM2 (8) and RRM3 (6,7) are unusual, for they each have an extra (fifth) β strand, connected by a long loop to β4 to form a five-stranded antiparallel β-sheet. This loop sits across the face of the β-sheet, and so occupies what is usually the RNA binding surface. Third, RRM1 and RRM2 are tethered by an ~30 amino acid linker (6), RRM2 and RRM3 are separated by a long and apparently flexible linker, and the twenty amino acid linker separating RRM3 and RRM4 forms part of their protein:protein interface (7). Assuming that RNA binds to the β-sheet surface of each RRM (8,9), the display of those surfaces is dramatically different in each RRM, and so could determine the sequence, length, and structural context of their preferred RNA targets.
The contribution of each RRM to RNA binding has been examined in several studies, and the conclusions have varied. For example, Perez et al., (10) concluded that PTB1 could bind to polynucleotides poly(rU), poly(rA), and poly(rC). PTB is also associated with viral and cellular IRES structures (11,12). The protein has been most efficiently crosslinked to a 5 nucleotide loop of StemLoop H in the EMCV IRES, which has the sequence UCUUU (13,14). Willis’ work on cellular IRES sequences shows that PTB is a required component of active translation from these sites (14). Lui et al. (15) used PTB1 without RRM4 in pre-mRNA splicing reactions with GABAA γ2 pre-mRNA, and concluded that binding was unperturbed but splicing regulation was lost. The Black lab has extensively studied regulation of neuronal c-src pre-mRNA by PTB and nPTB (5, 16–19). They conclude that it has several means to effect exon repression, including binding upstream and downstream of an excluded exon. That observation supports the model of exon looping out by PTB (20). Smith and colleagues have also compared PTB and nPTB repression (21) and the functions of PTB1 RRMs (22). Auweter et al., (9) have shown that RNA hexamers weakly associate with each RRM. PTB also binds to the 3′ UTR of mRNAs in the cytoplasm, where it has been linked to control of mRNA stability (23). The different structural presentations of polypyrimidine tracts confuse formulation of a consistent description of the PTB binding mechanism.
Here we compare PTB1 binding to pyrimidine sequences displayed in the context of structured RNAs with PTB1 binding to a single-stranded polypyrimidine tract. We conclude that the RRMs do have preferred target structures: PTB1:12 (containing RRM1 and RRM2) binds to short (U/C) tracts, but PTB1:34 (containing RRM3 and RRM4) does not (or only very weakly) and instead preferentially binds to longer single-stranded (U/C) tracts. A division of labor based on RNA structures helps to explain how PTB is capable of binding pyrimidine sequences in many contexts.
Experimental Procedures
Cloning
The human PTB1 gene cloned in pET 28A vector was a generous gift from Professor Doug Black (UCLA), and has an N-terminal his6, and an N-terminal extension that includes a thrombin cleavage site (MGSSHHHHHHSSGLVPRGSHMASMTGGQQMGRGSEFPRGSLPCAM1…) where M1 is the natural start site of the protein. A second PTB1 construct lacks the sequence between his6 and M1; the RNA binding properties of the two constructs are indistinguishable (data not shown). The starting sequence of PTB1:12 is identical to that of PTB1; it terminates at Val301. PTB1:34 begins at amino acid [Met334]G335Asn336 and ends at Ile531 (PTB1 numbering (3)). Proteins were overexpressed in E coli BL21 DE3 cells under control of the tac promoter. Tryptophan residues were introduced into proteins using standard site directed mutagenesis protocols in Quick-Change (Stratagene) and DNA constructs were sequenced to verify the substitution. Proteins include F98W on β2 in RRM1, F371W in the loop between β2 and β3 in RRM3, and F487W on β2 in RRM4.
Protein purification
Individual clones were grown overnight at 37 °C in LB/Kanamycin (50 µg/ml) followed by 50 fold dilution into terrific broth and grown at 37° C to an OD600 of 0.7. IPTG was added to a final concentration of 1 mM and growth continued for 4 hours. Cells were pelleted by centrifugation at 6,500 rpm for 15 min and frozen at − 80° C. The cell pellet was resuspended in 3 volumes of buffer A (50 mM sodium phosphate, 300 mM NaCl and 10% glycerol, pH 8). For lysis, cells were incubated at 4° C for 30 min with 5 mg/ml of lysozyme, 1 mM PMSF, 5 µg/ml Pepstatin and protease inhibitor cocktail (Sigma P2714) containing 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), trans-epoxysuccinyl-L-leucylamino(4-guanidino)butane(E-64), bestatin, leupeptin, aprotinin and EDTA. Cells were then pulse-sonicated for 30 s five times with cooling between. Cellular debris was removed by centrifugation at 10,000 ×g for 30 min at 4° C. The supernatant of His-tagged proteins was mixed with the pre-equilibrated Ni-NTA resin (Qiagen) in buffer A and rocked at 175 rpm for 1.5 hours on ice. The resin was then washed 4 times for 10 min each with 25 ml of buffer A plus 20 mM imidazole followed by elution of the protein with a gradient of imidazole (0 mM–250 mM). Peak fractions were collected, concentrated and exchanged into storage buffer (20 mM sodium cacodylate, 100 mM KCl, 0.5 mM DTT, 0.5 mM EDTA, 10% glycerol) using a Vivaspin with the appropriate molecular weight cut off. Proteins without the his-tag were purified by column chromatography as described (24).
Protein purifications were assessed by SDS PAGE. Protein concentrations were determined by the absorption spectrum, using ε280 = 12800 cm−1 M−1 (without Trp) or 18400 cm−1 M−1 (with Trp) according to the method of Gill & von Hippel (25). Protein secondary structure was measured by CD spectroscopy.
RNA transcription and purification
Most RNAs were transcribed from oligodeoxynucleotides, using the method of Milligan et al., (26). Oligonucleotides shown are the promoter DNA strand, with the promoter sequence in italics.
c-src pre m-RNA Exon NI:
5′-TTAATACGACTCACTATAGGGATCGGTGCCATCGATGTCAGGTGTGTACCCCTTAGGAGG3′
c-src pre m-RNA 39:
5′ – TTAATACGACTCACTATAGGATCCTTCTCTCTGCTTCTCTCTCGCTGGCCCTTAGGAGG-3′
c-src pre m-RNA 78:
5′ –TTAATACGACTCACTATAGGCAACCCTCTCCTTCTCTCTGCTTCTCTCTCGCTGGCCCTTA GGAGGAAGGTGCCATCGATGTCAGGTGTGTACCGAGG-3′
c-src pre m-RNA 3′ end (synthesized from PCR oligonucleotides):
5′ –TTAATACGACTCACTATAGGGATCGCTTCGCTGAGGCTGGGGGCTGCTCTCTGCATGTGCTTCCTCCACCGCCCCTGTGTGTTTCCAGCTCTCTCCCCGTCCCTTTAGCTTACCCTGCATCCCACCTGTATGAGCCGTACCC-3′
GABAA pre-mRNA Exon 4:
5′ – TTAATACGACTCACTATAGGGCTTCTTCGGATGTTTTCCTTCAAG-3′
HCV 3′ NTR X-SL2:
5′ – TTAATACGACTCACTATAGGGTCACGGCTAGCTGTGAAAGGTCCGTGAG-3′
HCV 3′ NTR X-SL3:
5′ - TTAATACGACTCACTATAGGAGACTTGGTGGCTCCATCTTAGCCCTA -3′
GABA RNA11
5′- TTAATACGACTCACTATAGGGACTACGCAATTCTCTTTTCTGTCTACAAATCCAAAG-3′
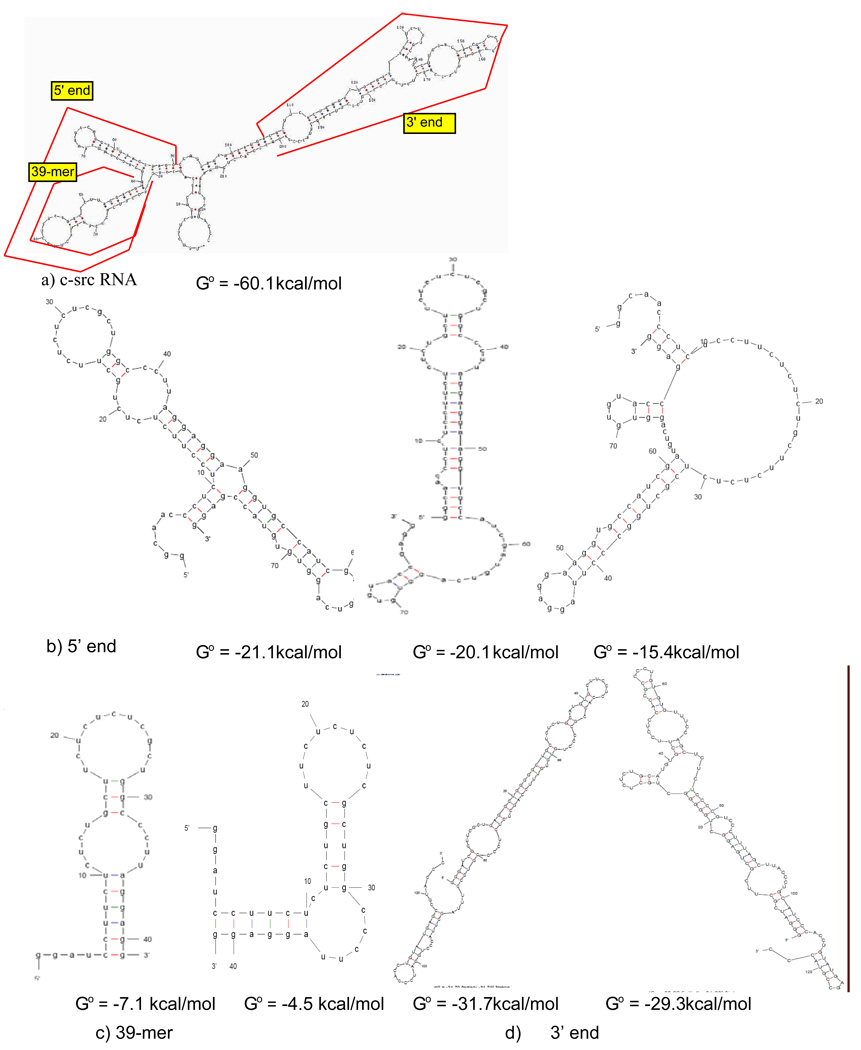
The 224 nt c-src intron/exon/intron (Figure 2) was transcribed from a PCR product into which a GGGACT start site was introduced at the 5’end. The locations of the sub-fragments are indicated on the predicted structure. In vitro transcription reactions used T7 RNA polymerase in 40 mM Tris, pH 8.0, 1 mM spermidine, 6 mM MgCl2. After phenol extraction and ethanol precipitation, RNAs were purified from denaturing polyacrylamide gels. The RNA was eluted from the gel by incubation in 0.3 M sodium acetate (pH 5.3) at 37 °C overnight. For large-scale transcriptions, RNA concentrations were determined spectrophotometrically using the approximation of 1 A260 = 40 µg/ml of RNA.
Figure 2.
Sequence and predicted structures of c-src RNAs that bind to PTB1. Structure predictions and folding free energies are by mfold (28,29).
Gel mobility shift assays (EMSA)
Electrophoretic mobility shift assays (EMSA) were typically used to quantify binding, since the stoichiometry of the association was not known. [α-32P]RNA in 10 mM KCl was heated to 95° C for three minutes and quenched on ice; yeast tRNA (Boehinger) was added to 10 µg/mL and the solution added to protein (typically 1 nM to 5 µM) in binding buffer (10 mM sodium cacodylate pH 7.5, 1 mM MgCl2, 100–250 mM NaCl, 20 µg/mL BSA). After binding at room temperature for twenty minutes, glycerol was added and the samples loaded on 8% polyacrylamide gels, 40:1 acrylamide:bis in 50 mM Tris/50 mM Glycine pH 8.3 and run in the cold at 8 V/cm for 3–4 hours. Experiments were repeated at least twice. Data were analyzed with a phosphorimager, and the fraction RNA bound was entered into the expression for a partition function (P) for association using Origin 7.5 (OriginLab, Northampton, MA). P = 1 + (Ki[P])ni + (KiKj[P]2)nj … where [P] is the known protein concentration, and the equilibrium association constants (Ki) and the Hill coefficients (ni) were allowed to float. Where only one complex is observed, there is only one ni = n1 = n; where there are two complexes, ni = n1, nj = n2, etc. The Hill coefficients are typically interpreted in terms of cooperativity, but because the binding stoichiometry within each complex observed in EMSA experiments is not known, the physical significance of ni is not clear.
Filter binding stoichiometry
Non-radioactive RNA was mixed with trace radioactive RNA to a final concentration of 500 nM to 1 µM RNA. The RNA was folded in solution by heating in water to 95° C, quenching on ice, then addition of KCl to a final concentration of 10 mM and yeast tRNA to a final 10 µg/mL tRNA. Increasing amounts of recombinant PTB1 proteins were incubated with the fixed concentration of RNA in 20 mM sodium cacodylate, 100 mM NaCl, 1 mM MgCl2, 20 µg/mL BSA, pH 7.5. The reaction was incubated at room temperature for 30 minutes in a polypropylene microtitre plate (96 wells). Reactions were filtered through presoaked Schleicher and Schuell 0.45 µm nitrocellulose filters, and read on a phosphorimager. Data were plotted as the bound RNA vs [protein]/[RNA]; the break point of the plot indicates the stoichiometry. Both components must be present at concentrations above the KD for this experiment to be accurate, so stoichiometry experiments use micromolar concentrations of RNA and protein.
Fluorescence measurements
Steady state tryptophan fluorescence was measured in the L-format with an SLM 8100 spectrofluorometer. Steady-state anisotropy values were calculated using the expression r = (I║ −GI ⊥)/(I║ + 2G I⊥), where I║ and I⊥ are the parallel and perpendicular intensities of the polarized fluorescence, G is the instrument correction factor (G = 1 for our instrument). For tryptophan, the excitation wavelength was 300 nm with a bandwidth of 2 nm and an emission bandwidth of 4 nm. Polarized fluorescence was detected at the emission maximum of each tryptophan.
Time-resolved fluorescence was measured using a home-built time correlated single photon counting instrument. The Coherent Titanium-sapphire laser was tuned to 900 nm, and the output light was tripled to 300 nm with a UOplaz tripler, and a Neos pulse picker was adjusted to a 7.6 MHz repetition rate. The instrument response function of the PMT was 150 ps fwhh. Tryptophan fluorescence decays were processed using FluoFit (PicoQuant). The goodness of fit was estimated by the shape and boundaries of the support planes of each parameter and the value of χ2 r.
For time-resolved anisotropy decays, the emission polarizer was alternated between the vertical (parallel) and horizontal (perpendicular) position with respect to the incident vertically polarized light. For each orientation, the polarization components of the decays were collected for equal time. Anisotropy decays curves were calculated from the equation:
where vv(t) is the parallel component and vh(t) is the perpendicular component. The G factor was calculated by tail matching in the Fluofit software. The anisotropy decays were described by two exponential decays as a function of time:
where βf is the amplitude associated with the fast anisotropy component Φf and βs is the amplitude associated with the slow anisotropy component Φs.
RESULTS
The PTB proteins
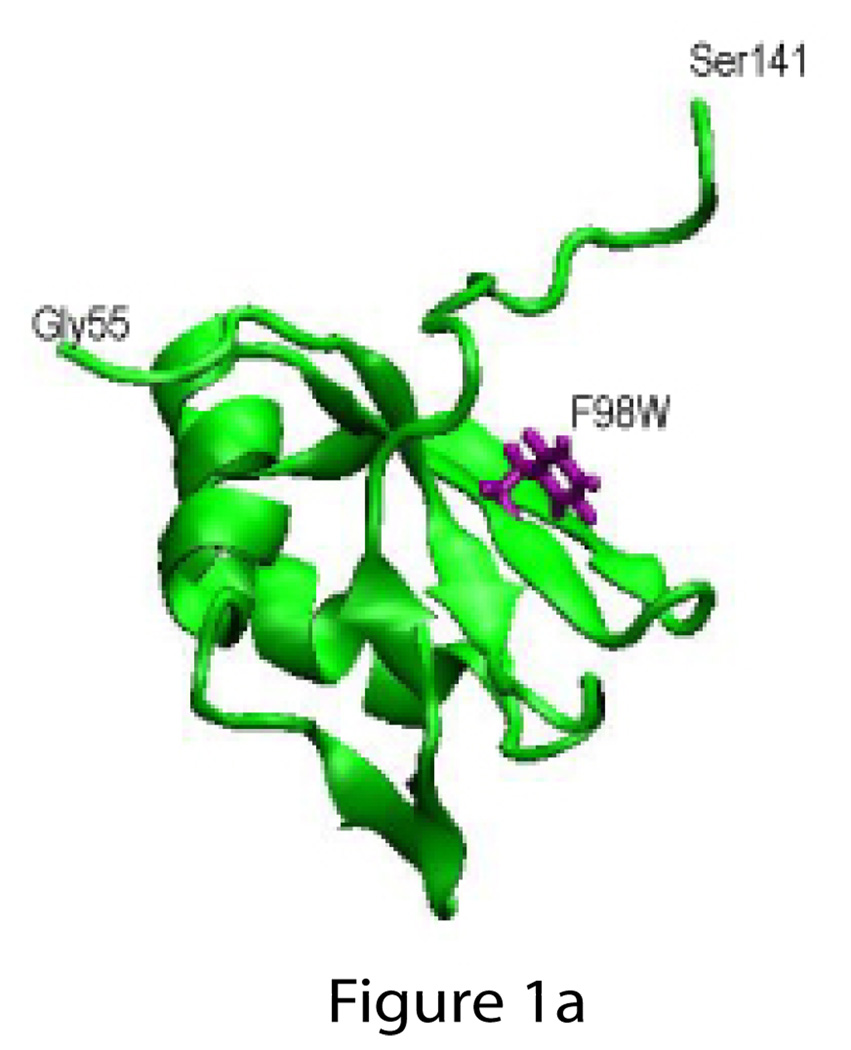
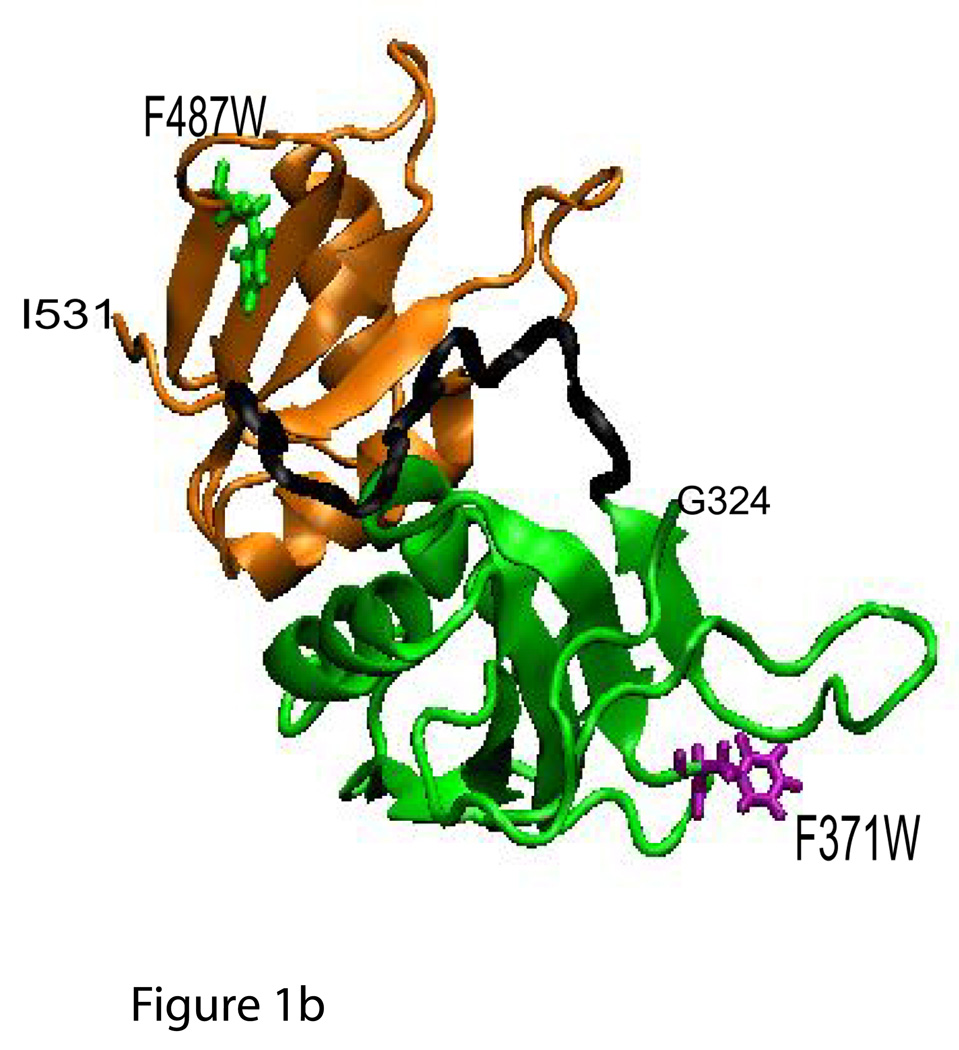
Full length PTB1 includes all four RRMs, and three mutant PTB1 constructs contain single tryptophan substitutions for aromatic residues. PTB1/W1 has W98 in RRM1, PTB1/W3 has W371 in RRM3, and PTB1/W4 has W487 in RRM4. These positions within the RRMs are shown in Figure 1. Three other sub-clones were also constructed: RRM1-W is the first RRM with the F98W substitution; PTB1:12 contains the first two RRMs of the protein (residues 1– 301), and PTB1:34 contains RRM3 and RRM4 (residues 334–531).
Figure 1.
a) Structures of PTB1 RRM1 (1sjq; Simpson et al., 2004) and b) RRM3/RRM4 complex (2evz; Vitali et al., 2006) showing the position of the tryptophan substitutions F98W in RRM1, F371W in RRM3, and F487W in RRM4.
The RNAs
For these experiments, six RNAs from known targets of PTB were bound to PTB1 and its constructs. Hepatitis C Virus (HCV) X RNA contains three stemloops and was subdivided into HCV X SL2 and SL3. Other RNAs come from c-src pre-mRNA (17) and the GABAA γ2 pre-mRNA exon 4 (27). Structures were predicted by mfold (28,29), and the lowest energy folds are illustrated.
RNAs not bound
PTB1 binds to the GABAA γ2 (GABA) pre-mRNA and excludes exon 4 during alternative splicing (27). In the context of an in vitro GABA intron/exon construct, exon 4 region is structured, while the upstream intron is unstructured (24). A short 27 nucleotide transcript of the exon 4 sequence is predicted to adopt several folds, each of which displays a UUCG tetraloop. Of the predicted GABA exon 4 structures, three are shown (Supplemental Figure 1). This GABA exon 4 RNA was not bound by PTB1, PTB1:12, or PTB1:34 in EMSA experiments.
PTB1 binds c-src pre-mRNA to exclude the short 24 nucleotide exon N1 (16). Unlike the GABA intron RNA, the c-src intron/exon construct is predicted to have more stable structures, and a transcript of the N1 exon region is predicted to adopt several structures that display two or three U/C sequences in bulges or hairpin loops (Supplemental Figure 1). This short 32 nucleotide RNA containing the N1 exon sequence was not bound by any PTB1 construct in EMSA experiments.
PTB binding to the 3′ end of the HCV genome is part of the viral replication machinery (30). The terminus is highly structured, with three hairpin loops comprising the 98 nucleotide X domain (31, 32, 24). HCV X SL2 has only two or three contiguous U/C bases, and an SL2 RNA synthesized here is predicted to adopt several nearly isoenergetic structures (Supplemental Figure 1). It does not bind to the PTB1 proteins.
EMSA experiments were run in salt concentrations as low as 50 mM KCl to favor electrostatic interactions, but neither HCV X SL2, c-src N1, nor GABA exon 4 RNAs were bound by PTB1 in these conditions. We conclude that these RNAs alone are not functional targets of PTB. However, in the context of the complete RNA sequence, these sites could be associated with the protein as a result of upstream or downstream polypyrimidine tracts that are high affinity binding sites. In fact, the GABA exon 4 is protected from nuclease digestion when the intron/exon4/intron fragment is bound to PTB1 (15, 24), as is SL2 of HCV X RNA (24). Alternatively, when flanking sequences are present the RNAs could adopt another structure that allows PTB access to their short (U/C) sequences.
Bound RNAs
c-src RNAs
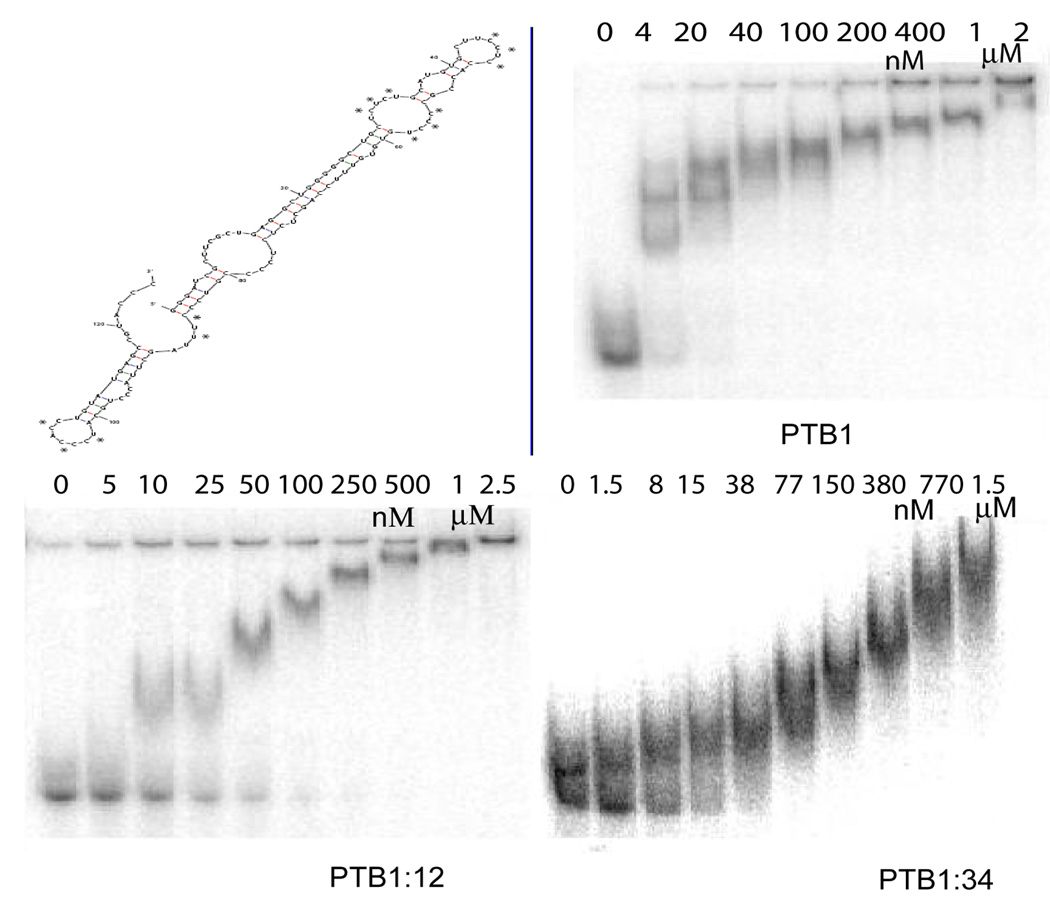
Predicted secondary structures of the c-src N1 intron/exon region suggest that this RNA can form very stable stemloops (Figure 2). To separate the upstream and downstream introns, which are both reported binding sites of PTB (18), the RNA was divided into segments: a 39 nucleotide 5′ intron sequence that flanks the N1 exon, a 78 nucleotide 5′ RNA that includes N1, and a 124 nucleotide portion of the 3′ intron. The smaller RNAs are mapped onto the predicted fold of the largest c-src construct in Figure 2.
The 5′ intron sequence including the N1 exon was predicted to have alternative folds (Figure 2) and nuclease probing data are consistent with one of them. Footprinting experiments of this 78 nucleotide RNA (RNA78) bound to 10 nM PTB1 show little change in the accessibility of the RNA to the nucleases, but at 1 µM PTB1, one large pyrimidine tract of RNA78 shows reduced cleavage (Supplementary Figure 2). Notably, there is no change in the overall pattern of cleavage with/without PTB, indicating that PTB has not remodeled the RNA structure. Stoichiometry experiments at 1 µM RNA show that the largest complex includes three PTB1 proteins bound to the 78-mer RNA (Figure 3), thus the footprinting experiments using 1 µM PTB1 correspond to saturating conditions. EMSA experiments give three distinct bands as PTB1 is titrated into RNA78: the first complex has an apparent affinity of 80 nM, the second 110 nM, and the third 450 nM (Figure 3).
Figure 3.
Protein binding to c-src 78-mer 5′ RNA. top: PTB1 + 78-mer 5′ RNA. Stoichiometry experiment at 1 µM RNA ♦ 39-mer 5′ RNA. ○ 78-mer 5′ + N1 RNA. 100 mM NaCl, 10 mM sodium cacodylate pH 7.5, 1 mM MgCl2, 10 mM DTT, 22° C. EMSA with PTB1 and 78-mer 5′ RNA. bottom: EMSA of PTB1:12 and PTB1:34 + 78-mer 5′ RNA. Gel mobility shift experiments at 4° C in 100 mM NaCl, 10 mM sodium cacodylate pH 7.5, 1 mM MgCl2, 10 mM DTT, 10 µg/mL tRNA, 20 µg/mL BSA.
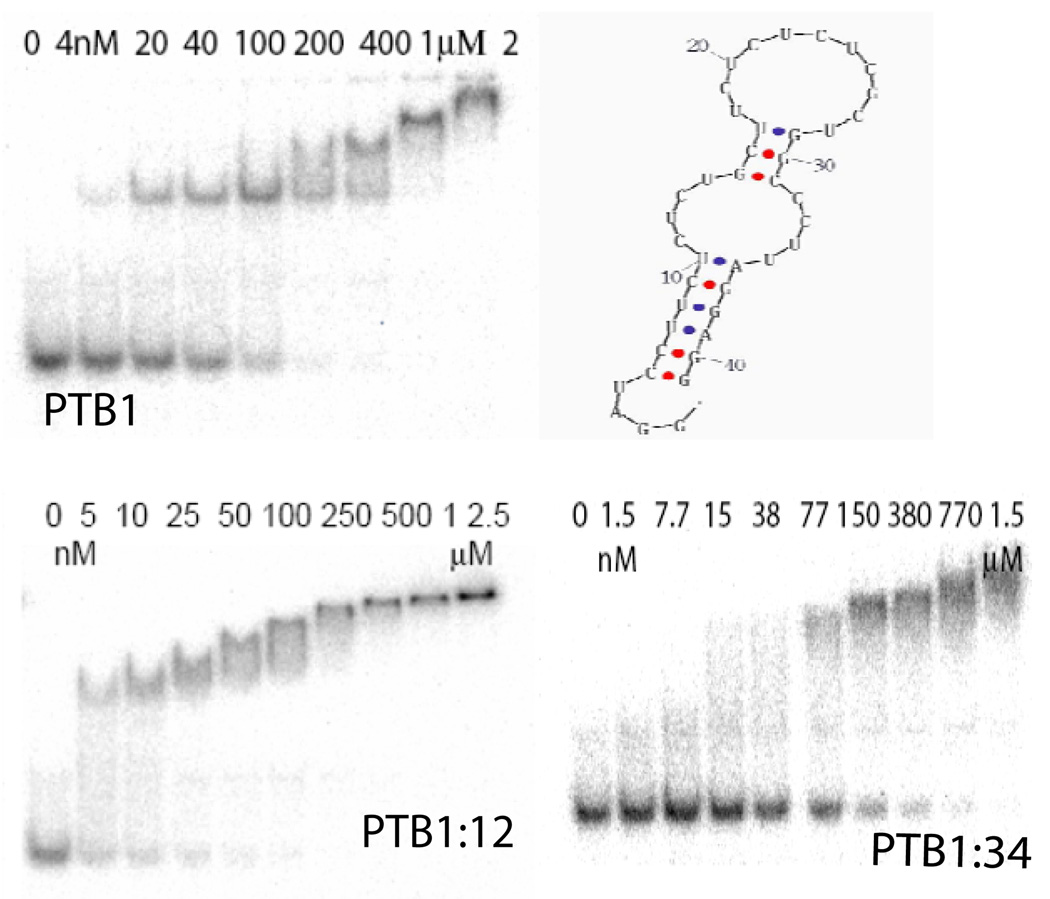
A shorter 39 nucleotide portion of the 5′ intron without the N1 sequences is predicted to display eight (U/C) nucleotides in a hairpin loop, as well as four on either side of an unpaired bulge (Figure 2). The most stable predicted structure has a folding free energy of −7.1 kcal/mol; alternative folds are less stable by at least 3 kcal/mol, and most contain the hairpin loop and bulged pyrimidines. EMSA results shows that PTB1 forms two complexes on this short 5′ intron RNA, the first with an affinity ~45 nM, and the second ~150 nM (Figure 4). Stoichiometry experiments at 1 µM RNA indicate three bound proteins (Figure 3) but whether all are in direct contact with RNA is not known.
Figure 4.
Protein binding to c-src 39-mer 5′ RNA. top: PTB1 EMSA experiment and most stable predicted structure of RNA. bottom: PTB1:12 and PTB1:34 EMSA. 4° C in 100 mM NaCl, 10 mM sodium cacodylate pH 7.5, 1 mM MgCl2, 10 mM DTT, 10 µg/mL tRNA, 20 µg/mL BSA.
The 3′ flanking intron sequence is bound by 4 – 5 PTB1 proteins at 1 µM RNA, and the first complex is formed at 4 nM PTB1 in EMSA experiments (Figure 5). Nuclease probing of this RNA indicates there are single-stranded polypyrimidine tracts, consistent with the structure shown in Supplementary Figure 2, but that much of the RNA is double-stranded. The longest internal bulge of polypyrimidines appears to be about 8 nucleotides. Footprinting experiments (Supplementary Figure 2) show that binding of PTB1 protects most of these sites, although the sequences between G81 and G90 remain somewhat sensitive to RNase A.
Figure 5.
Protein binding to c-src 3′ RNA. RNA structure consistent with nuclease probing. EMSA experiments at 4° C in 100 mM NaCl, 10 mM sodium cacodylate pH 7.5, 1 mM MgCl2, 10 mM DTT, 10 µg/mL tRNA, 20 µg/mL BSA.
X RNA hairpins
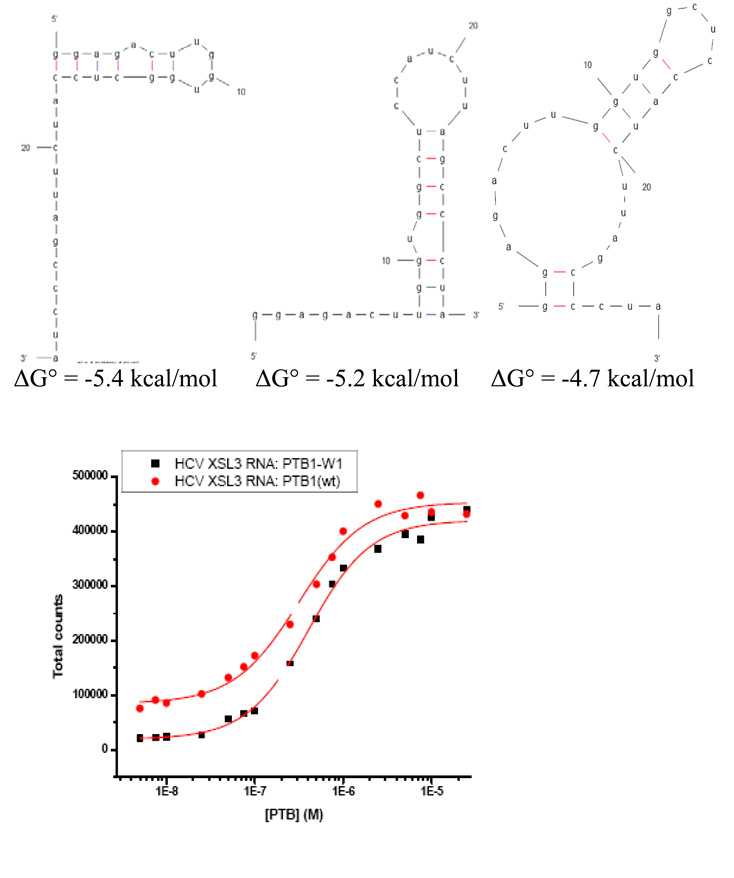
PTB1 binds to the 98 nucleotide X RNA, forming several complexes in EMSA experiments that are fit by two dissociation constants, and a final stoichiometry of two proteins/RNA (24). PTB1 binds to HCV X SL3 RNA with KD = 285 ± 120 nM in 100 mM NaCl calculated from EMSA (data not shown), 410 ± 30 nM in nitrocellulose filter binding experiments (Figure 6) and a stoichiometry of 1:1.
Figure 6.
HCV X SL3 binding. Three lowest energy predicted RNA structures and nitrocellulose filter binding experiment with PTB1 and PTB1/W1 proteins. Binding data are fit by a 1:1 complex with KD(WT) = 4.1 × 10−7 ± 3 × 10−8 M and KD(W1) = 3.1 × 10−7 ± 4 × 10−8 M. Binding in 100 mM NaCl, 10 mM sodium cacodylate pH 7.5, 1 mM MgCl2, 10 mM DTT, 10 µg/mL tRNA, 20 µg/mL BSA. HCV X SL3 RNA is bound by PTB1:12, so these experiments are important to measure the effect of tryptophan substitution in RRM1.
Separation of binding functions
PTB proteins can be separated into two parts via to the long linker that separates RRM2 and RRM3. For these experiments, PTB1:12 (RRM1 and RRM2) terminates at amino acid 301 and PTB1:34 (RRM3 and RRM4) begins with residue 334. Linker sequences from 301 to 334 are not included in either construct. Within the two domains, the RRM domains are displayed in very different geometries. RRM1 and RRM2 are separated by a 30 amino acid linker, and appear to be independent of each other as measured in sedimentation equilibrium experiments and in NMR (8). In contrast, RRM3 and RRM4 interact with each other, using their ~20 amino acid linker as part of the interface between the two domains (7). The RNA binding modes of these two parts of PTB are predicted to be very different, and as the data in Table 1 show, there is clearly a preference in their RNA targets.
Table 1.
Binding and stoichiometry data for PTB1:RNA complexesa
| RNA | PTB1 | PTB1:12 | PTB1:34 |
|---|---|---|---|
| HCV X§ | [P]/[R] = 2 | [P]/[R] = 2 | No binding |
| KD1 = 800 nM, n=1 | KD1 = 800 nM, n=1 | ||
| KD2 = 1 µM, n=1 | KD2 = 1 µM, n=1 | ||
| HCV X SL3 | [P]/[R] = 1 | [P]/[R] = 1 | No binding |
| KD=285 ± 120 nM. | KD = 1.6 ± 0.6 µM | ||
| GABA RNA11 | [P]/[R] = 2 | No binding | [P]/[R] = 4 ± 1 |
| KD= 15 nM ± 2 nM, | KD= 50 nM ± 3 nM | ||
| n=1.5 | n=3 | ||
| GABA exon4 | No binding | No binding | No binding |
| c-src exon N1 | No binding | No binding | No binding |
| c-src 5′ SL | [P]/[R] = 3 ± 1 | Binding | Binding |
| KD1= 45 nM ± 2, n=1.5 | |||
| KD2= 150 nM ± 7, n=1.5 | |||
| c-src 5′ SL + N1 | [P]/[R] = 3±1 | Binding | Binding |
| KD1= 80 ± 2 nM, n=1 | |||
| KD2 = 110 ± 6 nM, n=1 | |||
| KD3 = 450 ± 90 nM, n=2 | |||
| c-src 3′ end | Binding | Binding | Binding |
Binding in 100 mM NaCl, 10 mM sodium cacodylate, pH 7.5, (10 mM DTT for PTB1 and PTB1:12) 1 mM MgCl2.
HCV X data from Clerte & Hall (2006).
c-src
The 39 nucleotide 5′ end c-src RNA construct is bound by PTB1:12 and PTB1:34, but protein affinities differ by nearly an order of magnitude and the complexes show different patterns in EMSA experiments (Figure 4). PTB1:12 binding to the 5′ end c-src construct is first observed at 5 nM protein, and formation of a second shifted species is apparent at 25 nM protein. Curiously, PTB1:12 binding is tighter than that of PTB1 to c-src 5′ end RNA.
As is typical of PTB1:34 binding in EMSA experiments, the bands of the c-src 5′ RNA:protein complexes are diffuse (figure 4). PTB1:34 binding does not give a discernable band until nearly 80 nM protein, although the decrease in the intensity of the free RNA band indicates that some association occurs at lower protein concentrations.
The 78-mer [5′ end + N1] c-src RNA containing the N1 exon is designed to probe the influence of the N1 sequence in the context of the upstream intron RNA sequences. On this RNA, PTB1:12 binds with significantly weaker affinity than does PTB1, and PTB1:34 binding is not observed until about 150 nM protein (Figure 3). The presence of the N1 sequence appears to decrease the affinity of all PTB1 constructs for this RNA relative to the shorter 5′ construct.
The 3′ intron RNA is bound by both PTB1:12 and PTB1:34, but the complexes have dramatically different mobility in EMSA experiments (Figure 5). No distinct bands are observed in the PTB1:34 experiment, although binding is first clearly detected at 15 nM protein. By contrast, binding of PTB1:12 produces a series of shifted bands that more closely resemble those of the full length protein, although its affinity for the RNA is weaker. Binding by PTB1:12 does not shift all the RNA into complexes until it exceeds 50 nM, whereas by 20 nM PTB1, all the RNA is shifted.
HCV X Stem-loop 3
Only one PTB1:12 protein binds detectably to this RNA stemloop, and even then, its affinity is weak (~1.6 µM; data not shown). This is a 5-fold loss of binding affinity compared to that of PTB1, suggesting that other regions of the protein participate. PTB1:34 binding was not detected by EMSA, even at 5 µM protein. It is possible that the linker between RRM2 and RRM3 could interact with RNA in the context of PTB1, and so account for its observed increase in binding affinity.
Fluorescent probes of hairpin binding
PTB1/W sites in solution
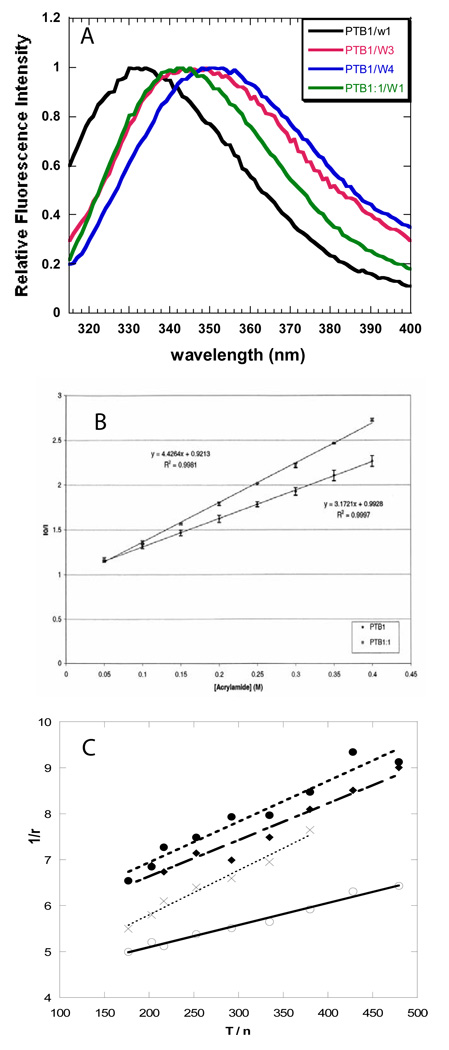
To hopefully identify the RRMs directly involved in PTB1 binding to RNAs, a single tryptophan residue was introduced into RRM1, RRM3, or RRM4 replacing a phenylalanine (Figure 1). The CD spectrum of each mutant PTB1 protein was identical to that of the wild type protein, with the exception of a small induced CD signal at the tryptophan wavelength. As shown in Figure 7, the fluorescence emission spectrum of each PTB1/W protein is distinctive as a result of different environments of each tryptophan sidechain. Also included in Figure 7 is the emission spectra of RRM1-W1 (the single domain expressed alone). The blue shift of PTB1/W1 fluorescence emission indicates that its environment is more nonpolar than in the free RRM1-W domain. Acrylamide quenching experiments indicate reduced solvent accessibility of W98 in PTB1 (Figure 7) and together these data suggest that the conformation of PTB1 leads to protection of the β-sheet surface of RRM1. Steady state fluorescence parameters for all proteins are summarized in Table 2.
Figure 7.
Fluorescence properties of PTB1/W proteins. A. Steady-state fluorescence emission spectra of PTB1/W (tryptophan) mutants. Excitation at 300 nm, 4 nm bandwidth. Intensity is normalized for comparison. B. Acrylamide quenching of W98 in PTB1/W1 and RRM1/W. C. Perrin plots of the steady-state anisotropy of the PTB1/W mutants and RRM1/W as a function of temperature (4° to 45° C). The viscosity of the buffer was determined by reference to that of water at a given temperature. The data were fit to the expression 1/r = 1/r0 + [(τ) / (r0θ)], where τ=Σαiτi2/Σαiτi . The limiting anisotropy for RRM1/W1 is r0(apparent) = 0.23 ; PTB1/W1 r0(apparent) = 0.24; PTB1/W3 r0(apparent) = 0.2 and PTB1/W4 r0(apparent) = 0.19. Protein concentrations were 4 µM in 10 mM sodium cacodylate pH 7.5, 100 mM NaCl. ▪▪▪X▪▪ PTB1/W1 y = 3.9 + 0097x, R = 0.988. ○ PTB1/W1, y = 4.14 + 0.0048x, R = 0.99. ♦ RRM1/W3, y = 5.05 + 0.0079x, R = 0.97. ▪▪●▪▪ PTB1/W4 y = 5.16 + 0.0089, R = 0.97.
Table 2.
Steady-state fluorescence properties of free and bound PTB/W proteinsa
| Protein | Fluorescence Emission λmax, |
Anisotropy 20 °C, 350 nm |
% quench |
|---|---|---|---|
| RRM1/W | 343 nm | 0.15 | |
| PTB1/W1 | 333 nm | 0.18 | |
| +GABA RNA11 | 338 nm | 25% (25%) | |
| + HCV X RNA | 338 nm | 80% | |
| + HCV X SL3 | Nd | 100% | |
| PTB1/W3 | 348 nm | 0.145 | |
| +GABA RNA11 | 348 nm | 60% (25%) | |
| + HCV X RNA | 348 nm | 20% | |
| + HCV X SL3 | 348 nm | 50% | |
| PTB1/W4 | 344 nm | 0.135 | |
| +GABA RNA11 | 344 nm | 40% (11%) | |
| + HCV X RNA | 344 nm | 20% | |
| + HCV X SL3 | 344 ns | 20% |
RNA concentration is 1 µM, protein concentration is adjusted to match stoichiometry (e.g. PTB1/W1 is 1 µM with HCV X SL3 and 2 µM with GABA RNA11 and HCV X). 100 mM NaCl, (250 mM) 10 mM sodium cacodylate pH 7.5, 10 mM DTT, room temperature.
To further characterize each tryptophan in the RRMs, their time-resolved fluorescence spectra were measured at 25° C. Tryptophan fluorescence is complex, due to its La and Lb absorption bands, and time-resolved spectra typically show several components. In each PTB1/W domain, the time-resolved fluorescence decays are best fit by three components, but the amplitudes and lifetimes are not identical (see Supplementary Table 1).
PTB1 properties in solution
SAXS experiments on PTB1 have described it as a relatively extended protein, the implication being that its RRMs can move independently (8,33). As another measure of the mobility of the individual RRMs and to estimate the protein hydrodynamics, the steady-state fluorescence anisotropy of the protein was measured as a function of temperature from 5° to 45° C. Data were fit to the Perrin Equation : 1/r = 1/r0 + [(τRT ) / ( r0Vη )] where r is the measured anisotropy of the protein, θR = (6 D)−1= (Vη) / (RT) is the global rotational correlation time of the protein, η the solution viscosity, τ is the intensity-weighted fluorescence lifetime and r0 is the limiting anisotropy. There are several assumptions in this formalism that directly apply to its use here for PTB1. The first is that the fluorophore has a single exponential fluorescence lifetime decay, which is not true for any of the three tryptophans in PTB1. Also, θR = (6 D)−1 is rigorously correct only for a spherical (globular) molecule, which PTB1 is certainly not. The latter limitation prevents any quantifiable description of the shape of PTB1 from these experiments, but it does not preclude a comparative study of the PTB1 constructs.
The anisotropy of the protein reported by the tryptophan sidechains can be an accurate measure of overall tumbling time θR if the sidechain is immobile. When the sidechain moves, then its segmental motion will contribute to the measured apparent anisotropy, and the value of the overall tumbling will be less accurately determined. The steady-state anisotropy data provide a measure of the local motions of each tryptophan, through calculation of the order parameter. The order parameter S2 = r0/r(0), where a high order parameter (0 ≤ S2 ≤ 1) indicates restricted mobility, due to a physical environment that limits its motion. For example, W98 in RRM1 appears somewhat restricted, with S2 = 0.74 in the single RRM1 and S2 = 0.77 in PTB1/W1 (see the Perrin plot in Figure 7). In PTB1/W1, W98 has more restricted segmental motion so its anisotropy will more accurately report on the overall tumbling of the domain, keeping in mind that tumbling occurs about molecular axes that are defined by the shape of the molecule (e.g. the molecule could be a sphere, rod or dumbbell). The rotational correlation times calculated for RRM1/W (θR = 6 ns) and PTB1/W1 (θR = 12 ns) reflect the differences in segmental motion of the tryptophans as well as the molecular dimensions.
In PTB1/W3 and PTB1/W4, the sidechains are more mobile (S2 = 0.64 and S2 = 0.61, respectively). The overall correlation times of PTB1/W3 and PTB1/W4 calculated from the steady-state experiment are similar (θR = 8 and 10 ns, respectively). NMR relaxation data of PTB1:34 in 100 mM KCl, 10 mM potassium phosphate pH 6.8, give an overall correlation time of 8.5 ns, while the separate RRM3 and RRM4 domains have tumbling times from 5–6 ns (data not shown). Taking into account the fluorescence and NMR data, it appears that (RRM3+RRM4) tumbles as a unit, but independently of the full-length protein. These fluorescence data clearly show that there is varying extents of flexibility between the RRMs in PTB1, which will impact how they interact with proximal RNA sites.
Time-resolved fluorescence anisotropy provides another measure of the mobility of the tryptophans as well as another indicator of the overall tumbling time of the RRMs. Each tryptophan in the protein shows characteristic fast depolarization attributed to internal photophysical processes and solvent relaxation; the values here range from 0.11 to 0.38 ns (Table 3). Each Trp also exhibits a slow component: Φs(PTB1/W1) = 7.3 ns, Φs(PTB1/W3) = 5.4 ns, Φs(PTB1/W4) = 8.4 ns. These slow anisotropy decay times should correspond to the correlation time of the domain, but the difference in the values of Φs for PTB1/W3 and PTB1/W4 indicates that photophysical processes are contributing to the values of r0 and τ. Recalling that r = r0/[1 + τ/θR], when τ « θR then the molecule will appear immobile during the fluorescence lifetime, but when τ ≈ θR then the anisotropy will be sensitive to factors affecting rotational motion. For free PTB1, all its tryptophans correspond to the latter case. Also worth noting is the relationship between the fluorescence lifetime of the tryptophan and the tumbling time of the protein: the fluorescence gives a window onto those processes that are on the same timescale, which here is about 5–7 ns.
Table 3.
Time-resolved anisotropy of free and bound PTB/W proteinsa
| Protein | βf | Φf (ns) | βs | Φs (ns) | Χ2 | <τ> (ns) |
|---|---|---|---|---|---|---|
| PTB1/W1 | 0.62 | 0.11 ± 0.08 | 0.38 | 7.3 ± 1.2 | 1.2 | 4.4 |
| + GABA RNA11@ |
0.31 | 0.10 ± 0.07 | 0.69 | 7.2 ± 0.52 | 1.2 | 4.0 |
| + HCV X RNA | 0.72 | 0.12 ± 0.07 | 0.28 | 7.9 ± 1.6 | 1.1 | 5.0 |
| PTB1/W3 | 0.53 | 0.23 ± 0.12 | 0.46 | 5.4 ± 1.2 | 1 | 3.9 |
| + GABA RNA11 |
0.67 | 0.25 ± 0.21 | 0.33 | 4.7 ± 1.5 | 1.08 | 4.3 |
| + HCV X RNA | 0.87 | 0.33 ± 0.11 | 0.13 | 9.1 ± 1.7 | 1.04 | 4.3 |
| + HCV X SL3 | 0.90 | 0.14 ± 0.04 | 0.10 | 21.6 ± 5.9 | 1.04 | 3.8 |
| PTB1/W4 | 0.63 | 0.38 ± 0.19 | 0.37 | 8.4± 2.2 | 1.07 | 5.6 |
| + GABA RNA11 |
0.62 | 0.34 ± .23 | 0.38 | 9.8 ± 3.3 | 1.02 | 5.3 |
| + HCV X RNA | 0.71 | 0.5± 0.2 | 0.29 | 17.5 ± 3.8 | 1.2 | 5.4 |
| + HCV X SL 3 | 0.73 | 0.23 ± 0.16 | 0.27 | 9.4 ± 3.2 | 1.1 | 5.5 |
Protein alone concentrations range from 1 to 5 µM in 100 mM NaCl, 10 mM sodium cacodylate pH 7.5, 10 mM DTT, room temperature.
250 mM NaCl only. RNA concentrations were 1 µM, and protein concentrations were adjusted to saturate the binding ([P]/[R] = 1 for HCV X SL3; [P]/[R] = 2 or HCV X and GABA RNA 11). Data acquired on a time-correlated single photon counting instrument, and analyzed with PicoQuant FluoFit. Steady state fluorescence spectra were collected before and after time-resolved experiments. <τ> values from Supplementary Table.
RNA binding
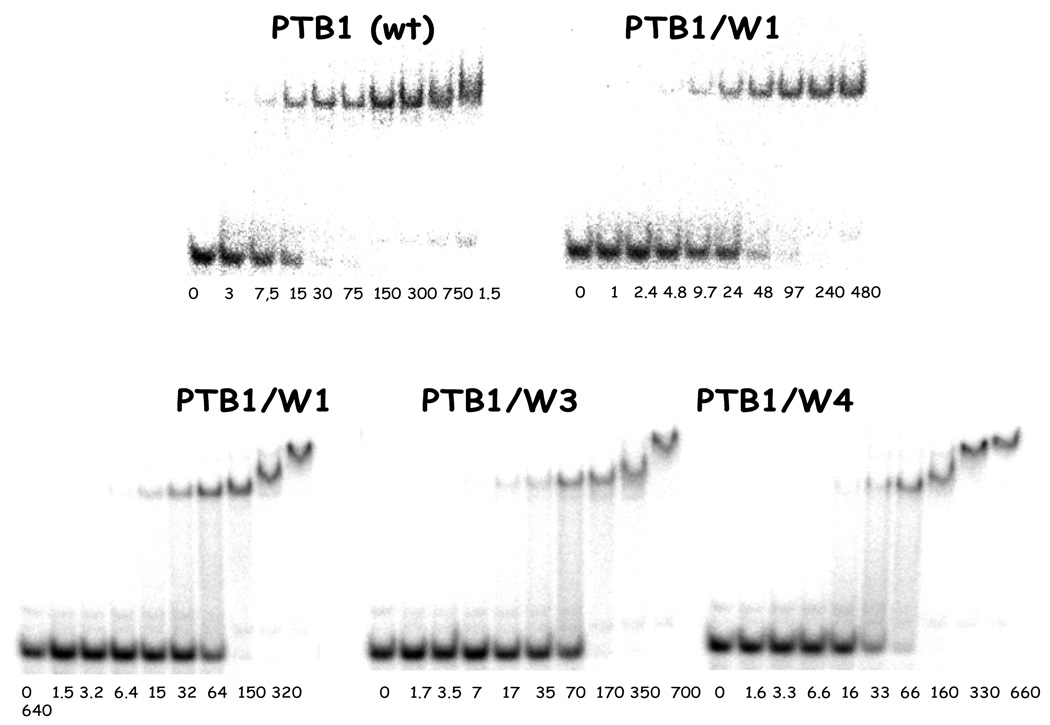
Introduction of a tryptophan into an RRM has the potential to alter the structure of the protein as well as its functional properties. The CD spectra show that the secondary structures of the proteins are not detectably altered, and from those data we infer that the proteins are folded. The RNA binding properties of each PTB1/W mutant were compared to that of wild type PTB1 in EMSA experiments using GABA RNA 11. This RNA is single-stranded and binds to PTB1 and to PTB1:34 with high affinity in these solution conditions (Table 1). EMSA experiments using GABA RNA11 and the PTB1/W mutants show that all proteins bind the RNA with similar affinities, and there appears to be about a two-fold reduction in affinity compared to the wild-type protein for this RNA (Figure 8).
Figure 8.
PTB1/W mutants binding to RNA. Top: EMSA of PTB1 and PTB/W1 binding to GABA 11 RNA. Bottom: all three tryptophan mutant proteins bound to GABA 11 RNA. This RNA is bound by PTB1:34, so these experiments are important to measure the effect of tryptophan substitution in those domains. We note that bands arising from higher order complexes are more distinct in the lower panel of gels, but this type of variability is occasionally observed for the wild type protein as well. EMSA experiments at 4° C in 100 mM NaCl, 10 mM sodium cacodylate pH 7.5, 1 mM MgCl2, 10 mM DTT, 10 µg/mL tRNA, 20 µg/mL BSA.
Binding of PTB1 and PTB1/W1 to HCV X SL3 was compared in nitrocellulose filter binding experiments. The calculated KD = 4.0 × 10−7 M ± 3 × 10−8 M and KD = 3.1 × 10−7 M ± 3 × 10−8 M for PTB1 and PTB1/W1, respectively, is the same within error (see Figure 6). HCV X SL3 RNA is bound by PTB1:12, so these experiments should assess the effect of the mutation at F98W on that association as well. We conclude that RNA binding of PTB1/W1 to this RNA is identical to that of wild type protein.
PTB1 Fluorescence upon binding RNA
Each PTB1/W protein was bound to HCV X, HCV X SL3, and the GABA RNA 11 used in EMSA experiments. Fluorescence experiments were carried out at 1 µM RNA, due to the relatively weak tryptophan signal. As such, these conditions correspond to those of the stoichiometry experiments.
HCV X RNAs
The steady-state fluorescence intensity at 25° C was measured to determine the environment of the Trp in the presence of the HCV X RNAs. These RNAs are bound by PTB1 and PTB1:12, but not by PTB1:34. The most striking response is the complete quenching of fluorescence intensity of PTB1/W1 upon binding to HCV X SL3 (this is a 1:1 complex), and its 80% quenching when bound to X RNA (a 2:1 complex) (Table 2). Loss of W98 fluorescence intensity could be caused by stacking with the RNA, a conformational change of the protein, or protein:protein interactions. The lifetime of the residual fluorescence of PTB1/W1 bound to X RNA increases from 5.3 to 7.6 ns (the longest component) and its average lifetime increases from <τ> = 4.4 to 5, a slight but significant change (supplementary Table 1).
The fluorescence changes of PTB1/W3 and PTB1/W4 bound to HCV X and HCV X SL3 were unanticipated, given that PTB1:34 does not bind to X or to X SL3 RNAs. In addition, the binding affinity of PTB1 and PTB1:12 for X RNA is identical, indicating that the C-terminal half of PTB1 does not contribute. However, HCV X RNA binding reduced the fluorescence intensity of both PTB1/W3 and PTB1/W4 by 20%. Since <τ> of W3 and W4 show little change in the HCV X complex (Table 3), static quenching is the most obvious mechanism responsible for the loss of fluorescence intensity, and stacking interactions are the most common source of static quenching. Either RNA nucleobase:tryptophan stacking or protein:protein stacking interactions could be responsible.
Those tryptophans that are not quenched in the complex show changes in other fluorescence parameters when X RNA is bound. For PTB1/W3, its time-resolved anisotropy has an increase in the amplitude of the short decay, and an increase in the slow decay time from 5.4 to 9.1 ns, which reports on the increase in the size of the complex. PTB1/W4 fluorescence changes more dramatically: binding to X RNA results in a 17.5 ns anisotropy decay time, even though its average lifetime does not change from that of the free protein. Since the fluorescence lifetime of W4 is longer than that of W3 in PTB1, W4 should be a more accurate indicator of the size of the complex (via its slower tumbling time). These data indicate that PTB1:34 does respond to binding of the X RNA, and based on the binding data, we suggest that there is a change in the conformation and relative flexibility of the two RRMs.
Binding of HCV X SL3 quenched PTB1/W3 fluorescence by 50% which again is a very curious result since PTB1:34 does not bind to HCV X SL3. The time-resolved anisotropy values change dramatically, for 90% of the decay is rapid, but 10% is very slow (21 ns). The average lifetime of PTB1/W3 free and bound to X SL3 is identical, however, indicating that its photophysics is unchanged, and that static quenching is the mechanism for its loss of fluorescence intensity. PTB1/W4 fluorescence intensity is quenched by 20% when HCV X SL3 is bound, but its average lifetime is that of free protein and its time-resolved anisotropy shows a modest increase in its long decay component. It is possible that the HCV X SL3 RNA exists in several conformations in solution, one of which is a better target for RRM3/RRM4 binding and these data report on that small population of complexes. An alternative explanation of the data is that the changes in the local environments of the two tryptophan sidechains upon X SL3 RNA binding are due to a global change in the shape of PTB1 protein within the complex that produces a more elongated rod-like shape with rotation about the small axis (fast anisotropy decay) and around the long axis (Φs = 21.6 ns).
GABA RNA 11
Binding to the single stranded GABA RNA 11 has the most significant effect on W3 and W4 fluorescence intensity. PTB1 and PTB1:34 bind with high affinity to this RNA, while PTB1:12 does not detectably bind. Both PTB1/W3 and PTB1/W4 fluorescence intensity is quenched by 40 – 50% upon complex formation. Since the stoichiometry of binding is 2:1, it is possible that in half of the bound PTB1/W3 and PTB1/W4 proteins, their fluorescence is completely quenched by direct binding to the RNA, while the remaining protein continues to emit with no change in the fluorescence lifetime or emission maxima. Time-resolved fluorescence parameters of PTB1/W3 and PTB1/W4 bound to GABA RNA 11 are only modestly altered from those of the free protein, further supporting this hypothesis. The fluorescence quenching upon binding the single-stranded GABA RNA 11 was salt-dependent, becoming less pronounced in both W3 (25%) and W4 (11%) at higher salt (250 mM NaCl), indicating that there is an electrostatic component to association, and that binding does indeed involve these two RRMs.
PTB1 binding affinity to GABA RNA 11 is higher than that of the PTB1:34 construct, indicating that other parts of the protein are contributing to binding. Indeed, PTB1/W1 fluorescence intensity is also reduced when GABA RNA 11 is bound, but by only 25%. Its time-resolved fluorescence parameters in the complex are nearly identical to those of the free PTB1/W1. Although PTB1:12 does not detectably bind to this RNA in EMSA experiments (24), the fluorescence data show that its environment does change when the RNA is bound. One interpretation of the observed quenching of PTB1/W1 fluorescence is that different conformers of the sidechain that are favored by a conformational change of the protein (RNA binding does alter the fluorescence emission maximum).
It would be very useful to know the geometry of these complexes, to know if they are extended or compact. That information would help to interpret the binding and fluorescence data, which now suggest that RNA binding either utilizes several RRMs or alters the structure of the free protein. It is certainly possible that the locally high RNA concentration leads to binding by RRMs that would naturally have low affinity for a particular RNA structure, but it is equally possible that the relative orientations of the RRMs and global flexibility of PTB1 changes when RNA is bound. Since the fluorescence experiments were carried out under conditions of maximum protein binding, fluorescence changes could also be due to protein:protein interactions. What the fluorescence data clearly show is that the binding modes of PTB1 to HCV X RNA, HCV X SL3 RNA, and GABA RNA 11 are very different.
DISCUSSION
The interaction of PTB1 with RNA can occur through any or all of the four RRMs, and the specific contacts could control the geometry and composition of the final complex. Regulatory decisions, such as exclusion of an exon or translational initiation, are likely to depend on these complexes, so it is important to identify which RRMs are involved in recognizing these RNAs and which RRMs can form protein/protein interactions.
Our previous experiments had suggested that RRM1 and RRM2 were capable of binding RNAs which PTB1:34 could bind only with weak affinity (24). Now, these experiments go on to show that the structural context of a polpyrimidine tract is a critical parameter for participation of each RRM. Although each RRM is capable of binding short RNA oligonucleotides (8,9), only the PTB1:12 construct binds with high affinity to short tracts displayed within a structured RNA, such as the hairpin RNAs used in these experiments. We suggest that in PTB1, RRM1 (and possibly RRM2, although we have no direct binding data for this domain) are selectively used to bind to short tracts such as those displayed in c-src RNAs and also in IRES elements.
Further, we postulate that the RRM3:RRM4 domain is reserved for binding longer single-stranded tracts. Indeed, Oberstrass et al., (34) showed that a RNA sequence of at least 15 nucleotides was necessary to span the distance between the β-sheet surfaces of RRM3 and RRM4, and in our experiments, this criterion is met only by GABA RNA 11 which is indeed bound tightly by PTB1:34.
Several reports of in vivo occurrences of truncated PTB proteins suggest that they are able to confer specific biological advantages to different RNAs. Back et al., (35) reported that the polio virus protease 3Cpro was capable of cleaving PTB in the linker between RRM2 and RRM3. Perhaps this separation is actively used by the polio IRES to control translation and ribosome recruitment through selective binding of PTB1:12 to its short U/C tracts. A report of an alternative splice-site that leads to a novel PTB1:34 suggests that this half of the protein could have autonomous regulatory functions (36).
A division of labor allows the protein to bind different sites on a single RNA molecule or different RNA molecules. Stoichiometry of binding of PTB1 and its halves show that these complexes are too large to form through only RNA:protein interactions (e.g. four PTB1:34 proteins ultimately bind to GABA RNA 11; see (24)). PTB1 is certainly a monomer in solution (8,37), but when it binds to RNA, sites could be exposed that lead to PTB:PTB or PTB:protein interactions (e.g. 38,39). Since there is typically more than one potential binding site for PTB in those RNAs that it regulates (2), and since multiple PTB proteins have been found associated with a single polypyrimidine tract (24), these complexes could be very large and diverse in their composition.
Several studies have already indicated the involvement of other proteins in PTB-mediated regulation. RRM2 and flanking sequences of its C-terminal linker were shown to be responsible for recruiting other proteins, in a ‘tethered function’ assay that bypassed its RNA binding activity (40). RRM2 was subsequently shown to bind (weakly) to a peptide from the Raver-1 protein and also to a nonamer RNA, forming a ternary complex that is easy to imagine forming in vivo (41). In the work of Cooper and colleagues (42), cell specific proteins have been identified that compete with PTB, one example being the alternative splicing of α-actinin (38). Alternative splicing of α-tropomyosin involves PTB bound to Raver-1 protein (39) and splicing of fas is regulated by PTB and TIA-1 (43). Specific ternary complexes could well be the heart of PTB regulatory activity in different cell types, and through its ability to recognize short and long, structured and unstructured, polypyrimidine tracts, PTB is well suited to be a versatile adaptor for RNP assembly.
Supplementary Material
Supporting information available: Figures of RNA folding, structure probing and PTB1 footprinting, and a table of time-resolved fluorescence decay components is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
We thank W. Tom Stump for excellent technical support and Professor Doug Black for the c-src RNA clones.
This work was supported by the National Institutes of Health R01 GM077231.
Abbreviations
- PTB1
polypyrimidine tract binding protein isoform 1
- RRM
RNA recognition motif
- HCV
Hepatitis C virus
- IRES
internal ribosome entry sites
- UTR
untranslated region
- EMSA
Electrophoretic mobility shift assays
References
- 1.Gama-Carvalho M, Barbosa-Morais NL, Brodsky AS, Silver PA, Carmo-Fonseca M. Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic-shuttling mammalian splicing factors. Genome Biol. 2006;7:R113. doi: 10.1186/gb-2006-7-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 3.Gil A, Sharp PA, Jamison SF, Garcia-Blanco MA. Characterization of cDNAs encoding the polypyrimidine tract binding protein. Genes Dev. 1991;5:1224–1232. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- 4.Ghetti A, Piňol-Roma S, Michael WM, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acid Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markovtsov V, Nikolic J, Goldman JA, Tyrck CW, Chou M-Y, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homologue of PTB. Mol Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conte MR, Grune T, Ghuman J, Kelly G, Ladas A, Matthews S, Curry S. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 2000;19:3132–3141. doi: 10.1093/emboj/19.12.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitali F, Henning A, Oberstrass FC, Hargous Y, Auweter SD, Erat M, Allain FH. Structure of the two most C-terminal RNA recognition motifs of PTB using segmental isotope labeling. EMBO J. 2006;25:150–162. doi: 10.1038/sj.emboj.7600911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson PJ, Monie TP, Szendroi A, Davydova N, Tyzack JK, Conte MR, Read CM, Cary PD, Svergun DI, Konarev PV, Curry S, Matthew S. Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure. 2004;12:1631–1643. doi: 10.1016/j.str.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Auweter SD, Oberstrass FC, Allain FHT. Solving the structure of PTB in complex with pyrimidine tracts: an NMR study of protein–RNA complexes of weak affinities. J. Mol. Biol. 2007;367:174–186. doi: 10.1016/j.jmb.2006.12.053. [DOI] [PubMed] [Google Scholar]
- 10.Perez I, McAfee JG, Patton JG. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry. 1997;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- 11.Kolupaeva VG, Hellen CUT, Shatsky IN. Structural analysis of the interaction of the pyrimidine tract binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell SA, Spriggs KA, Bushell M, Evans JR, Stoneley M, le Quesne JPC, Spriggs RV, Willis AE. Identification of a motif that mediates polypyrimidine tract binding protein dependent internal ribosome entry. Genes Dev. 2005;19:1556–1571. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witherell GW, Gil A, Wimmer E. Interaction of polypyrimidine tract binding protein with the encephalomyocarditis virus mRNA internal ribosomal entry site. Biochemistry. 1993;32:8268–8275. doi: 10.1021/bi00083a030. [DOI] [PubMed] [Google Scholar]
- 14.Hellen CUT, Witherall GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosome entry is identical to the nuclear pyrimidine tract binding protein. Proc. Natl. Acad. Sci. USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Zhang W, Reed RB, Liu W, Grabowski PJ. Mutations in RRM4 uncouple the splicing repression and RNA-binding activities of polypyrimidine tract binding protein. RNA. 2002;8:137–149. doi: 10.1017/s1355838202015029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan RC, Black DL. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou M-Y, Underwood JG, Nikolic J, Luu MHT, Black DL. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- 18.Amir-Ahmady B, Boutz PL, Markovtsov V, Phillips ML, Black DL. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol. 2008;15:183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spellman R, Llorian M, Smith CWJ. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Molec Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 23.Tillmar L, Welsh N. Hypoxia may increase rat insulin mRNA levels by promoting binding of the polypyrimidine tract-binding protein (PTB) to the pyrimidine-rich insulin mRNA 3′-untranslated region. Mol. Med. 2002;8:263–272. [PMC free article] [PubMed] [Google Scholar]
- 24.Clerte C, Hall KB. Characterization of multimeric complexes formed by the human PTB1 protein on RNA. RNA. 2006;12:457–475. doi: 10.1261/rna.2178406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill SC, von Hippell PH. Calculation of protein extinction coefficients from amino acid sequence data. Analytical Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 26.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashiya M, Grabowski PJ. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 28.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthews DH, Sabina J, Zuker M, Turner DH. Expanded Sequence Dependence of Thermodynamic Parameters Improves Prediction of RNA Secondary Structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 30.Gontarek RR, Gutshall LL, Herold KM, Tsai J, Sathe GM, Mao J, Prescott C, Del Vecchio AM. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3'NTR of the HCV RNA genome. Nucleic Acids Res. 1999;27:1457–1463. doi: 10.1093/nar/27.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolykhalov AA, Feinstone SM, Rice CM. Identification of a highly conserved sequence element at the 3' terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blight KJ, Rice CM. Secondary structure determination of the conserved 98-base sequence at the 3' terminus of hepatitis C virus genome RNA. J Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petoukhov MV, Monie TP, Allain FHT, Matthews S, Curry S, Svergun DI. Conformation of polypyrimidine tract binding protein in solution. Structure. 2006;14:1021–1027. doi: 10.1016/j.str.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FHT. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 35.Back SH, Kim YK, Kim WJ, Cho S, Oh HR, Kim JE, Jang SK. Translation of poliovirus mRNA is inhibited by cleavage of polypyrimidine tract binding proteins executed by poliovirus 3Cpro. J. Virol. 2002;76:2529–2542. doi: 10.1128/jvi.76.5.2529-2542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamilton BJ, Genin A, Cron RQ, Rigby WFC. Delineation of a novel pathway that regulated CD154 (CD40 ligand) expression. Mol Cell Biol. 2003;23:510–525. doi: 10.1128/MCB.23.2.510-525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monie TP, Hernandez H, Robinson CV, Simpson P, Matthews S, Curry S. The polypyrimidine tract binding protein is a monomer. RNA. 2005;11:1803–1808. doi: 10.1261/rna.2214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gromak N, Matlin AJ, Cooper TA, Smith CW. Antagonistic regulation of α-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA. 2003a;9:443–456. doi: 10.1261/rna.2191903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gromak N, Rideau A, Southby J, Scadden AD, Gooding C, Huttelmaier S, Singer RH, Smith CW. The PTB interacting protein raver1 regulates α-tropomyosin alternative splicing. EMBO J. 2003b;22:6356–6364. doi: 10.1093/emboj/cdg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson F, Smith CWF. A splicing repressor domain in polypyrimidine tract-binding protein. J Biol Chem. 2006;281:800–806. doi: 10.1074/jbc.M510578200. [DOI] [PubMed] [Google Scholar]
- 41.Rideau AP, Gooding C, Simpson PJ, Monie TP, Lorenz M, Huttelmaier S, Singer RH, Matthews S, Curry S, Smith CWJ. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat. Struct. Mol. Biol. 2006;13:839–848. doi: 10.1038/nsmb1137. [DOI] [PubMed] [Google Scholar]
- 42.Charlet-B N, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 43.Izquierdo JM, Majós N, Bonnal S, Martínez C, Castelo R, Guigó R, Bilbao D, Valcárcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information available: Figures of RNA folding, structure probing and PTB1 footprinting, and a table of time-resolved fluorescence decay components is available free of charge via the Internet at http://pubs.acs.org.