Abstract
Pre-mRNA splicing is regulated through combinatorial activity of RNA motifs including splice sites and splicing regulatory elements (SREs). Here, we show that the activity of the G-run class of SREs is ∼4-fold higher when adjacent to intermediate strength 5'ss relative to weak 5'ss, and ∼1.3-fold higher relative to strong 5'ss. This dependence on 5'ss strength was observed in splicing reporters and in global microarray and mRNA-Seq analyses of splicing changes following RNAi against heterogeneous nuclear ribonucleoprotein (hnRNP) H, which crosslinked to G-runs adjacent to many regulated exons. An exon’s responsiveness to changes in hnRNP H levels therefore depends in a complex way on G-run abundance and 5'ss strength, and other splicing factors may function similarly. This pattern of activity enables G-runs and hnRNP H to buffer the effects of 5'ss mutations, augmenting the frequency of 5'ss polymorphism and the evolution of new splicing patterns.
Keywords: alternative splicing, genetic buffering, hnRNP H, CLIP-Seq, mRNA-Seq
Introduction
Genetic changes that perturb pre-mRNA splicing are commonly associated with human genetic diseases, while other splicing alterations have contributed to evolutionary innovations1–4. Splicing may be disrupted either by mutation of sequence motifs present in every intron, namely the core 5' splice site (5'ss), 3' splice site (3'ss) or branch point, or by mutation of exonic or intronic SREs. Such changes frequently result in skipping of exons or other major alterations to the mRNA and the encoded protein, but may be compensated for during evolution by strengthening of other elements5. In a recent study, reciprocal compensatory evolution was observed for most pairs of splicing elements in human/mouse, with weakening of element A associated with strengthening of element B and vice versa, suggesting that most elements defining exons may contribute additively to exon recognition6. However, for the pair of the 5'ss and "G triplet" intronic splicing enhancers (ISEs; see below), compensatory evolution was unidirectional, suggesting that this pair of elements might have a special functional relationship6.
Poly-guanine sequences ("G-runs") play central roles in splicing of a number of important cellular and viral genes, commonly functioning through recruitment of splicing regulators of the heterogeneous nuclear ribonucleoprotein (hnRNP) F/H gene family7–15. Just three consecutive guanines, a "G triplet", are required for binding of hnRNP F/H proteins and for splicing activity16. G triplets are extremely abundant in mammalian introns, where they commonly function as ISEs, increasing the usage of adjacent splice sites. G triplets are most highly enriched in the ∼70 bases downstream of the 5'ss (Fig. 1a, Supplementary Fig. 1, [CP1]and refs 11,17). The extremely high density of G triplets located just 20–30 base pairs (bp) from the 5'ss, and the asymmetric coevolutionary relationship between these motifs suggested that strong functional links might exist between the 5'ss motif and adjacent G-run ISEs. Here, we explored this possibility using a battery of classical and high-throughput molecular genetic approaches[CP2] in human cells, uncovering an unexpected but highly consistent pattern of functional interdependency that has important genetic and evolutionary implications.
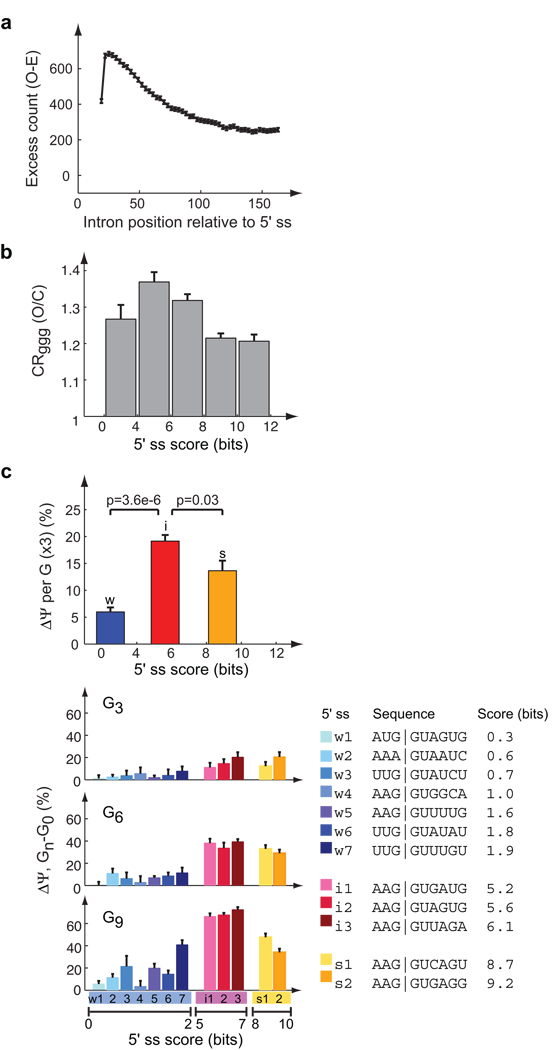
Figure 1. Abundance, conservation and 5’ss strength-dependent activity of G-run intronic splicing enhancers (ISEs).
(a) Excess count of GGG in introns downstream of human constitutive exons. Excess count is defined as the difference between the observed count and expected count (Supplementary Methods). Each point represents the center of a 30-nt window, with a 3-nt offset between successive windows. Black bars show the standard errors (SEM). (b) Mean and SEM of the conservation rate (CR) ratio, calculated as O/C (observed/control) conservation for GGG in positions +11–70 downstream of exons conserved between human and mouse. The intermediate 5’ss bins (4–6, 6–8 bits) had significantly higher CR and the very strong bin (10–12 bits) significantly lower CR (all P <0.05, Bonferroni corrected) than control sets sampled from the other 5’ss strength bins to match GGG counts. (c) ISE activity of G-runs in modular splicing reporters (Methods). Lower panels: change in “percent spliced in” (PSI or Ψ) (qRT-PCR) of the test exon of selected splicing reporters (Supplementary Table 1) with different 5’ss (listed at right) containing runs of 3, 6, or 9 G’s inserted in the downstream intron relative to the corresponding reporters with control inserts. Error bars indicate range of replicated experiments. Top: increase in Ψ of test exon with inserted G-runs relative to control insert (mean, SEM), normalized by number of G’s in inserted G-runs. Splicing reporters were grouped into weak (w), intermediate (i) and strong (s) bins according to 5’ss score of test exon. P-values calculated by Wilcoxon rank sum test.
Results
G triples are more conserved near intermediate 5'ss
The 5'ss sequences of mammalian introns vary greatly in the degree of complementarity to U1 small nuclear RNA (snRNA) and in their intrinsic activity in pre-mRNA splicing18. Using statistical models that capture mono- and di-nucleotide composition at pairs of 5'ss positions, log-odds scores can be assigned to 5'ss motifs that reliably predict function19. Using the MaxEnt model, scores of natural 5'ss typically range between 0 (occasionally below zero) and 12 bits, with the median around 9 bits. Increased density of G-rich and C-rich sequences adjacent to mammalian exons with weak 5'ss or weak 3'ss has been observed previously11,20.
Grouping orthologous pairs of human and mouse introns by their 5'ss scores, we observed that G triples in the downstream intron were more conserved than control trinucleotides (3mers) in all splice site strength groups, consistent with common ISE activity (Fig. 1b). However, significantly [CP4]greater conservation was seen for G triplets located adjacent to intermediate strength 5'ss (4–8 bits) than for those adjacent to strong (> 8 bit) or weak (< 4 bit) 5'ss (P < 0.05; Fig. 1b). In these and subsequent analyses, the boundaries between higher versus lower activity of intermediate versus weak or strong 5'ss appeared to fall at scores of 4 and 8 bits, respectively, corresponding to the 4th and 33rd percentiles of constitutive exon splice site scores (i.e. 1/3 of 5'ss are weaker than 8 bits). Here, our analyses included only G-runs located in the region +11 to +70 relative to the 5'ss, where G triples are most enriched. The region +1 to +10 was excluded, since G-runs that overlap with the 5'ss motif tend to suppress rather than activate splicing of the upstream exon21. Weaker exons are expected to be more dependent on enhancers. Therefore, the more pronounced conservation of G triplets adjacent to intermediate 5'ss relative to weak 5'ss was surprising, and suggested the hypothesis that the ISE activity of G-runs might vary depending on 5'ss strength, and that constitutive introns with weak 5'ss might depend more heavily on other types of ISEs.
G-run ISE activity depends on 5'ss strength
To test this hypothesis, G-run ISE activity was assessed as a function of 5'ss strength and sequence using splicing reporter minigenes transfected into cultured human cells (Fig. 1c). MaxEnt 5'ss scores correlated well with splicing activity, assessed by the fractional inclusion of a test exon (Supplementary Fig. 2). Here, we use "percent spliced in" (PSI or Ψ), the fraction of mRNAs that include an exon as a proportion of mRNAs that contain the flanking exons (see ref.22), determined by qRT-PCR. Insertion of G-runs totaling 3, 6 or 9 nucleotides (nt) downstream of the test exon consistently enhanced exon inclusion, with increased enhancement associated with longer G-runs. Splicing activation was particularly pronounced for intermediate strength 5'ss: Ψ values increased by 70%, from ∼20% to ∼90%, following insertion of G9 in three reporters with 5–7 bit 5'ss (Fig. 1c, Supplementary Table 1). Splicing enhancement by runs of G6 or G9 was more modest for exons with weaker (P = 3.6 × 10−6) or stronger (P = 0.03) 5'ss. (Enhancement did not differ significantly for G3). Considering all of the data, an increase in Ψ value of ∼20% per inserted G triplet was observed on average for intermediate 5'ss, approximately 1.3-fold greater than the mean enhancement for strong 5'ss, and some 4-fold higher than the mean for weak 5'ss (Fig. 1c, above).
ISE activity was much more dependent on 5'ss strength than specific sequence, with similar ISE activity observed for different 5'ss sequences of similar score (Fig. 1c; Supplementary Table 1), The dependence of ISE activity on 5'ss strength was robust to differences in starting Ψ value, i.e. Ψ value prior to insertion of G-runs (Supplementary Fig. 3a). No consistent pattern of dependence of ISE activity on 3'ss strength was seen (Supplementary Fig. 4a). These observations suggested that G-run ISEs located downstream of an exon recruit factor(s) that enhance splicing at a step closely associated with 5'ss function, such as 5'ss recognition by U1 small nuclear ribonucleoprotein (snRNP), or progression from U1:5'ss recognition to exon definition complex formation23 (see Discussion).
Intermediate 5'ss exons are more responsive to hnRNP H
HnRNP H is the most highly expressed member of the G-run-binding hnRNP F/H protein family in 293T cells24. RNAi directed against hnRNP H resulted in substantial (∼3–4-fold) reductions in target mRNA and protein levels by qRT-PCR and Western analysis 72 hours after initial siRNA transfection (Supplementary Fig. 5). Compensatory upregulation of closely related factors25 was not observed: expression of hnRNP F was also reduced by the siRNA used, while hnRNP H' (expressed ∼5-fold lower than H) was unaffected and hnRNP 2H9 was not detectably expressed (Methods).
To assess the activity of G-runs in regulation of endogenous exons, changes in exon inclusion were assessed following hnRNP H knockdown by deep sequencing of mRNAs (mRNA-Seq) using the Illumina platform, and by Affymetrix all-exon microarrays. The Ψ values of exons were estimated from mRNA-Seq read densities as described26. Analysis of mRNA-Seq read densities identified 214 exons whose Ψ values changed significantly, at a cutoff corresponding to a 5% false discovery rate (FDR). Of these, 79% (169 out of 214 ) had ≥3 G's in G-runs and 61% (131 out of 214) had ≥6 G's in G-runs within 70 bp of the 5'ss, both significantly higher than control exons whose Ψ values did not change (P < 1.7e-8 and P < 1.2e-11, respectively, Fisher's exact test). Furthermore, GGG was the most enriched 3mer within 70 bp 3' of the 214 exons (not shown), consistent with widespread reduction in ISE activity of G-runs following RNAi against hnRNP H.
Similar or greater Ψ value changes were associated with intronic GGGG motifs than with other 4mers containing GGG, with no other significant differences observed between GGGN and NGGG 4mers (Supplementary Fig. 6), suggesting that G-run length rather than flanking nucleotide context is the primary determinant of ISE activity in this system. Similar Ψ value changes were observed for exons flanked by G-runs independent of initial Ψ value or 3'ss strength (Supplementary Fig. 3b, Supplementary Fig. 4d), and for GGGs located at different positions within the range +11 to +70 relative to the 5'ss, to the extent that this variable could be assessed using the available data (Supplementary Fig. 6).
Larger changes in Ψ value were associated with larger numbers of G’s in intronic G-runs in both the mRNA-Seq and exon array analyses (Fig. 2a, Supplementary Fig. 7b), with better fit to a linear (additive) rather than multiplicative model of G-run ISE activity (Supplementary Figs. 7d, 7e). This relationship paralleled that observed for the splicing reporters (Fig. 1c, Supplementary Fig. 3a). Grouping expressed exons with downstream G-runs by 5'ss strength, the largest decreases in Ψ value following RNAi were observed for exons with intermediate (4–8 bit) 5'ss (Fig. 2b, P < 0.05). Thus, three independent lines of evidence – evolutionary conservation, splicing reporter analyses, and RNAi mRNA-Seq and exon array analyses (Supplementary Fig. 7c) – all supported the conclusion that G-run ISE activity is quite sensitive to 5'ss strength, with higher activity for exons containing intermediate-strength 5'ss.
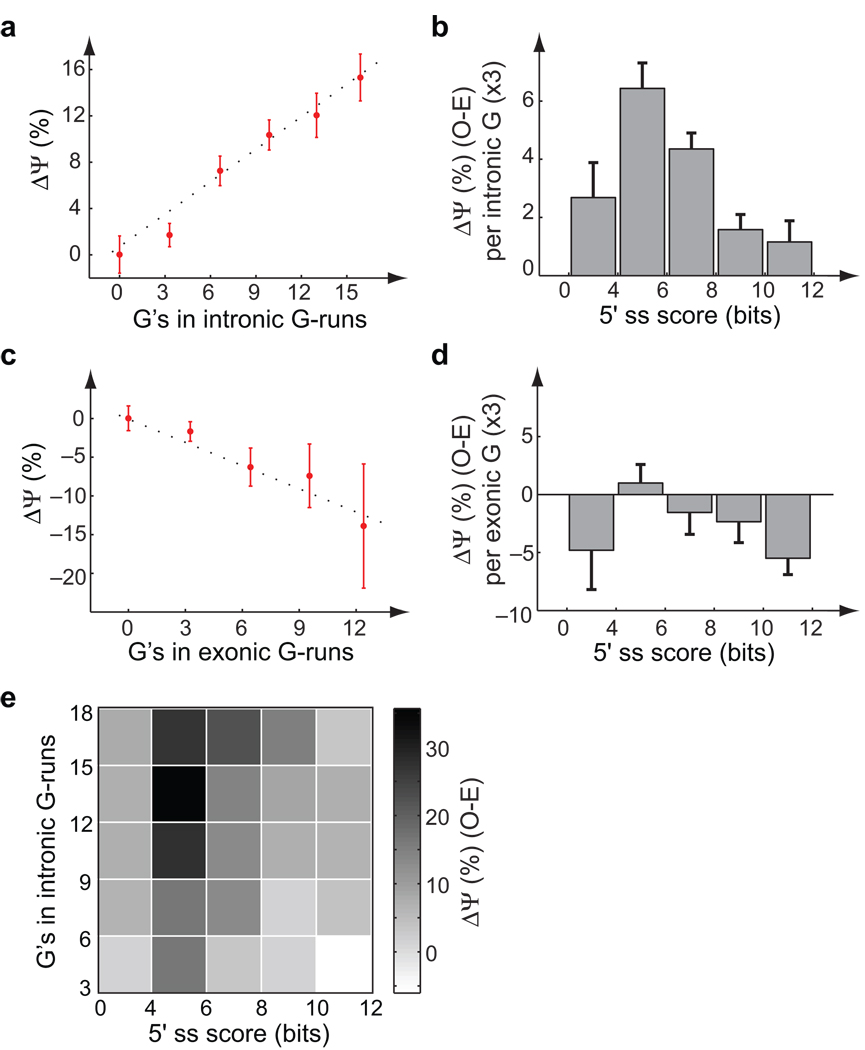
Figure 2. hnRNP H knockdown/mRNA-Seq analysis of G-run activity.
(a) Change in “percent spliced in” (PSI or Ψ) (mean, SEM; control - H knockdown) of exons grouped by number of G’s in downstream intronic G-runs (e.g., bin labeled 6 includes exons with 6–8 G’s in G-runs). Based on mRNA-Seq analysis of total RNA following RNAi directed against hnRNP H. (b) Change in Ψ (mean, SEM) of exons with ≥6 G’s in G-runs in downstream introns grouped by 5’ss score relative to control exons lacking exonic or intronic G-runs (O–E). The relative change was significantly higher for the intermediate (4–6 bit) and significantly lower for the strong (8–10 bit) 5’ss bins (both P < 0.05, Bonferroni corrected) relative to control sets sampled from the other 5’ss bins to match the distribution of numbers of G’s in runs G3 or longer. No significant differences in flanking nucleotide frequency were detected between 5’ss bins. (c) Same as (a) for exons with different numbers of G’s in G-runs in the exons, excluding those with upstream or downstream intronic G-runs. (d) Same as (b) for exons with ≥6 G’s in exonic G-runs. The relative change was significantly lower for the strong (10–12 bit) 5’ss bin relative to control sets sampled from the other 5’ss bins (P < 0.05, Bonferroni corrected). (e) Change in Ψ (O-E, same as (b)) of all exons grouped both by number of G’s in downstream intronic G-runs and 5’ss score.
Conversely, Ψ values of exons with internal G-runs tended to increase following hnRNP H knockdown, consistent with previous observations that exonic G-runs commonly function as ESSs27,28. Again, the change in Ψ value increased proportionally to total G-run length (Fig. 2c). [CP5] Effects of 5'ss strength were also observed for exons containing internal G-runs, with highest inferred ESS activity for exons with strong or weak 5'ss, and little or no ESS activity detected in the context of intermediate-strength 5'ss (Fig. 2d), a relationship inverse to that observed for the ISE activity of intronic G-runs. Measurement of Ψ values for a subset of exons by qRT-PCR yielded reasonably good correlation with Ψ values estimated by mRNA-Seq (Supplementary Fig. 8), and identified a high-confidence set of hnRNP F/H-responsive exons, including exons in the ATXN2, MADD and TARBP2 genes (Supplementary Fig. 8, Supplementary Table 2).
For the RNAi/mRNA-Seq experiment, it was possible to map the full spectrum of G-run ISE activity, as inferred from change in Ψ value, for exons with varying 5'ss strength, yielding a smoothly varying pattern (Fig. 2e[CP6]). It is clear from this representation that an exon's responsiveness to hnRNP H is not just a function of the density of G-runs, but is actually a function of both G-run length and 5'ss strength. The bivariate nature of this function is expected to result in finer regulatory discrimination between subsets of exons (e.g., between exons with strong, intermediate and weak 5'ss) in their responsiveness to changes in hnRNP H levels. Such changes may occur under developmental or physiological conditions or in disease states in which hnRNP H activity is altered such as myotonic dystrophy29,30.
The concordance between the activities of G-runs observed in the splicing reporter assays and in the hnRNP H knockdown experiment suggested that a substantial proportion of the effects observed in these systems were the result of direct effects of hnRNP H protein bound to intronic G-runs. Data from cross-linking/immunoprecipitation/sequencing (CLIP-Seq) experiments using antibodies against hnRNP H in 293T cells further supported this idea. The CLIP-Seq dataset, generated as part of a separate study of UTR-associated functions of hnRNP H, constituted 3.6 million 32-bp CLIP tag sequences that could be mapped uniquely to the human genome. In these CLIP tag sequences guanine was highly enriched, and GGG was the most abundant 3mer, enriched more than 5-fold relative to the average 3mer (Supplementary Table 3). Thus, these transcriptome-wide in vivo binding data were consistent with the high affinity of hnRNP H for runs of 3 or more guanines observed previously in vitro. Grouping introns by G-run density downstream of the 5'ss, we observed an approximately linear increase in CLIP tag density (normalized by gene expression) as a function of the number of guanines in G-runs (Supplementary Fig. 9a). This linear increase in binding paralleled the approximately linear increase in ISE activity as a function of G-run density observed in the splicing reporter and hnRNP H knockdown experiments. Exons whose expression changed following hnRNP H knockdown were substantially more likely to have associated CLIP tags than control exons (Supplementary Fig. 9b). Thus, both the overall pattern of linear increase in binding and activity associated with total G-run length and the association between binding and splicing change following knockdown provided further support for direct effects of hnRNP H being of primary importance in the observed pattern of G-run ISE activity. The set of exons whose Ψ values changed following hnRNP H knockdown and associated CLIP tag counts are provided in Supplementary Table 4.
Genetic buffering of 5'ss mutations by G-runs
The 5'ss strength-dependent activity of G-run ISEs and ESSs uniquely equips these elements to serve as "genetic buffers" capable of suppressing the phenotypes of 5'ss-weakening mutations that would otherwise cause substantial exon skipping. For example, in the absence of intronic G-runs, a mutation altering a strong (9.2 bit) 5'ss to intermediate (6.1 bit) strength reduced reporter exon inclusion from 56% to 21% (Fig. 3a). However, insertion of a G9 run in the downstream intron, in addition to enhancing exon inclusion, made inclusion of the exon tolerant to the same 5'ss-altering mutation as a result of the increased ISE activity in the presence of an intermediate rather than a strong 5'ss, with Ψ value actually increasing marginally from 90% to 93%. Presence of a downstream G-run ISE can therefore make an exon much less sensitive to 5'ss-altering mutations, with only the most drastic changes (e.g., reducing strength to < 4 bits) likely to result in substantially increased exon skipping. Large numbers of human exons are potentially affected by this mechanism. For example, more than 14,000 constitutive human exons (∼17% of the dataset used) had 5'ss > 8 bits and at least 6 G's in G-runs within 70 bp downstream of the 5'ss, and approximately one-third of randomly generated point mutations of these 5'ss reduced strength to the 4–8 bit range (not shown). This buffering mechanism is therefore applicable to a substantial proportion of 5'ss mutations in many thousands of human exons. Additional exons are likely buffered by G-run ESSs, since the splice site strength-dependence of G-run ESS activity also acts in a direction tending to buffer the effects of mutations from strong to intermediate 5'ss.
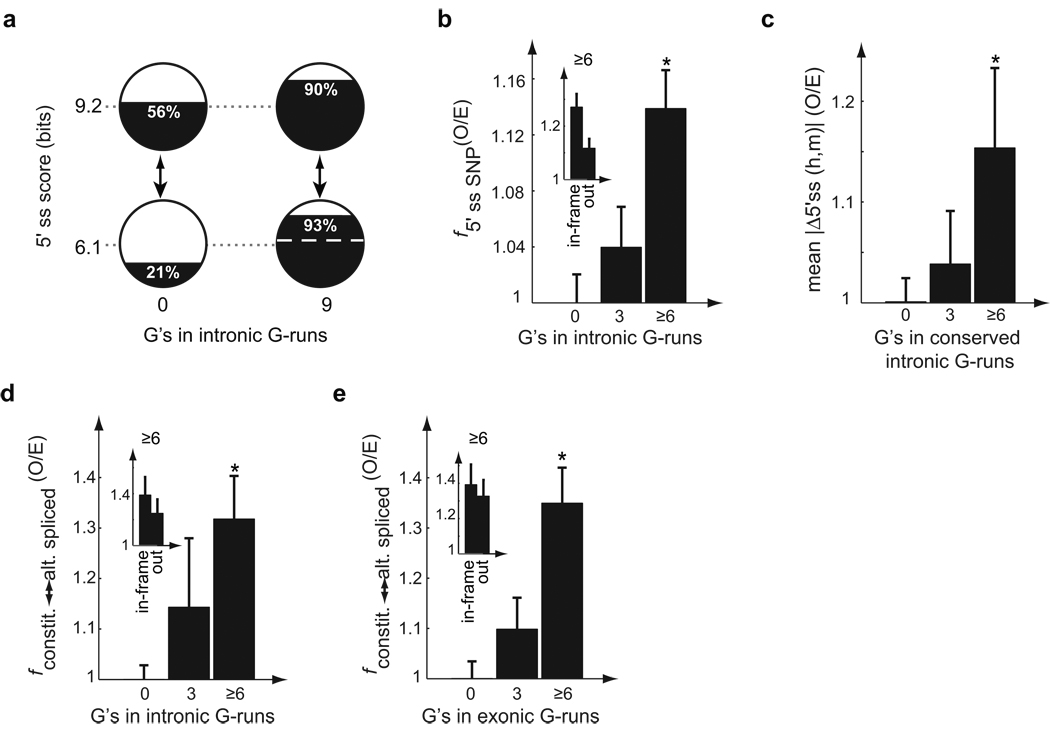
Figure 3. Genetic buffering by G-runs.
(a) G-runs as an evolutionary buffer to 5’ss mutations. Exon inclusion data from reporters s2 and i3 in Fig. 1c. Dashed white line represents percent inclusion that would result if the G-run activity were no greater for intermediate 5’ss exon than for strong 5’ss exon. (b) Higher SNP density in 5’ss of exons with downstream (+11–70) G-runs. Mean and SEM are shown of the ratio (O/E) between the fraction with 5’ss SNPs among exons with G-runs and that in control exons lacking G-runs but with matched compositional bias (*: P < 0.001, rank sum test). Inset shows results for in-frame and out-of-frame exons separately for exons with ≥6 G’s in G-runs. (c) Larger change in 5’ss strength between orthologous human/mouse exons with conserved downstream G-runs. O/E ratio was defined similarly as in (b), but absolute change in MaxEnt 5’ss scores was calculated (*: P < 0.001). (d) Higher fraction of exons with different splicing phenotypes in human and mouse (constitutive in human, alternative in mouse) in exons with ancestral downstream G-runs. Data were grouped by number of G’s in ancestral G-runs in positions +11–70. Mean and SEM are shown of the ratio (O/E) of the fraction with different splicing phenotypes among exons with ancestral G-runs and that of control exons lacking G-runs but matched for base composition, conservation and EST coverage in mouse (*: P < 0.001). Inset shows in-frame and out-of-frame exons separately. (e) Same as (d) for exons with ancestral exonic G-runs.
Equilibrium models of the evolution of cis-elements affecting exon splicing confirmed the intuitive expectations that presence of ISEs tends to relax constraints on the 5'ss, and that the sort of 5'ss strength-dependent ISE activity observed for G-runs relaxes selective pressure on 5'ss more than would 5'ss-independent ISE activity (Supplementary Fig. 10, Supplementary Methods). These models predict that the "flux" (i.e. number of changes occurring in the population per unit time) of neutral 5'ss mutations should be higher in constitutive exons flanked by G-run ISEs, and that these exons should therefore accumulate increased (neutral) genetic variation in their 5'ss sequences. Consistent with this prediction, a significantly higher frequency of single nucleotide polymorphisms (SNPs) was observed within the 5'ss consensus motifs of constitutive human exons with downstream G-runs of total length ≥ 6 than for control exons (Fig. 3b). This observation suggested that downstream intronic G-runs have buffered, i.e. suppressed the phenotypic effects of, a substantial fraction of 5'ss mutations in recent human evolution.
Orthologous human and mouse exons flanked by conserved G-runs diverged more in their 5'ss scores than control pairs of orthologous exons (Fig. 3c). Presence of intronic G-runs was therefore associated also with longer-term evolutionary change in 5'ss strength, as expected from the genetic buffering model.
An important but poorly understood evolutionary process is the evolution of alternative splicing patterns31. New alternative exons may sometimes derive from exons that previously were constitutively spliced or vice versa. Given the effects of G-runs on 5'ss variation, we asked whether presence of G-runs accelerated evolutionary changes in splicing patterns.
When G-runs totaling ≥ 6 G's were present ancestrally in the downstream intron, a ∼30% higher frequency of alternative splicing was observed in the mouse orthologs of constitutively spliced human exons than in control mouse exons (Fig. 3d, Supplementary Table 5). Acceleration of splicing level evolution was also observed when the conserved G-runs were located in the exon rather than the downstream intron (Fig. 3e). Some of these mouse-specific exon skipping events are expected to generate severely truncated proteins likely to lack function (e.g., in the MYEF2 gene) but may downregulate expression, while others are expected to generate isoforms missing one or more specific domains, e.g., an isoform of BMP-binding endothelial regulator protein (BMPER) that is predicted to lack just the central VWD domain, suggesting altered interaction properties (these and other examples are shown in Supplementary Fig. 11).
Regulation by hnRNP H and G-runs
Genes rich in intronic G-runs were more likely than control genes to encode proteins involved in a number of gene ontology (GO) categories related to development, membrane localization and signal transduction; genes containing hnRNP H-responsive exons were enriched for similar functions (Supplementary Table 6).
Cell type- and tissue-specific regulation of alternative splicing is thought to involve both highly tissue-specific factors such as Nova-1/Nova-2, and tissue-specific differences in the levels or activities of ubiquitously expressed factors such as hnRNPs. Because exons with intermediate strength 5'ss are more responsive to changes in hnRNP H levels than other exons, we expected that bioinformatic analyses of tissue-specific G-run enrichment should have greater statistical power in the subset of exons with intermediate 5'ss. This expectation was confirmed by analysis of G-run enrichment in sets of tissue-specifically-expressed exons (Supplementary Fig. 12), suggesting increased activity of hnRNP H in testis, consistent with Western analysis32, and also in adipose and MB435 cells
Intronic sequence conservation varies with 5'ss strength
Whether the activities of other SREs are similarly sensitive to splice site strength remains largely unexplored, with only a handful of reports addressing this issue (e.g., ref 33). Grouping exons by 5'ss strength, striking differences in patterns of evolutionary conservation were observed (Fig. 4). Notably, increased sequence conservation was observed adjacent to exons with weak 5'ss compared to those with stronger 5'ss. This pattern was observed both for exons constitutively spliced in human and mouse (“included-conserved exons” or ICEs; Fig. 4a), and for exons alternatively spliced in both species (“alternative-conserved exons” or ACEs; Fig. 4d), which exhibited much higher intronic conservation overall than ICEs34. These observations suggested that 5'ss strength fundamentally alters exon recognition and regulation, with intronic SREs playing a far greater role in splicing of exons with weak or intermediate 5'ss than in splicing of strong 5'ss exons. This idea is consistent with the very high conservation of 5'ss strength observed in ACEs35.
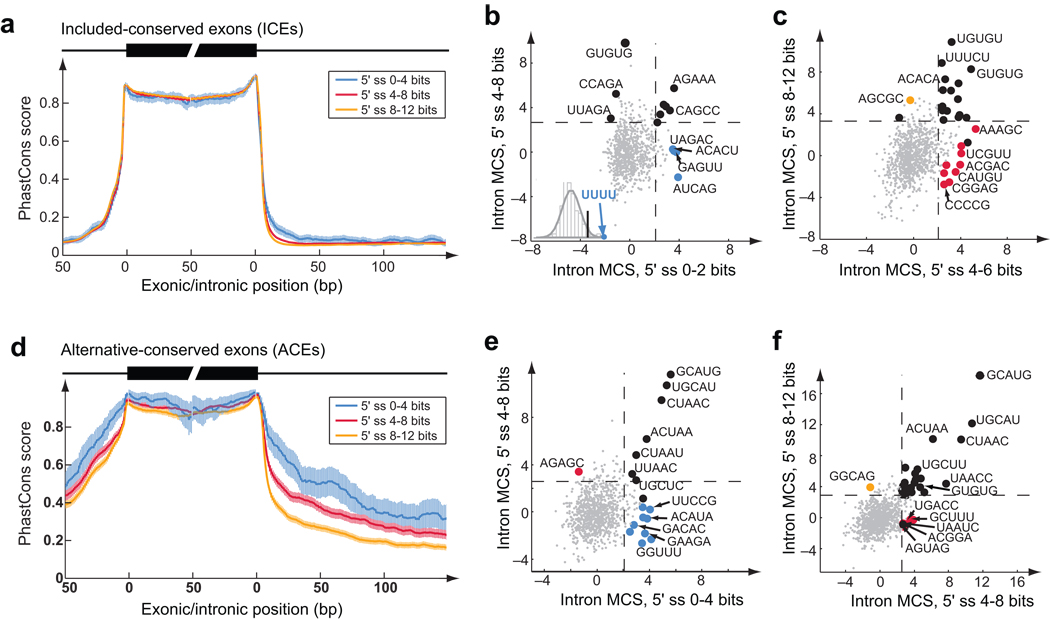
Figure 4.
Sequence conservation flanking exons dependent on 5’ss strength. (a) Conservation profile (mean and 95% confidence interval of PhastCons scores49) of exons and flanking introns for orthologous human/mouse included-conserved exons (ICEs) grouped by 5’ss score (bits). (b) Motif conservation scores (MCS, Supplementary Methods) of 5mers in downstream introns (11–70 nt from 5’ss) of ICEs with indicated 5’ss scores. Dashed lines show cutoff of significant MCS determined based on randomly shuffled data. Black dots represent motifs that are significantly conserved in more than one 5’ss groups. Motifs with more significant MCS in one group than all other groups are represented by colored dots in the same color scheme as in (a). Inset shows the histogram of the t-statistic of 4mers between the two indicated 5’ss groups. UUUU was the most significant in the 0–2 bits group. (c) Same as (b) for the 5’ss groups of 4–6 and 8–12 bits. (d, e, f) same as (a, b, c) for alternative-conserved exons (ACEs). Scatter plots are shown for the downstream intronic region 11–200 nt from the 5’ss. Because the number of exons was smaller in this analysis (∼3,000), only 3 bins of 5’ss strength were used (0–4, 4–8, 8–12 bits).
Some sequence motifs were highly conserved in intronic regions irrespective of 5'ss strength, suggesting that their activity does not depend on splice site strength (Fig. 4). This pattern was observed for 5mers matching the consensus binding motifs of the Fox-1/Fox-2 and STAR families of splicing factors (UGCAUG and ACUAAC, respectively36) and a few others.
Other motifs including UUUU were highly conserved only when adjacent to ICEs with very weak (0–2 bit) 5'ss, suggesting increased activity specifically in splicing of this class of exons. Consistent with this expectation, increased activity of U-run ISEs (which may act through the TIA-1 and/or TIAR splicing factors37) was observed in splicing of reporter exons with very weak 5'ss (Supplementary Fig. 13). Only one exonic motif was identified as differentially conserved dependent on 5'ss strength (Supplementary Table 7), suggesting that 5'ss strength-dependent activity is more common for intronic SREs. Previous studies of exonic motifs have observed increased density of certain exonic splicing enhancers (ESEs) in exons with weaker splice site sites38, a pattern expected even if ESE activity does not vary depending on splice site strength.
In addition, a diverse set of motifs were preferentially conserved adjacent to strong, intermediate, or weak 5'ss ICEs (Supplementary Table 7). Besides G triples (Fig. 1b), these motifs included GUGUG and UGUGU, which resemble the binding motifs of CELF family splicing factors39 and were conserved adjacent to ICEs and ACEs with intermediate and strong but not very weak 5'ss (Figs. 4b,c,f).
Discussion
Here, we present the first comprehensive study of the relationship between the strength of the 5'ss and splicing regulatory activity. The sensitivity of the splicing regulatory activity of G-runs to 5'ss strength suggests that G-run ISEs recruit factor(s) that enhance splicing at the step of initial 5'ss recognition by U1 snRNP or soon thereafter. Both U1:5'ss recognition and subsequent exon definition complex formation are important points of regulation40.
Several scenarios can be imagined that could account for the 5'ss strength-dependent activity of G-run ISEs. One possibility ("differential binding") is that the factor(s) responsible for splicing activation might bind more strongly to G-runs adjacent to intermediate strength 5'ss than to those near weak or strong 5'ss, with stronger binding leading to increased splicing activation. A weakness of this scenario is that how G-run binding would be affected by a motif located tens of bases away is not clear.
Another possibility ("differential activation") is that it is not binding to the pre-mRNA but activity in promoting splicing that varies for G-run-binding proteins depending on 5'ss strength, e.g., resulting from differences in the pathway of spliceosome assembly dependent on 5'ss strength. For example, if activation occurred through interaction with U1 snRNP, and if exons which have weak 5'ss and therefore low affinity for U1 snRNP were often spliced in a manner independent of U1 snRNP binding41–43. G-run activity might also vary depending on 5'ss strength for exons whose splicing is regulated kinetically, if activation occurred at a step which is rate-limiting for intermediate 5'ss exons, but a distinct step became rate-limiting for weak 5'ss exons. In-depth biochemical analyses are clearly needed to distinguish among these or other possible mechanisms.
The observed pattern of 5'ss strength-dependent ESS activity of exonic G-runs could potentially be explained through a combination of two competing activities of hnRNP H when bound to exonic G-runs: (i) a splicing inhibitory activity (e.g., involving inhibition of exon definition complex formation between the downstream 5'ss and upstream 3'ss) that occurs independently of 5'ss strength; and (ii) a splicing activating function that is similar or identical to that which is associated with intronic G-runs. Combined, these two activities might yield a pattern like that observed in Fig. 2d, with the inhibitory activity dominant in the case of weak or strong 5'ss, but roughly balanced by the more potent activating activity in the context of an intermediate 5'ss. Again, there are other possible scenarios.
The increased frequency of 5'ss polymorphism observed adjacent to G-run ISEs supports a common role for this motif as a buffer of genetic variation in the 5'ss. Such a buffering role could protect genes (presumably including disease genes) from some mutations that would otherwise disrupt their function, analogous to the buffering by some chaperones of mutations that would otherwise cause protein misfolding44.
Increased accumulation of neutral 5'ss polymorphisms, e.g., involving intermediate and strong 5'ss allele pairs, might contribute to evolution of alternative splicing. A straightforward pathway would involve reduction in the expression or activity of hnRNP H, e.g., through mutation of the hnRNP H locus. The 5'ss strength-dependence observed for G-run ISEs and ESSs (Fig. 1, Fig. 2) will tend to magnify differences between strong and intermediate 5'ss alleles when hnRNP H activity is reduced, thereby unmasking previously latent 5'ss variation as alternative splicing alleles, providing a substrate for natural selection. In the event that an allele generating an alternative splice of a formerly constitutive exon were advantageous or neutral, subsequent selection could act to tune the regulation, e.g., to bring it under the control of appropriate cell type- or condition-specific factors. Such a model would be directly analogous to the model of "evolutionary capacitance" by which the chaperone Hsp90 is proposed to accelerate evolutionary change45.
Changes in alternative splicing have been proposed as a major driver of phenotypic change in the mammalian lineage46,47, and G-runs and/or other motifs with potential to act as evolutionary capacitors of splicing change are likely to have accelerated these changes. Presence of intronic G-runs was not associated with an increase in the relative rate of non-synonymous substitutions (Supplementary Fig. 14) as would occur under the alternative "reduced selection pressure" model48.
Preferential conservation of a range of motifs adjacent to intermediate and weak 5'ss suggests that the activities of a number of different splicing factors may also exhibit 5'ss strength-dependent activity, as seen for G-runs and hnRNP H. In addition to potential roles in genetic buffering and effects on alternative splicing evolution, sensitivity to 5'ss strength may provide a general mechanism for tuning the responsiveness of distinct sets of exons to changes in the levels of a splicing factor, contributing to tissue-specific or environmentally regulated splicing.
Methods
Library Preparation for Illumina Sequencing
We used Poly-T capture beads to isolate mRNA from 10 ug of total RNA. First strand cDNA was generated using random hexamer-primed reverse transcription, and subsequently used to generate second strand cDNA using RNAase H and DNA polymerase. Sequencing adapters were ligated using the Illumina Genomic DNA sample prep kit. Fragments of ∼200 bp long were isolated by gel electrophoresis, amplified by 16 cycles of PCR, and sequenced on the Illumina Genome Analyzer. Further details regarding the mRNA-SEQ protocol can be found in50.
Splicing reporter constructs
To assess the activity of intronic splicing enhancers (ISEs) in the context of different splice sites, we used a ‘modular’ splicing reporter system described previously6. This reporter contains three exons with the test exon in the middle flanked by two GFP exons28. Splice site sequences were altered by site-directed mutagenesis at each splice site using primers covering the corresponding splice site (Supplementary Table 1). To insert sequences into the second intron of the splicing reporter, we used a reverse primer containing a SalI site and the desired insert sequence (e.g., G9) at its 5' end and a forward primer at the beginning of the upstream intron containing a HindIII site to mutate and amplify the test exon and its downstream intron via PCR. The sequences of the reverse primer were: CACGTCGACNNNNNNNNNNNNGTTGGAAAACAATAAAGAC (SalI site underlined and ISE region represented by N), and the forward primer was GAAACAAGGATGCTGTTAGAG. The resulting PCR product contains an ISE (or control) sequence 25 nt downstream from the 5'ss of the test exon and is inserted into the reporter backbone digested with SalI and HindIII. The control sequence used was CGTGCAAATCAA (designated G0 because it lacks G-runs). Nucleotides in the control sequence were replaced with different numbers of G runs to generate ISE sequences CGTGCGGGTCAA (designated G3), CGGGCGGGTCAA (G6), and CGGGGGGGGGAA (G9). All constructs were sequenced to confirm correct insert before transfection.
Cell culture, transfection, RNA purification and qRT-PCR
We cultured 293T cells with D-MEM medium supplemented with 10% (v/v) fetal bovine serum. The splicing reporter constructs were transfected (0.8 µg per well) with Lipofectamine 2000 (Invitrogen) in 12-well culture plates according to manufacturer instructions. Total RNA was purified from transfected cells using trizol / chlorophorm extraction followed by isopropanol precipitation and RNeasy column purification (Qiagen). The reverse transcription (RT) reaction was carried out using 2 µg total RNA with SuperScript III (Invitrogen). One tenth of the product from the RT reaction was used for PCR (20 cycles of amplification, with trace amount of α-32P-dCTP in addition to non-radioactive dNTPs). Quantitation of splicing isoforms was conduced as described previously51.
RNAi
We conducted knockdown of hnRNP H (H-KD) in four biological replicates (no. 1–3 for Exon array experiments and no. 4 for mRNA-SEQ, see below), as was the control knockdown using control siRNA. The dsRNA used for H-KD (IDT DNA) had sequences52: 5'-/5Phos/rArArCrUrUrGrArArUrCrArGrArArGrArUrGrArArGrUrCAA-3' 5'-rUrUrGrArCrUrUrCrArUrCrUrUrCrUrGrArUrUrCrArArGrUrUrCrA-3'
As a control we used the dsRNA Negative Control (DS ScrambledNeg) provided by IDT: 5'-/Phos/rCrUrUrCrCrUrCrUrCrUrUrUrCrUrCrUrCrCrCrUrUrGrUGA-3' 5'-rUrCrArCrArArGrGrGrArGrArGrArArArGrArGrArGrGrArArGrGrA-3'
The siRNA sequence for hnRNP H is partially complementary (at bases 1 to 19 from the 5' end of the siRNA with a mismatch at position 7) with the mRNA of the related gene, hnRNP F. In 293T cells, hnRNP H is known to be expressed at much higher levels than F by Western analysis24. From the analysis of mRNA-SEQ (see Supplementary Methods), we detected down-regulation at the mRNA level of both hnRNP H and F (∼3 fold) following siRNA transfection, considering only reads specific for each of these two closely-related genes. Similar analyses using Affymetrix exon arrays (see Supplementary Methods) yielded ∼2 fold down-regulation for both hnRNP H and F.
We used two different protocols in the knockdown experiments. Protocol 1, used for H-KD 1 and control 1 was as follows. Day 0: plate cells in 10cm dishes. Day 1: transfect 20nM siRNA with Lipofectamine 2000 (Invitrogen). Day 3: harvest cells. Protocol 2, used for H-KDs 2, 3 and 4 and controls 2, 3 and 4 was as follows. Day 0: plate cells in 10cm dishes. Day 1: transfect 20nM siRNA using Dharmafect 1 (Dharmacon) as transfection reagent. Day 2: transfect 50nM siRNA using Dharmafect 1 (Dharmacon) as transfection reagent. Day 4: harvest cells. Transfections were conducted using protocols suggested by the manufacturer of the transfection reagent. After cell harvest, three quarters of each dish were used for RNA extraction (using trizol / chlorophorm extraction followed by isopropanol precipitation and RNeasy column purification (Qiagen)) and one quarter was used for protein extraction. The quality of recovered RNA was verified by Bioanalyzer analysis (Agilent). Samples for mRNA-SEQ were processed and sequenced at Illumina Inc. Samples for exon array analysis were labeled, hybridized to the Affymetrix GeneChip® Human Exon 1.0 ST exon microarrays and scanned at the MIT BioMicrocenter following the manu facturer's instructions. The extent of H-KD was assessed both at the mRNA (real-time PCR) and protein levels (Western), as described in Supplementary Methods.
Analyses of organism- and tissue-specific alternative splicing
For Fig. 3e, we considered changes in splicing pattern where constitutive splicing was observed in human and alternative splicing in mouse rather than the reverse because the higher coverage of the human transcriptome in available expressed sequence tags (EST) and mRNA-Seq datasets enabled more confident identification of constitutive exons in human than in mouse. For Supplementary Fig. 12, we observed significant enrichment of G-runs downstream of exons with high Ψ values in three tissues (adipose, testis and the cell line MB435) in the set of intermediate 5'ss exons, while no significant enrichment for any tissue was observed in the strong 5'ss exon set, despite its larger size, and G-run enrichment was reduced to slightly below the Bonferroni-corrected P-value cutoff in the complete set of exons. This analysis suggested higher activity of hnRNP H in testis, consistent with the high levels of hnRNP H protein detected by Western analysis32, and also in adipose and MB435 cells. More generally, these observations suggest that subdividing exons based on splice site strength will provide greater statistical power to detect activity when considering SREs that have splice site strength-dependent activity.
Supplementary Material
Acknowledgments
We thank F. Allain and K. Lynch for helpful discussions, M. McNally, C. Nielsen, R. Sandberg, P. A. Sharp and members of the Burge lab for helpful comments on this manuscript, and G. P. Schroth and his research group for high-throughput cDNA sequencing.
Financial Disclosure
This work was supported by postdoctoral fellowships from the American Heart Association (X. X.) and the Human Frontiers Science Foundation (R. N.), by a training grant from the NIH (E. T. W.), by NSF equipment grant DBI-0821391, and by grants from the NIH (C. B. B.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Accession Codes
We have submitted the mRNA-SEQ reads to the GEO short read archive (accession no. GSE16642) and the Microarray data to GEO (accession no. GSE12386).
Supplementary Material
Supporting Information includes Methods, 7 Supplementary Tables, and 14 Supplementary Figures
Competing Interest Statement
The authors declare that they have no competing financial interests with respect to this work.
References
- 1.Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 2.Lu ZX, Peng J, Su B. A human-specific mutation leads to the origin of a novel splice form of neuropsin (KLK8), a gene involved in learning and memory. Hum Mutat. 2007;28:978–984. doi: 10.1002/humu.20547. [DOI] [PubMed] [Google Scholar]
- 3.Pan Q, et al. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 5.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao X, Wang Z, Jang M, Burge CB. Coevolutionary networks of splicing cis-regulatory elements. Proceedings of the National Academy of Sciences. 2007 doi: 10.1073/pnas.0707349104. 0707349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCullough AJ, Berget SM. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fogel BL, McNally MT. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J Biol Chem. 2000;275:32371–32378. doi: 10.1074/jbc.M005000200. [DOI] [PubMed] [Google Scholar]
- 9.Hastings ML, Wilson CM, Munroe SH. A purine-rich intronic element enhances alternative splicing of thyroid hormone receptor mRNA. Rna. 2001;7:859–874. doi: 10.1017/s1355838201002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H’/F/2H9 family. J Biol Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 11.Yeo G, Hoon S, Venkatesh B, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc Natl Acad Sci U S A. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNally LM, Yee L, McNally MT. Heterogeneous nuclear ribonucleoprotein H is required for optimal U11 small nuclear ribonucleoprotein binding to a retroviral RNA-processing control element: implications for U12-dependent RNA splicing. J Biol Chem. 2006;281:2478–2488. doi: 10.1074/jbc.M511215200. [DOI] [PubMed] [Google Scholar]
- 13.Kralovicova J, Vorechovsky I. Position-dependent repression and promotion of DQB1 intron 3 splicing by GGGG motifs. J Immunol. 2006;176:2381–2388. doi: 10.4049/jimmunol.176.4.2381. [DOI] [PubMed] [Google Scholar]
- 14.Marcucci R, Baralle FE, Romano M. Complex splicing control of the human Thrombopoietin gene by intronic G runs. Nucl. Acids Res. 2007;35:132–142. doi: 10.1093/nar/gkl965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauger DM, Lin C, Garcia-Blanco MA. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol Cell Biol. 2008;28:5403–5419. doi: 10.1128/MCB.00739-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez C, Allain FH. NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition. Nucleic Acids Res. 2006;34:3634–3645. doi: 10.1093/nar/gkl488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang XH, Leslie CS, Chasin LA. Dichotomous splicing signals in exon flanks. Genome Res. 2005;15:768–779. doi: 10.1101/gr.3217705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roca X, Sachidanandam R, Krainer AR. Determinants of the inherent strength of human 5' splice sites. Rna. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 20.Murray JI, Voelker RB, Henscheid KL, Warf MB, Berglund JA. Identification of motifs that function in the splicing of non-canonical introns. Genome Biol. 2008;9:R97. doi: 10.1186/gb-2008-9-6-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venables JP, et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol. 2008;28:6033–6043. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izquierdo JM, et al. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 24.Alkan SA, Martincic K, Milcarek C. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem J. 2006;393:361–371. doi: 10.1042/BJ20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CD, Kobayashi R, Helfman DM. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, et al. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH, et al. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 2005;33:3866–3874. doi: 10.1093/nar/gki698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul S, et al. Interaction of muscleblind, CUG-BP1 and hnRNP H proteins in DM1-associated aberrant IR splicing. Embo J. 2006;25:4271–4283. doi: 10.1038/sj.emboj.7601296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondrashov FA, Koonin EV. Evolution of alternative splicing: deletions, insertions and origin of functional parts of proteins from intron sequences. Trends Genet. 2003;19:115–119. doi: 10.1016/S0168-9525(02)00029-X. [DOI] [PubMed] [Google Scholar]
- 32.Honore B, Baandrup U, Vorum H. Heterogeneous nuclear ribonucleoproteins F and H/H' show differential expression in normal and selected cancer tissues. Exp Cell Res. 2004;294:199–209. doi: 10.1016/j.yexcr.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Modafferi EF, Black DL. Combinatorial control of a neuron-specific exon. Rna. 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Garg K, Green P. Differing patterns of selection in alternative and constitutive splice sites. Genome Res. 2007;17:1015–1022. doi: 10.1101/gr.6347907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 37.Del Gatto-Konczak F, et al. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5' splice site. Mol Cell Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 39.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith DJ, Query CC, Konarska MM. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crispino JD, Blencowe BJ, Sharp PA. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science. 1994;265:1866–1869. doi: 10.1126/science.8091213. [DOI] [PubMed] [Google Scholar]
- 42.Tarn WY, Steitz JA. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 1994;8:2704–2717. doi: 10.1101/gad.8.22.2704. [DOI] [PubMed] [Google Scholar]
- 43.Fukumura K, Taniguchi I, Sakamoto H, Ohno M, Inoue K. U1-independent pre-mRNA splicing contributes to the regulation of alternative splicing. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maisnier-Patin S, et al. Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nat Genet. 2005;37:1376–1379. doi: 10.1038/ng1676. [DOI] [PubMed] [Google Scholar]
- 45.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 46.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 47.Jin L, et al. The evolutionary relationship between gene duplication and alternative splicing. Gene. 2008;427:19–31. doi: 10.1016/j.gene.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Xing Y, Lee C. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc Natl Acad Sci U S A. 2005;102:13526–13531. doi: 10.1073/pnas.0501213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroth GP, Luo S, Khrebtukova I. Methods Mol. Biol. 2008 in press. [Google Scholar]
- 51.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim DH, et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.