Summary
Previous studies have suggested that the activity of the mammalian origin recognition complex (ORC) is regulated by cell-cycle-dependent changes in its Orc1 subunit. Here, we show that Orc1 modifications such as mono-ubiquitylation and hyperphosphorylation that occur normally during S and G2-M phases, respectively, can cause Orc1 to accumulate in the cytoplasm. This would suppress reassembly of pre-replication complexes until mitosis is complete. In the absence of these modifications, transient expression of Orc1 rapidly induced p53-independent apoptosis, and Orc1 accumulated perinuclearly rather than uniformly throughout the nucleus. This behavior mimicked the increased concentration and perinuclear accumulation of endogenous Orc1 in apoptotic cells that arise spontaneously in proliferating cell cultures. Remarkably, expression of Orc1 in the presence of an equivalent amount of Orc2, the only ORC subunit that did not induce apoptosis, prevented induction of apoptosis and restored uniform nuclear localization of Orc1. This would promote assembly of ORC-chromatin sites, such as occurs during the transition from M to G1 phase. These results provide direct evidence in support of the regulatory role proposed for Orc1, and suggest that aberrant DNA replication during mammalian development could result in apoptosis through the appearance of ‘unmodified’ Orc1.
Keywords: Cell division cycle, CHO, HeLa, ORC, Phosphorylation, Ubiquitylation
Introduction
One universal feature of eukaryotic DNA replication is that the genome is duplicated once and only once each time a cell divides. This is accomplished by inactivating the existing pre-replication complexes (pre-RCs) after S phase begins, yet concomitantly preventing assembly of new pre-RCs until mitosis is complete and a nuclear membrane is present. The importance of this principle is highlighted by the fact that endoreduplication (multiple rounds of DNA replication in the absence of an intervening mitosis) is a rare event. In mammals, it occurs only in the terminally differentiated trophoblast giant cells and in megakaryocytes.
Eukaryotic DNA replication begins with the binding of an origin recognition complex (ORC) to DNA, an event that is regulated by cell-cycle-dependent changes in one or more of the six ORC subunits (DePamphilis, 2005). In mammals, these changes appear to be specific for the largest ORC subunit, Orc1. The mammalian ORC consists of a stable core complex of ORC subunits Orc2, Orc3, Orc4 and Orc5 [referred to here as ORC(2–5)] that associate weakly with Orc1 and Orc6 (Dhar et al., 2001; Giordano-Coltart et al., 2005; Kneissl et al., 2003; Vashee et al., 2001). Interaction of Orc1 with the ORC core complex in the presence of ATP is essential for binding ORC to DNA and for assembly of pre-RCs in vitro (Giordano-Coltart et al., 2005). Selective inhibition of Orc1 synthesis in cultured cells confirms that it is also required for assembly of pre-RCs in vivo (Ohta et al., 2003), and the selective loss of Orc1 from chromatin during mitosis accounts for the fact that metaphase mammalian chromatin will not replicate in ORC-depleted Xenopus egg extracts (Li et al., 2000; Natale et al., 2000; Yu et al., 1998).
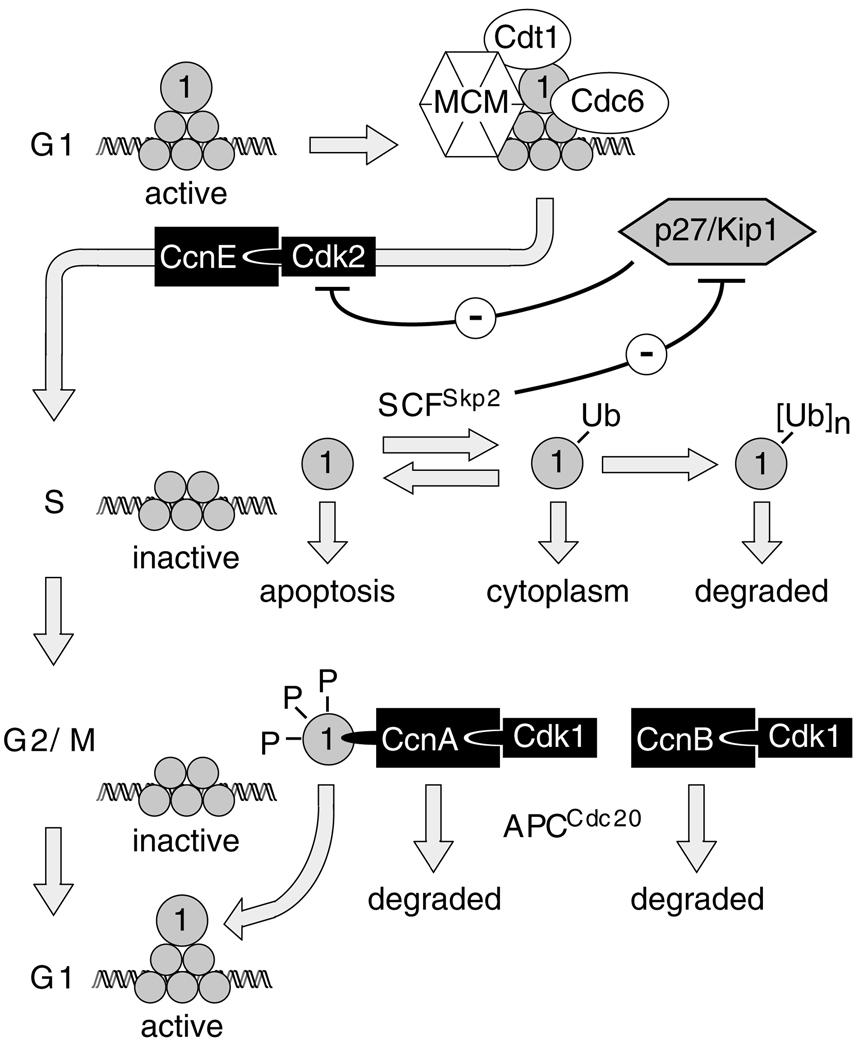
ORC activity in mammalian cells appears to be regulated by cell-cycle-dependent changes in the association of Orc1 with ORC-chromatin sites (DePamphilis, 2005). The so-called mammalian ‘ORC cycle’ involves three specific changes in Orc1 (see diagram in Fig. 10). First, the affinity of Orc1 for chromatin is selectively decreased as mammalian cells enter S phase (Kreitz et al., 2001; Li and DePamphilis, 2002; Ohta et al., 2003), a change that is reflected in the fact that Orc1 can no longer be crosslinked to DNA in S phase cells (Abdurashidova et al., 2003; Ladenburger et al., 2002). By contrast, the ORC(2–5) core remains stably bound to chromatin throughout cell division (discussed in DePamphilis, 2005). Second, Orc1 is subject to ubiquitylation during S phase (Fujita et al., 2002; Li and DePamphilis, 2002; Mendez et al., 2002; Ritzi et al., 2003; Tatsumi et al., 2003). In transformed human cells, 70–90% of the HsOrc1 is selectively degraded during S phase by an ubiquitin-dependent mechanism (Fujita et al., 2002; Mendez et al., 2002; Ritzi et al., 2003; Tatsumi et al., 2003) (A.V., unpublished data). HsOrc1 is then resynthesized and bound to chromatin during the transition from M to G1 phase. Finally, Orc1 is hyperphosphorylated during G2-M phase, as a result of its association with CcnA-Cdk1 (Li et al., 2004; Tatsumi et al., 2000; Thome et al., 2000). Since inhibition of CDK activity in metaphase cells causes rapid binding of Orc1 (Li et al., 2004) and minichromosome maintenance (MCM) proteins (Ballabeni et al., 2004) to chromatin, and premature initiation of DNA replication (Itzhaki et al., 1997), hyperphosphorylation of Orc1 during the G2-M phase presumably prevents its stable association with ORC(2–5)–chromatin sites. This would account for the absence of functional ORC-chromatin sites on metaphase chromatin (Li et al., 2000; Natale et al., 2000; Yu et al., 1998). Orc1 reassociates tightly with chromatin during the transition from M to G1 phase (Li and DePamphilis, 2002; Mendez et al., 2002; Natale et al., 2000) followed closely by the appearance of pre-RCs at specific genomic sites (Li et al., 2000; Natale et al., 2000).
Fig. 10.
The mammalian ‘ORC cycle’. All six ORC subunits (shaded circles) are bound tightly to chromatin during G1 phase to provide sites for initiation of pre-replication complex (pre-RC) assembly. DNA synthesis does not begin until pre-RCs are activated by CcnE-Cdk2, a protein kinase whose activity is inhibited during G1 phase by the CDK-specific inhibitor p27/Kip1. With the onset of S phase, the affinity of Orc1 for chromatin is selectively reduced, and Orc1 is targeted for ubiquitylation by SCFSkp2. Since SCFSkp2-dependent degradation of p27 is essential for cell-cycle progression (Kossatz et al., 2004), CcnE-Cdk2 activation is accompanied by Orc1 inactivation, resulting in initiation of S phase with concomitant suppression of pre-RC assembly. Orc1 is selectively degraded during S phase in human cells, but not in hamster cells. However, even a single ubiquitin adduct can inactivate Orc1 by transporting it to the cytoplasm. In M phase, Orc1 is hyperphosphorylated and bound tightly to CcnA-Cdk1, but weakly to chromatin.
Hyperphosphorylation favors cytoplasmic localization. When CcnA and CcnB are degraded during the transition from M to G1 phase, Cdk1 is inactivated, and Orc1 is dephosphorylated and rebound tightly to chromatin. If Orc1 is not associated with ORC, and if it is not ubiquitylated or hyperphosphorylated, then Orc1 can induce apoptosis.
Although these studies suggest that ORC activity in mammalian cells is regulated by cell-cycle-dependent changes in Orc1, other studies have suggested that this might not always be the case. In contrast to HeLa cells, the intracellular level of Orc1 in Chinese hamster ovary (CHO) cells remains constant throughout cell division (Li and DePamphilis, 2002; Li et al., 2004; Natale et al., 2000; Okuno et al., 2001), and this difference has been confirmed in parallel studies under identical conditions (S.G., unpublished data). Furthermore, although both hamster and human Orc1 can be ubiquitylated in vivo and in vitro (Li and DePamphilis, 2002; Mendez et al., 2002) (S.G., unpublished data), only a mono-ubiquitylated form of Orc1 has been detected during S phase in some studies (Li and DePamphilis, 2002), but not in others (Okuno et al., 2001). Moreover, there is no evidence that mono-ubiquitylation of Orc1 interferes with its function. Similarly, the effects of hyperphosphorylation on the ability of Orc1 to participate in DNA replication are implied from the inhibition of protein kinase activities in metaphase cells; direct evidence is lacking.
To determine whether or not mono-ubiquitylation and hyperphosphorylation of Orc1 can, in fact, suppress Orc1 function, modified and unmodified ORC1 genes were transiently expressed in CHO and HeLa cells, and their effects compared with those of transiently expressed Orc2. The results demonstrated that the cell-cycle-dependent changes in Orc1 described above could, in fact, affect ORC activity; activating it during the transition from M to G1 phase, and suppressing it during the transition from S to M phase. Unexpectedly, these experiments also revealed that unmodified Orc1 could induce apoptosis that was similar to apoptotic cells that had arisen spontaneously during cell proliferation. Moreover, the same modifications that prevented Orc1 from participating in DNA replication also neutralized its ability to induce apoptosis, suggesting that failure to regulate ORC activity during mammalian development could result in cell death.
Results
Orc1 can induce loss of cell adhesion
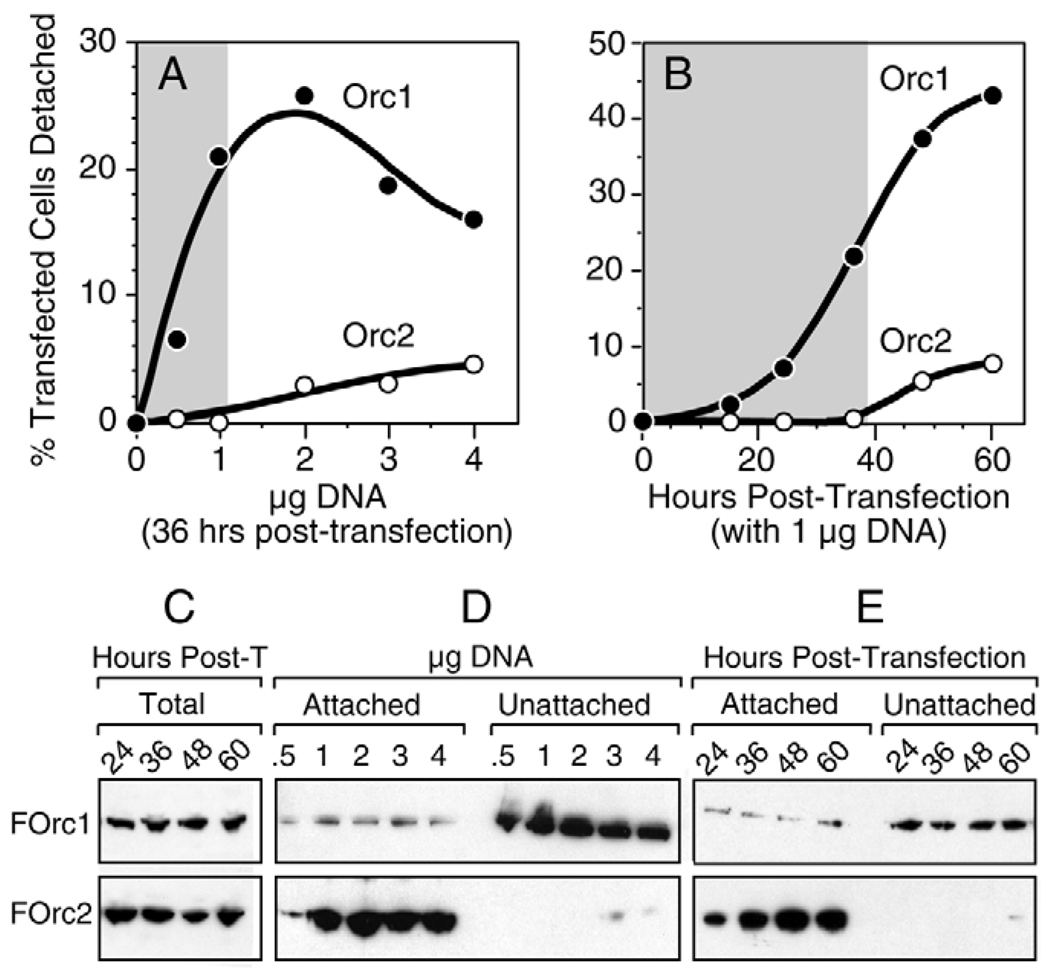
To determine whether or not changes in the cellular levels of Orc1 can affect cell viability, the Chinese hamster ovary cell line CHO C400 and the human tumor cell line HeLa were transfected with plasmid DNA expressing either their cognate Orc1 or Orc2 protein. DNA replication in these cell lines has been characterized extensively, and their Orc1 proteins have been reported to exhibit different sensitivities to ubiquitin-dependent degradation in vivo (see Introduction). When CHO cells were transfected with a plasmid that expressed FLAG-tagged Chinese hamster (Cg) Orc1 (pFCgOrc1), a fraction of them were observed to round up and lift off the dish. This fraction increased either with the amount of DNA transfected (Fig. 1A) or with the time elapsed following transfection (Fig. 1B). By contrast, comparatively few cells were released from the dish following transfection with pFCgOrc2, and this number was only slightly greater than the fraction of cells released by transfection with pCI, the parent plasmid (Fig. 1A,B). At 36 hours post-transfection with 1 µg DNA, the number of unattached cells was >20-fold greater with pFCgOrc1 relative to either pFCgOrc2 or pCI (shaded areas in Fig. 1). Similar results were obtained using wild-type CgOrc1 and CgOrc2 proteins, and by transfecting HeLa cells with the same vectors expressing the corresponding human proteins (data not shown). These results suggested that ectopic expression of CgOrc1 induced loss of cell adhesion.
Fig. 1.
Ectopic expression of CHO (Cg) Orc1 induced loss of cell adhesion under conditions where ectopic expression of CgOrc2 did not. (A) CHO cells were transfected with the amount of either pCgOrc1 (●) or pCgOrc2 (○) indicated and then incubated for 36 hours before determining the fraction of unattached cells. (B) CHO cells were transfected with 1 µg of either pCgOrc1 (●) or pCgOrc2 (○) and then incubated for the times indicated before determining the fraction of unattached cells. Data with pCI alone was subtracted from data for pCgOrc1 and pCgOrc2. Shaded areas indicate the conditions under which pCgOrc2 and pCI gave indistinguishable results. (C) CHO cells were transfected with 1 µg of either pFCgOrc1 (FOrc1) or pFCgOrc2 (FOrc2) for the times indicated in hours post-transfection (post-T). FOrc1 and FOrc2 were quantified in the total cell population (attached plus unattached) by immunoblotting using anti-FLAG antibody. A single band was detected that migrated as ~100 kDa for FOrc1 and ~65 kDa for FOrc2. (D) CHO cells, transfected as in panel A, were separated into those that lifted off the dish (unattached) and those that remained attached. Half of the unattached cells and 104 attached cells were assayed for either FOrc1 or FOrc2. (E) CHO cells, transfected as in panel B, were separated into attached and unattached cells before assaying for FOrc1 and FOrc2 for the times indicated. In each experiment, attached and unattached cell proteins were fractionated in the same gel, transferred to the same immunoblot, and detected with the same anti-FLAG antibody to ensure accurate comparison.
To test this hypothesis, ratios of ectopically expressed Orc protein were determined in attached cells versus unattached cells. Comparison of aliquots of the total cell population (attached plus unattached cells) revealed that FCgOrc1 and FCgOrc2 were the only proteins detected by anti-FLAG antibody, and that the total amount of FCgOrc2 expressed in hamster cells was about twice that of FCgOrc1 (Fig. 1C). Unattached cells were greatly enriched for FCgOrc1 whereas attached cells were greatly enriched for FCgOrc2, regardless of whether cells had been transfected with different amounts of DNA and then cultured for 36 hours (Fig. 1D), or transfected with 1 µg DNA and then cultured for increasing amounts of time (Fig. 1E). To ensure accurate comparisons, the same amounts of attached and unattached cell lysates were present on the same immunoblot where Orc1 and Orc2 were detected with the same anti-FLAG antibody. At 36 hours post-transfection with 1 µg DNA, the ratio of FCgOrc1 in unattached cells to FCgOrc1 in attached cells was 6.8, whereas the same ratio for FCgOrc2 was <0.01. Thus, ectopic expression of Orc1 induced loss of cell adhesion whereas ectopic expression of Orc2 did not.
Only Orc2 did not induce loss of cell adhesion
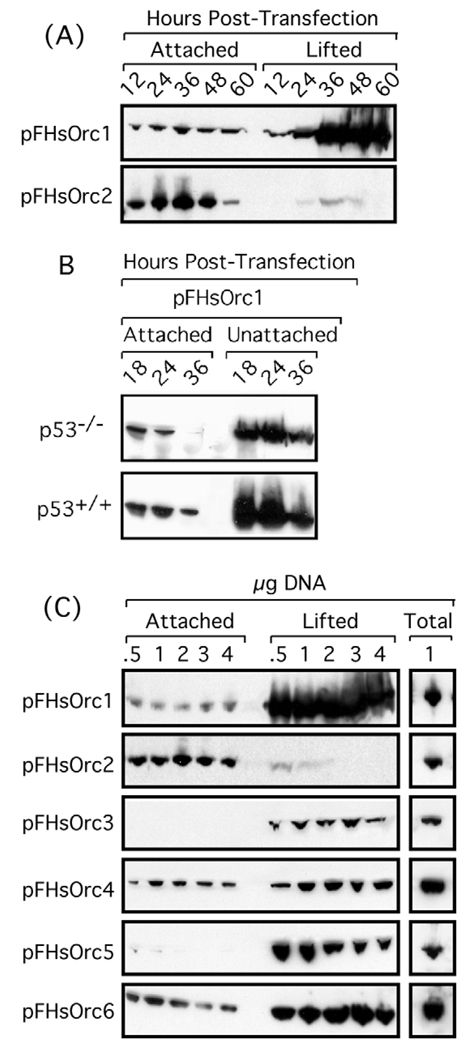
In contrast to CHO cells where the steady-state level of Orc1 is constant during cell division, HeLa cells selectively degrade Orc1 during S phase. Therefore, we determined whether HsOrc1 would also induce loss of cell adhesion in a HeLa cell line that proliferated as a monolayer. The results (Fig. 2) were essentially the same as with CgOrc1 and CgOrc2 in CHO cells (Fig. 1).
Fig. 2.
Ectopic expression of human (Hs) Orc1, Orc3, Orc4, Orc5 and Orc6 induced loss of cell adhesion under conditions where ectopic expression of HsOrc2 did not. (A) HeLa cells were transfected with 1 µg of the plasmid indicated (pFHsOrc1 or pFHsOrc1) and then incubated for the times indicated before determining the fraction of unattached cells. (B) HCT116 cells (±TP53 gene, as indicated by p53−/− and p53+/+) transfected with 1 µg of pFHsOrc1 for the times indicated. (C) CHO cells were transfected with the amount of plasmid DNA indicated and then incubated for 36 hours before determining the fraction of unattached cells.
The ability of ORC subunits to induce loss of cell adhesion appeared to be independent of Tp53, because both CHO (Hu et al., 1999; Lee et al., 1997) and HeLa cells (Hoppe-Seyler and Butz, 1993) contain little Tp53 activity. In fact, transfecting Tp53+/+ HCT116 cells and Tp53−/− HCT116 cells with a plasmid that expressed FLAG-tagged human (Hs) Orc1 (pFHsOrc1) gave results (Fig. 2B) indistinguishable from those with either CHO (Fig. 1) or HeLa cells (Fig. 2). Therefore, loss of cell adhesion induced by Orc1 was not dependent on a functional TP53 gene.
To determine whether either Orc1 or Orc2 was unique among ORC subunits, all six human ORC subunits were tested in parallel for their ability to induce loss of cell adhesion. Results with HsOrc3, 4, 5 and 6 were essentially the same as those with HsOrc1 (Fig. 2B). Only Orc2 did not induce loss of cell adhesion.
Orc1 can induce changes in cell morphology
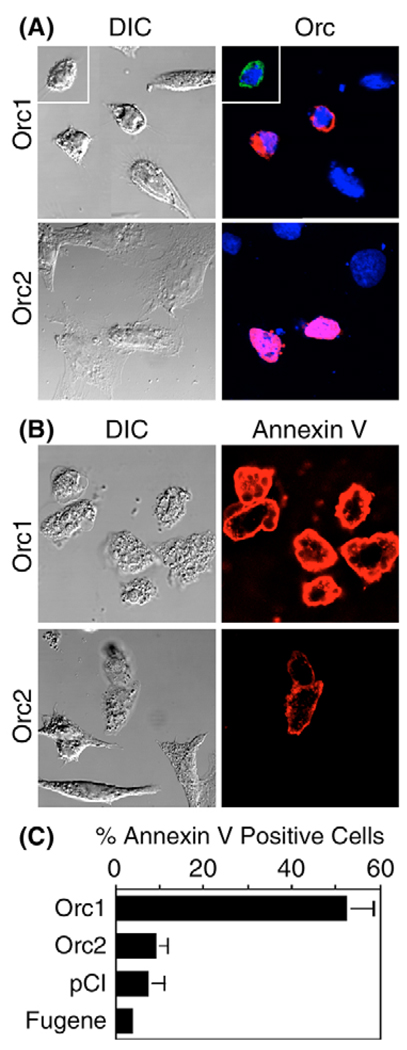
A second characteristic of apoptosis is a rounded cell morphology with blebbing (Otsuki et al., 2003). Such morphology was routinely observed in fixed cells that contained FLAG-tagged Orc1 (FOrc1), but not in those that contained FOrc2 (Fig. 3A). A third characteristic of apoptosis is the translocation of phosphatidylserine from the inner to the outer leaflet of the plasma membrane, a change that can be detected by the calcium-dependent binding of annexin-V to living cells (Vermes et al., 1995). Approximately half of the cells transfected with Orc1 expression vectors bound annexin-V, and these cells also exhibited rounded morphology and blebbing (Fig. 3B,C). By contrast, only ~9% of the cells transfected with Orc2 expression vectors bound annexin-V (Fig. 3C), and these cells appeared morphologically normal. Moreover, the intensity of the stain was significantly less with Orc2 transfectants than with Orc1 transfectants. Cells transfected with either pCI or Fugene alone were similar to those transfected with Orc2 expression vectors. Similar results were obtained with human cells transfected with pFHsOrc1 or pFHsOrc2 (data not shown). Thus, ectopic expression of Orc2 had little effect on cell morphology.
Fig. 3.
Ectopic expression of Orc1 induced exposure of phosphatidylserine and blebs on cell surfaces, but Orc2 did not. (A) CHO cells were transfected with 1 µg pFCgOrc1 (Orc1) or pFCgOrc2 (Orc2) DNA. At 24 hours post-transfection, they were fixed and then stained with anti-FLAG antibody (red). Inset: cells transfected with pCgOrc1 [stained with anti-CgOrc1 antibody (green)] gave results comparable with those transfected with pFCgOrc1. (B) Live transfected cells were stained with annexin-V to reveal cells undergoing apoptosis. (C) The fraction of cells that bound annexin-V was determined for the indicated plasmid or with Fugene alone. DIC, differential interference contrast.
Both necrotic cells and late-stage apoptotic cells exhibit a loss of membrane integrity that can be detected by staining with a non-vital dye such as BOBO-1. Approximately half of the annexin-V+ cells in the pFHsOrc1-transfected population also stained with BOBO-1 at 36 hours post-transfection (data not shown). By contrast, <3% of the cells transfected with pFCgOrc2, pCI or Fugene alone were stained with BOBO-1.
Orc1 can induce DNA fragmentation
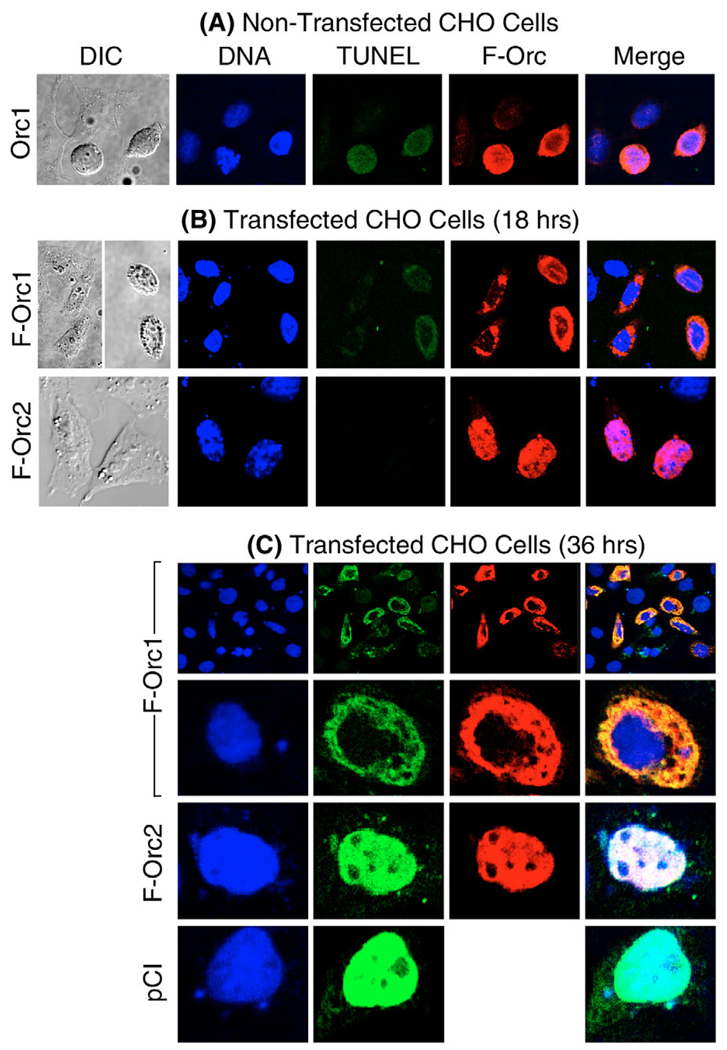
A fourth characteristic of apoptosis is nuclear DNA fragmentation. Apoptotic endonucleases generate free 3′-OH groups at the ends of DNA fragments that can be labeled with fluorescein-conjugated dUTP by terminal deoxynucleotidyl transferase [TUNEL assay (Otsuki et al., 2003)]. The same cells can then be stained with anti-FLAG antibody to identify the ectopically expressed protein and with Hoechst dye to identify nuclear DNA.
When this protocol was carried out with non-transfected CHO cells attached to the dish, ~1% or fewer exhibited unusually high levels of endogenous Orc1 protein that tended to accumulate perinuclearly (around the genomic DNA, Fig. 4A). The rounded, blebbed morphological appearance of these cells and the clear presence of fragmented nuclear DNA (TUNEL-positive) cells suggested that they were undergoing apoptosis.
Fig. 4.
Ectopic expression of Orc1 induced DNA fragmentation similar to non-transfected cells that spontaneously over-expressed Orc1. (A) Non-transfected CHO cells were stained with Hoechst to stain DNA (blue), with TUNEL assay to label double-stranded DNA breaks (green), and with anti-FLAG antibody to label FCgOrc1 (red; F-Orc). DIC, differential interference contrast. (B) CHO cells were transfected with 1 µg of either pFCgOrc1 (F-Orc1) or pFCgOrc2 (F-Orc2). At 18 hours post-transfection, the attached cells were treated as in panel A. (C) Same as in panel B, except that attached cells were analyzed at 36 hours post-transfection, and pCI-transfected cells were included. In the merged images, yellow indicates sites where DNA breaks and Orc were co-localized, and white indicates sites where DNA and Orc1 were co-localized.
Similar results could be induced in CHO cells by ectopic expression of FCgOrc1. Following transfection, ~40% of the attached cells expressed FCgOrc1, and all of these cells contained fragmented DNA, although the intensity of the TUNEL stain was significantly greater at 36 hours post-transfection (Fig. 4C) than at 18 hours post-transfection (Fig. 4B). Moreover, the fragmented DNA and the FCgOrc1 were coincident when the two images were merged; both were perinuclear. The same perinuclear staining pattern appeared when pFCgOrc1-transfected cells were stained only with the TUNEL assay and with Hoechst dye. Similar results were obtained with human cells (data not shown).
In contrast to ectopic expression of Orc1, ectopic expression of Orc2 was not tightly linked to apoptosis. At 18 hours post-transfection, approximately 40% of the cells contained FCgOrc2 distributed throughout the nucleus, none of these cells was clearly TUNEL positive and all of them exhibited a normal morphology (Fig. 4B). By 36 hours post-transfection, approximately 10% of the cells that contained FCgOrc2 also contained fragmented DNA, and the FCgOrc2 and TUNEL stains, which were coincident, were distributed throughout the nucleus (Fig. 4C). Since results with cells transfected with pCI (which expressed no protein) were indistinguishable from those with pFCgOrc2 (Fig. 4C), the DNA fragmentation observed in Orc2-expressing cells resulted from the transfection protocol and not from ectopic expression of Orc2. Similar results were obtained with human cells (data not shown).
Loss of cell adhesion preceded DNA fragmentation
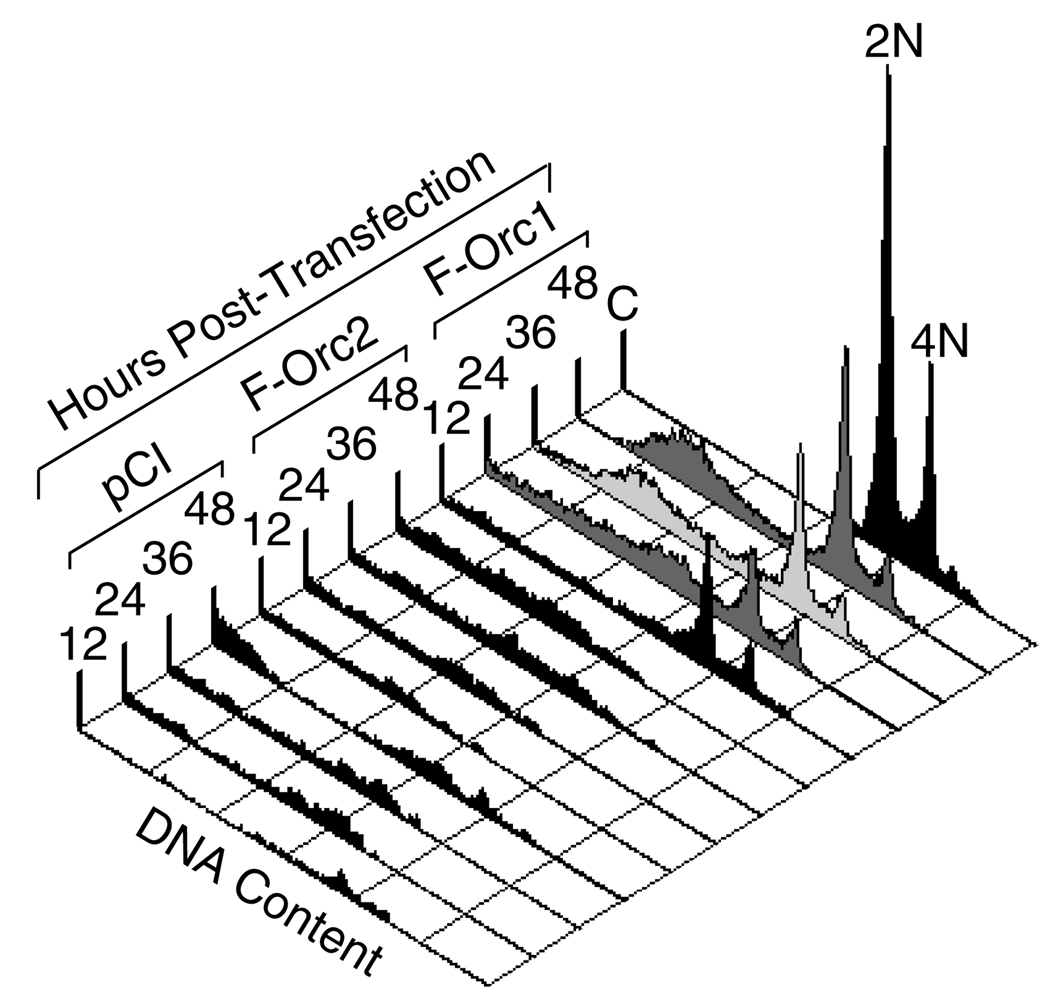
To determine whether or not ectopic expression of ORC proteins induced re-replication of genomic DNA as well as DNA fragmentation, the unattached cells in a transfected cell culture were subjected to fluorescence activated cell sorting (FACS) at various times after transfection (Fig. 5). These results revealed that cells in which Orc1 expression had induced a loss of adhesion to the dish consisted of two populations, one with a normal DNA content [i.e. distributed between 2N (G1 phase) and 4N (G2-M phase)] and one with degraded genomic DNA (<2N). Since all of the Orc1-expressing cells that remained attached to the dish (Fig. 4) were TUNEL positive (enriched in damaged DNA), these FACS data revealed that loss of cell adhesion preceded extensive DNA fragmentation. By contrast, the comparatively small number of unattached cells from either pFCgOrc2 or pCI transfections contained only degraded genomic DNA, consistent with necrosis rather than apoptosis. Similar results were obtained with human cells, as well as when cells were transfected with wild-type Orc1 and Orc2 proteins.
Fig. 5.
Ectopic expression of Orc1 induced loss of cell adhesion prior to extensive DNA fragmentation, but not endoreduplication. Unattached CHO cells at the indicated times post-transfection with pCI, pFCgOrc2 (F-Orc2) or pFCgOrc1 (F-Orc1) were analyzed by fluorescence activated cell sorting (FACS). Control (C) was non-transfected cells. Cells at G1 phase (2N) and G2/M phase (4N) are indicated. Similar results were obtained with HeLa cells transfected with DNA expressing human Orc proteins (data not shown).
Orc1 did not induce endoreduplication
Cells containing >4N DNA content were not detected at any time following transfection with pFCgOrc1, revealing that ectopic expression of Orc1 did not induce endoreduplication (Fig. 5).
Low levels of Orc1 can rapidly induce caspase-3 activity
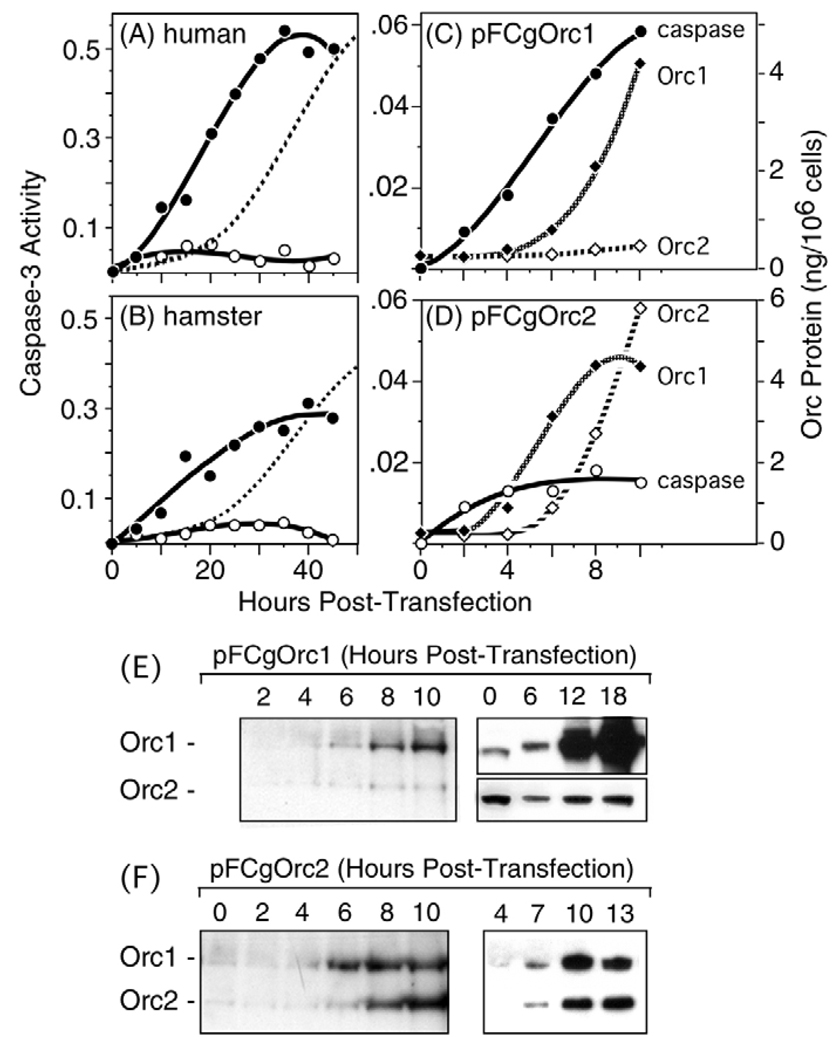
A sixth characteristic of apoptosis is the rapid appearance of caspase-3 activity, a protease that is indispensable for apoptotic DNA fragmentation (Otsuki et al., 2003). Caspase-3 activity increased immediately following transfection of cells with Orc1 expression vectors (Fig. 6A,B). By contrast, the amount of caspase-3 activity induced by ectopic expression of Orc2 was only marginally greater than with pCI alone. Moreover, pretreatment of cells with 50 µM Z-VAD-fmk, a cell-permeable caspase inhibitor, revealed that about 90% of the ‘caspase-3 activity’ observed with pCI was not inhibited by Z-VAD-fmk. Therefore, the Z-VAD-fmk-resistant activity observed with pCI-transfected cells was subtracted from the activity observed with Orc1 and Orc2 expression vectors. The results revealed that induction of caspase-3 preceded the loss of cell adhesion by 15–20 hours (Fig. 6A,B; dashed line). By 36 hours post-transfection, CgOrc1 was approximately sixfold more effective than CgOrc2 and about sevenfold better than pCI in inducing caspase-3 activity. In human cells, these differences were ~10-fold and ~12-fold, respectively.
Fig. 6.
Low levels of ectopic Orc1 induced caspase-3 activity. (A) HeLa cells were transfected with 1 µg of either pCI, pFHsOrc1 (●) or pFHsOrc2 (○). At the indicated times post-transfection, extracts from ~106 cells were prepared by the freeze-thaw method and assayed for Z-VAD-fmk-sensitive caspase-3 activity using the colorimetric CaspACE™ Assay System (Promega). Activities observed with pCI were subtracted from those observed with Orc expression vectors. The fraction of unattached transfected cells is indicated by broken line (0.5=50%). (B) CHO cells transfected with pCI, pFCgOrc1 (●) or pFCgOrc2 (○) were analyzed as in panel A. (C) Amounts of Orc1 (◆) and Orc2 (◇) proteins in CHO cells transfected with pFCgOrc1 were determined by densitometry of data such as those in panel E. Comparisons were made only where images were not saturating. These results were compared with the appearance of caspase-3 activity (●) at the beginning of transfection. Comparison with standard amounts of recombinant Orc protein fractionated in parallel was used to determine the cellular concentration of Orc. (D) Amounts of Orc1 (◆) and Orc2 (◇) in CHO cells transfected with pCgOrc2 were determined from data such as those in panel E, and then compared with caspase-3 activity (○) at the beginning of transfection. (E) Total CgOrc1 (Orc1) and CgOrc2 (Orc2) in CHO cells following transfection with pFCgOrc1 was detected by immunoblotting with both anti-CgOrc1 and anti-CgOrc2 IgG. Two independent experiments are shown. In the second, different exposures of the same gel were required to visualize both Orc1 and Orc2. (F) Total CgOrc1 and CgOrc2 in CHO cells following transfection with pFCgOrc2 was determined as in panel E. Two independent experiments are shown.
Immunoblotting with anti-CgOrc1 antibody revealed that the amount of FCgOrc1 protein produced during the first 6 hours post-transfection, the time when caspase-3 activity was induced, was equivalent to the endogenous level of CgOrc1 (Fig. 6C,E). To quantify the cellular concentration of CgOrc1, total cell lysates were fractionated in parallel with known amounts of recombinant CgOrc1, and the resulting gels subjected to immunoblotting using anti-CgOrc1 antibody (data not shown). Purified Orc1 protein was quantified by direct comparison with known amounts of bovine serum albumin. The results were then compared with caspase-3 activity as a function of hours post-transfection (Fig. 6C).
Untransfected CHO cells contained ~2.3×104 molecules CgOrc1/cell [(3.69 ng CgOrc1/106 cells)(mole/9.5954×104 gm)(6.02×1023 molecules/mole)(gm/109 ng)=2.32×104 molecules CgOrc1/cell], which is consistent with a previous estimate of ~3×104 molecules CgOrc1/ cell (Natale et al., 2000). Following transfection with pFCgOrc1, the amount of Orc1 did not increase significantly until 6 hours post-transfection when the cells contained, on average, 2.8×104 molecules CgOrc1 each. Since 40% of the cells in these experiments expressed FLAG-tagged Orc1, each transfected cell contained ~3.6×104 molecules CgOrc1/cell at 6 hours [(2.8×104 molecules/cell) –0.6(2.3×104 molecules/untransfected cell)]/0.4=3.6×104 molecules/transfected cell]. By 12 hours, each transfected cell contained 24.6×104 molecules CgOrc1. Thus, 6 hours after transfection, when caspase-3 activity was clearly induced, each transfected cell contained only ~1.6-fold more Orc1 than each untransfected cell.
Orc2 can suppress induction of apoptosis by Orc1
Ectopic expression of Orc2 generally exhibited a small but detectable induction of caspase-3 activity above that produced by the DNA vector alone (Fig. 6D). This appeared to result from the fact that ectopic expression of Orc2 stimulated endogenous expression of Orc1 (Fig. 6D,F). Orc1 accumulated faster than Orc2, which could account for the rapid, but limited, induction of caspase-3 activity. Once the level of Orc2 was equivalent to that of Orc1, no further induction of caspase-3 activity was observed (Fig. 6F), suggesting that Orc2 suppressed induction of apoptosis by Orc1.
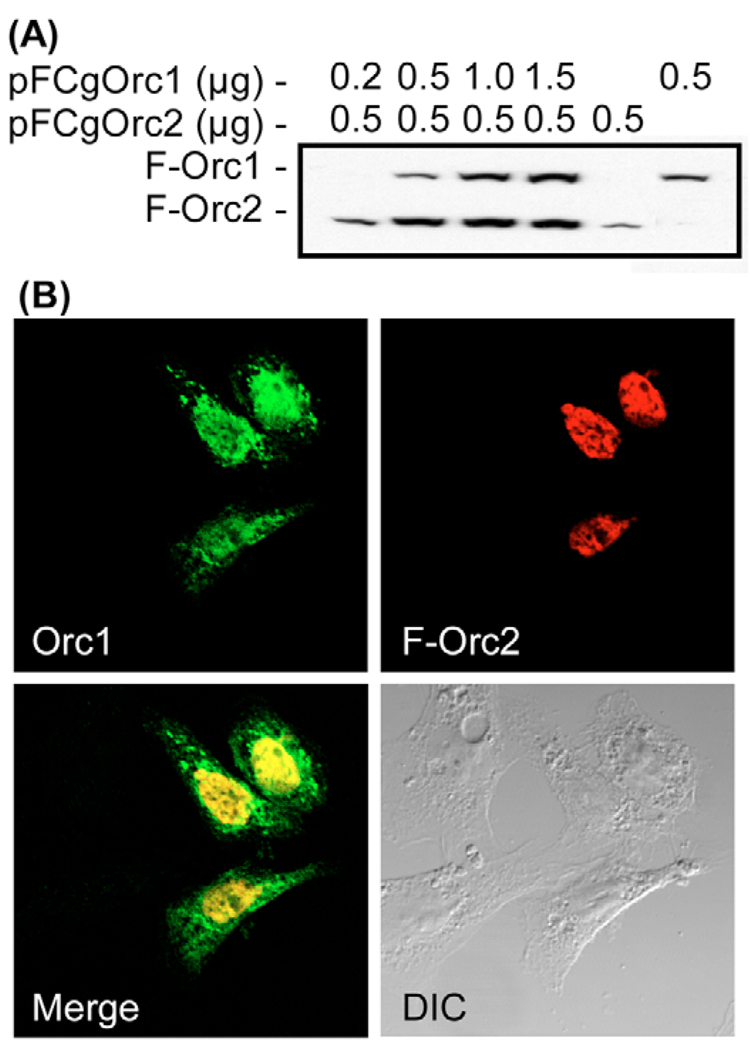
To test this hypothesis, CHO cells were co-transfected with pFCgOrc1 and pFCgOrc2. When CHO cells ectopically expressed either FCgOrc1 or CgOrc1 for 24 hours, Orc1 accumulated perinuclearly (Fig. 3A and Fig. 4B). By contrast, when CHO cells ectopically expressed FCgOrc2 for 24 hours, FCgOrc2 accumulated uniformly throughout the nucleus (Fig. 3A and Fig. 4B). However, when CHO cells were co-transfected with 1.5 µg pCgOrc1 and 0.5 µg pFCgOrc2 [the ratio of expression vectors that produced equivalent amounts of Orc1 and Orc2 during co-transfection (Fig. 7A)], then both Orc1 and Orc2 were distributed uniformly throughout the nucleus of transfected cells (Fig. 7B, Orc1, F-Orc2) with some Orc1 spreading into the cytoplasm (Fig. 7B, Merge). Furthermore, the transfected cells exhibited normal morphology (Fig. 7B, DIC), and <10% of the cells bound annexin-V (the fraction routinely observed when cells were transfected with pCI). Thus, induction of apoptosis by Orc1 is suppressed in the presence of Orc2.
Fig. 7.
Co-expression of Orc2 with Orc1 suppressed induction of apoptosis by Orc1. (A) CHO cells were transfected with the indicated amounts of pFCgOrc1 and pFCgOrc2, and cell lysates were subjected to immunoblotting at 24 hours post-transfection using anti-FLAG antibody. (B) CHO cells were co-transfected with 1.5 µg pCgOrc1 and 0.5 µg pFCgOrc2. At 24 hours post-transfection, cells were stained with both anti-CgOrc1 (Orc1) and anti-FLAG (F-Orc2) antibodies. Merged images revealed common areas of localization (yellow). DIC, differential interference contrast.
Ubiquitylation can suppress Orc1 induction of apoptosis
Endogenous CgOrc1 has been reported to undergo mono-ubiquitylation during S phase (Li and DePamphilis, 2002). However, since CgOrc1 is not selectively degraded during S phase, one can only speculate that this post-translational modification serves to prevent Orc1 from forming active ORC-chromatin sites. To determine whether or not mono-ubiquitylated forms of Orc1 are inactivated, the C-terminal amino acid in FHsOrc1 and FCgOrc1 was fused either to a single human ubiquitin (Ub), or to a single HsUb in which all seven lysine residues had been replaced by arginines (UbKO). UbKO cannot be extended by polyubiquitylation. Data are shown for HeLa cells; similar results were obtained with CHO cells.
Expression levels for FHsOrc1-Ub and FHsOrc1-UbKO in HeLa cells were about a third to a half of expression levels for FHsOrc1 (Fig. 8A). At 12 and at 24 hours after transfecting HeLa cells with pFHsOrc1, 30–40% of the cells expressed FHsOrc1 in their nuclei, and these cells exhibited morphological changes consistent with apoptosis (Fig. 3). Moreover, a similar fraction of cells exhibited strong annexin-V binding (Fig. 8B), and the transfected cell population exhibited increased caspase-3 activity (Fig. 8C).
Fig. 8.
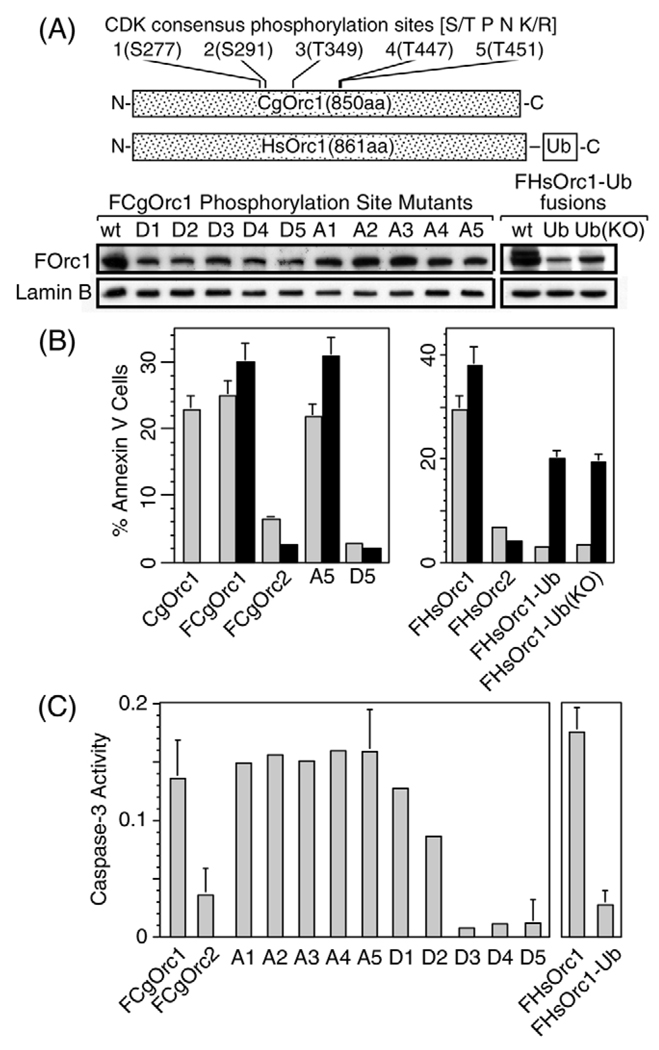
Post-translational modifications of Orc1 suppressed its ability to induce exposure of phosphatidylserine on cell surfaces and caspase-3 activity. (A) The five CDK consensus phosphorylation sites in hamster Orc1 were mutated cumulatively, either (S/T→D) or (S/T→A). Thus, D1 contains a single amino acid change (S277D), whereas D5 contains five amino acid changes, one at each site. FCgOrc1 and FHsOrc1 also were modified by addition of either a single human ubiquitin (Ub) fused to their C-terminus, or a single ubiquitin in which all seven lysines had been ‘knocked-out’ by changing them to arginines [Ub(KO)]. CHO cells were transfected with 1 µg of the indicated phosphorylation site mutant, and HeLa cells were transfected with 1 µg of the indicated Ub-fusion protein. Cells were incubated for 12 hours before staining the expressed protein with anti-FLAG antibody to determine their relative expression levels; wt, wild type. (B) The fraction of annexin-V-positive cells was determined at 12 hours (light bars) and 24 hours (dark bars) post-transfection for the indicated plasmid. (C) Caspase-3 activity was determined at 12 hours post-transfection for the indicated plasmid, and corrected for activity produced by pCI transfection.
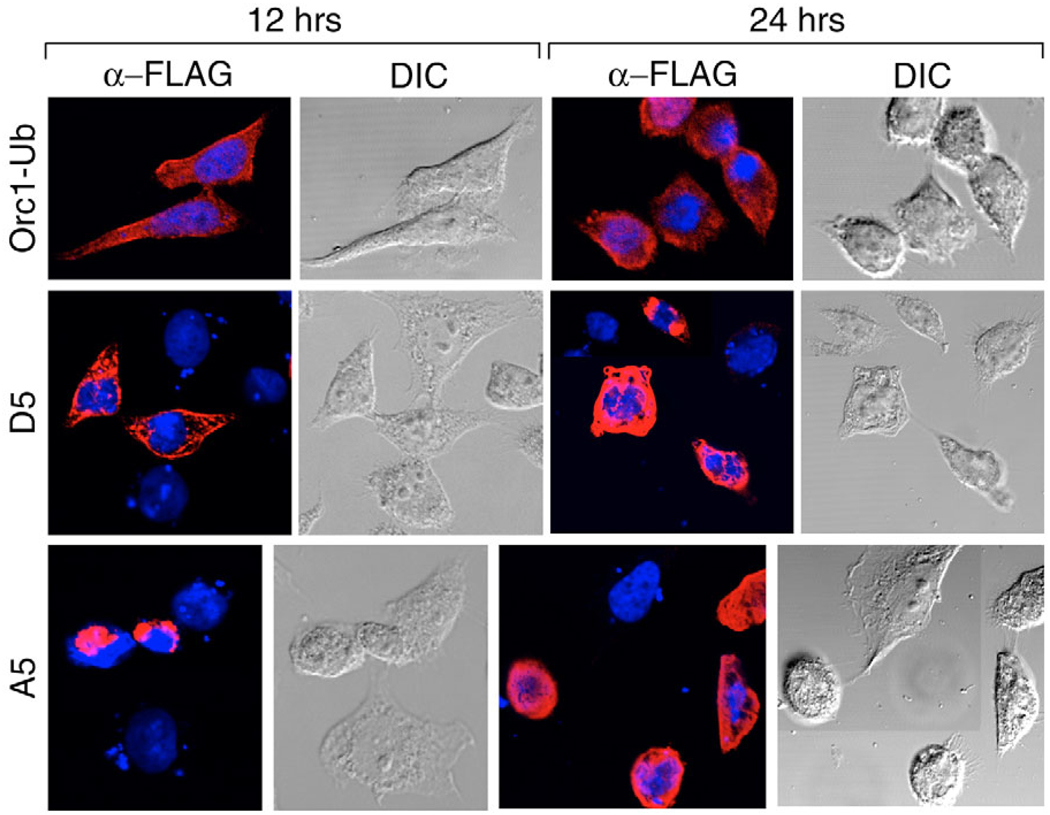
In contrast to unmodified Orc1, 12 hours after transfecting cells with either pFHsOrc1-Ub or pFHsOrc1-UbKO, about 40% of the cells expressed the mono-ubiquitylated protein, but its location was clearly cytoplasmic, and the morphology of the cell remained normal (Fig. 9, Orc1-Ub). Furthermore, the fraction of cells that stained with annexin-V (Fig. 8B) and that expressed caspase-3 (Fig. 8C) were reduced to background levels. Similar results were obtained at 24 hours post-transfection (Fig. 9), although the fraction of cells that bound annexin-V had increased (Fig. 8B). Results with pFHsOrc1-UbKO were indistinguishable from those with pFHsOrc1-Ub. Cytoplasmic accumulation of HsOrc1-Ub was specific for the ubiquitin adduct, because fusion of the C-terminus of HsOrc1 to either FLAG (1 kDa) or green fluorescent protein (59 kDa) results in nuclear accumulation (Lidonnici et al., 2004). Thus, a single ubiquitin can simultaneously prevent Orc1 from participating in DNA replication by sequestering it in the cytoplasm and can suppress the ability of Orc1 to induce apoptosis.
Fig. 9.
Post-translational modifications of Orc1 affected the subcellular localization of Orc1 and suppressed changes in cell morphology characteristic of apoptosis. HeLa cells were transfected with 1 µg pFHsOrc1-Ub (Orc1-Ub). CHO cells were transfected with 1 µg of either pFCgOrc1(A5) or pFCgOrc1(D5). At 12 and 24 hours post-transfection, cells were stained with anti-FLAG antibody to detect FCgOrc1 (red) and with Hoechst to detect nuclear DNA (blue). Merged images revealed FCgOrc1 localization relative to DNA, and DIC images revealed the morphology of the cell. DIC, differential interference contrast.
Hyperphosphorylation can suppress Orc1 induction of apoptosis
Previous studies have implicated hyperphosphorylation of Orc1 in preventing Orc1 from forming functional ORC-chromatin complexes during M phase (see Introduction). Therefore, to test this hypothesis and to determine whether or not hyperphosphorylation of Orc1 could prevent Orc1-induced apoptosis, each of the five CDK-dependent phosphorylation consensus sites in CgOrc1 (Fig. 8A) were mutated either to mimic phosphorylation (S/T→D) or to prevent phosphorylation (S/T→A). Expression of these mutant proteins was similar to wild-type protein (Fig. 8A).
FCgOrc1 containing five S/T→A mutations (A5) behaved like the parental FCgOrc1 protein. It accumulated perinuclearly as early as 12 hours post-transfection (Fig. 9) and induced changes in cell morphology (Fig. 9), annexin-V binding (Fig. 8B) and caspase-3 activity (Fig. 8C) that were consistent with induction of apoptosis. By contrast, FCgOrc1 containing five S/T→D mutations (D5) mimicked Orc1-Ub fusion proteins in that it rapidly accumulated in the cytoplasm (Fig. 9, 12 hours post-transfection), did not alter cell morphology (Fig. 9), did not induce annexin-V binding (Fig. 8B) and did not induce caspase-3 activity (Fig. 8C). Interestingly, loss of the ability to induce caspase-3 activity was associated with the sequential conversion of the first three CDK-dependent phosphorylation sites to a quasi-phosphorylated state (Fig. 8C, D1 to D3). Thus, as previously reported for HsOrc1 (Laman et al., 2001), quasi-phosphorylation of CgOrc1 caused it to localize to the cytoplasm where it cannot participate in DNA replication. In addition, multiple quasi-phosphorylated sites suppressed the ability of Orc1 to induce apoptosis.
Discussion
The results presented here demonstrate that low levels of unmodified Orc1 can trigger apoptosis in a manner similar to apoptotic cells that arise spontaneously in proliferating cell cultures. Moreover, induction of apoptosis by Orc1 can be suppressed by cell-cycle-specific modifications of Orc1 such as association with Orc2, ubiquitylation and hyperphosphorylation (Fig. 10). Furthermore, a single ubiquitin adduct or quasi-hyperphosphorylation of Orc1 caused it to accumulate in the cytoplasm where it could not participate in DNA replication. These results provide direct evidence that mono-ubiquitylation and hyperphosphorylation of Orc1 are, in fact, capable of inhibiting assembly of functional ORC-chromatin sites during the S, G2 and M phases of cell division, as previously suggested (see Introduction and Fig. 10). Finally, these results suggest that aberrant DNA replication during mammalian development could result in apoptosis through the appearance of ‘unmodified’ and ‘unbound’ Orc1. In brief, mammalian cells are sensitive to the physical state of Orc1. When Orc1 is part of a functional ORC complex, it is benign, but when Orc1 disengages from the ORC complex, it becomes a potential threat to the cell. Thus, association with Orc2 during G1 phase, ubiquitylation during S phase, and phosphorylation during M phase are three cell-cycle-dependent Orc1 modifications that not only restrict the assembly of functional ORC-chromatin sites during G1 phase, they prevent Orc1 from inducing apoptosis when it is not functionally engaged with ORC-chromatin sites during the transition from S to M phase.
Induction of apoptosis
Cells transfected with an expression vector for either wild-type Orc1 or FLAG-tagged Orc1 rapidly underwent five changes that are established characteristics of apoptosis and a sixth change, the perinuclear localization of Orc1, that may be unique to induction of apoptosis by Orc1. The cellular concentration of Orc1 began to increase by 6 hours post-transfection, at which time a distinct increase in caspase-3 activity could be detected even though the total amount of Orc1 had not exceeded twice its endogenous level. Induction of caspase-3 activity is an early event in apoptosis, and it is required for the appearance of DNA fragmentation (Otsuki et al., 2003, and references therein). By 18 hours post-transfection, Orc1 was observed to accumulate in a perinuclear location and 3′-ends of broken DNA were detected by the TUNEL assay. The exposure of phosphatidylserine on the surface of living cells was also detected at this time by the binding of annexin-V to cell surfaces. The intensity of annexin-V reached saturation by 24 hours post-transfection, at which time cells that expressed ectopic Orc1 exhibited a rounded shape and blebbing. This process was accompanied by a loss of cell adhesion that was detected by the release of Orc1-expressing cells into the culture medium. About half of the cells ectopically expressing Orc1 lost cell adhesion before their genomes were significantly fragmented. Similar results were obtained with both CHO and HeLa cells transfected with their cognate Orc1.
The results presented here provide a strong correlation between the perinuclear localization of Orc1 and its ability to induce apoptosis. Without exception, all Orc1 molecules that induced apoptosis also accumulated perinuclearly; by contrast, without exception, all Orc1 molecules that did not induce apoptosis did not localize perinuclearly. Either ubiquitylation or hyperphosphorylation of Orc1 suppressed its ability to induce apoptosis and caused it to localize to the cytoplasm. In fact, the perinuclear localization of ectopically expressed Orc1 mimicked the localization pattern for endogenous Orc1 in proliferating cells that had spontaneously undergone apoptosis (Fig. 4). In both cases, higher than normal levels of Orc1 accumulated perinuclearly and were associated with fragmented DNA as visualized by the TUNEL assay. This observation might reflect reports that endogenous HsOrc1 is selectively bound to a nuclease-resistant component of the nucleus (Kreitz et al., 2001; Ohta et al., 2003).
Orc2 was unique among the ORC subunits in that it did not induce apoptosis. The frequency and intensity of apoptotic symptoms observed with Orc2-transfected cells were virtually the same as those observed with the parent plasmid pCI. The slight induction of apoptosis above this background observed with ectopic expression of Orc2 resulted from a rapid increase in the level of endogenous Orc1. When Orc2 levels eventually reached those of Orc1, no further increase in caspase-3 activity was observed, suggesting that Orc2 suppressed the induction of apoptosis by increased levels of endogenous Orc1. This conclusion was confirmed by the facts that ectopic expression of equivalent amounts of Orc1 and Orc2 did not induce apoptosis, and that Orc1 produced under these conditions was localized throughout the nucleus rather than perinuclearly. Thus, Orc2 can neutralize the toxic effects of Orc1. Orc1 does not form a dimeric complex with Orc2 in vitro (Vashee et al., 2001), but does bind to the ORC(2–5) core complex (Dhar et al., 2001; Giordano-Coltart et al., 2005; Kneissl et al., 2003; Vashee et al., 2001). Therefore, suppression of Orc1 toxicity by co-expression with Orc2 would require that Orc2 stimulate synthesis of other ORC subunits. In fact, Orc2 does regulate expression of some, if not all, of the other ORC subunits. Specific inhibition of Orc2 expression inhibits expression of Orc3 and, in some cases, the other four ORC subunits (Machida et al., 2005; Prasanth et al., 2004), and conversely, ectopic expression of Orc2 stimulates expression of Orc1 (Fig. 6F) and presumably the remaining ORC subunits. Given the fact that the remaining ORC subunits were also toxic when expressed alone, we suggest that Orc2 plays a central role during cell proliferation by rapidly assembling newly synthesized ORC subunits into chromatin-bound ORC cores, thereby preventing induction of apoptosis by individual ORC subunits.
Regulation of Orc1 activities during cell proliferation
Of the six mammalian ORC subunits, Orc1 appears to be unique in its role as a regulator of ORC activity. Thus, the fact that levels of ectopic Orc1 comparable with those of endogenous Orc1 could induce apoptosis underscores the importance of Orc1 modifications in preventing apoptosis. This is accomplished during G1 phase by the association of Orc1 with Orc2 and the other ORC subunits when the bulk of endogenous Orc1 is bound to chromatin (DePamphilis, 2005; Li et al., 2004). With the onset of S phase, the association between Orc1 and chromatin becomes salt sensitive, and Orc1 becomes subject to ubiquitylation.
In hamster cells, Orc1 ubiquitylation is limited to one or two ubiquitin adducts but, in HeLa cells, Orc1 undergoes ubiquitin-dependent degradation (see Introduction). Whereas degradation of Orc1 would clearly prevent it from inducing apoptosis, as well as from forming active ORC-chromatin sites during S phase, the function of mono-ubiquitylation has not been clear. The results presented here show that even a single ubiquitin adduct is sufficient to prevent Orc1 from initiating assembly of pre-RCs by translocating it to the cytoplasm. The addition of other polypeptides to either the N- (this report) or C-terminus (Lidonnici et al., 2004) of Orc1 did not have this effect. In addition, a single ubiquitin was sufficient to suppress Orc1 induction of apoptosis. Since the same results were obtained using either Ub or UbKO molecules, the presence of a single ubiquitin on Orc1 does not target it for polyubiquitylation and subsequent degradation. In fact, previous studies have shown that fusion of a single ubiquitin to the C-terminus of either yeast α-factor receptor or human Tp53 causes the subcellular redistribution of these proteins to mimic the effects of their normal mono-ubiquitylation pattern (Li et al., 2003; Terrell et al., 1998). For example, both Tp53-Ub and Tp53-UbKO localize to the cytoplasm of human cells (Li et al., 2003). Since ubiquitin hydrolases can cleave the isopeptide bonds that link ubiquitin to its target protein, mono-ubiquitylation provides a reversible reaction that can prevent apoptosis without requiring the cell to expend enormous energy to degrade and then resynthesize Orc1. The difference between HeLa cells and CHO cells might simply reflect differences in their ratios of Orc1 to SCF, the protein complex that targets Orc1 for ubiquitylation (Mendez et al., 2002). For example, low levels of Mdm2 activity, the enzyme that ubiquitylates Tp53, induce mono-ubiquitylation and nuclear export of Tp53, whereas high levels promote polyubiquitylation and nuclear degradation. Thus, either pathway could protect mammalian cells from induction of apoptosis by Orc1 during the transition from G1 to S phase.
With the onset of G2-M phase, Orc1 becomes hyperphosphorylated by CcnA-Cdk1 and is selectively disengaged from ORC-chromatin sites in metaphase cells (Li et al., 2004). Here, we show that CgOrc1 in which at least three of the five CDK-phosphorylation sites were changed from S/T to D in order to mimic phosphorylation also induced Orc1 to localize in the cytoplasm, consistent with the behavior of the endogenous CgOrc1 in metaphase CHO cells. In addition, this quasi-phosphorylated Orc1 failed to induce apoptosis, whereas Orc1 in which the same S/Ts were changed to A in order to prevent phosphorylation marginally stimulated the ability of Orc1 to induce apoptosis. These results show that hyperphosphorylation of Orc1 can, in fact, prevent association of Orc1 with chromatin. This is consistent with previous studies (Li et al., 2004) suggesting that CDK-dependent phosphorylation of Orc1 during mitosis prevented stable association of Orc1 with chromatin, and with the report that CDK phosphorylation of Drosophila Orc1 inhibits its ability to bind DNA (Remus et al., 2005). In addition, these results reveal that hyperphosphorylation can also prevent the disengaged Orc1 from inducing apoptosis.
Orc1 as an indicator of aberrant DNA replication
Previous studies have revealed that inactivation of proteins involved either in the assembly (Cdc6, Mcm2) or activation (Cdc7, Cdc45) of pre-RCs can trigger apoptosis, as does inappropriate expression of geminin, a specific inhibitor of pre-RC assembly (Feng et al., 2003; Kim et al., 2002; Shreeram et al., 2002; Yim et al., 2003). Moreover, apoptosis induced by a variety of stimuli coincides with the destruction of Cdc6 (Blanchard et al., 2002). The destruction of Cdc6 appears to have a causal role in apoptosis, because stable Cdc6 mutants that cannot be cleaved by caspases inhibit apoptosis (Pelizon et al., 2002), perhaps by inhibiting the production of a caspase cleavage fragment of Cdc6 that has dominant-negative effects on Cdc6 function (Yim et al., 2003). Thus, it is clear that disproportionate changes in the levels of specific proteins that regulate initiation of DNA replication can induce apoptosis, and that at least one protein, Cdc6, is part of the induction pathway.
These studies also concluded that over-expressing Cdc6 or Cdt1, or repressing expression of geminin, induces apoptosis by triggering the ATM/ATR-dependent DNA damage checkpoint pathways, because these events induced re-replication of DNA. In adult tissues, this pathway requires Tp53 activity. Tp53 functions as a tumor suppressor by activating downstream genes that either induce apoptosis or arrest cell division (Ding and Fisher, 2002). In fact, the absence of Tp53 activity in cancer cells is primarily responsible for their ability to escape apoptosis and survive (Vogelstein et al., 2000). Thus, it is surprising that Tp53-dependent apoptosis is not required for suppression of the early onset of spontaneous tumors during mouse development; only the ability of Tp53 to induce cell-cycle arrest and retain chromosome stability is required (Liu et al., 2004). Moreover, embryonic stem cells that sustain DNA damage undergo Tp53-independent apoptosis (Aladjem et al., 1998). Part of the explanation might be that Orc1 provides a mechanism for detecting aberrant DNA replication that does not rely on either re-initiating DNA replication or on Tp53 activity, and therefore it can function from fertilized egg to adult.
Materials and Methods
Expression plasmids
HsORC1, HsORC2, HsORC3, HsORC4, HsORC5 and HsORC6 genes, and CgORC1 (AF254572) and CgORC2 (AF254573) were isolated from genomic DNA and cloned into pCI (Promega). HsORC1 (NM_004153) was also taken from pKG28 (Mendez et al., 2002), and HsORC2 (NM_006190) was also provided by A. Dutta (University of Virginia Health Sciences System, Charlottesville, VA). Where indicated, a single copy of the FLAG epitope (DYKDDDDK) with a spacer (EFKGLRR) was linked to the N-terminus of Orc. Single amino acid changes were made in FCgOrc1 using QuikChange® Site-Directed Mutagenesis Kit (Stratagene, Cat #200518). Primer sequences can be provided upon request. Mutations were introduced sequentially beginning with S277. The C-terminus of F-Orc1 was fused to a single ubiquitin (Ub) molecule as described for Tp53 by inserting the gene into either pCDNA3.1(TOPO, His-HA-Ub) or pCDNA3.1(TOPO, His-HA-UbKO) (Li et al., 2003). The accuracy of these constructions was confirmed by DNA sequencing. Plasmids were purified using a Qiagen plasmid purification kit followed by CsCl dye-density equilibrium centrifugation.
Cells
HeLa cells (ATCC #CCL-2) and CHO C400 cells (Leu and Hamlin, 1989) were cultured as monolayers in 185 cm2 flasks using Dulbecco’s modified Eagle’s medium supplemented with either 10% (HeLa cells) or 5% (CHO cells) fetal bovine serum and 1% non-essential amino acids at 37°C in a humidified incubator with 5% CO2. HCT116 cells (Bunz et al., 1998) were cultured in McCoys 5A medium supplemented with 10% fetal bovine serum.
DNA transfection
Cells were seeded (1–2×105/well) in a six-well cluster plate (Costar), and then transfected the following day (~75% confluent) using the Roche Fugene 6 protocol. Unattached cells were recovered from the cell culture medium by centrifugation (4000 g, 10 minutes, ambient temperature), resuspended in 30 µl SDS sample buffer and incubated for 10 minutes at 80°C. The attached cells were washed with 10 ml phosphate buffered saline (PBS), lysed by addition of 300 µl/well of 2× SDS sample buffer plus 5% β-mercaptoethanol, and incubated for 10 minutes at 80°C. Proteins in 20 µl aliquots (~104 cell equivalents) were detected by immunoblotting. Transfection efficiency, as determined by immunostaining for the expressed protein, was 30–40%.
Immunoblotting
Cell lysates were fractionated by electrophoresis (150 V constant, ambient temperature) in a 4–12% NuPage Bis-Tris polyacrylamide gel in NuPage MOPS-SDS running buffer (Invitrogen). Proteins were detected by immunoblotting using the Invitrogen protocol with a nitrocellulose membrane (Protran BA83, Schleicher & Schuell). FLAG-tagged proteins were detected with mouse anti-FLAG monoclonal antibody (Sigma). Orc1 and Orc2 were detected with rabbit anti-CgOrc1 and anti-CgOrc2 (Li et al., 2004), and goat anti-HsOrc1 and anti-HsOrc2 IgG (Santa Cruz) antibodies. Primary antibodies were detected with either antimouse, anti-rabbit or anti-goat IgG conjugated to horseradish peroxidase (Pierce). Antibodies were diluted in 5% non-fat dry milk, 0.1% Tween-20 and PBS to minimize background. Chemiluminescence was detected using either SuperSignal West Pico or West Duro Chemiluminescent substrate (Pierce).
Microscopic analyses
Cells were cultured on chamber slides (Lab-Tek). For immunofluorescence, cells were washed with PBS, fixed with freshly prepared 4% paraformaldehyde in PBS for 20 minutes at ambient temperature, washed once with PBS, fixed with ice-cold 100% methanol for 10 minutes, blocked with 3% BSA, 0.1% Tween 20 in PBS for 30 minutes, and then incubated with the appropriate antibody for up to 2 hours at 4°C. Cells were then washed twice in PBS for 10 minutes, and incubated either with anti-rabbit IgG coupled with Cy2 (Molecular Probes) or with anti-mouse IgG coupled with Cy3 for 1 hour at room temperature. DNA was stained for 5 minutes with 5 µg/ml Hoechst (Molecular Probes). Slides were mounted in Dako Fluorescent Mounting Medium (DAKO Cytomation) and viewed with a confocal laser scanning microscope (LEICA TCS-SP2). For non-transfected cells, the signal from the primary antibody was increased by incubating with a second biotin-labeled antibody that was detected by Streptavidine conjugated with fluorescent Cy3 dye (Jackson Laboratories).
For annexin-V binding, cells were stained with annexin-V Alexa Fluor 568 and BOBO-1 (Molecular Probes) using the Roche protocol. For DNA fragmentation (TUNEL assay), DNA ends were labeled with fluorescein-dUTP using an In Situ Cell Death Detection Kit (Roche). Cells were then stained with mouse anti-FLAG monoclonal antibody and anti-mouse IgG-Cy3 secondary antibody.
Fluorescence activated cell sorting (FACS)
Unattached cells were collected by centrifugation, washed twice with ice-cold PBS, fixed in 70% ethanol at 4°C for 3 hours, washed twice with ice-cold PBS, incubated in 0.5 ml PBS supplemented with 100 µg/ml RNase A (Roche) at 37°C for 30 minutes, and then washed again with PBS. Cells were stained with 0.5 ml of 50 µg/ml propidium iodide (Roche) for 30 minutes at 4°C. Propidium-stained DNA fluorescence was quantified by FACScan flow cytometry (Becton Dickinson).
Caspase-3 activity
Extracts from ~106 cells were prepared by the freeze-thaw method and assayed for caspase-3 activity according to the manufacturer’s instructions (CaspACE™ Assay System, Colorimetric, Promega). Lysates were centrifuged at 15,000 g for 20 minutes at 4°C. The supernatant was incubated with enzyme-specific colorimetric substrates Ac-DEVD-pNA at 37°C for 4 hours. Cleavage of the colorimetric substrate and release of pNA (yellow) was measured as absorbance at 405 nm. Caspase-3 activity detected in non-transfected cells maintained in parallel with the transfected cells was subtracted from the transfected cell data as background activity. The error bars indicate the s.e.m. for each set of assays in a particular experiment and were compiled from a minimum of three assays. Since transfection efficiency varied from one experiment to the next, our conclusions were based on the ratio of Orc1 annexin-positive cells to Orc2 annexin-positive cells in each experiment.
Acknowledgments
We thank B. Stillman for pKG28, A. Dutta for HsOrc2, B. Vogelstein for HCT116 cells, and A. Dey and T. Tomahico for help with confocal microscopy and FACS analyses.
References
- Abdurashidova G, Danailov MB, Ochem A, Triolo G, Djeliova V, Radulescu S, Vindigni A, Riva S, Falaschi A. Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J. 2003;22:4294–4303. doi: 10.1093/emboj/cdg404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- Ballabeni A, Melixetian M, Zamponi R, Masiero L, Marinoni F, Helin K. Human Geminin promotes pre-RC formation and DNA replication by stabilizing CDT1 in mitosis. EMBO J. 2004;23:3122–3132. doi: 10.1038/sj.emboj.7600314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard F, Rusiniak ME, Sharma K, Sun X, Todorov I, Castellano MM, Gutierrez C, Baumann H, Burhans WC. Targeted destruction of DNA replication protein Cdc6 by cell death pathways in mammals and yeast. Mol. Biol. Cell. 2002;13:1536–1549. doi: 10.1091/mbc.02-02-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle. 2005;4:70–79. doi: 10.4161/cc.4.1.1333. [DOI] [PubMed] [Google Scholar]
- Dhar SK, Delmolino L, Dutta A. Architecture of the human origin recognition complex. J. Biol. Chem. 2001;276:29067–29071. doi: 10.1074/jbc.M103078200. [DOI] [PubMed] [Google Scholar]
- Ding HF, Fisher DE. Induction of apoptosis in cancer: new therapeutic opportunities. Ann. Med. 2002;34:451–469. doi: 10.1080/078538902321012405. [DOI] [PubMed] [Google Scholar]
- Feng D, Tu Z, Wu W, Liang C. Inhibiting the expression of DNA replication-initiation proteins induces apoptosis in human cancer cells. Cancer Res. 2003;63:7356–7364. [PubMed] [Google Scholar]
- Fujita M, Ishimi Y, Nakamura H, Kiyono T, Tsurumi T. Nuclear organization of DNA replication initiation proteins in mammalian cells. J. Biol. Chem. 2002;277:10354–10361. doi: 10.1074/jbc.M111398200. [DOI] [PubMed] [Google Scholar]
- Giordano-Coltart J, Ying CY, Gautier J, Hurwitz J. Studies of the properties of human origin recognition complex and its Walker A motif mutants. Proc. Natl. Acad. Sci. USA. 2005;102:69–74. doi: 10.1073/pnas.0408690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe-Seyler F, Butz K. Repression of endogenous p53 transactivation function in HeLa cervical carcinoma cells by human papillomavirus type 16 E6, human mdm-2, and mutant p53. J. Virol. 1993;67:3111–3117. doi: 10.1128/jvi.67.6.3111-3117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Miller CM, Ridder GM, Aardema MJ. Characterization of p53 in Chinese hamster cell lines CHO-K1, CHO-WBL, and CHL: implications for genotoxicity testing. Mutat. Res. 1999;426:51–62. doi: 10.1016/s0027-5107(99)00077-9. [DOI] [PubMed] [Google Scholar]
- Itzhaki JE, Gilbert CS, Porter AC. Construction by gene targeting in human cells of a ‘conditional’ CDC2 mutant that rereplicates its DNA. Nat. Genet. 1997;15:258–265. doi: 10.1038/ng0397-258. [DOI] [PubMed] [Google Scholar]
- Kim JM, Nakao K, Nakamura K, Saito I, Katsuki M, Arai K, Masai H. Inactivation of Cdc7 kinase in mouse ES cells results in S-phase arrest and p53-dependent cell death. EMBO J. 2002;21:2168–2179. doi: 10.1093/emboj/21.9.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneissl M, Putter V, Szalay AA, Grummt F. Interaction and assembly of murine pre-replicative complex proteins in yeast and mouse cells. J. Mol. Biol. 2003;327:111–128. doi: 10.1016/s0022-2836(03)00079-2. [DOI] [PubMed] [Google Scholar]
- Kossatz U, Dietrich N, Zender L, Buer J, Manns MP, Malek NP. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev. 2004;18:2602–2607. doi: 10.1101/gad.321004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitz S, Ritzi M, Baack M, Knippers R. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 2001;276:6337–6342. doi: 10.1074/jbc.M009473200. [DOI] [PubMed] [Google Scholar]
- Ladenburger EM, Keller C, Knippers R. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol. 2002;22:1036–1048. doi: 10.1128/MCB.22.4.1036-1048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laman H, Peters G, Jones N. Cyclin-mediated export of human Orc1. Exp. Cell Res. 2001;271:230–237. doi: 10.1006/excr.2001.5360. [DOI] [PubMed] [Google Scholar]
- Lee H, Larner JM, Hamlin JL. Cloning and characterization of Chinese hamster p53 cDNA. Gene. 1997;184:177–183. doi: 10.1016/s0378-1119(96)00592-6. [DOI] [PubMed] [Google Scholar]
- Leu TH, Hamlin JL. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol. Cell. Biol. 1989;9:523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CJ, DePamphilis ML. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol. 2002;22:105–116. doi: 10.1128/MCB.22.1.105-116.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CJ, Bogan JA, Natale DA, DePamphilis ML. Selective activation of pre-replication complexes in vitro at specific sites in mammalian nuclei. J. Cell Sci. 2000;113:887–898. doi: 10.1242/jcs.113.5.887. [DOI] [PubMed] [Google Scholar]
- Li CJ, Vassilev A, DePamphilis ML. Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex’s largest subunit (Orc1) from binding to chromatin during mitosis. Mol. Cell. Biol. 2004;24:5875–5886. doi: 10.1128/MCB.24.13.5875-5886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono-versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- Lidonnici MR, Rossi R, Paixao S, Mendoza-Maldonado R, Paolinelli R, Arcangeli C, Giacca M, Biamonti G, Montecucco A. Subnuclear distribution of the largest subunit of the human origin recognition complex during the cell cycle. J. Cell Sci. 2004;117:5221–5231. doi: 10.1242/jcs.01405. [DOI] [PubMed] [Google Scholar]
- Liu G, Parant JM, Lang G, Chau P, Chavez-Reyes A, El-Naggar AK, Multani A, Chang S, Lozano G. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat. Genet. 2004;36:63–68. doi: 10.1038/ng1282. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Teer JK, Dutta A. Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J. Biol. Chem. 2005;280:27624–27630. doi: 10.1074/jbc.M502615200. [DOI] [PubMed] [Google Scholar]
- Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell. 2002;9:481–491. doi: 10.1016/s1097-2765(02)00467-7. [DOI] [PubMed] [Google Scholar]
- Natale DA, Li CJ, Sun WH, DePamphilis ML. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G(1) transition in mammals. EMBO J. 2000;19:2728–2738. doi: 10.1093/emboj/19.11.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S, Tatsumi Y, Fujita M, Tsurimoto T, Obuse C. The ORC1 Cycle in Human Cells: II. Dynamic changes in the Human ORC complex during the cell cycle. J. Biol. Chem. 2003;278:41535–41540. doi: 10.1074/jbc.M307535200. [DOI] [PubMed] [Google Scholar]
- Okuno Y, McNairn AJ, den Elzen N, Pines J, Gilbert DM. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 2001;20:4263–4277. doi: 10.1093/emboj/20.15.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki Y, Li Z, Shibata MA. Apoptotic detection methods – from morphology to gene. Prog. Histochem. Cytochem. 2003;38:275–339. doi: 10.1016/s0079-6336(03)80002-5. [DOI] [PubMed] [Google Scholar]
- Pelizon C, d’Adda di Fagagna F, Farrace L, Laskey RA. Human replication protein Cdc6 is selectively cleaved by caspase 3 during apoptosis. EMBO Rep. 2002;3:780–784. doi: 10.1093/embo-reports/kvf161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzi M, Tillack K, Gerhardt J, Ott E, Humme S, Kremmer E, Hammerschmidt W, Schepers A. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 2003;116:3971–3984. doi: 10.1242/jcs.00708. [DOI] [PubMed] [Google Scholar]
- Shreeram S, Sparks A, Lane DP, Blow JJ. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene. 2002;21:6624–6632. doi: 10.1038/sj.onc.1205910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi Y, Tsurimoto T, Shirahige K, Yoshikawa H, Obuse C. Association of human origin recognition complex 1 with chromatin DNA and nuclease-resistant nuclear structures. J. Biol. Chem. 2000;275:5904–5910. doi: 10.1074/jbc.275.8.5904. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C. The ORC1 cycle in human cells: I. Cell cycle-regulated oscillation of human ORC1. J. Biol. Chem. 2003;278:41528–41534. doi: 10.1074/jbc.M307534200. [DOI] [PubMed] [Google Scholar]
- Terrell J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- Thome KC, Dhar SK, Quintana DG, Delmolino L, Shahsafaei A, Dutta A. Subsets of human origin recognition complex (ORC) subunits are expressed in non-proliferating cells and associate with non-ORC proteins. J. Biol. Chem. 2000;275:35233–35241. doi: 10.1074/jbc.M005765200. [DOI] [PubMed] [Google Scholar]
- Vashee S, Simancek P, Challberg MD, Kelly TJ. Assembly of the human origin recognition complex. J. Biol. Chem. 2001;276:26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin-V. J. Immunol. Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Yim H, Jin YH, Park BD, Choi HJ, Lee SK. Caspase-3-mediated cleavage of Cdc6 induces nuclear localization of p49-truncated Cdc6 and apoptosis. Mol. Biol. Cell. 2003;14:4250–4259. doi: 10.1091/mbc.E03-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wu JR, Gilbert DM. Analysis of mammalian origin specification in ORC-depleted Xenopus egg extracts. Genes Cells. 1998;3:709–720. doi: 10.1046/j.1365-2443.1998.00224.x. [DOI] [PubMed] [Google Scholar]