Abstract
Refining phenotypes for the study of neuropsychiatric disorders is of paramount importance in neuroscience. Poor phenotype definition provides the greatest obstacle for making progress in disorders like schizophrenia, bipolar disorder, Attention Deficit/Hyperactivity Disorder (ADHD), and autism. Using freely available informatics tools developed by the Consortium for Neuropsychiatric Phenomics (CNP), we provide a framework for defining and refining latent constructs used in neuroscience research and then apply this strategy to review known genetic contributions to memory and intelligence in healthy individuals. This approach can help us begin to build multi-level phenotype models that express the interactions between constructs necessary to understand complex neuropsychiatric diseases.
Keywords: gene, SNP, review, cognition, phenomics
In the wake of mapping the human genome, the most significant challenge for Neuroscience is mapping the human phenome—the expression of a complex interaction of genes and environment observed at molecular, cellular, systems, and behavioral levels [(Freimer and Sabatti, 2003; Bearden and Freimer, 2006); see also Bilder et al., 2009 for more detailed exposition of the “phenomics” strategy]. Without links from genome to multiple levels of phenomic data, advances in genetics will not be fully utilized. The challenge of finding genetic determinants for latent phenotypic constructs depends first on our ability to adequately define and refine these phenotypes. Once this occurs, we can begin to build complex multivariate phenotypic maps that may more adequately address the complex interaction between phenome and genome currently being investigated in the field of neuroscience. The first generation of genome-wide association studies (GWAS) makes it clear that psychiatric diagnostic phenotypes may not offer the most traction for gene discovery. This has prompted many investigators in the field to pursue endophenotypes, phenotypes presumably “intermediate” between the overt expression of the syndromes themselves, and more basic levels of gene expression. In this review, we propose that a next step, multivariate multi-level models that flesh out the relationships between numerous intermediate phenotypes, will be essential to identify the complex unfolding of biological paths from genome to syndrome.
The discovery of the key components of the phenome that reflect critical paths to human disease is of paramount importance, but further and often ignored, we must recognize that the labeling and operational definition of these phenotypes poses a high hurdle. Only through objective construct definition can we hope to determine the validity of these constructs, and so far there is often argument even about the labels of constructs, much less their full operational definitions. The lack of an agreed upon phenotype lexicon undermines our progress. While genomics has begun to develop agreed-upon lexica, such as the terminology adopted for the Gene Ontology project [(Ashburner et al., 2000); http://www.geneontology.org/], researchers in neuroscience are left to speculate about the relationships between different processes and concepts. Developing a phenotype ontology or lexicon is a major undertaking, but is necessary for our field to make progress in connecting the genome to the phenome.
Latent phenotypes used in neuroscience are often ill-defined in the literature and reflect a combination of folk psychology and popular buzz-words, often lacking detailed construct validity. For instance, we previously found that usage of the term “cognitive control” in the literature grew exponentially over the last 10 years, even while it was still being measured with the same cognitive tasks used to describe other cognitive concepts (such as working memory, task switching/set shifting, response inhibition, and response selection). These findings suggest that the latent construct had been re-framed without equal advances in paradigm development (Sabb et al., 2008). A poor literature definition for a construct can pose challenges when investigators seek to link that phenotype to other phenotypes (in order to better define a multivariate phenotype) or to genotype (in order to define a genetic association). Poorly defined phenotypes can lead to negative results and failures to replicate findings, as is frequently seen in psychiatric research.
While poorly defined phenotypes are most clearly evident at the level of cognitive concepts, ill-defined phenotypes occur between all levels of inquiry, including neural systems (e.g. mirror neuron’s role in language) and signaling pathways (e.g. RAS role in learning). Neuroscience is a sufficiently new field, where discovery requires some speculative inference. In an effort to explain results, correlations or causal links are suggested where they may not exist (e.g. “reverse inference,” in functional neuroimaging (Poldrack, 2006). New tools will be necessary to highlight relationships in models that are tenuous and can be further tested empirically.
The benefit of good phenotype definition can be seen in the recent successes of GWAS outside the field of neuroscience. For instance, in studies of type 1 diabetes, recent advances in technology and knowledge have allowed researchers to determine four new genes from GWAS, by examining a clear (endo)phenotype in immune system cell histocompatibility antigen molecules (Nepom, 1995; Ueda et al., 2003; Barratt et al., 2004; Vella et al., 2005; Bottini et al., 2006; Lowe et al., 2007; Qu et al., 2007). Thus even with a polygenic disease, a coherent phenotype for diabetes has allowed the genetic contributions to be fully realized in these association studies.
In order to achieve adequate phenotype definitions for genetic studies, we must construct multivariate phenotypic models. Current approaches to phenotype definition lack an ability to differentiate between disagreements that result from (a) different theoretical interpretations of data versus, (b) non-comparable datasets that are inappropriately compared. Explicitly modeling phenotypes can expose quantitative relationships between phenotypic elements to allow consensus building. These rich phenotypic networks that contain quantitative relationships between network nodes (i.e. a quantology [Parker et al., submitted for publication]) can then be compared to genotypic data. Here we provide a framework and tools including http://pubatlas.org for (PubMed) knowledge representation and the http://phenowiki.org knowledge base in order to support this phenotypic modeling of quantitative effects.
In order to identify phenotype constructs that may ultimately be successful in genetic association studies, the field needs to move beyond the now traditional endophenotype approach and begin to build and refine multivariate multilevel phenotype models. These models may help expose the complex interactions between phenotypes and their relationship to underlying genetic architecture (Sabb et al., 2008). As neuropsychiatric syndromes are most likely caused by many genes with many small effects, understanding the relationships between multiple phenotypes is even more important.
Using more sophisticated bioinformatics tools to examine patterns in the literature through text mining can lead to a more objective definition of the components (“nodes”) of a model and the general patterns of interaction between nodes (“edges”). The current literature in neuroscience is too vast for the reader to objectively digest based on the results simple PubMed searches. Performing data reduction from a single perspective can lead to bias. For instance, we found that a number of papers use the Mini-Mental State Exam (MMSE) as a measure of memory. While most researchers may disagree with this operational definition, searching the literature for “genes and memory” would provide hits for these manuscripts. In conducting a meta-analysis or review, it is then up to the personal perspective of the author as to whether to include these findings. Using bio-informatics literature-based approaches can provide an objective account of the literature, which will serve as an initial lexicon from which the field can build consensus on what operational definitions are relevant for particular latent constructs. A common criticism of bioinformatics is referred to as the “garbage in/garbage out” problem, which states that it is difficult to draw conclusions from inadequate data. We propose, however, that a larger problem in neuroscience is the inability to digest the relevant information to draw any conclusions. Rather than using our approach to reach conclusions, we provide a way of organizing and visualizing the data that allows researchers to draw the conclusions. In essence, we think it is important to draw the entire map, but not tell the field how to get from point A to point B. These tools can provide better methods for culminating current findings and progressively building on previous research (i.e. meta-analyses), which, as noted by Rosenthal (1984), is a significant challenge in research.
Building these models, we feel, provides significant value to the neuroscience community. Using these techniques can improve our ability to digest the large amounts of data generated every day in neuroscience journals, share knowledge by developing free online tools to build collaborative meta-analytic models, build consensus among currently debated latent constructs like cognitive control, and expose interactions between phenotypes that may mediate correlations. Ultimately, these models may expose weak links in current hypotheses and drive hypothesis testing of particularly tenuous relationships.
In this meta-analytic review, we begin to build a multivariate multilevel model of two complex phenotypes (memory and intelligence) and examine current cognitive, neural, and genetic associations. Our methods detail a strategy for using informatics procedures to help refine and build consensus for poorly defined phenotypes, which can ultimately drive hypothesis testing to pursue novel genetic associations. Through collaborative use of these free tools, we ultimately aim to build a neuroscience knowledge base. This proof of concept example reveals overlap in genes, pathways, and neural systems between these two putatively disparate latent constructs.
METHODS
“Memory” and “intelligence” are broad phenotypes that are used in a variety of ways in the literature. Rather than impose a single perspective on each of these phenotypes by having the authors define the construct, we used a more unbiased approach that considers the current landscape of the literature to aid in our definition and construction of these phenotypes. Using similar methods described previously (Sabb et al., 2008), we constructed each phenotype to include its co-occurrence with other terms in the literature. This provides a construct definition based on existing knowledge across the field, and not solely from the perspective of these authors, which still represents a crucial obstacle in building consensus in neuroscience.
CNP lexica
For both constructs we used two lexica under development by the CNP and freely available online (http://phenomics.ucla.edu) in order to help drive our phenotype definition and paper selection.
In order to build the most comprehensive lexica, we started with a large list (2387 concepts, and 3824 tasks) acquired from index sections of textbooks (Handel, 1989; Levelt, 1993; Hunt and Ellis, 2004; Medin et al., 2005; Reisberg, 2005; Sternberg and Mio, 2006), the World Wide Web (WWW), and previous experience by the authors. While not exhaustive, we hope these lexica can provide the groundwork, which can be further refined through collaborative editing by the entire field.
Lexica were refined using stemming tools (Lovins, 1968), with which we grouped syntactically similar terms together and ordered them by their prefixes and suffixes. Such regrouping greatly eases review by domain experts by helping them identify and categorize the latent cognitive concepts or tasks. Initial domain expert review centered on identifying those terms that reflected “concepts” and “tasks” separately, while dropping terms that referred to “effects” or “theories.” The terms that were categorized as concepts by two of the three authors (R.M.B., R.A.P., F.W.S.) were selected as concept lexicon candidates. Further, these candidate terms were examined for hits in the PubMed database. Those that had at least one hit were kept in the lexicon, yielding 900 terms. A similar procedure was used to create a list of candidate cognitive tasks. It should be emphasized that these lexica are meant to serve as starting points rather than comprehensive or finished products. Given the aim of these projects is to develop collaborative tools for cognitive phenotype definition, it is hoped that investigators seeking to use these tools will add the concept terms and specific tasks of interest to them, and that some of the existing terms may ultimately be “pruned” if these are not used.
PubAtlas
Using PubAtlas, a freely available web-based tool for knowledge representation of the PubMed database, these terms and tasks were then examined for their relationship to the phenotype of interest (memory or IQ). PubAtlas (see http://pubatlas.org) interrogates the PubMed literature and provides “heat maps” of Jaccard coefficients (a co-occurrence statistic based on the union and intersection of terms) for any set of terms in an easy to use and flexible web-interface.
We selected the top five concepts most frequently used in conjunction with our phenotypes to provide cohesive and manageable sets for review (Table 1). These collections of concepts refined our phenotype selection (enriched concept phenotype) and were then compared with our task lexicon. We chose the top six tasks from this expanded concept phenotype to establish an enriched task phenotype description (see methods flow chart, Fig. 1, Table 1). Several strong Jaccard coefficients for items initially in our task lexicon were not considered in this review, including “Naming” and “Digit-Span.” “Naming” was judged to be a family of tasks and not a specific task, while we have previously shown “Digit-Span” was more strongly correlated with “working memory” (Sabb et al., 2008).
Table 1.
Co-occurrence statistics for concepts and tasks
| Task | Total hits | Intersection | Jaccard | |
|---|---|---|---|---|
| Memory (4237) | Fluency | 1871 | 81 | −4.31 |
| Paired Associates | 2735 | 78 | −4.48 | |
| AVLT | 360 | 34 | −4.9 | |
| CVLT | 441 | 34 | −4.92 | |
| MMSE | 3650 | 42 | −5.23 | |
| WMS | 702 | 27 | −5.23 | |
| Intelligence (7730) | WAIS | 2528 | 1862 | −3.74 |
| VIQ | 915 | 915 | −4.44 | |
| FSIQ | 874 | 854 | −4.51 | |
| WISC | 702 | 675 | −4.74 | |
| Stanford-Binet | 475 | 381 | −5.31 | |
| WCST | 1204 | 321 | −5.5 |
Output from PubAtlas reflecting co-occurrence strength for concepts and tasks. Gives total hits for each concept, total hits for each task, the number of hits for the intersection of concept and task, and the Jaccard coefficient, a measure based on the intersection divided by the union of sets.
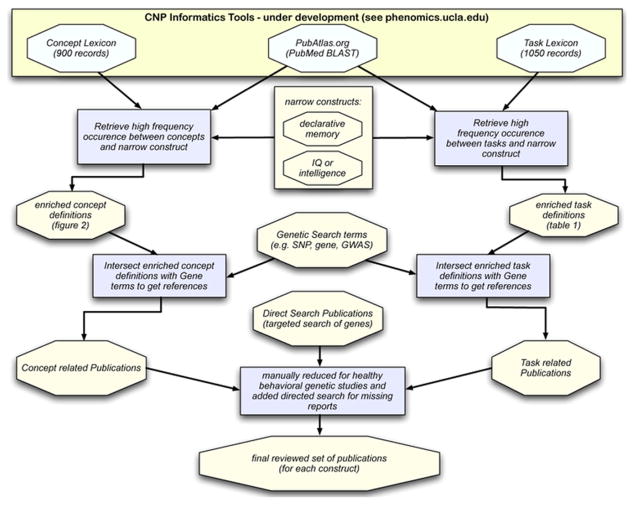
Fig. 1.
Methods flow chart. We used two lexica underdevelopment as well as the PubAtlas web-tool to arrive at our final reviewed set of publications. We performed two steps in parallel for concept (depicted on the left) and task (on the right) to find concepts and tasks that frequently co-occur with the narrow phenotype definitions of memory and intelligence (in blue boxes). These were then intersected with common gene search terms. Finally, with step 3, we reduced this list by excluding references that did not mention genetic or behavior focus, or did not contain a sample of healthy individuals.
Finally, tasks and concepts were used in PubMed searches with keywords related to genetics in order to find the best recall and precision to evaluate the genetic findings for these phenotypes (it may be noted that in literature mining and web search, the term “recall” is used to signify “sensitivity” or complete retrieval of all relevant material, while “precision” signifies “specificity” or retrieval of only relevant material, without irrelevant material). Along with the relevant cognitive/behavioral terms (related to either memory or IQ), we added a conjunction (AND) for genetic keywords: “haplotype” OR “SNP” OR “single nucleotide polymorphism” OR “gene” OR “linkage disequilibrium” OR (“genetic” AND (“association” OR “linkage”)) OR “GWAS” OR heritab*.
Construct operational definitions
In this manuscript, we made several choices given the breadth of the literature that has been investigated related to these topics. We decided to look at genes related only to healthy function. We also look only at the human behavioral studies; we excluded neuroimaging studies, as other contributions for this special issue feature neuroimaging. Specific constructs are defined in each section below.
Phenowiki
Phenowiki (http://phenowiki.org) is an online collaborative database for phenotype interrogation, annotation, evaluation, and selection. Phenowiki currently provides a collaborative web site for description, background, validity, and other important characteristics about tasks used in neuroscience. Further, it provides a semi-structured database for user-inputted quantitative data about relationships between tasks and other phenotypes related to neuropsychiatry. Quantitative data including effect sizes used in this review have been added to the Phenowiki database. These data are used to build a “hypothesis web” of the memory and intelligence phenotypes. This “quantology,” as we describe them, allows visualization of quantitative relationships between related entities (Parker et al., submitted for publication).
Methods caveats
Ideally, these methods provide a framework for using informatics to identify, examine, and refine, eventually engaging in an iterative process of task refinement to optimize measurement of a specific phenotype. There do remain some current limitations to this approach. Firstly, our lexica are works in progress. They are not exhaustive, and without help from the community can never reach that point. Improved lexica will provide better representation of the literature and improve recall on PubMed searches. Our second caveat is that the current literature mining was done on titles and abstracts in the PubMed database. While the PubMed database is extensive, it is not exhaustive, and similarly, titles and abstracts do not contain the richness of full text. Expanding to richer datasets is currently under development. Finally, while we aim to provide a fully objective literature-based representation of these constructs, as we develop this proof of concept example some subjective decisions must be made. We chose arbitrary Jaccard thresholds and similarly decided not to review procedural or working memory. These were done largely to reduce the scope of this review, but future work could expand this scope to yield fuller definition of specific constructs.
“Candidate” genes vs. GWAS
There will soon be results from large GWAS to provide richer knowledge about genetic associations with intelligence and memory constructs. Although some commentary suggests that these studies of cognitive phenotypes may be disappointing (Wade N, see NYT article September 2008), it remains unclear to what extent this may just reflect problems with sample size or problems with phenotype definition as proposed here. Indeed, Maher et al. (2008) very recently noted two new “adequately powered” GWAS studies for bipolar disorder and schizophrenia with sample sizes above 10,000, which may provide a “boost” for neuropsychiatry genetics work.
A majority of the existing literature for genetic association reflects studies of specific “candidate” genes, usually interrogated based on putative association with a disorder or signaling pathway. The possibility that evidence for association in these genes represents false-positive findings has been reviewed in detail elsewhere (Glatt and Freimer, 2002; Flint and Munafo, 2007); in brief, so far none of these genetic loci passes significance testing at the genome-wide level. Of note, the “adequately powered” GWAS studies noted above did not illuminate the usual cast of characters seen from typical candidate gene work (Maher et al., 2008). Below we review specific genes raised by our PubMed query and their relationship to our constructs of interest. Thus this review undoubtedly contains some inappropriate genetic targets, but we believe nevertheless this multivariate strategy will be important in detecting which genes (or neural systems, concepts, etc.) are inappropriate and which require greater efforts. We recognize the irony in reviewing this work after being critical of its approach; however, we feel the aggregation of these data is important in building models that represent the current state of knowledge in neuroscience. In this review, we remain agnostic as to the current strength of the finding as more than a few studies still have very small sample sizes and report very weak associations making it easier to understand why some of these studies were difficult to replicate. Recent studies using whole-genome suggest P-values at least on the order of 10−7 may be needed in order to survive correction across the genome, which should make them easier to replicate (Glatt and Freimer, 2002). This highlights another significant problem of the “candidate” gene approach, demonstrating the inertia in neuropsychiatric genetics as the same loci are tested time and time again, without correcting for genome-wide significance.
INTELLIGENCE
Intelligence is a poorly defined and often disputed construct, yet it is arguably the most well-studied behavioral phenotype in humans. Here we will avoid questions about controversy/dispute of its theoretical (construct) validity with respect to genetic findings, but point the reader to an excellent discussion of some of these issues (Sternberg et al., 2005).
Behaviorally, intelligence is a complex phenotype involving many well-validated tests (e.g. WAIS), however apparent shared variance among a variety of tests in all areas of cognition led researchers to propose “g.” The concept of g (for “general cognitive ability”) was proposed nearly a century ago to acknowledge the substantial covariance among diverse psychometric tests of cognitive ability (Spearman, 1904). In a meta-analysis of 322 studies, the average correlation among diverse tests (e.g. abstract reasoning, spatial abilities, and verbal fluency) is about 0.30 (Carroll, 1993), suggesting support for a single underlying latent factor such as “g.” The construct of “intelligence” continues to be measured today following paradigms largely established before 1918 [(Boake, 2002); see also Bilder et al. (in press)]. There remains controversy about the most appropriate definition of the intelligence construct broadly, whether it is best represented as “g,” as “g” with secondary “specific” intelligences, or best conceptualized as a “positive manifold” of multiple discrete sub-constructs (van der Maas et al., 2006). A full review of the behavioral manifestation of the construct is beyond the scope of this review, but we point the reader to a number of recent reviews that consider both the behavioral constructs (Neisser et al., 1996; Gottfredson, 1997).
Latent construct definition
Our review method yielded 56 hits for the enriched task definition of intelligence (see Fig. 1) and 123 hits for the enriched concept definition that served as our operational definition (see Figs. 1 and 2, Table 2). For the intelligence construct there was significant overlap between hits in the task and concept suggesting a fairly common set of tasks used to examine this construct. This resulted in the final list of reviewed papers for the intelligence construct of 75 articles. From these, we reduced the list to only include healthy behavioral studies, leading us to tabulate 34 different effects of gene function on intelligence.
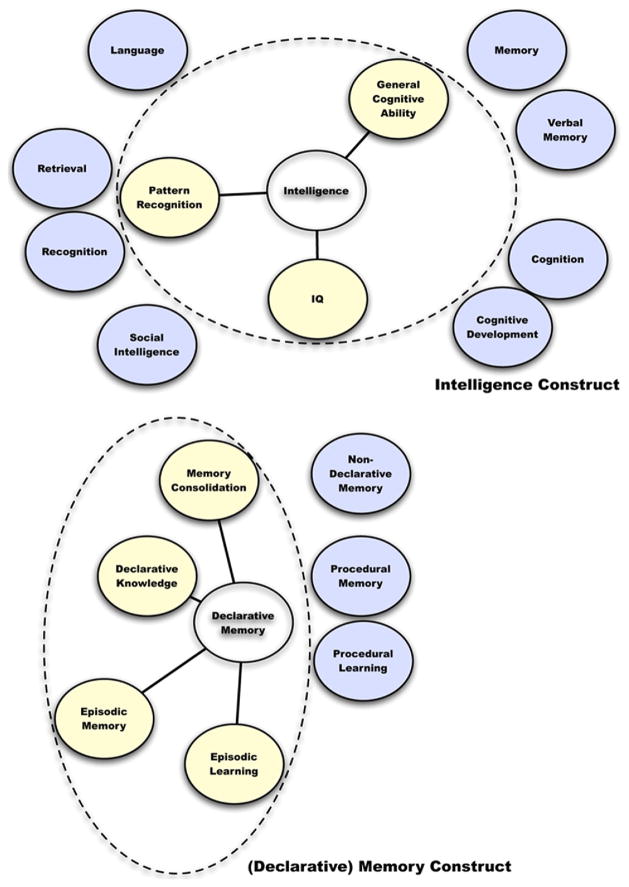
Fig. 2.
Phenotype construct maps. Graphical depictions of our enriched concept phenotype definitions. Distance from the center suggests co-occurrence strength. Dotted line represents concepts chosen to define the construct. Blue circles represent other frequent concepts that were not included in our phenotype definition.
Table 2.
Parameters for memory and intelligence effect (sizes)
| # | 1st Author | PMID | Source | Gene | SNP | Task | Indicator (condition) | Statistic | Stat value |
P-value | N | Cohen’s d |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Memory effects | ||||||||||||
| 1 | Gondo | 16095668 | Task | 5-HTT | S/L allele | MMSE | Cognitive function | Not reported | 265 | |||
| 2 | Reynolds | 17564514 | Concept | 5-HTT | U79746 [exon 1B −925 T/G] | Thurston Picture Memory | Quadratic change score at age 65 | F(,) | 3.13 | 0.05 | 112 | 0.31 |
| 3 | Roiser | 16893493 | Concept | 5-HTT | S/L allele/tryptophan depletion | CANTAB pattern recognition | Percent correct | P-value only | 0.05 | 30 | 0.62 | |
| 4 | Strange | 18614671 | Concept | 5-HTT | S/L allele | Word Perception Task | # Items recalled | F(1,26) | 4.865 | 0.036 | 28 | 0.71 |
| 5 | Mannie | 18775088 | Direct | 5-HTT | S/L allele | RAVLT | Delayed recall | No stats, ES derived from means/SD | 31 5-HT avg |
0.32 0.41 |
||
| 6 | Albert | 17402816 | Concept | APOE | Presence of e4 | composite | Episodic memory | Mixed model | 0.89 | 0.001 | 327 | 0.34 |
| 7 | Andersson | 17124416 | Concept | APOE | Presence of e4 | RAVLT | Delayed recall score used for group assignment | Not reported | 124 | |||
| 8 | Andersson | 17124416 | Concept | APOE | Presence of e4 | MMSE | Cognitive status | P-value only | 0.01 | 124 | 0.42 | |
| 9 | Deary | 15222832 | Task | APOE | Presence of e4 | WMS-R logical memory | Delayed recall | F(1,457) | 7.84 | 0.005 | 462 | 0.24 |
| 10 | Ferguson | 12502505 | Task | APOE | Presence of e4 | Borkowski verbal fluency | # Of items in 60 s | P-value only | 0.063 | 96 | 0.31 | |
| 11 | Fiocco | 18374494 | Concept | APOE | Presence of e4 | Unrelated word pairs (not normed) | Total recall of word pairs | Not reported | 63 | |||
| 12 | de Frias | 17379671 | Concept | APOE | Presence of e4 | Episodic memory (from factor structure −nyberg03) | Recall composite score | t () | 5.69 | 0.001 | 524 | 0.27 |
| 13 | de Frias | 17379671 | Concept | APOE | Presence of e4 | episodic memory (from factor structure −nyberg03) | Recognition composite score | t () | 6.2 | 0.001 | 524 | 0.27 |
| 14 | de Frias | 17379671 | Concept | APOE | Presence of e4 | Episodic memory (from factor structure −nyberg03) | Fluency composite score | t () | 4.65 | 0.001 | 524 | 0.27 |
| 15 | Halkala | 7644133 | Task | APOE | Presence of e4 | Buschke’s list learning | Total recalled over six trials | P-value only | 0.01 | 916 | 0.15 | |
| 16 | Halkala | 7644133 | Task | APOE | Presence of e4 | Visual Reproduction Test | Reproduce three figures | Not reported | 916 | |||
| 17 | Harwood | 12075916 | Task | APOE | Presence of e4 | MMSE | Age and education adjusted score | OR | 9.5 | 0.004 | 57 | 0.74 |
| 18 | Helkala | 8938259 | Concept | APOE | Presence of e4 | Buschke’s list learning; figure recall | Sum of word list recall over six trials | P-value only | 0.003 | 632 | 0.22 | |
| 19 | Jorm | 17201525 | Concept | APOE | Presence of e4 | CVLT | Delayed recall | P-value only | 0.91 | 6560 | 0.03 | |
| 20 | Mondadori | 17077159 | Concept | APOE | Presence of e4 | Associative encoding | Delayed recall | Odds ratio | 1.6 | 0.009 | 340 | 0.26 |
| 21 | Nilsson | 17100509 | Concept | APOE | Presence of e4 | Composite | Composite score for recall data | F(4,1542) | 6.48 | 0.005 | 657 | 0.20 |
| 22 | Reynolds | 17564514 | Concept | APOE | Presence of e4 | Thurston Picture Memory | Quadratic change score at age 65 | F(,) | 1.22 | 150 | ||
| 23 | Schultz | 18235080 | Concept | APOE | Presence of e4 | WMS-III | Logical memory subtest, immediate and delayed recall | F(1,617) | 6.79 | 0.009 | 626 | 0.19 |
| 24 | Strandburg | 15748778 | Task | APOE | Presence of e4 | MMSE | Cognitive impairment | P-value only | 0.02 | 357 | 0.22 | |
| 25 | Tagarakis | 17606532 | Task | APOE | Presence of e4 | MMSE | Cognitive impairment | Not reported | 137 | |||
| 26 | Tagarakis | 17606532 | Task | APOE | Presence of e4 | WMS-R | Cognitive impairment | Not reported | 137 | |||
| 27 | van Munster | 17728664 | Task | APOE | Presence of e4 | MMSE | Cognitive impairment | P-value only | 0.057 | 72 | 0.37 | |
| 28 | Yip | 12205106 | Task | APOE | Presence of e4 | MMSE | Dementia | Adjusted OR | 1.5 | 2034 APOE avg |
0.12 |
|
| 29 | Egan | 12553913 | Concept | BDNF | val66met | WMS-R | Recall of items from two stories after 30 min delay | F(2,591) | 3.89 | 0.02 | 641 | 0.16 |
| 30 | Dempster | 15719396 | Direct | BDNF | val66met | WMS-R logical memory | Delayed recall | t() | −2.65 | 0.01 | 114 | 0.44 |
| 31 | Hariri | 12890761 | Concept | BDNF | val66met | Scene Recognition Task | Old/new judgment— percent correct | F(1,26) | 9.44 | 0.005 | 64 | 0.67 |
| 32 | Harris | 16446742 | Task | BDNF | val66met | Verbal fluency | # Words in 1 min | F(,) | 1.3 | 0.28 | 460 | 0.05 |
| 33 | Harris | 16446742 | Task | BDNF | val66met | Logical memory | Recall of items in story | F(,) | 1.7 | 0.19 | 460 | 0.08 |
| 34 | Harris | 16446742 | Task | BDNF | val66met | Moray House Test (MHT) | Test of verbal reasoning taken at age 11 | F(,) | 4.2 | 0.016 | 460 | 0.20 |
| 35 | Strauss | 15626819 | Concept | BDNF | val66met | Verbal declarative memory | Composite score | F(4,54) | 1.4 | 0.25 | 19 BDNF avg |
0.31 0.23 |
| 36 | Apud | 17063156 | Concept | COMT | val158met | Free recall of word list | # Correct/15 words | Not reported | 47 | |||
| 37 | Frias | 15319576 | Concept | COMT | val158met | Recall test; recognition tests | # Correct/16 items over three tests | F(2,277) | 3.29 | 0.04 | 286 | 0.21 |
| 38 | Harris | 15979789 | Concept | COMT | val158met | WMS-R logical memory portion | Recall of items in story | F(,) | 3.6 | 0.028 | 460 | 0.18 |
| 39 | Strauss | 15626819 | Concept | COMT | val108/158met | WMS | Logical memory (delayed) subtest | F(4,54) | 0.49 | 0.15 | 49 | 0.30 |
| 40 | Strauss | 15626819 | Concept | COMT | val158met | Paired assoc learning | Delayed score | F(4,54) | 0.49 | 0.79 | 49 COMT avg |
0.23 0.17 |
| 41 | Nacimas | 18378080 | Concept | KIBRA | T/C allele (rs 17070145) | 24-h Delayed Five Words Recall | Recall of five and paired words 24 h delay | P-value only | 0.04 | 70 | 0.42 | |
| 42 | Nacimas | 18378080 | Concept | KIBRA | T/C allele (rs 17070145) | Short Story | Immediate recall | P-value only | 0.03 | 70 | 0.45 | |
| 43 | Need | 18205171 | Task | KIBRA | T/C allele (rs 17070145) | CANTAB verbal recall | Not provided | P-value only | 0.63 | 319 | 0.04 | |
| 44 | Need | 18205171 | Task | KIBRA | T/C allele (rs 17070145) | Story recall (Green, 2005) | Not provided | P-value only | 0.39 | 319 | 0.03 | |
| 45 | Need | 18205171 | Task | KIBRA | T/C allele (rs 17070145) | AVLT | Delayed recall | P-value only | 0.56 | 365 | 0.02 | |
| 46 | Schaper | 17353070 | Concept | KIBRA | T/C allele (rs 17070145) | Verbal Learning and Memory Test | Total recall | Effect size d | 0.74 | 0.004 | 64 | 0.74 |
| 47 | Almeida | 18194457 | Direct | KIBRA | T/C allele (rs 17070145) | CERAD | Delayed recall | F(,) | 3.91 | 0.049 | 312 KIBRA avg |
0.19 0.19 |
| 48 | Luciano | 19077176 | DTNBP1 | rs 742105 English cohort | Verbal declarative memory | Memory cumulative recall | t | −2.67 | 0.01 | 745 | 0.168 | |
| 49 | Luciano | 19077176 | DTNBP1 | rs 742105 English cohort | Verbal declarative memory | Delayed recall | t | −1.93 | 0.05 | 745 | 0.119 | |
| 50 | Luciano | 19077176 | DTNBP1 | rs 742105 English cohort | Verbal declarative memory | Memory factor | t | −2.32 | 0.02 | 745 | 0.149 | |
| 51 | Luciano | 19077176 | DTNBP1 | 6-SNP Burdick risk haplotype ACT in Scottish cohort | Verbal declarative memory | Logical memory immediate | F(1,1022) | 4.55 | 0.03 | 1054 DTNBP1 avg |
0.114 0.14 |
|
| Intelligence effects | ||||||||||||
| 1 | Turic | 11166947 | Con | APOE | apoE/−491 AT | WISC | High g (136)>avg g (103) | Chi | 2.599 | 0.761 | 202 | 0.10 |
| 2 | Turic | 11166947 | Con | APOE | apoE/Th1/e47cs | WISC | High g (136)>avg g (103) | Chi | 3.364 | 0.498 | 202 | 0.00 |
| 3 | Turic | 11166947 | Con | APOE | Presence of e4 | WISC | High g (136)>avg g (103) | Chi | 2.877 | 0.238 | 202 | 0.23 |
| 4 | Pendleton | 11983298 | Con | APOE | Presence of e4 | Heim | Ah4 part 1 score | t () | −0.7 | 0.48 | 767 | 0.05 |
| 5 | Ferguson | 12502505 | Task | APOE | Presence of e4 | WAIS | PIQ raw score | P-value only | 0.037 | 96 | 0.33 | |
| 6 | Puttonen | 12886039 | Con | APOE | Presence of e4 | Mental Arithmetic | Correct trials | t () | 57 | −0.42 | ||
| 7 | Puttonen | 12886039 | Con | APOE | Presence of e4 | Mental Arithmetic | Adjusted correct trials | t () | 57 | −0.51 | ||
| 8 | Puttonen | 12886039 | Con | APOE | Presence of e4 | Mental Arithmetic | # Of trials | t () | 57 | −0.46 | ||
| 9 | Deary | 15222832 | Task | APOE | Presence of e4 | Ravens | Correct completed | F(1,457) | 1.46 | 0.23 | 1462 APOE avg |
0.11 0.09 |
| 10 | Harris | 16446742 | Con | BDNF | val66met | Ravens | Correct completed | F(,) | 7 | 0.001 | 904 | 0.20 |
| 11 | Harris | 16446742 | Con | BDNF | val66met | Moray House Test | F(,) | 4.2 | 0.016 | 904 | 0.14 | |
| 12 | Tsai04 | From savitz | BDNF | WAIS | Piq | P-value only | 0.046 | 114 | 0.30 | |||
| 13 | Goldberg | 17988784 | Task | BDNF | matched IQ | Not applicable |
BDNF avg |
0.21 |
||||
| 14 | Comings | 12556901 | Con | CHRM2 | a->t 1890 | WAIS-R | Fsiq | F(,) | 4.12 | 0.017 | 828 | 0.15 |
| 15 | Dick | 17160701 | Task | CHRM2 | 8 SNPS <0.05 | WAIS-R | Piq | 8 SNPs P<05 | 0.05 | 1113 | 0.10 | |
| 16 | Gosso | 17996044 | Con | CHRM2 | rs 2061174 | WAIS-R | Piq | Chi | 9.14 | 0.003 | 391 | 0.28 |
| 17 | Gosso | 17996044 | Con | CHRM2 | rs 323650 | WISC | Viq | Chi | 9.5 | 0.002 | 371 CHRM2 avg |
0.30 0.17 |
| 18 | Barnett | 17202556 | Direct | COMT | Val158Met | WISC | Viq | F(2,1763) | 9.97 | 0.001 | 8707 | 0.21 |
| 19 | Barnett | 17202556 | Direct | COMT | Val158Met | WISC | Fsiq | F(2,1639) | 4.28 | 0.05 | 8707 | 0.15 |
| 20 | Malhotra | 11925305 | Task | COMT | Val158Met | WCST | Perseverative errors | F(2,70) | 4.43 | 0.02 | 73 | 0.71 |
| 21 | Reuter | 16026865 | Con | COMT | Val158Met | Not reported | 96 | |||||
| 22 | de Frias | 16102234 | Con | COMT | Val158Met | WAIS | Block design | F(1,285) | 4.59 | 0.03 | 292 | 0.25 |
| 23 | Starr | 17548151 | Task | COMT | Val158Met | Ravens | Correct completed | F(2935.7) | 7.92 | 0.001 | 473 | 0.26 |
| 24 | Tsai03 | 12566168 | Savitz | COMT | Val158Met | WCST | Perseverative errors | P-value only | 0.755 | 120 | 0.12 | |
| 25 | Tsai03 | 12566168 | Savitz | COMT | Val158Met | WAIS | Viq | P-value only | 0.431 | 120 | 0.03 | |
| 26 | Tsai03 | 12566168 | Savitz | COMT | Val158Met | WAIS | Piq | P-value only | 0.658 | 120 | 0.07 | |
| 27 | Tsai03 | 12566168 | Savitz | COMT | Val158Met | WAIS | Fsiq | P-value only | 0.511 | 120 COMT avg |
0.00 0.21 |
|
| 28 | Ball | 9507981 | Task | DRD2 | TaqI A1/A2 | WISC | High g (136)>avg g (105) | 0.41 | 102 | 0.04 | ||
| 29 | Rodriguez–Jimenez | 17230034 | Task | DRD2 | C957T | WCST | Perseverative errors | F | 5.849 | 0.018 | 83 | 0.73 |
| 30 | Han | 18555060 | Task | DRD2 | TaqI A1/A2 | WCST | Not reported | 40 | ||||
| 31 | Tsai02 | 11979061 | Task | DRD2 | TaqI A1/A2 | WISC | Piq | 0.036 | 112 | 0.34 | ||
| 32 | Tsai02 | 11979061 | Task | DRD2 | TaqI A1/A2 | WISC | Viq | 0.289 | 112 | 0.10 | ||
| 33 | Petrill | 9145541 | Savitz | DRD2 | TaqI A1/A2 | WISC | High g (51)>avg g (51)>low g (35) | Not significant, not reported | 137 | |||
| 34 | Berman | 7755518 | Savitz | DRD2 | TaqI A1/A2 | WISC | Fsiq | No stats, ES derived from means/sd | 73 DRD2 avg |
0.07 0.29 |
||
| 35 | Burdick | 16415041 | DTNBP1 | CTCTAC risk haplotype | General cognitive ability | Composite factor score | F(1,125) | 4.29 | 0.04 | 126 | 0.311 | |
| 36 | Luciano | 19077176 | DTNBP1 | rs 1047631 English cohort | Cattell | Culture fair | t | −2.12 | 0.03 | 745 | 0.136 | |
| 37 | Luciano | 19077176 | DTNBP1 | 3-SNP Donohoe risk haplotype ACT in English cohort | General ability | Alice heim 2 | F(1,728) | 3.73 | 0.05 | 745 | 0.119 | |
| 38 | Luciano | 19077176 | DTNBP1 | 3-SNP Donohoe risk haplotype ACT in English cohort | General ability | G factor (Wave 1) | F(1,688) | 5.67 | 0.02 | 745 | 0.149 | |
| 39 | Luciano | 19077176 | DTNBP1 | rs 742105 English cohort | General ability | G factor (Wave 2) | t | −2.19 | 0.03 | 745 | 0.136 | |
| 40 | Luciano | 19077176 | DTNBP1 | 6-SNP Burdick risk haplotype ACT in Australian cohort | Verbal ability | Nart | 0.04 | 1806 | 0.081 | |||
| 41 | Luciano | 19077176 | DTNBP1 | 3-SNP Donohoe risk haplotype ACT in English cohort | Verbal ability | Mill hill a | F(1,724) | 11.82 | 0.001 | 745 | 0.224 | |
| 42 | Luciano | 19077176 | DTNBP1 | rs 2619539 English cohort | Verbal ability | Mill hill a | t | 2.09 | 0.04 | 745 DTNBP1 avg |
0.127 0.13 |
|
Cohen’s d effect sizes were calculated from t or F statistics when degrees of freedom were provided. Otherwise, P-value and sample size were used to calculate (Cohen’s d) effect sizes. This table contains incomplete information, only what we were able to glean from the article. All effects are available online (http://phenowiki.org), and we encourage the authors to fill in missing data for their studies. To compute averages we only chose one effect per (independent) sample. We chose the strongest evidence for implication of that gene for the average, but this table contains additional relevant effects. “From” column, notes what method was used to find that paper (either from our task, concept, or direct search strategy, or cross-pollination from a paper already in the list).
Despite the ambiguity in the validity of this construct, there is relatively consistent evidence that IQ has demonstrated moderate heritability. Meta-analyses based on more than 10,000 twin pairs, 8000 parent-offspring pairs and 25,000 sibling pairs yield heritability of about 50% (McGuffin et al., 2001). This is an important, yet sometimes surprisingly overlooked, characteristic of cognitive phenotypes if they are to be investigated in studies for genetic determinants.
Intelligence shows strong covariance with almost every aspect of cognition. This makes literature-based mining for term-frequency measures somewhat more challenging as often studies report controlling for IQ. This challenge, however, reinforces the fact that it is difficult to dissociate intelligence from any aspect of cognition.
Neural systems
The neural correlates of intelligence have not been clearly elucidated. There are, however, a number of brain regions that have been consistently implicated in functional and structural imaging studies of intelligence in humans (e.g. Shaw, 2007; Thompson et al., 2002). Using our PubMed BLAST tool, PubAtlas, to extract the co-occurrence of brain regions and cognitive concepts, we show that the most common brain regions implicated in intelligence are the corpus callosum, the prefrontal cortex, the hippocampus, and the basal ganglia. These brain regions also correspond well to neural systems targets implicated in the genetic expression and signaling pathways for downstream effects of candidate genes implicated in intelligence [e.g. Kovas and Plomin (2006) present an excellent theoretical review of this work]. Thus, converging evidence from molecular and behavioral work highlights the importance of the PFC, HC, and BG.
A full review of the neural systems evidence for the genes, signaling pathways, behaviors, and diseases discussed here is beyond the scope of this meta-analysis. Further work will be required to quantify the specific effect sizes for these relationships, but until that work is completed we have pointed the reader to valuable reviews of specific evidence for the involvement of the basal ganglia, hippocampus, and prefrontal cortex.
Genes
Interrogating the PubMed literature using the search criteria described above revealed several associations using a “candidate” gene approach. Indeed there are a number of excellent review articles detailing the genetic findings of intelligence (Plomin and Petrill, 1997; de Geus et al., 2001; Plomin and Spinath, 2004). For this special issue, we summarize genetic association findings as part of our multivariate modeling approach. The full PubMed reference list returned from our search and additional details about each article are available on our website (http://www.phenowiki.org). As to be expected with a “candidate” gene approach, a number of well-known genes have been investigated across a variety of disorders and linked to a number of “endophenotypes.” These associations are generally weak, but exposing all the interactions together may provide additional clarity.
APOE
The apolipoprotein E gene and associated protein are involved in the breakdown of beta-amyloid deposits and has been implicated in Alzheimer’s disease (Bertram and Tanzi, 2008). Three relatively large studies report no significant effect of the e-4 allele on IQ using different tests (Turic et al., 2001; Pendleton et al., 2002; Deary et al., 2004). In contrast, Ferguson and colleagues (2003), reported worse performance on WAIS-R-test for those with a copy of e-4, however, they studied 96 women all with type I diabetes. One report even suggests that e-4 allele conferred better mental arithmetic, which the authors used as a measure of general cognitive ability (e-4 was better). Together these studies show weak evidence for the role of APOE in intelligence (Cohen’s d effect size = 0.09).
Brain derived neurotrophic factor (BDNF)
The BDNF gene encodes for a secretory protein involved in a number of important functions throughout the CNS, from glutamate signaling to cell survival to plasticity. Abnormal BDNF expression has been observed in patients with schizophrenia (Lu and Martinowich, 2008) and mood disorders (Martinowich et al., 2007; Martinowich and Lu, 2008). A previous review (Savitz et al., 2006) of BDNF and cognition noted one study that examined the WAIS-R in 114 Chinese women (Tsai et al., 2004). Separately, Harris and colleagues (2006) report a significant effect of BDNF genotype on Raven’s progressive matrices measures of intelligence in two large healthy cohorts. Thus, BDNF shows a small effect on intelligence (Cohen’s d effect size = 0.21).
Catechol-O-methyltransferase (COMT)
COMT is one of the most studied genetic polymorphisms in relation to human cognition. The allure of the val158met polymorphism is the apparent fourfold differences in COMT enzyme activity associated with that substitution. In an older cohort, Starr and co-authors (2007) found that COMT has an effect on overall cognition using a battery of tests. de Frias and co-workers (2005) report poorer performance on block design WAIS-R in those with a Val allele. Malhotra et al. (2002) report WCST errors was impacted by COMT. A recent review by Savitz et al. (2006) reveals one study of healthy subjects, who report no differences in a very small sample (six met/met), making interpretation difficult (Tsai et al., 2003). Overall, there is a small effect for the role of COMT in intelligence (Cohen’s d = 0.21).
Dopamine receptor d2 (DRD2)
The Taq1 allelic variant of the DRD2 is important for dopamine signaling, especially in the striatum (Bowirrat and Oscar-Berman, 2005). Two early studies in a healthy population from the Plomin group found no difference in allelic (A1/A2) frequency using WISC-R (Petrill et al., 1997; Ball et al., 1998). The review by Savitz et al. (2006) notes a study (Berman and Noble, 1995), that found no difference in WISC IQ associated with Taq A1 allele in the control group (in a study of alcoholism). More recently, Tsai et al.(2002) tested 112 female Chinese participants using the WAIS-R and found differences in PIQ but not FIQ or VIQ.
Several researchers have also used the Wisconsin Card Sorting Task as a measure of general functioning. This task is very complex and often shows significant correlation with more traditional measures of intelligence [e.g.; Ardila et al., 2000], and while it is unclear how well it overlaps with the construct, it is frequently associated with general ability (in the literature). Rodriguez-Jimenez and colleagues (2006) used the WCST as a measure of abstract reasoning ability and found more perseverative errors. Han and colleagues (2008) also found no effect in WCST errors in a control group (vs. meth abuse), but did not report any statistics for the control sample.
CHRM2
CHRM2 is a gene that encodes a cholinergic/muscarinic type 2 receptor. Located on chromosome 7q, it contains a number of SNPs studied in neuropsychiatry. This receptor is primarily a presynaptic auto-receptor that modulates release (Luo et al., 2007). Three studies from two different groups (Gosso et al., 2006b; Dick et al., 2007b) report several SNPs on the CHRM2 gene are associated with intelligence. In relatively large samples, both Gosso et al. (2006b) and Dick et al. (2007b) found modulation in this gene on WAIS-R at a number of different SNP locations. More recent fine mapping of the CHRM2 gene by Gosso et al. (2006b) confirmed two SNPs from their previous study and found two new ones. These studies suggest a very small effect size (Cohen’s d = 0.17) for the role of this gene in intelligence.
Dysbindin-1 (DTNBP1)
DTNBP1 encodes dystrobrevin-binding protein, which is expressed downstream in synaptic terminals in a number of subcortical and cortical regions including the hippocampus, the frontal lobe, the temporal lobe, and the midbrain. There is also growing evidence that suggests increased risk for schizophrenia is associated with dysbindin’s influence on cognitive and pre-frontal function (Posthuma et al., 2005; Burdick et al., 2006; Donohoe et al., 2007). Burdick and colleagues (2006) first reported an association between DTNPB1 six-locus haplotype (CTCTAC) and (g) in two independent samples of patients with schizophrenia and healthy controls. Another group found a relationship between WAIS-III IQ scores and several single SNPs in healthy controls, patients with schizophrenia, and their siblings but did not report statistics for these groups separately (Zinkstok et al., 2007). A very recent report that employed a diverse range of cognitive tasks and SNPs in three large independent samples of healthy individuals composed of Australian, English, and Scottish descent provides a large number of association results for dysbindin (Luciano et al., 2009). These associations differed across study population and cognitive measure, but generally supported a relationship between DTNBP1 genotype and cognitive ability, although most effect sizes for memory and IQ were quite small. Luciano and colleagues (2009) test a large number of SNPs and cognitive measures, of which those that passed a nominal P-value threshold of P<0.05 are included in Table 2, and the remaining effects available online through http://phenowiki.org (http://www.phenowiki.org). Even when including just the significant effects in the report by Luciano et al. (2009), the average weighted effect size for DTNBP1 in intelligence is only 0.13 (Cohen’s d). These authors, however, did replicate the association between general cognitive ability and the original 6-SNP haplotype (Burdick et al., 2006). Thus, there is consistent but very weak evidence beyond the initial association study for the role of dysbindin in intelligence in the healthy population.
GWAS or GW linkage reports
Plomin’s group and his research colleagues have championed the QTL strategy. In a genome-wide allelic (or “pooled”) association method, pools of DNA are formed by combining samples from individuals differing in mean score on IQ, and then a comparison is made of the frequency of alleles for each marker between the comparison groups. Recently, this group reported association for general cognitive ability (g) taken at age 7 in a sample of 6000 twins (Butcher et al., 2005). Five of the 10,000 typed single nucleotide polymorphisms (SNPs) (located on chromosomes 2, 6, 7, 11, and 18) showed replicable association but together accounted for less than 1% of ability variance. Though these associations were confirmed by individual genotyping, a meta-analysis across six population samples did not support an association between the 5-SNP set and (g); (Luciano et al., 2008). Of course, at the time this study was conducted, the prevailing technology was far less developed than today, thus prompting both the use of the pooled DNA method and genotyping at “only” 10,000 SNPs, while today GWAS routinely genotype each individual case at more than 350,000 SNPs.
Five genome-wide studies converge to identify regions on chromosomes 2, 6, and 14 as explaining some of the variance in intelligence (Luciano et al., 2005; Wainwright and Jordan, 2005; Buyske et al., 2006; Dick et al., 2007a). Most recently, Butcher et al. (2008) further explored a genome-wide association scan employing a 500,000 SNP microarray (Butcher et al., 2005). They found six SNPs that yielded significant associations with (g), although again the effect size explained less than 1% of the phenotypic variance. This has led some to suggest that effect sizes may be much smaller than previously considered (Meaburn et al., 2006).
Plomin and Kovas (2005) have argued for a “generalist gene” hypothesis, to explain how the phenotypic relationship with (g) is largely mediated genetically, where the genes for individual variation in intelligence are often general in their effects. This hypothesis is grounded in considerable multivariate genetic research showing that there is substantial genetic overlap between broad areas of cognition such as language, reading, mathematics, and general cognitive ability [see; Deary et al., 2006 for review]. One analysis (Luo et al., 1994) using 17 ability measures from two intelligence test batteries found that all the tests were influenced by genetic sources common to all tests. Another study (Rijsdijk et al., 2002) investigated correlations using the Raven’s Progressive Matrices and the WAIS in 194 twins. The authors found that the covariation among the WAIS subtests and between the subtests and Raven’s was predominantly influenced by a second-order genetic factor, lending support to the notion of a biological basis for (g). These data substantiate the notion that specific cognitive abilities are influenced by a pervasive genetic factor whose contribution to each phenotypic measure does not differ significantly.
Intelligence construct conclusions
Candidate genes have not provided us much traction, as they explain little of the variance in this phenotypic construct. If we look at the average effect size for each of the candidate genes provided here and what percentage of variance that explains, we can calculate a liberal approximation of how much variance we may currently be able to explain. This is assuming a very simple additive model with no interaction between genes, so it is unlikely to be as strong as suggested here. These five genes might explain 5.5% of the variance in a construct that shows very strong heritability (at least 50%). Thus, a large portion of the variance has yet to be explained.
There are other genes that may warrant consideration once evidence for their association is replicated by independent groups. These genes may shed additional light on the complex interaction of the multivariate representation of intelligence. Cathepsin D, a protease implicated in AD, has been studied in a large cohort of older adults using the Heim Intelligence Test (Payton et al., 2003). SNAP-25 codes for a membrane protein that is essential for vesicle docking at the presynaptic terminal. Two papers by Gosso and colleagues (2006a, 2008) show WISC scores differed for different SNAP-25 SNPs. Another interesting study by Reuter et al. (2005) found an interaction between COMT/D2 and the Stroop test. This highlights the challenges facing neuroscience going forward as we aim to understand the role of specific genes and the interactions between these genes in the pursuit of association with complex phenotypes.
MEMORY
Memory is a broad and multifactorial concept (Squire, 1992). The intuitive concept of memory, involving the recollection of previous events or facts, is most often identified with the declarative memory system, which is in turn often considered to rely upon the medial temporal lobe (hippocampus and related structures) as well as other brain regions including the prefrontal cortex, but clearly involves a broad range of cellular processes widely distributed throughout the brain. Declarative memory is often subdivided into three processes; encoding (the creation of memory traces based on experience), consolidation (the fixation of those memory traces into a longer-term store) and retrieval (the recollection of stored information). The construct (Fig. 2) defined by the results of our informatics procedures suggests a close relation between declarative memory and declarative knowledge, memory consolidation, episodic memory, and episodic learning in the literature. Although in the literature declarative memory is frequently also associated with procedural memory and learning, as well as non-declarative memory, we chose not to include those constructs here, as largely agreed to represent different forms of memory that have different tasks, disorders, and neural substrates.
Latent construct definition
For the declarative memory construct (see Fig. 2, Table 1), our procedure led to 50 hits for the enriched task definition and 118 hits for the enriched concept definition. As with the IQ construct, we reduced this list by excluding papers that did not include either genetics or behavior. We note here that we also did not consider “working memory” or “short-term memory,” as these may be more related to attention/control processes than memory (Sabb et al., 2008), and we did not consider “procedural memory” in an effort to reduce the scope of review. We did find that “declarative memory” and “episodic memory” showed a very high overlap in papers, suggesting these terms are used interchangeably in the literature. There was very little overlap in the task and concept lists, suggesting less uniformity in the tasks used to measure (declarative) memory, and consistent with the idea that “memory” has not been measured as a unitary construct. Somewhat surprisingly, several papers used the MMSE as a test of memory. Although widely accessible, this might not be the most accurate indicator of declarative memory, and it highlights the need for better consideration of phenotype measures for latent cognitive constructs. Overall, we report 47 effects of genes on memory performance in the healthy population. For each construct, a full list of hits for the original queries and the pruned lists of hits are available online at http://phenowiki.org.
As mentioned above, although sometimes overlooked in establishing phenotype definition, examining the heritability of the construct and especially the tasks that measure them is an important step (Gottesman and Gould, 2003). Several studies confirm the heritability of declarative memory. In 1997, McClearn reported the heritability of memory at ~50% using the WAIS (McClearn et al., 1997). In an aged population, Johansson and colleagues (1999) found the heritability of episodic memory ranged from 0.04 to 0.49, depending on the specific indicator used.
Neural level
The neural correlates of memory are well studied in neuropsychology, functional imaging, and molecular neuroscience. Seminal work by Knowlton and Squire (Squire et al., 1993) developed a model that dissociates declarative and non-declarative memory systems. These systems are served by different brain networks, with declarative putatively driven by the medial temporal lobe including the hippocampus, while non-declarative forms of memory are putatively served by the basal ganglia (Knowlton et al., 1996). These brain regions are consistent with those implicated in intelligence (especially when one considers that the PFC is largely implicated in working memory studies). As with intelligence, there is good convergence between different approaches that implicate the same brain regions, as these regions are presumed to be the focus of disorders like schizophrenia (Gothelf et al., 2000), and show molecular mechanisms putatively involved in memory [i.e. long term potentiation (LTP)] (Pelletier and Lacaille, 2008). We hope with further work in building models like these, the field can begin to quantitatively describe the specific relationships between these nodes. This may help to elucidate similarities and more importantly the differences in critical pathways between disease and genes.
Genes
As with IQ, our search criteria revealed several putative associations with candidate genes in the PubMed literature. A majority of the papers studying genetics effects of memory have focused on the APOE gene, with most of them reporting significant differences for those with the e4 allele. Here we review briefly review APOE and those with at least two reports in healthy samples as recovered using our search criteria. Again, the full PubMed reference list is available on our website (http://www.phenowiki.org).
APOE
The APOE e-4 allele is a known risk factor for AD but its role in normal memory is still debated. Of the 22 papers examined here, only five failed to report a significant effect on memory for carriers of the e4 allele (Yip et al., 2002; Jorm et al., 2007; Tagarakis et al., 2007b; van Munster et al., 2007; Fiocco et al., 2008). The remaining studies found that those with the APOE e-4 allele had poorer memory performance using a variety of different tasks.
Several studies used a rather tenuous operational definition of memory in the MMSE (Haan et al., 1999; Harwood et al., 2002; Yip et al., 2002; Strandberg et al., 2005; Andersson et al., 2007; Tagarakis et al., 2007a; van Munster et al., 2007). Across these studies, however, lower MMSE scores were seen for those with a copy of the e-4 allele than those without the e-4 allele. As mentioned above, the total MMSE score may not be the most reliable indicator of declarative memory performance. A study by Anderson et al. (2007) lends support to this, by using both the MMSE and the Rey Auditory Verbal Learning Test (RAVLT). They found e4 carriers performed worse on the MMSE but not on RAVLT.
Other studies used well-established memory batteries (WMS, Buschke) to assess the relationship between APOE and memory. Two studies from the same group used Buschke’s tests. Helkala studied a large sample of non-demented elderly subjects and found APOE e4 carriers scored significantly lower on Buschke’s list learning test (Helkala et al., 1995), and in the same cohort found those with the e-4 allele were poorest at a three year follow-up on Buschke’s selective reminding test (Helkala et al., 1996). Deary et al. (2004) reported poor WMS (logical memory portion) scores, and Schultz et al. (2008) found significantly lower scores across WMS-III measures for e4 carriers after controlling for IQ.
Some studies used more general memory tasks, like Mondadori et al. (2007), who found better episodic memory in a young sample of e4 carriers on an associative encoding task, which led the authors to propose that e4 may be associated with better neural efficiency in younger persons. Ferguson et al. (2003) reported a trend towards significantly worse performance on Borkowski’s verbal Fluency task in e-4 carriers. Reynolds et al. (2007) found significant relationships between the e4 allele of APOE as well as 5HTT on the Thurston Picture Memory Test.
Finally, a number of studies examined composite measures or batteries of tests. de Frias et al. (2007) found those e-4 carriers with higher cholesterol levels had greater declines in episodic memory over 10 years on composite scores for tests of recall, recognition, and fluency. Similarly, Nilsson et al. (2006) found significantly poorer performance on tests of free recall, cued recall, word/face/name recognition. Albert et al. (2007) examined longitudinal change in cognitive performance among individuals with mild cognitive impairment and found individuals with APOE e4 allele scored significantly lower on a battery of tasks [CVLT; Free & Cued Selective Reminding Test; Rey-O; Delayed Word Recall; WMS].
BDNF
BDNF was important for both intelligence and memory constructs, with respect to its role in memory, Egan and colleagues (2003) reported poorer episodic memory as well as abnormal hippocampal activation using fMRI. Two other studies included in this review also found an effect of this genetic variant on declarative memory (Hariri et al., 2003; Harris et al., 2006), while one study found no effect (Strauss et al., 2004).
COMT
COMT was also shared between memory and intelligence construct. Four studies examined the role of COMT in declarative memory. Two of these studies report no effect of the val158met genotype on verbal episodic memory (Apud et al., 2007), or logical memory and paired associate word learning (Strauss et al., 2004). Two other studies report positive findings where one found met/met subjects performed best on recall and recognition tests of episodic memory (de Frias et al., 2004) and one found heterozygotes had significantly higher scores on the logical memory portion of the WMS test (Harris et al., 2005).
Kidney and brain expressed protein (KIBRA)
Initially discovered in a whole genome study, the KIBRA gene encodes a phosphoprotein. It has subsequently been shown to be involved in memory retrieval, and is expressed in hippocampus (Papassotiropoulos et al., 2006). Our search revealed two studies that concurred with original reports of the KIBRA gene’s role in memory (Need et al., 2008; Schaper et al., 2008). Another study, however, indicated the opposite effect; a positive effect on memory performance for the C-allele (Nacmias et al., 2008). The authors suggest the KIBRA genotype could affect subjects complaining of memory deficits differentially from subjects who do not.
5-HT transporter gene (5-HTT)
5-HTT and protein are involved in the uptake of 5-HT from the synapse and thus in modulating serotonergic signaling (for review see; Aleman et al., 2008). Three studies in our review directly investigated short and long allelic variants in the 5-HTT gene with different operational definitions for memory. One group found a significant effect of the s-allele on emotion-induced retrograde amnesia, but not emotional memory encoding (Strange et al., 2008). Another study found no genetic effect of this allele on MMSE performance (Gondo et al., 2005). Finally, Mannie and colleagues (2008) demonstrated a moderately strong effect for the RAVLT. The largest study of 5-HTT did not report finding an association, but failed to report enough statistics to be included in the average. The evidence for association in this gene highlights the need for the approach presented here. There are multiple positive reports in the literature using tenuous phenotype definitions, and one large study that did not present enough information for proper meta-analysis. Building these multilevel models will expose the weak links in the chains of evidence.
Memory construct conclusions
Current genetic associations provide very limited success. One GWAS success has come in the identification of KIBRA, however, further studies have not replicated that finding as successfully. More work is also needed to establish the pathway between gene and expression of memory phenotypes. Several other genes that may warrant further investigation include MTHR, 5HT2a, CAMTA1, SOAT and GCLM. Overall, if one takes these average effect sizes at face value, a very small proportion of the variance has been explained in this construct (~7%), in comparison to its heritability of around 50%.
GENERAL DISCUSSION
Using an informatics approach, we propose a framework for building complex phenotypic models that provides a more objective definition of constructs. To highlight the benefits of this approach, we reviewed the genetic contributions to memory and intelligence in the healthy population.
“Intelligence” and “memory” functions are appealing targets for genetic study, given that these are widely appreciated in their lay sense, and disorders of each have enormous public health significance. Indeed cognitive disorders that impact general intelligence and memory may be among the greatest causes of disability we face in mental health. Both constructs have long been measured, and psychometric properties of many measurements (i.e. tests) are well established. There continues to be controversy, however, about how best to measure these constructs, and what their components may be. The challenge of identifying the components of these constructs is highlighted by current studies, which in attempts to discern genetic bases of behavioral phenotypes, place a focus more clearly on connecting abstract concepts such as “intelligence” and “memory” to specific biological processes. The initial hope that mapping these phenotypes to genetic targets might be easy was based on the substantial heritability of these constructs. Estimates of heritability in each of these constructs suggest that approximately 50% of phenotype variance is explained by genetic variation, providing a large target for genetic analyses. Given, however that only small proportions of variance have been explained by the currently identified genes, it may be that this initial hope was unrealistic, and that a large number of genes are likely to contribute.
This demonstrates that current phenotype definitions lack the specificity and complexity necessary for the study of genetic determinants. We know neither memory nor intelligence is a unitary construct, and their representations in literature co-occurrence confirms this. One important step towards improving phenotype definitions is the development of neuroscience lexica. Using two lexica (one for cognitive tasks, another for cognitive concepts), we found differences between the literatures that represent task and concept definitions of the constructs, especially in the memory construct, further suggesting there could be stronger agreement on what tasks are best used to measure this concept. In particular, several studies used MMSE as an indicator of declarative memory function. Better definition of latent concepts in neuroscience, and their relationship to the indicators, the actual measurement assessed, may help to provide researchers with more prudent ways to select appropriate measures for their phenotypes of interest.
The literature is replete with genes putatively associated with these two constructs. Many of these potential targets were not surprising as they were linked to clinical disorders (e.g. APOE and AD), or important transmitter systems (e.g. COMT, DRD2 and dopamine signaling), or molecular neuroscience hypotheses (e.g. BDNF and LTP) that have been foci of cognitive research. Our review also revealed that many of the same genes are implicated in both memory and intelligence constructs. This may suggest several potential biases in looking for genetic determinants.
Overall, we found relatively small portions of variance have been explained for either intelligence (~5.5%) or memory (~7%) constructs. With both constructs putatively 50% heritable, this suggests the candidate gene approach may not be the most fruitful process in examining genetic targets for complex phenotypic constructs like memory or intelligence.
This demonstrates one potential bias is highlighted in this review. The candidate gene approach creates a low barrier for researchers to investigate a particular gene (so far few “candidates” in neuropsychiatry have genome-wide significance), and the field has so far lacked any systematic method to evaluate the relative validity of hypotheses supporting candidacy (i.e. there remain no clear figures of merit regarding the strength of the putative causal pathway between genotype and phenotype). This is another form of instrumentation inertia (Bilder, 2002), in which some initial observation leads to continued investigation without regard to the actual strength of the findings. The number of papers on a topic sometimes seems more influential than the strength of these findings, both in phenotype and genotype selection. This problem may be compounded by the “file drawer problem,” where positive results are reported widely, but negative results are less frequently published which could reveal weaker relations of putative candidate genes with their phenotypic targets.
One scenario that is often mentioned but not investigated is that our phenotype constructs are not defined enough to extract differences between IQ and memory. It remains unclear whether efforts to define more discrete processes that contribute to intelligence and memory can be identified and linked more directly to gene action. However, if this is to happen, it is unlikely to be at the level of parsing cognitive concepts, and will require firmer links to underlying biological processes. For example, it may well be that we will require intermediate links at the level of protein structure, markers of neurodevelopmental processes, and more discrete markers of cellular and signaling system function to forge paths from genome to behavioral phenotypes. This highlights the need for improved translational models of intelligence and memory functioning. Informatics strategies can help support more appropriate modeling of these translational constructs.
Multi-level models of phenotype constructs are necessary to represent the complexity of many gene interactions (see Fig. 3). The framework provided here builds the necessary platform needed to construct multi-level models of complex phenotypes that can help build consensus by exposing quantitative relationships between phenotypic concepts used in genetic research. Until recently, examination of genetic studies involved looking at a syndrome level phenotype (e.g. schizophrenia) and its relationship to a particular gene (e.g. COMT). More recent focus on endophenotypes has driven researchers to examine phenotypes theoretically “closer” to the genome, including everything from symptoms down to proteins. A multi-level approach subscribed to by the CNP, involves examining a complex web of phenotypes and their inter-relationships, as well as the overall phenotype’s relationship to the genome. We have described this as a “quantology,” a visual representation of quantitative relationships between any set of entities [Parker et al. (submitted for publication)].
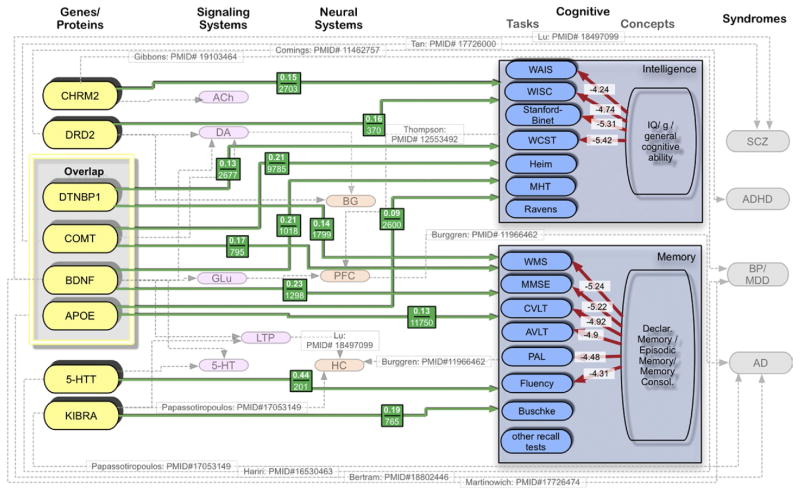
Fig. 3.
Multi-level model of memory and intelligence. Complex phenotype model showing overlap in genetic targets as well as signaling and neural systems between disparate latent constructs of memory and intelligence. Shows five levels of phenotype complexity (gene, signaling, neural, cognitive, and syndrome). Grey lines represent widely held belief and references that support those relationships but not quantitatively reviewed. Red lines show cumulative literature co-occurrence with Jaccard coefficient labels. Green lines represent Cohen’s d effect size estimates determined from this review. Green line labels give average weighted effect size and cumulative sample size that contributed to the effect.
Here we provided a proof of concept example for our multi-level approach. We provide a framework for building more objective literature-based models that can then be examined using meta-analytic approaches. This provides the field with a larger frame of reference for hypothesis testing of critical pathways between gene and phene, and helps the field build consensus in phenotype definition. We hope the next step is to expand and connect these models to create an effect-size knowledge base for neuroscience. Through visualization of these models, we can better capture the wide breadth of research results in our field and expose points of weakness that require further study in the pathway between gene and syndrome.
CONCLUSION
In summary, in order to make progress in linking the genome to complex neuropsychiatric disorders, we need better phenotype definitions. This will require large collaboration and consensus building from the field. It will require new tools and new approaches that include multi-level phenotype constructs using the phenomics approach (Bilder et al., this issue). We have proposed a novel informatics strategy to encourage objective definitions of phenotypes as a starting point and demonstrated this approach for “memory” and “intelligence.” Using these tools, the field of neuroscience can make progress in understanding the phenome, which may ultimately unlock the genetic determinants of neuropsychiatric diseases.
Acknowledgments
This work is supported by the following grants: UL1DE019580 (Bilder), RL1LM009833 (Parker), RO1MH082795 (Poldrack), RL1MH083269 (Cannon), RL1MH083268 (Freimer), and a NARSAD Young Investigator Award (Sabb).
Abbreviations
- BDNF
brain-derived neurotrophic factor
- COMT
catechol-O-methyltransferase
- DRD2
dopamine receptor d2
- DTNBP1
dysbindin-1
- GWAS
genome-wide association studies
- KIBRA
kidney and brain expressed protein
- LTP
long term potentiation
- MMSE
Mini-Mental State Exam
- RAVLT
Rey Auditory Verbal Learning Test
- 5-HTT
5-HT transporter gene
References
- Albert M, Blacker D, Moss MB, Tanzi R, McArdle JJ. Longitudinal change in cognitive performance among individuals with mild cognitive impairment. Neuropsychology. 2007;21:158–169. doi: 10.1037/0894-4105.21.2.158. [DOI] [PubMed] [Google Scholar]
- Aleman A, Swart M, van Rijn S. Brain imaging, genetics and emotion. Biol Psychol. 2008;79:58–69. doi: 10.1016/j.biopsycho.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Andersson C, Blennow K, Johansson SE, Almkvist O, Engfeldt P, Lindau M, Eriksdotter-Jonhagen M. Differential CSF biomarker levels in APOE-epsilon4-positive and -negative patients with memory impairment. Dement Geriatr Cogn Disord. 2007;23:87–95. doi: 10.1159/000097354. [DOI] [PubMed] [Google Scholar]
- Apud JA, Mattay V, Chen J, Kolachana BS, Callicott JH, Rasetti R, Alce G, Iudicello JE, Akbar N, Egan MF, Goldberg TE, Weinberger DR. Tolcapone improves cognition and cortical information processing in normal human subjects. Neuropsychopharmacology. 2007;32:1011–1020. doi: 10.1038/sj.npp.1301227. [DOI] [PubMed] [Google Scholar]
- Ardila A, Pineda D, Rosselli M. Correlation between intelligence test scores and executive function measures. Arch Clin Neuropsychol. 2000;15:31–36. [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball D, Hill L, Eley TC, Chorney MJ, Chorney K, Thompson LA, Detterman DK, Benbow C, Lubinski D, Owen M, McGuffin P, Plomin R. Dopamine markers and general cognitive ability. Neuroreport. 1998;9:347–349. doi: 10.1097/00001756-199801260-00031. [DOI] [PubMed] [Google Scholar]
- Barratt BJ, Payne F, Lowe CE, Hermann R, Healy BC, Harold D, Concannon P, Gharani N, McCarthy MI, Olavesen MG. Remapping the insulin gene/IDDM2 locus in type 1 diabetes. Diabetes. 2004;53:1884–1889. doi: 10.2337/diabetes.53.7.1884. [DOI] [PubMed] [Google Scholar]
- Bearden CE, Freimer NB. Endophenotypes for psychiatric disorders: ready for primetime? Trends Genet. 2006;22:306–313. doi: 10.1016/j.tig.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Berman SM, Noble EP. Reduced visuospatial performance in children with the D2 dopamine receptor A1 allele. Behav Genet. 1995;25:45–58. doi: 10.1007/BF02197241. [DOI] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Thirty years of Alzheimer’s disease genetics: the implications of systematic meta-analyses. Nat Rev. 2008;9:768–778. doi: 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- Bilder RM. Computerized neurocognitive assessment for schizophrenia: historical background and current trends. J Adv Schizophr Brain Res. 2002;4:74–80. [Google Scholar]
- Bilder RM, Sabb FW, Parker DS, Kalar D, Chu WW, Fox J, Poldrack RA. Cognitive ontologies for neuropsychiatric phenomics research. Cogn Neuropsychiatry. 2009 doi: 10.1080/13546800902787180. PMID: 19344640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boake C. From the Binet–Simon to the Wechsler–Bellevue: tracing the history of intelligence testing. J Clin Exp Neuropsychol. 2002;24:383–405. doi: 10.1076/jcen.24.3.383.981. [DOI] [PubMed] [Google Scholar]
- Bottini N, Vang T, Cucca F, Mustelin T. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Semin Immunol. 2006;18(4):207–213. doi: 10.1016/j.smim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Bowirrat A, Oscar-Berman M. Relationship between dopaminergic neurotransmission, alcoholism, and reward deficiency syndrome. Am J Med Genet B Neuropsychiatr Genet. 2005;132:29–37. doi: 10.1002/ajmg.b.30080. [DOI] [PubMed] [Google Scholar]
- Burdick KE, Lencz T, Funke B, Finn CT, Szeszko PR, Kane JM, Kucherlapati R, Malhotra AK. Genetic variation in DTNBP1 influences general cognitive ability. Hum Mol Genet. 2006;15(10):1563–1568. doi: 10.1093/hmg/ddi481. [DOI] [PubMed] [Google Scholar]
- Butcher LM, Davis OS, Craig IW, Plomin R. Genome-wide quantitative trait locus association scan of general cognitive ability using pooled DNA and 500K single nucleotide polymorphism microarrays. Genes Brain Behav. 2008;7(4):435–446. doi: 10.1111/j.1601-183X.2007.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher LM, Meaburn E, Knight J, Sham PC, Schalkwyk LC, Craig IW, Plomin R. SNPs, microarrays and pooled DNA: identification of four loci associated with mild mental impairment in a sample of 6000 children. Hum Mol Genet. 2005;14:1315–1325. doi: 10.1093/hmg/ddi142. [DOI] [PubMed] [Google Scholar]
- Buyske S, Bates ME, Gharani N, Matise TC, Tischfield JA, Manowitz P. Cognitive traits link to human chromosomal regions. Behav Genet. 2006;36:65–76. doi: 10.1007/s10519-005-9008-9. [DOI] [PubMed] [Google Scholar]
- Carroll JB. Human cognitive abilities: a survey of factor-analytic studies. Cambridge University Press; 1993. [Google Scholar]
- Deary IJ, Spinath FM, Bates TC. Genetics of intelligence. Eur J Hum Genet. 2006;14:690–700. doi: 10.1038/sj.ejhg.5201588. [DOI] [PubMed] [Google Scholar]
- Deary IJ, Whiteman MC, Pattie A, Starr JM, Hayward C, Wright AF, Visscher PM, Tynan MC, Whalley LJ. Apolipoprotein e gene variability and cognitive functions at age 79: a follow-up of the Scottish mental survey of 1932. Psychol Aging. 2004;19:367–371. doi: 10.1037/0882-7974.19.2.367. [DOI] [PubMed] [Google Scholar]
- de Frias CM, Annerbrink K, Westberg L, Eriksson E, Adolfsson R, Nilsson LG. COMT gene polymorphism is associated with declarative memory in adulthood and old age. Behav Genet. 2004;34:533–539. doi: 10.1023/B:BEGE.0000038491.06972.8c. [DOI] [PubMed] [Google Scholar]
- de Frias CM, Annerbrink K, Westberg L, Eriksson E, Adolfsson R, Nilsson LG. Catechol O-methyltransferase Val158Met polymorphism is associated with cognitive performance in nondemented adults. J Cogn Neurosci. 2005;17:1018–1025. doi: 10.1162/0898929054475136. [DOI] [PubMed] [Google Scholar]
- de Frias CM, Bunce D, Wahlin A, Adolfsson R, Sleegers K, Cruts M, Van Broeckhoven C, Nilsson LG. Cholesterol and triglycerides moderate the effect of apolipoprotein E on memory functioning in older adults. J Gerontol B Psychol Sci Soc Sci. 2007;62:112–118. doi: 10.1093/geronb/62.2.p112. [DOI] [PubMed] [Google Scholar]
- de Geus EJ, Wright MJ, Martin NG, Boomsma DI. Genetics of brain function and cognition. Behav Genet. 2001;31:489–495. doi: 10.1023/a:1013360909048. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Kramer J, Wang JC, Hinrichs A, Bertelsen S, Kuperman S, Schuckit M, Nurnberger J, Jr, Edenberg HJ, Porjesz B, Begleiter H, Hesselbrock V, Goate A, Bierut L. Association of CHRM2 with IQ: converging evidence for a gene influencing intelligence. Behav Genet. 2007a;37:265–272. doi: 10.1007/s10519-006-9131-2. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Kramer J, Wang JC, Hinrichs A, Bertelsen S, Kuperman S, Schuckit M, Nurnberger J, Jr, Edenberg HJ. Association of CHRM2 with IQ: converging evidence for a gene influencing intelligence. Behav Genet. 2007b;37:265–272. doi: 10.1007/s10519-006-9131-2. [DOI] [PubMed] [Google Scholar]
- Donohoe G, Morris DW, Clarke S, McGhee KA, Schwaiger S, Nangle JM, Garavan H, Robertson IH, Gill M, Corvin A. Variance in neurocognitive performance is associated with dysbindin-1 in schizophrenia: a preliminary study. Neuropsychologia. 2007;45(2):454–458. doi: 10.1016/j.neuropsychologia.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Ferguson SC, Deary IJ, Perros P, Evans JC, Ellard S, Hattersley AT, Frier BM. Apolipoprotein-e influences aspects of intellectual ability in type 1 diabetes. Diabetes. 2003;52:145–148. doi: 10.2337/diabetes.52.1.145. [DOI] [PubMed] [Google Scholar]
- Fiocco AJ, Poirier J, Joober R, Nair NP, Lupien SJ. Acute and long-term associations between ApoE genetic polymorphism, cortisol levels, and declarative memory performance in older adults. Psychoneuroendocrinology. 2008;33:625–633. doi: 10.1016/j.psyneuen.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychol Med. 2007;37(2):163. doi: 10.1017/S0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimer N, Sabatti C. The human phenome project. Nat Genet. 2003;34:15–21. doi: 10.1038/ng0503-15. [DOI] [PubMed] [Google Scholar]
- Glatt CE, Freimer NB. Association analysis of candidate genes for neuropsychiatric disease: the perpetual campaign. Trends Genet. 2002;18:307–312. doi: 10.1016/S0168-9525(02)02670-7. [DOI] [PubMed] [Google Scholar]
- Gondo Y, Hirose N, Arai Y, Yamamura K, Shimizu K, Takayama M, Ebihara Y, Nakazawa S, Inagaki H, Masui Y, Kitagawa K. Contribution of an affect-associated gene to human longevity: prevalence of the long-allele genotype of the serotonin transporter-linked gene in Japanese centenarians. Mech Ageing Dev. 2005;126:1178–1184. doi: 10.1016/j.mad.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Gosso MF, de Geus EJ, Polderman TJ, Boomsma DI, Heutink P, Posthuma D. Common variants underlying cognitive ability: further evidence for association between the SNAP-25 gene and cognition using a family-based study in two independent Dutch cohorts. Genes Brain Behav. 2008;7:355–364. doi: 10.1111/j.1601-183X.2007.00359.x. [DOI] [PubMed] [Google Scholar]
- Gosso MF, de Geus EJ, van Belzen MJ, Polderman TJ, Heutink P, Boomsma DI, Posthuma D. The SNAP-25 gene is associated with cognitive ability: evidence from a family-based study in two independent Dutch cohorts. Mol Psychiatry. 2006a;11:878–886. doi: 10.1038/sj.mp.4001868. [DOI] [PubMed] [Google Scholar]
- Gosso MF, van Belzen M, de Geus EJC, Polderman JC, Heutink P, Boomsma DI, Posthuma D. Association between the CHRM2 gene and intelligence in a sample of 304 Dutch families. Genes Brain Behav. 2006b;5:577–584. doi: 10.1111/j.1601-183X.2006.00211.x. [DOI] [PubMed] [Google Scholar]
- Gothelf D, Soreni N, Nachman RP, Tyano S, Hiss Y, Reiner O, Weizman A. Evidence for the involvement of the hippocampus in the pathophysiology of schizophrenia. Eur Neuropsychopharmacol. 2000;10:389–395. doi: 10.1016/s0924-977x(00)00097-3. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Gottfredson L. Mainstream science on intelligence: an editorial with 52 signatories, history, and bibliography. Intelligence. 1997;24:13–23. [Google Scholar]
- Haan MN, Shemanski L, Jagust WJ, Manolio TA, Kuller L. The role of APOE epsilon4 in modulating effects of other risk factors for cognitive decline in elderly persons. JAMA. 1999;282:40–46. doi: 10.1001/jama.282.1.40. [DOI] [PubMed] [Google Scholar]
- Han DH, Yoon SJ, Sung YH, Lee YS, Kee BS, Lyoo IK, Renshaw PF, Cho SC. A preliminary study: novelty seeking, frontal executive function, and dopamine receptor (D2) TaqI A gene polymorphism in patients with methamphetamine dependence. Compr Psychiatry. 2008;49:387–392. doi: 10.1016/j.comppsych.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Handel S. Listening: an introduction to the perception of auditory events. Cambridge, MA: MIT Press; 1989. [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SE, Fox H, Wright AF, Hayward C, Starr JM, Whalley LJ, Deary IJ. The brain-derived neurotrophic factor Val66Met polymorphism is associated with age-related change in reasoning skills. Mol Psychiatry. 2006;11:505–513. doi: 10.1038/sj.mp.4001799. [DOI] [PubMed] [Google Scholar]
- Harris SE, Wright AF, Hayward C, Starr JM, Whalley LJ, Deary IJ. The functional COMT polymorphism, Val 158 Met, is associated with logical memory and the personality trait intellect/imagination in a cohort of healthy 79 year olds. Neurosci Lett. 2005;385:1–6. doi: 10.1016/j.neulet.2005.04.104. [DOI] [PubMed] [Google Scholar]
- Harwood DG, Barker WW, Ownby RL, Mullan M, Duara R. Apolipoprotein E genotype and cognitive impairment in community-dwelling black older adults. Int J Psychiatry Med. 2002;32:55–67. doi: 10.2190/1W2E-190C-RG27-VHUJ. [DOI] [PubMed] [Google Scholar]
- Helkala EL, Koivisto K, Hanninen T, Vanhanen M, Kervinen K, Kuusisto J, Mykkanen L, Kesaniemi YA, Laakso M, Riekkinen P., Sr The association of apolipoprotein E polymorphism with memory: a population based study. Neurosci Lett. 1995;191:141–144. doi: 10.1016/0304-3940(95)11575-h. [DOI] [PubMed] [Google Scholar]
- Helkala EL, Koivisto K, Hanninen T, Vanhanen M, Kervinen K, Kuusisto J, Mykkanen L, Kesaniemi YA, Laakso M, Riekkinen P., Sr Memory functions in human subjects with different apolipoprotein E phenotypes during a 3-year population-based follow-up study. Neurosci Lett. 1996;204:177–180. doi: 10.1016/0304-3940(96)12348-x. [DOI] [PubMed] [Google Scholar]
- Hunt RR, Ellis HC. Fundamentals of cognitive psychology. Boston: McGraw-Hill Higher Education; 2004. [Google Scholar]
- Johansson B, Whitfield K, Pedersen NL, Hofer SM, Ahern F, McClearn GE. Origins of individual differences in episodic memory in the oldest-old: a population-based study of identical and same-sex fraternal twins aged 80 and older. J Gerontol B Psychol Sci Soc Sci. 1999;54:P173–P179. doi: 10.1093/geronb/54b.3.p173. [DOI] [PubMed] [Google Scholar]
- Jorm AF, Mather KA, Butterworth P, Anstey KJ, Christensen H, Easteal S. APOE genotype and cognitive functioning in a large age-stratified population sample. Neuropsychology. 2007;21:1–8. doi: 10.1037/0894-4105.21.1.1. [DOI] [PubMed] [Google Scholar]
- Knowlton BJ, Mangels JA, Squire LR. A neostriatal habit learning system in humans. Science. 1996;273:1399–1402. doi: 10.1126/science.273.5280.1399. [DOI] [PubMed] [Google Scholar]
- Kovas Y, Plomin R. Generalist genes: implications for the cognitive sciences. Trends Cogn Sci. 2006;10:198–203. doi: 10.1016/j.tics.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Levelt WJM. Speaking from intention to articulation. Cambridge, MA: MIT Press; 1993. p. xiv.p. 566. [Google Scholar]
- Lovins JB. Development of stemming algorithm. Mechanical translation and computational. Linguistics. 1968;11:22–31. [Google Scholar]
- Lowe CE, Cooper JD, Brusko T, Walker NM, Smyth DJ, Bailey R, Bourget K, Plagnol V, Field S, Atkinson M. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet. 2007;39:1074–1082. doi: 10.1038/ng2102. [DOI] [PubMed] [Google Scholar]
- Lu B, Martinowich K. Cell biology of BDNF and its relevance to schizophrenia. Novartis Found Symp. 2008;289:119–129. doi: 10.1002/9780470751251.ch10. discussion: 129–135, 193–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Lind PA, Deary IJ, Payton A, Posthuma D, Butcher LM, Bochdanovits Z, Whalley LJ, Visscher PM, Harris SE, Polderman TJ, Davis OS, Wright MJ, Starr JM, de Geus EJ, Bates TC, Montgomery GW, Boomsma DI, Martin NG, Plomin R. Testing replication of a 5-SNP set for general cognitive ability in six population samples. Eur J Hum Genet. 2008;16:1388–1395. doi: 10.1038/ejhg.2008.100. [DOI] [PubMed] [Google Scholar]
- Luciano M, Miyajima F, Lind PA, Bates TC, Horan M, Harris SE, Wright MJ, Ollier WE, Hayward C, Pendleton N, et al. Variation in the dysbindin gene and normal cognitive function in three independent population samples. Genes Brain Behav. 2009;8(2):218–227. doi: 10.1111/j.1601-183X.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- Luciano M, Posthuma D, Wright MJ, de Geus EJ, Smith GA, Geffen GM, Boomsma DI, Martin NG. Perceptual speed does not cause intelligence, and intelligence does not cause perceptual speed. Biol Psychol. 2005;70:1–8. doi: 10.1016/j.biopsycho.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Zhang H, Wang S, Gelernter J. CHRM2 variation predisposes to personality traits of agreeableness and conscientiousness. Hum Mol Genet. 2007;16:1557–1568. doi: 10.1093/hmg/ddm104. [DOI] [PubMed] [Google Scholar]
- Luo X, Petrill SA, Thompson LA. An exploration of genetic g: hierarchical factor analysis of cognitive data from the Western reserve twin project. Intelligence. 1994;18:335–347. [Google Scholar]
- Maher BS, Riley BP, Kendler KS. Psychiatric genetics gets a boost. Nat Genet. 2008;40:1042–1044. doi: 10.1038/ng0908-1042. [DOI] [PubMed] [Google Scholar]
- Malhotra AK, Kestler LJ, Mazzanti C, Bates JA, Goldberg T, Goldman D. A functional polymorphism in the COMT gene and performance on a test of prefrontal cognition. Am J Psychiatry. 2002;159:652–654. doi: 10.1176/appi.ajp.159.4.652. [DOI] [PubMed] [Google Scholar]
- Mannie ZN, Barnes J, Bristow GC, Harmer CJ, Cowen PJ. Memory impairment in young women at increased risk of depression: influence of cortisol and 5-HTT genotype. Psychol Med. 2008:1–6. doi: 10.1017/S0033291708004248. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Lu B. Interaction between BDNF and serotonin: role in mood disorders. Neuropsychopharmacology. 2008;33:73–83. doi: 10.1038/sj.npp.1301571. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat Neurosci. 2007;10:1089–1093. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Johansson B, Berg S, Pedersen NL, Ahern F, Petrill SA, Plomin R. Substantial genetic influence on cognitive abilities in twins 80 or more years old. Science. 1997;276:1560–1563. doi: 10.1126/science.276.5318.1560. [DOI] [PubMed] [Google Scholar]
- McGuffin P, Riley B, Plomin R. Genomics and behavior: toward behavioral genomics. Science. 2001;291:1232–1249. doi: 10.1126/science.1057264. [DOI] [PubMed] [Google Scholar]
- Meaburn E, Butcher LM, Schalkwyk LC, Plomin R. Genotyping pooled DNA using 100K SNP microarrays: a step towards genomewide association scans. Nucleic Acids Res. 2006;34:e27. doi: 10.1093/nar/gnj027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medin DL, Ross BH, Markman AB. Cognitive psychology. Hoboken, NJ: John Wiley & Sons; 2005. [Google Scholar]
- Mondadori CR, De Quervain DJ, Buchmann A, Mustovic H, Wollmer MA, Schmidt CF, Boesiger P, Hock C, Nitsch RM, Papassotiropoulos A, Henke K. Better memory and neural efficiency in young apolipoprotein E epsilon4 carriers. Cereb Cortex. 2007;17:1934–1947. doi: 10.1093/cercor/bhl103. [DOI] [PubMed] [Google Scholar]
- Nacmias B, Bessi V, Bagnoli S, Tedde A, Cellini E, Piccini C, Sorbi S, Bracco L. KIBRA gene variants are associated with episodic memory performance in subjective memory complaints. Neurosci Lett. 2008;436:145–147. doi: 10.1016/j.neulet.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KN, Wagoner AP, Gumbs CE, Giegling I, Moller HJ, Francks C, Muglia P, Roses A, Gibson G, Weale ME, Rujescu D, Goldstein DB. Failure to replicate effect of Kibra on human memory in two large cohorts of European origin. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:667–668. doi: 10.1002/ajmg.b.30658. [DOI] [PubMed] [Google Scholar]
- Neisser U, Boodoo G, Bouchard TJ, Boykin AW, Brody N, Ceci S, Halpern DF, Loehlin JC, Perloff R, Sternberg RJ, Urbina S. Intelligence: knowns and unknowns. Am Psychol. 1996;51:77–101. [Google Scholar]
- Nepom GT. Class II antigens and disease susceptibility. Annu Rev Med. 1995;46:17–25. doi: 10.1146/annurev.med.46.1.17. [DOI] [PubMed] [Google Scholar]
- Nilsson LG, Adolfsson R, Backman L, Cruts M, Nyberg L, Small BJ, Van Broeckoven C. The influence of APOE status on episodic and semantic memory: data from a population-based study. Neuropsychology. 2006;20:645–657. doi: 10.1037/0894-4105.20.6.645. [DOI] [PubMed] [Google Scholar]
- Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D, Wollmer MA, Aerni A, Coluccia D, Hanggi J, Mondadori CR, Buchmann A, Reiman EM, Caselli RJ, Henke K, de Quervain DJ. Science. Vol. 314. 2006. Common Kibra alleles are associated with human memory performance; pp. 475–478. [DOI] [PubMed] [Google Scholar]
- Payton A, Holland F, Diggle P, Rabbitt P, Horan M, Davidson Y, Gibbons L, Worthington J, Ollier WE, Pendleton N. Cathepsin D exon 2 polymorphism associated with general intelligence in a healthy older population. Mol Psychiatry. 2003;8:14–18. doi: 10.1038/sj.mp.4001239. [DOI] [PubMed] [Google Scholar]
- Pelletier JG, Lacaille JC. Long-term synaptic plasticity in hippocampal feedback inhibitory networks. Prog Brain Res. 2008;169:241–250. doi: 10.1016/S0079-6123(07)00014-3. [DOI] [PubMed] [Google Scholar]
- Pendleton N, Payton A, van den Boogerd EH, Holland F, Diggle P, Rabbitt PM, Horan MA, Worthington J, Ollier WE. Apolipoprotein E genotype does not predict decline in intelligence in healthy older adults. Neurosci Lett. 2002;324:74–76. doi: 10.1016/s0304-3940(02)00135-0. [DOI] [PubMed] [Google Scholar]
- Petrill SA, Plomin R, McClearn GE, Smith DL, Vignetti S, Chorney MJ, Chorney K, Thompson LA, Detterman DK, Benbow C, Lubinski D, Daniels J, Owen M, McGuffin P. No association between general cognitive ability and the A1 allele of the D2 dopamine receptor gene. Behav Genet. 1997;27:29–31. doi: 10.1023/a:1025659124405. [DOI] [PubMed] [Google Scholar]
- Plomin R, Kovas Y. Generalist genes and learning disabilities. Psychol Bull. 2005;131:592–617. doi: 10.1037/0033-2909.131.4.592. [DOI] [PubMed] [Google Scholar]
- Plomin R, Petrill SA. Genetics and intelligence: what’s new? Intelligence. 1997;24:53–77. [Google Scholar]
- Plomin R, Spinath FM. Intelligence: genetics, genes, and genomics. J Pers Soc Psychol. 2004;86:112–129. doi: 10.1037/0022-3514.86.1.112. [DOI] [PubMed] [Google Scholar]
- Poldrack RA. Can cognitive processes be inferred from neuro-imaging data? Trends Cogn Sci. 2006;10:59–63. doi: 10.1016/j.tics.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Posthuma D, Luciano M, Geus EJC, Wright MJ, Slagboom PE, Montgomery GW, Boomsma DI, Martin NG. A genomewide scan for intelligence identifies quantitative trait loci on 2q and 6p. Am J Hum Genet. 2005;77(2):318–326. doi: 10.1086/432647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu HQ, Montpetit A, Ge B, Hudson TJ, Polychronakos C. Toward further mapping of the association between the IL2RA locus and type 1 diabetes. Diabetes. 2007;56:1174–1176. doi: 10.2337/db06-1555. [DOI] [PubMed] [Google Scholar]
- Reisberg D. Cognition: exploring the science of the mind. New York: W. W. Norton; 2005. [Google Scholar]
- Reuter M, Peters K, Schroeter K, Koebke W, Lenardon D, Bloch B, Hennig J. The influence of the dopaminergic system on cognitive functioning: a molecular genetic approach. Behav Brain Res. 2005;164:93–99. doi: 10.1016/j.bbr.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Reynolds CA, Gatz M, Berg S, Pedersen NL. Genotype-environment interactions: cognitive aging and social factors. Twin Res Hum Genet. 2007;10:241–254. doi: 10.1375/twin.10.2.241. [DOI] [PubMed] [Google Scholar]
- Rijsdijk FV, Vernon PA, Boomsma DI. Application of hierarchical genetic models to Raven and WAIS subtests: a Dutch twin study. Behav Genet. 2002;32:199–210. doi: 10.1023/a:1016021128949. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Jimenez R, Hoenicka J, Jimenez-Arriero MA, Ponce G, Bagney A, Aragues M, Palomo T. Performance in the Wisconsin Card Sorting Test and the C957T polymorphism of the DRD2 gene in healthy volunteers. Neuropsychobiology. 2006;54:166–170. doi: 10.1159/000098652. [DOI] [PubMed] [Google Scholar]
- Rosenthal R. Meta-analytic procedures for social research. Sage Publications; 1984. [Google Scholar]
- Sabb FW, Bearden CE, Glahn DC, Parker DS, Freimer N, Bilder RM. A collaborative knowledge base for cognitive phenomics. Mol Psychiatry. 2008;13:350–360. doi: 10.1038/sj.mp.4002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitz J, Solms M, Ramesar R. The molecular genetics of cognition: dopamine, COMT and BDNF. Genes Brain Behav. 2006;5:311–328. doi: 10.1111/j.1601-183X.2005.00163.x. [DOI] [PubMed] [Google Scholar]
- Schaper K, Kolsch H, Popp J, Wagner M, Jessen F. KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol Aging. 2008;29:1123–1125. doi: 10.1016/j.neurobiolaging.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Schultz MR, Lyons MJ, Franz CE, Grant MD, Boake C, Jacobson KC, Xian H, Schellenberg GD, Eisen SA, Kremen WS. Apolipoprotein E genotype and memory in the sixth decade of life. Neurology. 2008;70:1771–1777. doi: 10.1212/01.wnl.0000286941.74372.cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P. Intelligence and the developing human brain. Bioessays. 2007;29:962–973. doi: 10.1002/bies.20641. [DOI] [PubMed] [Google Scholar]
- Spearman C. General intelligence, objectively determined and measured. Am J Psychol. 1904;15:201–293. [Google Scholar]
- Squire LR. Declarative and nondeclarative memory: multiple brain systems supporting learning and memory. J Cogn Neurosci. 1992;4:232–243. doi: 10.1162/jocn.1992.4.3.232. [DOI] [PubMed] [Google Scholar]
- Squire LR, Knowlton B, Musen G. The structure and organization of memory. Annu Rev Psychol. 1993;44:453–495. doi: 10.1146/annurev.ps.44.020193.002321. [DOI] [PubMed] [Google Scholar]
- Starr JM, Fox H, Harris SE, Deary IJ, Whalley LJ. COMT genotype and cognitive ability: a longitudinal aging study. Neurosci Lett. 2007;421:57–61. doi: 10.1016/j.neulet.2007.05.023. [DOI] [PubMed] [Google Scholar]
- Sternberg RJ, Grigorenko EL, Kidd KK. Intelligence, race, and genetics. Am Psychol. 2005;60:46–59. doi: 10.1037/0003-066X.60.1.46. [DOI] [PubMed] [Google Scholar]
- Sternberg RJ, Mio JS. Cognitive psychology. Belmont, CA: Thomson/Wadsworth; 2006. [Google Scholar]
- Strandberg TE, Pitkala K, Eerola J, Tilvis R, Tienari PJ. Interaction of herpesviridae, APOE gene, and education in cognitive impairment. Neurobiol Aging. 2005;26:1001–1004. doi: 10.1016/j.neurobiolaging.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Strange BA, Kroes MC, Roiser JP, Tan GC, Dolan RJ. Emotion-induced retrograde amnesia is determined by a 5-HTT genetic polymorphism. J Neurosci. 2008;28:7036–7039. doi: 10.1523/JNEUROSCI.0834-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J, Barr CL, George CJ, Ryan CM, King N, Shaikh S, Kovacs M, Kennedy JL. BDNF and COMT polymorphisms: relation to memory phenotypes in young adults with childhood-onset mood disorder. Neuromol Med. 2004;5:181–192. doi: 10.1385/NMM:5:3:181. [DOI] [PubMed] [Google Scholar]
- Tagarakis GI, Tsolaki-Tagaraki F, Tsolaki M, Diegeler A, Kazis D, Rouska E, Papassotiropoulos A. The role of SOAT-1 polymorphisms in cognitive decline and delirium after bypass heart surgery. Clin Res Cardiol. 2007a;96:600–603. doi: 10.1007/s00392-007-0539-3. [DOI] [PubMed] [Google Scholar]
- Tagarakis GI, Tsolaki-Tagaraki F, Tsolaki M, Diegeler A, Tsilimingas NB, Papassotiropoulos A. The role of apolipoprotein E in cognitive decline and delirium after bypass heart operations. Am J Alzheimers Dis Other Demen. 2007b;22:223–228. doi: 10.1177/1533317507299415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P, Cannon TD, Toga AW. Mapping genetic influences on human brain structure. Ann Med. 2002;34:523–536. doi: 10.1080/078538902321117733. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Hong CJ, Yu YW, Chen TJ. Association study of a brain-derived neurotrophic factor (BDNF) Val66Met polymorphism and personality trait and intelligence in healthy young females. Neuropsychobiology. 2004;49:13–16. doi: 10.1159/000075333. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Yu YW, Lin CH, Chen TJ, Chen SP, Hong CJ. Dopamine D2 receptor and N-methyl-D-aspartate receptor 2B subunit genetic variants and intelligence. Neuropsychobiology. 2002;45:128–130. doi: 10.1159/000054951. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Yu YWY, Chen TJ, Chen JY, Liou YJ, Chen MC, Hong CJ. Association study of a functional catechol-O-methyltransferase-gene polymorphism and cognitive function in healthy females. Neurosci Lett. 2003;338:123–126. doi: 10.1016/s0304-3940(02)01396-4. [DOI] [PubMed] [Google Scholar]
- Turic D, Fisher PJ, Plomin R, Owen MJ. No association between apolipoprotein E polymorphisms and general cognitive ability in children. Neurosci Lett. 2001;299:97–100. doi: 10.1016/s0304-3940(00)01789-4. [DOI] [PubMed] [Google Scholar]
- Ueda H, Howson JMM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KMD, Smith AN, Di Genova G. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- van der Maas HLJ, Dolan CV, Grasman R, Wicherts JM, Huizenga HM, Raijmakers MEJ. A dynamical model of general intelligence: the positive manifold of intelligence by mutualism. Psychol Rev. 2006;113:842. doi: 10.1037/0033-295X.113.4.842. [DOI] [PubMed] [Google Scholar]
- van Munster BC, Korevaar JC, de Rooij SE, Levi M, Zwinderman AH. The association between delirium and the apolipoprotein E epsilon4 allele in the elderly. Psychiatr Genet. 2007;17:261–266. doi: 10.1097/YPG.0b013e3280c8efd4. [DOI] [PubMed] [Google Scholar]
- Vella A, Cooper JD, Lowe CE, Walker N, Nutland S, Widmer B, Jones R, Ring SM, McArdle W, Pembrey ME. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:773–779. doi: 10.1086/429843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade N. A dissenting voice as the genome is sifted to fight disease. New York Times; 2008. September 15, [Google Scholar]
- Wainwright M, Jordan M. New directions in statistical signal processing: from systems to brain. Cambridge, MA: MIT Press; 2005. A variational principle for graphical models. [Google Scholar]
- Yip AG, Brayne C, Easton D, Rubinsztein DC. Apolipoprotein E4 is only a weak predictor of dementia and cognitive decline in the general population. J Med Genet. 2002;39:639–643. doi: 10.1136/jmg.39.9.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkstok JR, de Wilde O, van Amelsvoort TA, Tanck MW, Baas F, Linszen DH. Association between the DTNBP1 gene and intelligence: a case control study in young patients with schizophrenia and related disorders and unaffected siblings. Behav Brain Funct. 2007;3:19. doi: 10.1186/1744-9081-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]