Abstract
The mammalian Bombesin (Bn) peptides neuromedin B (NMB) and gastrin-releasing peptide (GRP) actions are mediated by two receptors (NMB-receptor, GRP-receptor) which are widely distributed in the GI tract and CNS. From primarily animal studies NMB/GRP-receptor activation has physiological/pathophysiological effects in the CNS and GI tract including stimulating of growth of cancers and normal tissues. Whereas these Bn receptor's effects have been extensively studied in nonhuman cells and animals, little is known of the physiological/pathological role(s) in humans, largely due of lack of potent antagonists. To address this issue we compared NMB-receptor/GRP-receptor affinity/potency of 10 chemical classes of putative antagonists (35 compounds) for human Bn-receptors by performing binding studies or assessing abilities to activate hGRP/hNMB-receptor[assessing phospholipase C activation] in 4 different cells containing native Bn receptors or transfected receptors. From binding studies 23 were GRP-receptor-preferring, 4 were NMB-receptor, and 8-nonselective. For the hGRP-receptor-preferring analogues none showed hGRP-receptor agonist activity, but 13 were full or-partial hNMB-receptor agonists at hNMB-receptors. For hNMB-receptor-preferring analogues none were agonists. Analogue #24([(3-Ph-Pr6),His7,D-Ala11,D-Pro13,Ψ(13-14), Phe14]Bn(6-14)NH2) and analogue #7[D-Phe6,Leu13,Ψ(CH2NH),Cpa14]Bn(6-14) were the most potent (0.2-1.4 nM) and selective ((>10, 000-fold) for the hGRP-receptor with analogue #7.5[D-Tpi6,Leu13, Ψ(CH2NH),Leu14]Bn(6-14) [RC-3095] (0.2-1.4 nM) slightly less selective. Analogue #34(PD168368) had the highest affinity for hNMB-receptor (1.32-1.58 nM) and the greatest selectivity (2298-6952-fold) for the hNMB-receptor. These results demonstrate numerous putative hGRP/hNMB-receptor antagonists identified in nonhuman cells and/or animals have agonist activity at the hNMB-receptor, limiting their potential usefulness. However, a number were identified which were potent/selective for human Bn receptors and should be useful for investigating their roles in human physiological/pathophysiological conditions.
1. Introduction
The mammalian Bombesin (Bn)-related peptides, neuromedin B (NMB) and gastrin-releasing peptide (GRP) share close structural similarity with identities of 7 of the 10 amino acids in the COOH-terminus [24] and their actions are mediated by two receptors, the GRP-receptor and NMB-receptor, which are also closely related, having 55% amino acid identities [24,29,32].
GRP/NMB-receptor are widely distributed in mammals in both the central nervous system (CNS) and peripheral tissues, including the gastrointestinal (GI) tract [24,29,30,32,44]. Animal studies suggest GRP/NMB-receptors may be involved in a broad spectrum of biological responses including in the CNS [circadian rhythm, TSH secretion, behavior control, thermo-regulation, satiety], in the immune system [effects on macrophages, lymphocytes, leukocytes, dendritic cells], endocrine [release of numerous hormones/neurotransmitters], GI tract [motility, secretion, growth], as well as the urogenital tract and respiratory system[17,19,24,29,32,44,65]. Not only GRP, but also to a lesser extent NMB, have important pathophysiological effects including a prominent effect on the growth and/or differentiation of a number of important human tumors [colon, prostrate, lung, head/neck squamous cell, CNS, pancreatic and some gynecologic cancers], and in some cases, function as autocrine growth factors [11,17,24,28,41,48,65]. Also, GRP/NMB-receptor's are one of the G protein-coupled receptor (GPCR) family most frequently ectopically expressed or overexpressed by a different tumors including prostate cancer, small cell lung cancer, breast cancer, CNS tumors (glioblastomas) and carcinoids (intestinal, thymic, bronchial) [17,22,24,28,41,48,50,51,65].
In general the role of GRP/NMB-receptor in human physiology or pathology is largely unknown [17,24,65], in large part due to a lack of knowledge about potentially useful receptor antagonists in humans. This has occurred for a number of reasons. First, marked species differences in the affinity and selectivity of a wide range of Bn-putative agonist/antagonist analogues for GRP/NMB-receptor have been described depending on the tissue/species studied [3,9,24,35,52,63,64]. Therefore, studies of receptor selectivity using non-human receptors may not be applicable to human studies. Second, studies showed that various putative Bn receptor antagonists can have marked differences in with the same analogue able to function as an antagonist or an agonist, ranging from functioning as a pure antagonist, partial agonist, or full agonist depending on the assay system and species studied [10,12,21,24,25,34,53,64]. Therefore, identification of a potential antagonist from animal studies may or may not be applicable in humans. Third, the search for high affinity receptor antagonists for the GRP-receptor has been very successful and a large number of different chemical classes of antagonists have been described primarily from nonhuman studies, including various bombesin analogues [D-Phe12 Bn analogues, des Met14 analogues, Bn pesudopeptides, Bn amides, alkylamides, esters, D-Pro13-psuedopeptides], substituted substance P and somatostatatin analogues, and peptoids][24,25]. In contrast, for the NMB-receptor only a few putative high affinity receptor antagonists have been described including peptoids and substituted somatostatin analogues [13,42,52]. The peptoids in particular have not been investigated in detail in human tissues. Peptoid antagonists, which were identified based on a strategy which resulted in not only Bn receptor antagonists, but also CCK and tachykinin receptor antagonists [20], involved identifying the molecular recognition interactions between the endogenous ligand and receptor of interest, and then making a small topographic analogue [1,13,20]. In general, with all of these putative Bn receptor antagonists, there are few studies with human Bn receptors, especially using cellular systems containing receptor expression densities that usually occur with native Bn receptors. The later point is crucial in assessing agonist activity of a possible putative Bn receptor antagonist, because Bn receptor density can have important effects on receptor interaction/activation, particularly if cells with high densities seen frequently with transfected cell systems, are studied [59], and therefore may not be applicable to native cells Bn receptor densities, seen in vivo.
The present study was performed to attempt to identify receptor antagonists for human bombesin receptors (i.e. hNMB-receptor, hGRP-receptor) that might be useful in human studies to define the role of these receptors in both physiological and pathological conditions. To address this issue, in the present study we evaluate the pharmacology of 35 putative antagonist compounds for all three human mammalian Bn-receptor subtypes GRP-receptor, NMB-receptor and BRS-3 using human cells expressing natively hGRP-receptor, hGRP-receptor transfected Balb 3T3-cells, hNMB-receptor transfected NCI-H1299, hNMBR-receptor transfected Balb 3T3, and hBRS-3 transfected Balb 3T3 cells. Our results identify a number of the potent and selective antagonists for hGRP-receptor and hNMB-receptor, which could be useful for investigating the role of GRP/NMB-receptor in human physiological/pathophysiological conditions.
2. Materials and methods
2.1. Materials
The following cells and materials were obtained from the sources indicated: Balb 3T3 (mouse fibroblast) cells and HuTu-80 (human duodenal cancer cell line) from the American Type Culture Collection, (Rockville, MD); NCI-H1299 (non-small cell lung cancer cells) were gift from Herb Oie (National Cancer Institute-Navy Medical Oncology Branch, Naval Medical Center, Bethesda, MD); the mammalian expression vectors, pCD2 and pcDNA3, Lipofectamine PLUS Reagent, OPTI-MEM I Reduced-Serum Medium and GENETICIN selective antibiotic (G418 Sulfate) from Invitrogen (Carlsbad, CA); Dulbecco's minimum essential medium (DMEM), RPMI 1640, phosphate-buffered saline (PBS), fetal bovine serum (FBS) and trypsin/versene solution from Biosource International (Camarillo, CA); PD176252 and PD168368 from Tocris Bioscience (Ellisville, MO); Na125I (2,200 Ci/mmol) from Amersham Biosciences (Piscataway, NJ); 1,3,4,6-tetrachloro-3α, 6α-diphenylglycouril (IODO-GEN) and dithiothreitol (DTT) from Pierce Biotechnology Inc (Rockford, IL); myo-[2-3H]Inositol (20 Ci/mmol) from Amersham Pharmacia Biotech (Piscataway, NJ); formic acid, ammonium formate, disodium tetraborate, soybean trypsin inhibitor, bacitracin and AG 1-X8 resin from Bio-Rad, (Richmond, CA); bovine serum albumin fraction V (BSA) and N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) from ICN Pharmaceutical, Inc. (Aurora, OH). All other chemicals were of the highest purity commercially available.
2.2. Methods
2.2.1. Preparation of peptides
The peptides were synthesized using standard solid-phase methods as described previously [39]. In brief, solid-phase syntheses of peptide amides were carried out using Boc chemistry on methylbenzhydrylamine resin (Advanced ChemTech, Louisville, KY) followed by hydrogen fluoride-cleavage of free peptide amides. The crude peptides were purified by preparative high-performance liquid chromatography on columns (2.5 × 50 cm) of Vydac C18 silica (10 μm), which was eluted, with linear gradients of acetonitrile in 0.1% (v/v) trifluoroacetic acid. Homogeneity of the peptides was assessed by analytical reverse-phase high-performance liquid chromatography, and the purity was usually 97% or higher. Amino acid analysis (only amino acids with primary amino acid groups were quantitated) gave the expected amino acid ratios. Peptide molecular masses were obtained by matrix-assisted laser desorption mass spectrometry (Thermo Bioanalysis Corp., Hemel, Helmstead, UK), and all corresponded well with calculated values.
2.2.2. Growth and maintenance of cells
HuTu-80 cells, which contain native hGRP-receptor [66], were grown in DMEM. Balb 3T3 cells stably expressing human BRS-3 receptors, human NMB-receptor, or human GRP-receptor and NCI-H1299 cell stably expressing human NMB-receptor were made as described previously [3,40,55] and grown in DMEM or RPMI 1640, respectively. NCI-N417 small cell lung carcinoma cells which contain native human BRS-3 receptors [55] were grown in RPMI-1640. All the cells were grown in their respective propagation media supplemented with supplemented 10% FBS and 300 mg/liter of G418 sulfate and incubated at 37°C in a 5% CO2 atmosphere.
2.2.3. Cell transfections and isolation of Stable Cell lines
Stable transfection: Fifteen μg of plasmid DNA [human epitope-tagged NMB-receptor cDNA in the mammalian expression vectors pcDNA3] was used for transfection of NCI-H1299 cells with 25 μl Lipofectamine. BALB 3T3 cells were transfected using the Ca3(PO4)2 precipitation method [3]. Three days after transfection cells were split in a ratio of 1:3 and the selection antibiotic G418 was added to the regular growth medium at a concentration of 800 μg/ml. Single colonies were isolated two weeks later and expanded in growth medium containing G418 (300 μg/ml).
Receptor density on the single colonies and the HuTu-80 cell line were determined using 0.75 nM 125I-[d-Tyr6, β-Ala11, Phe13, Nle14]Bn-(6-14). Total saturable binding was determined using 1 μM Bn and using the ligand specific activity the cell number or the protein present, the receptor densities were calculated. The results of the binding experiments were expressed as fmoles of 125I-[d-Tyr6, β-Ala11, Phe13, Nle14]Bn-(6-14) bound per 106 of cells or as fmoles bound/mg protein. hGRP-receptor containing HuTu-80 cells had a receptor density of 1.68 ± 0.08 fmoles/106 cells (8.13 ± 0.39 fmoles/mg protein) and the hGRP-receptor transfected Balb 3T3 cells had a receptor density of 15.16 ± 0.21 fmoles/106 cells (43.17 ± 1.0 fmoles/mg protein). The hNMB-receptor transfected Balb 3T3 cells had a receptor density of 77 ± 6-fmoles/106 cells (162.0 ± 13.4 fmoles/mg protein) and NCI-H1299 transfected with hNMB-receptor had a receptor density of 2.11 ± 0.20 fmoles/106 cells (4.65 ± 0.40 fmoles/mg protein).
2.2.4. Preparation of 125I-[Tyr4]Bn, 125I-[d-Tyr0]NMB and 125I-[d-Tyr6, β-Ala11, Phe13, Nle14]Bn-(6-14)
Preparation of 125I-[Tyr4]Bn, 125I-[d-Tyr0]NMB and 125I-[d-Tyr6, β-Ala11, Phe13, Nle14]Bn-(6-14) at a specific activity of 2,200 Ci/ mmol was prepared by a modification of methods described previously [37-40]. Briefly, 0.8 μg of IODO-GEN (in 0.01 μg/ml chloroform) was transferred to a vial, dried under a stream of nitrogen, and washed with 100 μl of KH2PO4 (pH 7.4). To the reaction vial, 20 μl of 0.5 M KH2PO4 (pH 7.4), 8 μg of peptide in 4 μL of water, and 2 mCi (20 μl) Na 125I were added, mixed gently, and incubated at room temperature for 6 min. The incubation was stopped by the addition of 100 μl of distilled water. Radiolabeled peptide was separated using a Sep-Pak (Waters Associates) and high-performance liquid chromatography as described previously [40,58]. Radioligand was stored with 0.5% BSA at -20°C.
2.2.6. Whole cell radioligand binding assays
Cells were incubated for 1 h at 21°C in 250 μl of binding buffer containing 24.5 mM HEPES (pH 7.4), 98 mM NaCl, 6 mM KCl, 2.5 mM KH2PO4, 5 mM sodium pyruvate, 5 mM sodium fumarate, 5 mM sodium glutamate, 2 mM glutamine, 11.5 mM glucose, 0.5 mM CaCl2, 1.0 mM MgCl2, 0.01% (w/v) soybean trypsin inhibitor, 0.2 % (v/v) amino acid mixture, 0.2% (w/v) BSA, and 0.05% (w/v) bacitracin with 50 pM 125I-[Tyr4]Bn, 125I-[d-Tyr0]NMB or 125I-[d-Tyr6, β-Ala11, Phe13, Nle14]Bn-(6-14) (2,200 Ci/mmol), respectively, in the presence of the indicated concentration of unlabeled peptides. Although receptor expression over the ranges for cells used in this study [59] has been shown not to alter receptor affinity or potency under the experimental conditions used in this study, as an added precaution to correct for any differences in ligand bound by different cell line, binding results with each receptor were compared only to results with receptor containing cells binding similar amounts of ligand. This was accomplished as described previously [16] by varying the cell concentration between 0.05 − 2 × 106 cells/ml for each receptor so that <20% of the total added radioactive ligand was bound during the incubation and the results compared to cells transfected with native receptor adjusted in concentration to bind a similar amount of ligand.
After the incubation, 100 μl aliquot were added to 400 μl microfuge tubes (PGC Scientific, Frederick, MD), which contained 100 μl of binding buffer to determine the total radioactivity. The bound tracer was separated from unbound tracer by pelleting the cells through the binding buffer by centrifugation at 10,000 × g in a Microfuge E (Beckman, Fullerton, CA) for 3 min. The supernatant was aspirated and the pelleted cells were rinsed twice with a washing buffer which contained 1% (w/v) BSA in PBS. The amount of radioactivity bound to the cells was measured in a Cobra II Gamma counter (Packard Instruments, Meriden, CT). Binding was expressed as the percentage of total radioactivity that was associated with the cell pellet. All binding values represented saturable binding (i.e., total binding minus nonsaturable binding). Nonsaturable binding was defined as the amount of binding that occurred with 1 μM Bn, 1 μM NMB or 1 μM [D-Tyr6, β-Ala11, Phe13, Nle14]Bn-(6-14) in the incubation solution. Nonsaturable binding was <15% of the total binding in all experiments. Each point was measured in duplicate, and each experiment was replicated at least four times. Calculation of affinity was performed by determining the IC50 using the curve-fitting program KaleidaGraph (Synergy Software). The Mann Whitney U test was used to determine the statistical significance of differences.
2.2.7. Measurement of inositol phosphates
Changes in total [3H]inositol phosphates ([3H]IP) was measured as described previously [2-4,16,54]. Briefly, hGRP-receptor-, hNMB-receptor- or hBRS-3, transfected Balb 3T3, hNMB-receptor-transfected NCI-H1299 or HuTu-80 cells were subcultured into 24-well plates (5.0 × 104 cells/well) in regular propagation media and then incubated for 24 h at 37°C in a 5% CO2 atmosphere. The cells were then incubated with 3 μCi/ml of myo- [2-3H] inositol in growth media supplemented with 2% FBS for an additional 24 hr. Before assay, the 24-well plates were washed by incubating for 30 min at 37°C with 1 ml/well of PBS (pH 7.0) containing 20 mM lithium chloride. The wash buffer was aspirated and replaced with 500 μl of IP assay buffer containing 135 mM sodium chloride, 20 mM HEPES (pH 7.4), 2 mM calcium chloride, 1.2 mM magnesium sulfate, 1 mM EGTA, 20 mM lithium chloride, 11.1 mM glucose, 0.05% BSA (w/v) and incubated with or without any of the peptides studied. After 60 min of incubation at 37°C, the experiments were terminated by the addition of 1 ml of ice cold 1% (v/v) hydrochloric acid in methanol. Total [3H]IP was isolated by anion exchange chromatography as described previously [2-4,54]. Briefly, samples were loaded onto Dowex AG1-X8 anion exchange resin columns, washed with 5 ml of distilled water to remove free [3H]inositol, then washed with 2 ml of 5 mM disodium tetraborate/60 mM sodium formate solution to remove [3H]glycerophosphorylinositol. Two ml of 1 mM ammonium formate/100 mM formic acid solution were added to the columns to elute total [3H]IP. Each eluate was mixed with scintillation cocktail and measured for radioactivity in a scintillation counter.
2.2.8. Species Comparison
The generation of Table 5, which compares the affinities and agonist/antagonist activities for human GRP/NMBR/BRS-3 receptors for the most important compounds tested in this study, to results for rat, mouse, and guinea pig, was made by reviewing papers using Medline in the literature and using data from our studies at the NIH and were primarily from the following papers [1,8-10,18,24-26,31,33,38,40,42,47,52,55,61,63,64].
Table 5.
Comparison of affinities/activities of antagonists for GRP-receptor and NMB-receptor in different species.
| Ki (nM) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mGRPR | rGRPR | gpGRPR | hGRPR | rNMBR | hNMBR | ||||||||
| Peptide # | Group | Peptide structure | |||||||||||
| IC50 | IC50 | Ag/Antag | IC50 | Ag/Antag | IC50 | Ag/Antag | IC50 | Ag/Antag | IC50 | Ag/Antag | |||
|
|
|
|
|
|
|
|
|
|
|||||
| #2 | I | [Tyr4, d-Phe12]Bn | >5,000 | >10,000 | Antag | 4,000 ± 300 | Antag | 912 ± 76 | Antag | 3,330 ± 240 | Antag | 1,860 ± 60 | pAg |
| #3 | II | [d-Arg1, d-Trp7,9,Leu11]SP | 2,700 ± 490 | >10,000 | pAg | 5,000 ± 500 | Antag | 1,780 ± 50 | Antag | 1,400 ± 800 | Antag | 1,590 ± 30 | No act |
| #4 | II | [d-Pro4, d-Trp7,9,10]SP-(4-11) | 7,690 ± 880 | >10,000 | pAg | >10,000 | Antag | >10,000 | No act | 1,410 ± 90 | Antag | >10,000 | No act |
| #5 | III | [Leu13,ψ(CH2NH),Leu14]Bn | 65 ± 8 | 434 ± 65 | pAg | 60 ± 6 | Antag | 8.12 ± 0.56 | Antag | >10,000 | No act | >10,000 | No act |
| #6 | III | [d-Phe6,Leu13,ψ(CH2NH), d-Phe14]Bn-(6-14) | 12.5 ± 1.8 | 62 ± 10 | pAg | 10 ± 2 | Antag | 12.6 ± 0.5 | Antag | >10,000 | ND | >10,000 | pAg |
| #7 | III | [d-Phe6,Leu13,ψ(CH2NH),Cpa14]Bn(6-14) | 4.9 ± 0.8 | 41 ± 5 | Antag | 7.2 ± 1.0 | Antag | 1.23 ± 0.09 | Antag | 2,700 ± 20 | Antag | >10,000 | No act |
| #8 | IV | [d-Phe6]Bn(6-13)NH2 | 23 ± 1 | 27 ± 6 | Antag | 96 ± 21 | Antag | 4.93 ± 0.39 | Antag | >10,000 | ND | >10,000 | No act |
| #10 | V | [d-Phe6]Bn(6-13)Ethylamide | 2.9 ± 0.4 | 27 ± 6 | Antag | 16 ± 4 | Antag | 2.22 ± 0.13 | Antag | 2,200 ± 473 | ND | >10,000 | pAg |
| #11 | V | [d-Phe6-Bn](6-13)Propylamide (PA) | 1.7 ± 0.2 | 6.4 ± 0.8 | pAg | 4.4 ± 0.9 | Antag | 0.39 ± 0.12 | Antag | 3,000 ± 130 | ND | 3,090 ± 70 | pAg |
| #12 | V | [d-Tyr6]Bn-(6-13)Propylamide (PA) | 1.8 ± 0.1 | 14 ± 1 | pAg | 8.7 ± 0.9 | Antag | 1.41 ± 0.06 | Antag | 3,000 ± 200 | ND | >10,000 | pAg |
| #14 | V | [d-Phe6]Bn(6-13)Butylamide (BA) | 14 ± 1 | 14 ± 1 | Ag | 28 ± 4 | pAg | 1.02 ± 0.09 | Antag | 2,300 ± 560 | ND | 1,780 ± 30 | pAg |
| #15 | V | [d-Phe6]Bn-(6-13)Hexylamide | 6.1 ± 1.1 | 103 ± 8 | Ag | 16 ± 2 | pAg | 2.19 ± 0.04 | Antag | >10,000 | ND | >10,000 | pAg |
| #16 | VI | [d-Phe6]Bn(6-13)Heptylamide | 10.8 ± 0.8 | 92 ± 12 | Antag | 19 ± 3 | Antag | 5.12 ± 0.33 | ND | 2,530 ± 150 | ND | 6,100 ± 630 | ND |
| #17 | VI | [d-Phe6]Bn-(6-13)ME | 1.0 ± 0.2 | 10.3 ± 1.4 | Antag | 6.7 ± 0.8 | Antag | 3.98 ± 0.80 | Antag | 7,500 ± 400 | ND | >10,000 | Ag(10μM) |
| #18 | VI | [d-F-5,Phe6, d-Ala11]Bn(6-13)ME | 0.2 ± 0.1 | 5.5 ± 1.4 | Antag | ND | ND | 0.12 ± 0.01 | Antag | 1,760 ± 284 | ND | >10,000 | pAg |
| #20 | VI | [d-Phe6]Bn-(6-13)Ethyl ester | 1.6 ± 0.4 | 5 ± 1 | Antag | 2.6 ± 0.4 | Antag | 0.14 ± 0.03 | Antag | 2,500 ± 150 | No act | >10,000 | pAg |
| #24 | VII | [(3-Ph-Pr6),His7, d-Ala11, d-Pro13,ψ(13-14), Phe14]Bn(6-14)NH2) | 0.001 ± 0.0004 | 0.7 ± 0.1 | Antag | ND | ND | 0.23 ± 0.01 | Antag | >10,000 | ND | > 10, 000 | Antag |
| #30 | VIII | [d-Nal,Cys,Tyr,D-Trp, Lys,Val, Cys, Nal]NH2 | >10,000 | >10,000 | No act | ND | ND | 2,460 ± 120 | No act | 60 ± 9 | Antag | 275 ± 7 | Antag |
| #33 | X | PD 176252 | ND | 1.0 ± 0.2 | Antag | ND | ND | 170 ± 11 | Antag | 16 | Antag | 0.08 ± 0.01 | Antag |
| #34 | X | PD 168368 | 2,151 ± 402 | 1,300 ± 190 | ND | ND | ND | 1,738 ± 105 | Antag | 39 ± 9 | Antag | 0.25 ± 0.02 | Antag |
3. Results
In this study, we have compared the ability of a number of members of various chemical classes of peptides/peptoids that are reported to function as Bn receptor antagonists [9,24,25,52,56,63,64], to interact with each of the human Bn-related receptors (Tables 1, 2). To be certain that the results obtained were reflective of the response in natural tissues, two controls were incorporated in this study. First, the hGRP-receptor and hNMB-receptor were both evaluated in Balb 3T3 cells, a cell line which when Bn receptors were stably expressed in, the transfected Bn receptors behaved both, pharmacologically and in terms of cell signaling in a similar manner to wild type GRP-receptor [2,3]. Second, the duodenal cancer cell line, HuTu-80, which natively expresses hGRP-receptor [66] was studied. Furthermore, because a human cell line expressing only native NMB-receptors in sufficient number to do pharmacologic studies, was not available, hNMB-receptor was over-expressed in NCI-H1299 cells, which natively have a very low density of hNMB-receptor [5,40], with the hNMB-receptor transfected NCI-H1299 having a receptor density in the same range as native hGRP-receptor in human HuTu-80 cells [i.e. HuTu-80 (GRP-receptor density=1.68 ± 0.08 fmoles/106 cells) vs hNMB- receptor transfected (NCI-H1299 cells 2.11 ± 0.20 fmoles/106 cells)].
Table 1.
Affinities of antagonists for hGRP-receptor, hNMB-receptor and hBRS-3 receptor in Balb-3T3, HuTu-80 and NCI-H1299 cell lines.
| IC50 | |||||||
|---|---|---|---|---|---|---|---|
| hGRP-R | hNMB-R | hBRS-3 | |||||
| Peptide number | Group | Peptide structure | |||||
| Balb-3T3 | HuTu-80 | Balb-3T3 | NCI-1299 | Balb-3T3 | |||
| GRP | GRP | 0.12 ± 0.02 | 0.11 ± 0.01 | 490 ± 40 | 295 ± 35 | >10,000 | |
| NMB | NMB | 38.0 ± 2.1 | 30.1 ± 2.6 | 0.063 ± 0.002 | 0.031 ± 0.001 | 4,800 ± 400 | |
| 35 | [d-Tyr6, β-Ala11,Phe13, Nle14]Bn-(6-14) | 0.07 ± 0.01 | 0.03 ± 0.01 | 0.31 ± 0.01 | 0.76 ± 0.03 | 0.55 ± 0.03 | |
| 2 | I | [Tyr4, D-Phe12]Bn | 1950 ± 160 | 912 ± 76 | 3090 ± 320 | 1860 ± 60 | >10,000 |
| 3 | II | [d-Arg1, D-Trp7,9, Leu11]SP | 794 ± 51 | 1780 ±50 | 4480 ± 860 | 1590 ± 30 | >10,000 |
| 4 | II | [d-Pro4, D-Trp7,9,10]SP-(4-11) | 3800 ± 270 | >10,000 | >10,000 | >10,000 | 2300 ± 400 |
| 5 | III | [Leu13,ψ(CH2NH), Leu14]Bn | 7.72 ± 0.31 | 8.12 ± 0.56 | >10,000 | >10,000 | >10,000 |
| 6 | III | [d-Phe6, Leu13,ψ(CH2NH), D-Phe14]Bn-(6-14) | 12.72 ± 0.61 | 12.62 ± 0.50 | 7200 ± 690 | >10,000 | >10,000 |
| 7 | III | [d-Phe6, Leu13,ψ(CH2NH), Cpa14]Bn(6-14) | 1.35 ± 0.20 | 1.23 ± 0.09 | >10,000 | >10,000 | >10,000 |
| 7.5 | III | [d-TPi6, Leu13,ψ(CH2NH), Leu14]Bn(6-14) | 1.58 ± 0.08 | 1.32 ± 0.07 | 871 ± 44 | 776 ± 39 | >10,000 |
| 8 | IV | [d-Phe6]Bn(6-13)NH2 | 2.70 ± 0.24 | 4.93 ± 0.39 | >10,000 | >10,000 | >10,000 |
| 10 | V | [d-Phe6]Bn(6-13)Ethylamide | 1.72 ± 0.14 | 2.22 ± 0.13 | >10,000 | >10,000 | >10,000 |
| 11 | V | [d-Phe6-Bn](6-13)Propylamide (PA) | 2.63 ± 0.15 | 0.39 ± 0.12 | 2440 ± 160 | 3090 ± 70 | 1910 ± 300 |
| 12 | V | [d-Tyr6]Bn-(6-13)Propylamide (PA) | 1.82 ± 0.10 | 1.41 ± 0.06 | >10,000 | >10,000 | 1900 ± 300 |
| 14 | V | [d-Phe6]Bn(6-13)Butylamide (BA) | 2.18 ± 0.19 | 1.02 ± 0.09 | 3110 ± 160 | 1780 ± 30 | 692 ± 27 |
| 15 | V | [d-Phe6]Bn-(6-13)Hexylamide | 3.89 ± 0.45 | 2.19 ± 0.04 | 8140 ± 400 | >10,000 | 3250 ± 60 |
| 17 | VI | [d-Phe6]Bn-(6-13)Methyl ester (ME) | 0.89 ± 0.03 | 3.98 ± 0.80 | >10,000 | >10,000 | 5290 ± 1990 |
| 18 | VI | [d-F-5, Phe6, D-Ala11]Bn(6-13)Metyl ester (ME) | 0.89 ± 0.08 | 0.12 ± 0.01 | >10,000 | >10,000 | >10,000 |
| 20 | VI | [d-Phe6]Bn-(6-13)Ethyl ester | 0.28 ± 0.04 | 0.14 ± 0.03 | >10,000 | >10,000 | 5300 ± 1400 |
| 22 | VII | [d-Phe6, His7, D-Ala11, D-Pro13,ψ(13-14), Phe14]Bn-(6-14)NH2 | 4.89 ± 0.34 | 3.16 ± 0.60 | >10,000 | 3310 ± 70 | >10,000 |
| 23 | VII | [d-Tyr6, His7, D-Ala11, D- Pro13,ψ(13-14), Phe14]Bn-(6-14)NH2 | 8.91 ± 0.64 | 9.33 ± 0.14 | >10,000 | >10,000 | >10,000 |
| 24 | VII | [(3-Ph-Pr6), His7, D-Ala11, D-Pro13, ψ(13-14), Phe14]Bn(6-14)NH2) | 0.22 ± 0.03 | 0.23 ± 0.01 | 4899 ± 282 | >10,000 | 6820 ± 860 |
| 30 | VIII | [d-Nal, Cys,Tyr, D-Trp, Lys,Val, Cys, Nal]NH2 | 4570 ± 230 | 2460 ± 120 | 605 ± 23 | 275 ± 7 | 338 ± 11 |
| 31 | VIII | [d-Nal, Cys, Tyr, D-Trp,Orn,Val, Cys, Nal]NH2 | >10,000 | 3470 ± 360 | 630 ± 18 | 468 ± 16 | 537 ± 26 |
| 32 | IX | [d-Cys6, D-Ala11, Leu13,ψ(CH2NH), Cys14]Bn(6-14) | 2460 ± 70 | 2090 ± 70 | >10,000 | >10,000 | >10,000 |
| 33 | X | PD 176252 | 213 ± 15 | 170 ± 11 | 0.53 ± 0.03 | 0.17 ± 0.01 | >10,000 |
| 34 | X | PD 168368 | 1172 ± 85 | 1738 ± 105 | 0.51 ± 0.02 | 0.25 ± 0.02 | >10,000 |
HuTu-80 (1.5 × 106 cells/ml) and hGRPR-Balb-3T3 transfected (0.5 × 106 cells/ml) were incubated with 50pM 125I-[Tyr4]Bn, hNMBR-NCI-H1299 transfected (1 × 106 cells/ml) and hNMBR-Balb-3T3 transfected (0.05 × 106 cells/ml) with 125I-[D-Tyr0]NMB and hBRS-3-Balb-3T3 transfected (0.8 × 106 cells/ml) with 50 pM 125I-[D-Tyr6, β-Ala11, Phe13, Nle14]Bn-(6-14) for 1 h at 22°C. Increasing concentrations of unlabeled peptide were added, and dose-response curves were analyzed using KaleidaGraph. Values are mean ± S.E. from at least four experiments. >10,000 means the affinity was greater than 10,000 nM.
Abbreviations: β-Ala, β-Alanine; Nle, norleucine; GRP, gastrin-releasing peptide; ψ; pseudopeptide bond, (i.e., CONH changed to CH2NH); Ph-Pr, phenylpropanolamine; PA, propylamide; BA, butylamide; ME, methyl ester; SP, substance P; D-F-5, pentafluoro; Nal, β-napthylalanine; Orn, ornithine; Cpa, chlorophenylalamine, PD176252, (3-(1H-Indol-3-yl)-N-[1-(5-methoxy-pyridin-2-yl)-cyclohexylmethyl]-2-methyl-2-[3-(4-nitro-phenyl)-ureido]-propionamide); PD168368, (3-(1H-Indol-3-yl)-2-methyl-2-[3(4-nitro-phenyl)-ureido]-N-(1-pyridin-2-yl-cyclohexylmethyl)-propionamide; TPI,2.3.4.9-tetrahydro-1H-pyrido[3,4-]indol-3-carboxylic acid).
Table 2.
Affinities of antagonists for hGRP-receptor, hNMB-receptor and hBRS-3 receptor in Balb-3T3 cell lines.
| IC50 | |||||
|---|---|---|---|---|---|
| hGRPR | hNMBR | hBRS-3 | |||
| Peptide number | Group | Peptide structure | |||
| Balb-3T3 | Balb-3T3 | Balb-3T3 | |||
| GRP | GRP | 0.12 ± 0.02 | 490 ±10 | >10,000 | |
| NMB | NMB | 38.0 ± 2.1 | 0.063 ± 0.002 | 4,800 ± 400 | |
| 1 | I | [d-Phe12, Leu14]Bn | >10,000 | >10,000 | >10,000 |
| 9 | IV | Bn(6-13)NH2 | 355 ± 37 | >10,000 | >10,000 |
| 13 | V | [d-Phe6,Thr10, Phe13]Bn(6-13)PA | 178 ± 16 | 1320 ± 30 | >10,000 |
| 16 | VI | [d-Phe6]Bn(6-13)Heptylamide | 5.12 ± 0.33 | 6100 ± 630 | >10,000 |
| 19 | VI | [d-F-5- Phe6, His7, D-Ala11, D-Pro13]Bn(6-13)ME | 240 ± 18 | >10,000 | >10,000 |
| 21 | VII | [N-AC-GRP(20-26)]Ethyl ester | 2.04 ± 0.20 | >10,000 | >10,000 |
| 25 | VII | [d-F-5-Phe6, His7, D-Ala11, D-Tic13,ψ (13-14), Phe14]Bn(6-14)NH2 | 8.81 ± 2.54 | >10,000 | >10,000 |
| 26 | VII | [d-F-5-Phe6, His7, D-Ala11, D-Pro13,ψ (13-14), Phe14]BN(6-14)NH2 | 1740 ± 40 | >10,000 | >10,000 |
| 27 | VII | [d-F-5-Phe6, His7, D-Ala11, D-Pro13,ψ (13-14),Tic14]Bn(6-14)NH2 | 19.42 ± 1.52 | >10,000 | >10,000 |
| 28 | VII | [d-F-5-Phe6, His7, D-Ala11, D-Tic13,ψ (13-14),Tic14]Bn(6-14)NH2 | >10,000 | >10,000 | >10,000 |
| 29 | VIII | [d-Nal, Cys,Tyr, D-Trp, Lys, Nal, Cys,Thr]-NH2 | 3290 ± 290 | 2490 ± 360 | >10,000 |
Abbreviations: D-F-5, pentafluoro; Tic, 1,2,3,4,-tetrahydroisoquinoline-3-carboxylic acid; ψ; pseudopeptide bond, (i.e., CONH changed to CH2NH); ME, methyl ester; N-AC, N-acetyl; PA, propylamide; Nal, β-napthylalalanine.
As described previously in other human and nonhuman tissues, the native GRP-receptor ligand, GRP, had high affinity for hGRP-receptor on both cell lines (i.e. 0.11-0.12 nM), a >3000-fold lower affinity for hNMB-receptor and a >10,000-fold lower affinity for hBRS3 (Table1)[2-4,24,29]. In contrast, similar to described previous in human and other nonhuman tissues, the native NMBR ligand, NMB, had very high affinity for the hNMB-receptor on both cell lines (0.033-0.063 nM) with a 600-fold lower affinity for the hGRP-receptor and a >10,000 fold lower affinity for the hBRS3 (Table 1)[2-4,24,29,45,60]. In contrast, similar to recent described in various human and nonhuman cells, the novel synthetic Bn analogue, [D-Tyr6, β-Ala11,Phe13, Nle14]Bn-(6-14)(#35, Table 1) had high affinity for all human Bn receptors, including hBRS-3 (Table 1) [24,38-40,43,49,51,55].
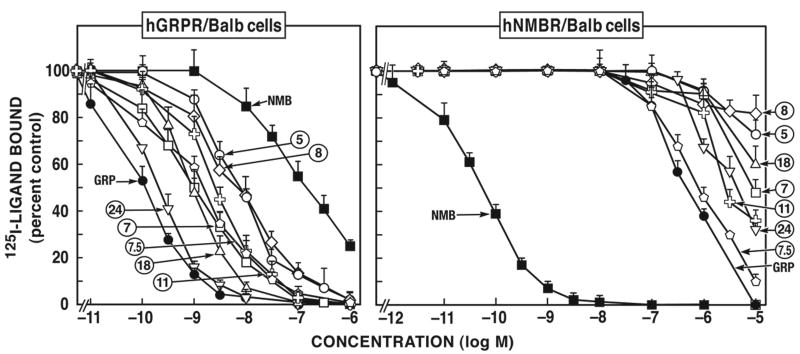
The different putative bombesin antagonists or ligands could be divided into 10 chemical categories (Tables 1, 2) including: Group I, [d-Phe12Bn-analogues] (#1, #2); Group II, [d-amino acid substituted substance P analogues] (#3, #4); Group III, [Ψ13-14 pseudo-bond Bn-peptides analogues] (#5-#7.5); Group IV, [des-Met14 amide analogue] (#8); Group V, [des-Met14 alkyl amide analogues] (#10-#12, #14-#16); Group VI, [des-Met14 ester analogues] (#17-#21); Group VII, [d-Pro13, Ψ13-14 pseudo-bond peptide analogues] (#22-#28); Group VIII, [d-amino acid substituted somatostatin (SS) analogues] (#29, #30, #31); Group IX, [cyclic Bn peptide analogues] (#32); and Group X, [peptoids] (#33, #34). For the hGRP-receptor in both hGRP-receptor transfected Balb 3T3 cells and for the native hGRP-receptor in HuTu-80 cells (Fig. 1, 2; Tables 1, 2) the relative affinities of the most potent antagonists in the different chemical classes were: the [d-Pro13, Ψ13-14]Bn-analogue [peptide #24 (IC50= 0.2 nM) > Bn(6-13)esters [#17, #18, #20 and #21 (IC50= 0.3-2.0 nM) = [Ψ13-14]Bn (6-14) analogues [peptides #5, #6, #7 and #7.5 (IC50= 1.4-13.0 nM) = Bn(6-13) amide and alkyl amides [peptides #8, #10-#16] > Peptoids [peptides #33 and #34 (IC50= 170-1,172 nM) > d-amino and SP substituted analogues [peptides #2 and #3 (IC50= 800-1,950 nM, Fig. 3; Table 1)] > cyclic Bn-analogues [#32] = [d-Phe12]Bn-analogues [peptides #1 and #2 (IC50= 1,000-10,000 nM, Fig. 1; Table 1 and 2)] = substituted SS analogues [peptides #29, #30 and #31 (IC50= 2,500-10,000 nM, Fig. 2; Table 1, 2)].
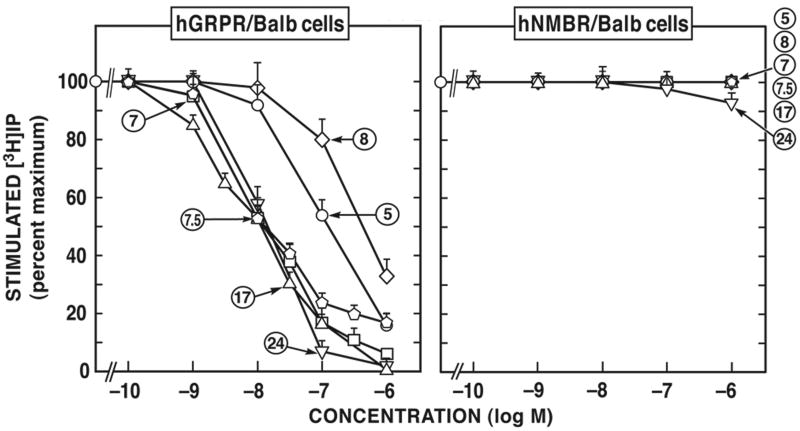
Figure 1.
Comparison of affinities of various selected human GRP receptor-preferring putative antagonists for BALB 3T3 cells transfected with hGRP-receptor (left) or hNMB-receptor (right). The experimental conditions were similar to those outlined in the legend to Table 1 and as described under Materials and Methods. Values are mean ± S.E.M. from at least four experiments. >10,000 means the affinity was greater than 10,000 nM. Numbers refer to the peptide number in Table 1.
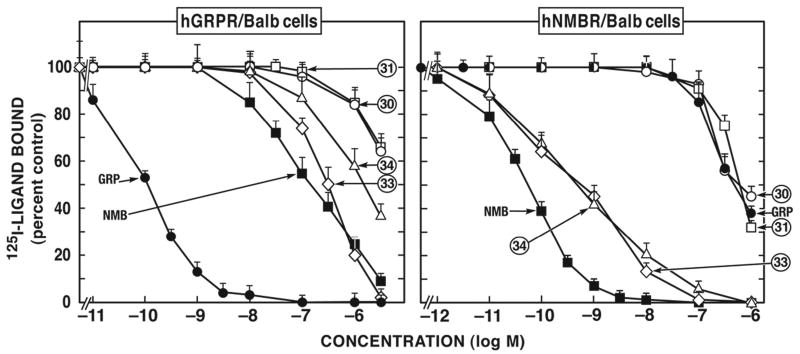
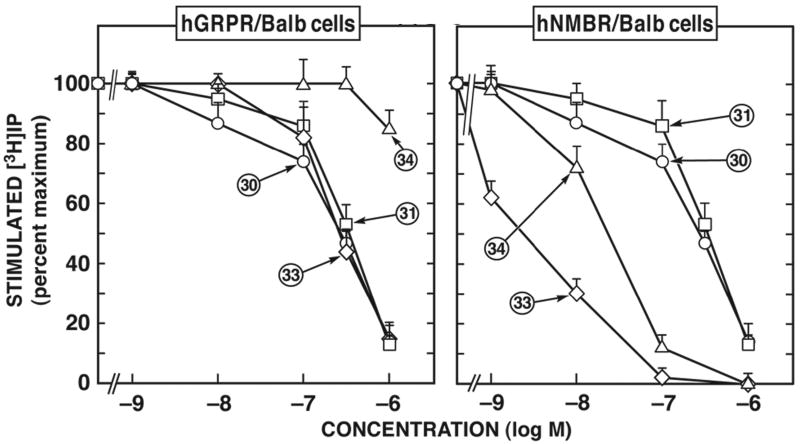
Figure 2.
Comparison of affinities of various selected human NMB receptor-preferring putative antagonists for BALB 3T3 cells transfected with hGRP-receptor (left) or hNMB-receptor (right). The experimental conditions were similar to those outlined in the legend to Table 1 and as described under Materials and Methods. Values are mean ± S.E.M. from at least four experiments. >10,000 means the affinity was greater than 10,000 nM. Numbers refer to the peptide number in Table 1.
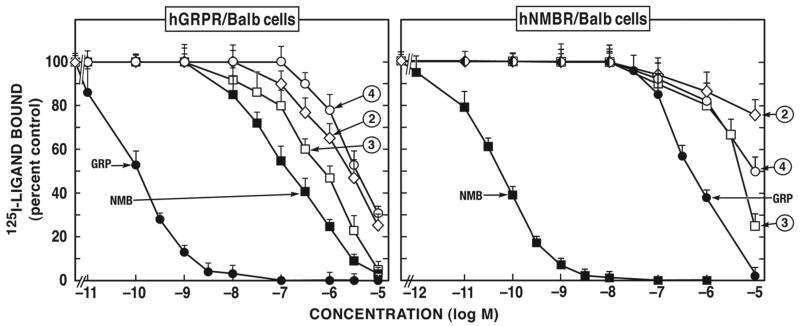
Figure 3.
Affinities of various non-selective putative antagonists for BALB 3T3 cells transfected with hGRP-receptor (left) or hNMB-receptor (right). The experimental conditions were similar to those outlined in the legend to Table 1 and as described under Materials and Methods. Values are mean ± S.E.M. from at least four experiments. Numbers refer to the peptide number in Table 1.
In contrast, with the hNMB-receptor expressed in either hNMB-receptor transfected Balb 3T3 cells or NCI-H1299 cells, only two putative antagonists, the peptoid analogues (#33, #34, Fig. 2; Table 1) had high affinities (IC50= 0.17-0.53 nM) (Table 1, 2). All of the other putative Bn receptor antagonists had an affinity >250 nM for the hNMB- receptor, with most having an affinity >1 μM for the hNMB-receptor (Fig. 2, Table 1, 2).
All of the antagonists had low affinity (>0.3 uM) for the hBRS-3 receptor in hBRS-3 transfected Balb 3T3 cells (Table 1). Previous studies have reported similar results for a few Bn-receptor antagonists with hBRS-3 transfected Balb 3T3 cells and with NCI-N417 lung cancer cells, which possess low density of native hBRS-3-receptors [52,55]. To confirm the low affinities for hBRS-3 were not due to the transfected cell used, 5 antagonists [8,14,17, 30, 31] were tested on both cells and similar affinities were obtained with each peptide on native and transfected hBRS-3 cell lines (data not shown)
In terms of the putative hGRP-receptor antagonist's affinity compared to the native agonist, GRP, some had an affinity almost equal to GRP's (0.12 ± 0.02 nM), such as the [d-Pro13, Ψ13-14, Phe14]Bn(6-14) analogue peptide #24 (0.22 ± 0.03 nM) and the [d-Phe6]Bn(6-13)ethyl ester analogue peptide #20 (0.28 ± 0.04 nM); whereas others were 5 to 10-fold less potent than GRP, but retained high affinity including the [d-F5-Phe6]Bn(6-13)methyl ester analogue peptide #18 (0.89 ± 0.08 nM), the Bn(6-13) propylamide analogue peptide #11 (2.63 ± 0.15 nM) and the Bn(6-13) heptylamide analogue, peptide #16 (5.12 ± 0.33 nM). Others had an affinity that was 50-100 lower than GRP, resulting in intermediate affinities including peptide #5 (7.72 ± 0.31 nM), peptide #25 (8.81 ± 2.54 nM) and peptide #6 (12.72 ± 0.61 nM) and finally a group had low affinity (>1,000) including peptides #3, #4, #28-30, #31, #32 and the peptoid, #34 (Table 1, 2). With the hNMB- receptor, only two putative antagonists the peptoid antagonists (#33, #34, Fig. 2; Table 1) had affinities (0.17-0.53 nM) within same range as the natural agonist NMB (0.03-0.06 nM, Fig. 1, 2; Table 2).
The different putative antagonists were grouped by their selectivity for hGRP-receptor or hNMB-receptor (Fig. 1). Members of 6 chemical groups [Groups III, IV, V, VI, VII, IX (Fig.1, Table 1)] showed selectivity for hGRP-receptor over hNMB-receptor. Specifically, in order of increasing selectivity the cyclic analogue (#32, Table 1) was >5 fold selective for the hGRP-receptor over the hNMB-receptor; the [Ψ13-14]Bn pseudopeptide analogue, peptide #5 and the Bn(6-13)propyl amide analogue, peptide #11 had ≥1,000-fold greater selectivity for hGRP-receptor over hNMB-receptor expressing in Balb 3T3 cells; the [Ψ13-14, Cpa14]Bn(6-14) analogue peptide #7, the [D-Tpi6, Ψ13-14, Leu13]Bn(6-14) analogue, peptide #7.5 and the [d-F5-Phe6]Bn(6-13)methyl ester analogue, peptide #18, were 7,400 and 11,000-fold more selective for hGRP-receptor over hNMB-receptor, respectively expressed in Balb 3T3 cells, and finally the [d-Pro13, Ψ13-14, Phe14]Bn(6-14), peptide #24, exhibited more than 30,000-fold selectivity for hGRP-receptor over hNMB-receptor in Balb 3T3 cells (Fig. 1, Table 1). Similar selectivity was seen in the hGRP-receptor containing Hutu-80 cells and in hNMB-receptor transfected NCI-H1299 cells.
In the case of hNMB-receptor, only members of two chemical groups (Groups VIII and X) were selective for hNMB-receptor over hGRP-receptor (Fig. 1, 2; Table 1). The peptoid analogues #33 and #34 belonging to group X (Fig. 2; Table 1) showed the greatest selectivity (400-6,952-fold) for hNMB-receptor over hGRP-receptor. In addition, members of Group VIII, the putative antagonist d-amino substituted somatostatin analogue peptide #30 and #31 (Fig. 2; Table 1), showed a 3-8-fold selectivity for hNMB-receptor over hGRP-receptor (Fig. 2; Table 1).
A number of the putative antagonists were non-selective and had low affinity for both hNMB-receptor and hGRP-receptor. Specifically, this included the group I peptides [d-Phe12]Bn-analogues #1 and #2 (Fig. 3; Table 1 and 2) and the Group VIII peptides, d-amino acid substituted SS analogue #29 (Table 2). The Group II d-amino substituted SP antagonist derivatives peptides #3 and #4, not only had low affinity for hGRP-receptor (800-3,800 nM) and for hNMB-receptor (4,500-10,000 nM) (Fig. 3; Table 1) but also, neither of them was selective for these receptors.
None of the putative Bn receptor antagonists were selective for the hBRS-3 receptor and except for the d-amino acid substituted analogues #30, #31 (IC50= 340-540 nM) and the [d-Phe6]Bn(6-13)butylamide analogue (IC50= 692 nM), all the other putative antagonists had had affinities >1 μM for hBRS-3, and many >10 μM (Table 1, 2).
To determine whether these putative Bn antagonists, functioned as Bn receptor antagonists or had agonist activity, their ability to stimulate or inhibit agonist-stimulated [3H]IP was determined. Activation of phospholipase C was assessed because the native, as well as the transfected hGRP-receptor [3,15,24,66], hNMB-receptor [3,24] and hBRS-3 [14,24,52,54,55] have all shown to activate phospholipase C and stimulate generation of inositol phosphates.
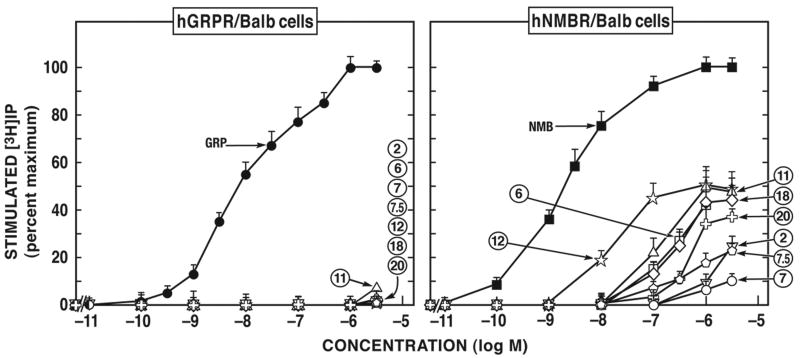
None of the putative antagonists tested in this study showed agonist activity for hGRP-receptor at concentrations up 10 μM in hGRP-receptor transfected Balb 3T3 or HuTu-80 cells (Fig. 4; Table 3). However, with the hNMB-receptor, twelve of the compounds (analogues #2, #6, #7, #7.5, #10-#12, #14, #15, #17, #18, #20) (Fig. 4; Table 4) had agonist activity at concentrations up to 10 μM in hNMB-receptor transfected Balb 3T3 cells and also in hNMB-receptor transfected NCI-H1299, with the exception of the [Ψ13-14]Bn pseudopeptide analogue, peptide #7; which was a partial agonist (10% of the efficacy of NMB at 10 μM) in hNMB-receptor transfected Balb 3T3 cells, but had no activity in hNMB-receptor transfected NCI-H1299 (Table 4). This group of twelve compounds with NMB-receptor agonist activity included the [d-Phe12]Bn-analogue, peptide #2, the (Ψ13-14]Bn (6-14) analogues, peptide #6, #7 and #7.5; the Bn(6-13) alkylamide analogues, peptides #10, #11, #12, #14, #15 and the Bn(6-13) ester analogues, peptides #17, #18, #20. For these twelve peptides with agonist activity at the hNMB-receptor, none was a fully efficacious agonist causing a similar full increase in [3H]IP seen with NMB (Fig. 4; Table 4). Peptides #6, #10, #11, #12, #14, #15, #18 and #20 were partial agonists (34-61% of the efficacy of NMB) (Fig. 4, Table 4) and four peptides, the d-amino SP substituted analogue peptide #2, the [Ψ13-14, Cpa14]Bn(6-14) analogue, peptide #7, [D-Tpi6, Ψ13-14, Leu13]Bn(6-14) analogue, peptide #7.5 and the Bn(6-13) ester analogue, peptide #17 had agonist activity when used up to 10 μM in hNMB-receptor transfected Balb 3T3 cells, but higher concentrations could not be used to determine whether they were full or partial agonists (Fig. 4; Table 4).
Figure 4.
Comparison of the ability of various selected putative human Bn receptor antagonists to stimulate [3H]IP generation in Balb 3T3 cells transfected with hGRP-receptor (left) or hNMB-receptor (right). hGRP-receptor-transfected BALB 3T3 and hNMB-receptor-transfected BALB 3T3 were incubated with each ligand at the indicated concentrations for 60 min. EC50 is the concentration of the indicated peptide causing half-maximal stimulation seen with 1 μM GRP or NMB. The results are the means ± S.E.M. from at least three experiments performed in duplicate. The control and maximal stimulated [3H]IP values for the hGRP-R-transfected BALB 3T3 cells were 2,547 ± 210 and 25,720 ± 1,785 dpm, respectively. The control and maximal stimulated [3H]IP values for the hNMBR-transfected BALB 3T3 cells were 1,895 ± 230 and 71,466 ± 3,010 dpm, respectively. Numbers refer to the peptide number in Tables 3 and 4.
Table 3.
Ability of GRP antagonists to stimulate [3H]IP generation in hGRP-receptor Balb 3T3 and HuTu-80 cells.
| [3H]IP | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| hGRPR-R-Balb-3T3 | HuTu-80 (hGRP-R) | |||||||||
| Peptide Number |
Group | Peptide structure | Ag/Ant ag |
% max | EC50 (nM) |
IC50 (nM) | Ag/Antag | % max | EC50 (nM) | IC50 (nM) |
| GRP | GRP | Ag | 100 | 6.8 ± 0.3 | Ag | Ag | 100 | 8.2 ± 0.2 | Ag | |
| 2 | I | [Tyr4, D-Phe12]Bn | No act | No act | No act | >10,000 | Antag | Antag | Antag | 8128 ± 226 |
| 3 | II | [d-Arg1, D-Trp7,9, Leu11]SP | Antag | Antag | Antag | 1580 ± 81 | Antag | Antag | Antag | 8313 ± 154 |
| 4 | II | [d-Pro4, D-Trp7,9,10]SP-(4-11) | No act | No act | No act | >3000 | No act | No act | No act | >3,000 |
| 5 | III | [Leu13,ψ(CH2NH), Leu14]Bn | Antag | Antag | Antag | 134 ± 2 | Antag | Antag | Antag | 100 ± 5 |
| 6 | III | [d-Phe6, Leu13,ψ(CH2NH), D-Phe14]Bn-(6-14) | Antag | Antag | Antag | 151 ± 13 | Antag | Antag | Antag | 240 ± 26 |
| 7 | III | [d-Phe6, Leu13,ψ(CH2NH), Cpa14]Bn(6-14) | Antag | Antag | Antag | 10.3 ± 1.2 | Antag | Antag | Antag | 20.0 ± 1 |
| 7.5 | III | [d-TPi6, Leu13,ψ(CH2NH), Leu14]Bn(6-14) | Antag | Antag | Antag | 11.6 ± 0.6 | Antag | Antag | Antag | 19.1 ± 1 |
| 8 | IV | [d-Phe6]Bn(6-13)NH2 | Antag | Antag | Antag | 416 ± 8 | Antag | Antag | Antag | 398 ± 11 |
| 10 | V | [d-Phe6]Bn(6-13)Ethylamide | Antag | Antag | Antag | 52.5 ± 4.3 | Antag | Antag | Antag | 43.6 ± 3.6 |
| 11 | V | [d-Phe6-Bn](6-13)PA | Antag | Antag | Antag | 31 ± 1 | Antag | Antag | Antag | 36.3 ± 1.0 |
| 12 | V | [d-Tyr6]Bn-(6-13)PA | Antag | Antag | Antag | 275 ± 13 | Antag | Antag | Antag | 40.7 ± 1.4 |
| 14 | V | [d-Phe6]Bn(6-13)BA | Antag | Antag | Antag | 21.3 ± 1.6 | Antag | Antag | Antag | 15.5 ± 2.0 |
| 15 | V | [d-Phe6]Bn-(6-13)Hexylamide | Antag | Antag | Antag | 380 ± 31 | Antag | Antag | Antag | 204 ± 14 |
| 17 | VI | [d-Phe6]Bn-(6-13)ME | Antag | Antag | Antag | 15.5 ± 1.0 | Antag | Antag | Antag | 10.9 ± 1.5 |
| 18 | VI | [d-F-5, Phe6, D-Ala11]Bn(6-13)ME | Antag | Antag | Antag | 3.6 ± 0.2 | Antag | Antag | Antag | 4.2 ± 0.9 |
| 20 | VI | [d-Phe6]Bn-(6-13)Ethyl ester | Antag | Antag | Antag | 2.0 ± 0.3 | Antag | Antag | Antag | 1.4 ± 0.8 |
| 22 | VII | [d-Phe6, His7, D-Ala11, D-Pro13,ψ(13-14), Phe14]Bn-(6-14)NH2 | Antag | Antag | Antag | 31 ± 3 | Antag | Antag | Antag | 25 ± 1 |
| 23 | VII | [d-Tyr6, His7, D-Ala11, D- Pro13, ψ(13-14), Phe14]Bn-(6-14)NH2 | Antag | Antag | Antag | 168 ± 20 | Antag | Antag | Antag | 158 ± 14 |
| 24 | VII | [(3-Ph-Pr6), His7, D-Ala11, D-Pro13,ψ(13-14), Phe14]Bn(6-14)NH2) | Antag | Antag | Antag | 15 ± 1 | Antag | Antag | Antag | 6.0 ± 0.3 |
| 30 | VIII | [d-Nal, Cys,Tyr, D-Trp, Lys,Val, Cys, Nal]NH2 | No act | No act | No act | >3,000 | No act | No act | No act | >3,000 |
| 31 | VIII | [d-Nal, Cys, Tyr, D-Trp,Orn,Val, Cys, Nal]NH2 | No act | No act | No act | >3,000 | No act | No act | No act | >3,000 |
| 32 | IX | [d-Cys6, D-Ala11, Leu13,ψ(CH2NH), Cys14]Bn(6-14) | Antag | Antag | Antag | 2,000 | No act | No act | No act | >10,000 |
| 33 | X | PD 176252 | Antag | Antag | Antag | 263 ± 14 | Antag | Antag | Antag | 275 ± 13 |
| 34 | X | PD 168368 | No act | No act | No act | >3,000 | Antag | Antag | Antag | 1778 ± 57 |
Ability of different compounds to stimulate increases in [3H]IP formation in hGRP-R-transfected Balb-3T3 cells and HuTu-80. The cells (5.0 × 104 cells/well) were loaded with myo-[2–3H]inositol as described under Materials and Methods, washed, and incubated with the indicated concentrations of compounds for 45 min at 37°C. Values are expressed as the percentage of the total [3H]IP release stimulated by 1 μM GRP. Results are the means ± S.E.M. from at least theree experiments, and each point was determined in duplicate.
Abbreviations: Ag, Agonist; Antag, Antagonist; No act, No agonist/antagonist activity at 10 μM.
Table 4.
Ability of antagonists to stimulate [3H]IP generation in hNMB-receptor Balb 3T3 and NCI-H1299 cells.
| [3H]IP | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| hNMB-R-Balb-3T3 | NCI-H1299 (hNMB-R) | |||||||||
| Peptide Number |
Group | Peptide structure | Ag/Antag | % max |
EC50 (nM) | IC50 (nM) | Ag/Antag | % max | EC50 (nM) | IC50 (nM) |
| NMB | NMB | Ag | 100 | 4.0 ± 0.2 | Ag | Ag | 100 | 1.0 ± 0.2 | Ag | |
| 2 | I | [Tyr4, D-Phe12]Bn | Ag (10μM) | 24 | Ag (10μM) | >10,000 | pAg | 34 | 103 ± 5 | pAg |
| 3 | II | [d-Arg1, D-Trp7,9, Leu11]SP | Antag | Antag | Antag | 1175 ± 98 | Antag | No act | No act | 2344 ± 80 |
| 4 | II | [d-Pro4, D-Trp7,9,10]SP-(4-11) | No act | No act | No act | >10,000 | Antag | No act | No act | 7080 ± 344 |
| 5 | III | [Leu13,ψ(CH2NH), Leu14]Bn | No act | No act | No act | >10,000 | No act | No act | No act | >10000 |
| 6 | III | [d-Phe6, Leu13,ψ(CH2NH), D-Phe14]Bn-(6-14) | pAg | 42 | 269 ± 21 | pAg | pAg | 32 | 528 ± 26 | pAg |
| 7 | III | [d-Phe6, Leu13,ψ(CH2NH), Cpa14]Bn(6-14) | Ag (10μM) | 10 | Ag (10μM) | >10,000 | No act | No act | No act | >10,000 |
| 7.5 | III | [d-TPi6, Leu13,ψ(CH2NH), Leu14]Bn(6-14) | Ag (10μM) | 24 | Ag (10μM) | >10,000 | Ag (10μM) | 17 | Ag (10μM) | >10,000 |
| 8 | IV | [d-Phe6]Bn(6-13)NH2 | No act | No act | No act | >10,000 | No act | No act | No act | >10,000 |
| 10 | V | [d-Phe6]Bn(6-13)Ethylamide | pAg | 50 | 275 ± 18 | pAg | pAg | 28 | 144 ± 11 | pAg |
| 11 | V | [d-Phe6-Bn](6-13)PA | pAg | 50 | 100 ± 4 | pAg | pAg | 35 | 218 ± 10 | pAg |
| 12 | V | [d-Tyr6]Bn-(6-13)PA | pAg | 51 | 16.2 ± 2.3 | pAg | pAg | 31 | 28.0 ± 1.5 | pAg |
| 14 | V | [d-Phe6]Bn(6-13)BA | pAg | 61 | 200 ± 14 | pAg | pAg | 60 | 21.4 ± 1.6 | pAg |
| 15 | V | [d-Phe6]Bn-(6-13)Hexylamide | pAg | 58 | 269 ± 12 | pAg | pAg | 45 | 144 ± 18 | pAg |
| 17 | VI | [d-Phe6]Bn-(6-13)ME | Ag (10μM) | 42 | Ag (10μM) | >10,000 | Ag (10μM) | 32 | Ag (10μM) | >10,000 |
| 18 | VI | [d-F-5, Phe6, D-Ala11]Bn(6-13)ME | pAg | 43 | 213 ± 9 | pAg | pAg | 34 | 316 ± 19 | pAg |
| 20 | VI | [d-Phe6]Bn-(6-13)Ethyl ester | pAg | 34 | 158 ± 10 | pAg | pAg | 15 | 59 ± 6 | pAg |
| 22 | VII | [d-Phe6, His7, D-Ala11, D-Pro13, ψ(13-14), Phe14]Bn-(6-14)NH2 | No act | No act | No act | >10,000 | No act | No act | No act | >10,000 |
| 23 | VII | [d-Tyr6, His7, D-Ala11, D- Pro13,ψ(13-14), Phe14]Bn-(6-14)NH2 | No act | No act | No act | >10,000 | No act | No act | No act | >10,000 |
| 24 | VII | [(3-Ph-Pr6), His7, D-Ala11, D-Pro13,ψ(13-14), Phe14]Bn(6-14)NH2) | No act | No act | No act | >10,000 | Antag | Antag | Antag | 955 ± 24 |
| 30 | VIII | [d-Nal, Cys,Tyr, D-Trp, Lys,Val, Cys, Nal]NH2 | No act | No act | No act | 216 ± 36 | Antag | Antag | Antag | 269 ± 12 |
| 31 | VIII | [d-Nal, Cys, Tyr, D-Trp,Orn,Val, Cys, Nal]NH2 | Antag | Antag | Antag | 363 ± 16 | Antag | Antag | Antag | 315 ± 15 |
| 32 | IX | [d-Cys6, D-Ala11, Leu13,ψ(CH2NH), Cys14]Bn(6-14) | No act | No act | No act | >10,000 | No act | No act | No act | >10,000 |
| 33 | X | PD 176252 | Antag | Antag | Antag | 3.1 ± 0.1 | Antag | Antag | Antag | 2.4 ± 0.1 |
| 34 | X | PD 168368 | Antag | Antag | Antag | 19.4 ± 0.7 | Antag | Antag | Antag | 24.5 ± 1.2 |
Ability of different compounds to stimulate increases in [3H]IP formation in hNMB-R-transfected Balb-3T3 cells and NCI-H1299. The cells (5.0 × 104 cells/well) were loaded with myo-[2–3H]inositol as described under Materials and Methods, washed, and incubated with the indicated concentrations of compounds for 45 min at 37°C. Values are expressed as the percentage of the total [3H]IP release stimulated by 1 μM NMB. Results are the means ± S.E.M. from at least theree experiments, and each point was determined in duplicate.
Abbreviations: Ag, Agonist; Antag, Antagonist; No act, No agonist/antagonist activity at 10 μM; pAg, partial Agonist; Ag, Agonist at 10 μM.
Of the twelve analogues with agonist activity at hNMB-receptor, the most potent in hNMB-receptor transfected Balb-3T3 or in NCI-H1299 cells was [d-Tyr6]Bn(6-13)propyl amide (peptide #12, EC50= 16.2 nM), which had a potency (6-17 fold) higher than the other partial agonists, peptides #6, #10, #11, #14, #15, #18 and #20, but was 81-fold less potent than the natural ligand, NMB in the activation of hNMB-receptor in Balb-3T3 or NCI-H1299 cells (Fig. 4; Table 4). Peptides #6, #10, #11, #14, #15, #18 and #20 had potencies 700-1,400-fold lower than NMB for the activation of the hNMB-receptor in both hNMB-receptor-containing cell lines (Fig. 4; Table 4).
Thirteen putative Bn-receptor antagonist peptides including analogues #3, #4, #5, #8, #23, #24, #30, #31, #32 and peptoids #33 and #34 did not have agonist activity in hNMB-receptor transfected Balb 3T3 or NCI-H1299 cells, at concentrations up to 10 μM (Fig. 4; Table 4). We examined their abilities to inhibit GRP- and NMB-stimulated [3H]IP in the different cells. Most of the twelve peptides inhibited GRP-stimulated [3H]IP increases in hGRP-receptor transfected Balb 3T3 cells and HuTu-80 cells (Fig. 5; Table 3). The highest potency for inhibiting GRP-stimulated [3H]IP in GRP-receptor-cells was seen with the des-Met14 esters, peptides #18 and #20 (IC50= 6-15 nM, Table 3) and the analogue, peptide #24 (IC50= 1-4 nM, Fig. 5; Table 3). Others were less potent, but still retained relatively high potency including the Ψ13-14 pseudo-bond peptide #7, [D-Tpi6, Ψ13-14, Leu13]Bn(6-14) analogue, peptide #7.5 and the [d-Phe6]Bn(6-13)methyl ester peptide #17 (IC50= 11-16 nM) (Fig.5, Table 3). Still others, showed intermediate potencies for inhibiting-hGRP-receptor activity (i.e. 50-400 nM, Table 3) including the Ψ13-14 pseudo-bond peptide analogue, peptide #5, the [d-Pro13, Ψ13-14, Phe14]Bn(6-14) derivate peptide #23 and the [d-Phe6]Bn(6-13)NH2 peptide #8. Finally, a group had low potency (IC50=1,600->10,000 nM) for inhibiting GRP-stimulated [3H]IP increases including the d-amino acid substituted somatostatin analogues (#30, #31) and peptoid #34 (Table 3).
Figure 5.
Comparison of the ability of various selected putative human GRP-preferring receptor antagonists without agonist activity, to inhibit the stimulation of [3H]IP generation by GRP in Balb 3T3 cells transfected with hGRP-receptor or NMB in cells transfected with hNMBR. hGRP-receptor-transfected BALB 3T3 and hNMB-receptor-transfected BALB 3T3 were incubated with 1 nM GRP (control: 4,210 ± 350 dpm; maximal: 25,020 ± 2,356 dpm) or 1 nM NMB (control: 2,280 ± 310 dpm; maximal: 33,288 ± 2,130 dpm) and each antagonist at indicated concentrations for 60 min. Results are expressed as the percentage of total [3H]IP released by 1 nM GRP or 1 nM NMB in the presence of various concentrations of each antagonist. The results are the means ± S.E.M. from at least three experiments performed in duplicate. Numbers refer to the peptide number in Tables 3 and 4.
For the hNMB-receptor-preferring peptoid analogues, peptide #33 and #34, each inhibited NMB-stimulated [3H]IP generation with higher potency (IC50= 2.4-25 nM) than they inhibited GRP-receptor-stimulated increases in [3H]IP (IC50= 263 to >3,000nM) and each was more potent for inhibiting hNMB-receptor stimulated activity than any other Bn-receptor putative antagonist studied (Fig. 6, Table 4).
Figure 6.
Comparison of the ability of various selected putative human NMB receptor preferring antagonists without agonist activity, to inhibit the stimulation of [3H]IP generation by GRP in Balb 3T3 cells transfected with hGRP-receptor or NMB in cells transfected with hNMB-receptor. hGRP-receptor-transfected BALB 3T3 and hNMB-receptor-transfected BALB 3T3 were incubated with 1 nM GRP (control: 4,210 ± 350 dpm; maximal: 25,020 ± 2,356 dpm) or 1 nM NMB (control: 2,280 ± 310 dpm; maximal: 33,288 ± 2,130 dpm) and each compound at indicated concentrations for 60 min. Results are expressed as the percentage of total [3H]IP released by 1 nM GRP or 1 nM NMB in the presence of various concentrations of each ligand. The results are the means ± S.E.M. from at least three experiments performed in duplicate. Numbers refer to the peptide number in Tables 3 and 4.
In terms of selectivity based on potency for inhibiting agonist activity, the pseudopeptide analogue, peptides #5, the des Met14 amide analogue, peptide #8 and the [d-Pro13, Ψ13-14, Phe14]Bn(6-14) analogue, peptide #24 functioned as highly selective hGRP-receptor antagonists in hGRP-receptor transfected Balb-3T3 and HuTu-80 cells, with no activity at the hNMB-receptor transfected Balb-3T3 or NCI-H1299 cells (Fig. 5, Table 3 and 4). However, the Cpa11 [Ψ13-14 pseudopeptide analogue], peptide #7, [D-Tpi6, Ψ13-14, Leu13]Bn(6-14) analogue, peptide #7.5 and the des-Met14 methyl ester analogue, peptide #17, although they were moderately potent antagonists in hGRP-receptor transfected Balb-3T3 or HuTu-80 cells, both had some agonist activity at a concentration of 10 μM with the hNMB-receptor transfected Balb 3T3 cells (Table 4). In contrast, each of the des-Met14 alkyl amides (#10, #11, #12, #14, #15), des-Met14 esters (#17, #18, #20) and the Ψ13-14 pseudo-bond analogue, peptide #6, which functioned as antagonists in hGRP-receptor transfected Balb 3T3 or HuTu cells (Table 3) were partial agonists at the hNMB-receptor transfected Balb 3T3 and NCI-H1299 cells at higher concentrations (10 μM, Fig. 4; Table 4).
In terms of potency and selectivity for the hNMB-receptor based on their ability to inhibit NMB-stimulated cell activation, the two peptoids, peptide #33 and #34 inhibited NMB-stimulated [3H]IP generation in hNMB-receptor transfected Balb 3T3 and NCI-H1299 cells with the highest potency (IC50= 2.4-24.5 nM, Fig. 6, Table 4) of all the Bn putative antagonists studied. While peptide #33 inhibited also GRP-stimulated [3H]IP generation (IC50= 275 nM, Fig. 6, Table 4) in hGRP-receptor transfected Balb-3T3 or HuTu-80 cells, peptide #34 was less potent at the hGRP-receptor (IC50= 1,778 to > 3,000 nM, Fig. 6, Table 4) making it 33 to 100-fold more selective for hNMB-receptor over hGRP-receptor. None of the d-amino acid substituted somatostatin analogues (peptides #30, #31), stimulated [3H]IP generation in hGRP-receptor transfected Balb-3T3 or HuTu-80 cells, and neither of them were able to inhibit GRP-stimulated [3H]IP with a potency higher than 3,000 nM in both cell lines (Fig. 6, Table 3). However, these two compounds (#30, #31) showed similar antagonist activity inhibiting NMB-stimulated [3H]IP generation (IC50= 216-363 nM. Fig.6, Table 4) in hNMB-receptor transfected Balb-3T3 or NCI-H1299 cells and therefore based on potency, they were 10 times more selective for hNMB-receptor over hGRP-receptor.
4. Discussion
This study was designed to assess the ability of previously described Bn receptor antagonists, primarily investigated in nonhuman cells, to function as possible receptor antagonist at human receptors and be possibly useful for human studies. Although 35 different compounds (10 different chemical groups) were tested, this study was not designed to provide detailed structure-function insights, because new, novel Bn analogues were not systematically made to provide information in this area. A number of our results support the conclusion that several members of the 10 different chemical groups of putative Bn-receptor antagonists have sufficient affinity for hGRP-receptors and a few for hNMB-receptors, to be potentially generally useful. For hGRP-receptors, based on affinities, the putative Bn-antagonists could be divided into four broad categories. Category-one has very-high affinity for hGRP-receptor (i.e.<1 nM) includes des-Met14-Bn-esters [#17-18,20] and [d-Pro13, Ψ13-14]Bn-analogue [#24]. Category-two has high-affinity for hGRP-receptors (i.e.1-5 nM), includes des-Met14Bn-alkylamides [#10-12,14] and a des-Met14Bn-amide [#8]. Category-three has moderate-affinity for hGRP-receptors (i.e.6-30 nM) includes some Ψ13-14pseudopeptide-bond Bn-analogues [#5,6]. Category-four has low-very low affinities (i.e.>30 nM) and includes [d-Phe12]Bn-analogues, D-amino acid substituted-substance P-analogues, d-amino acid-substituted-somatostatin (SS)-analogues, cyclic-Bn peptides and peptoids. In contrast to hGRP-receptors, only the peptoids [analogues#33-34], had very high affinity (<1 nM) for hNMB-receptors with all other analogues showing very low affinity (i.e.>250 nM). Our results with hGRP-receptors have similarities and differences from previous studies. In the very-high affinity group (IC50<1 nM), #18([d-F-5, Phe6, D-Ala11]Bn(6-13)methylester), had hGRP-receptor affinities of 0.12-0.89 nM which are similar to reported for mGRP-receptor and >5-times higher than for rGRP-receptors (Table-5). Similarly, analogue #20 ([d-Phe6]Bn(6-13)ethylester) showed a higher affinity (IC50=0.14-0.28 nM) than reported for the GRP-receptor from mouse, guinea pig or rat (Table-5). Finally, analogue #24, ([(3-Ph-Pr6), His7, D-Ala11, D-Pro13,(Ψ13-14), Phe14]Bn(6-14)NH2), had a hGRP-receptor affinity of 0.22-0.23 nM, similar to reported in rat pancreas, 10-18-fold higher than human lung carcinoma cells, or human T47D breast cancer cells (IC50=4-10 nM)[46], but 220-fold lower than mGRP-receptors (Table-5). Our finding that [D-Phe6]Bn(6-13)amide [analogue #8] and various alkylamides [analogues#10,11] had high affinity (Category-two, 1-5 nM) for the hGRP-receptor agree with some studies in the literature[63,66,67] and are similar to their affinity for the mGRP-receptors (Table-5), however, differ from results for guinea pig and rGRP- receptors for which they have lower affinities (Table-5). Similarly, our results are in agreement with several studies, which report various [Ψ13-14]Bn pseudopeptides [#5,6] have moderately-high (6-30 nM) affinity for hGRP-receptors in various cells[67], as well as for the mGRP-receptor[67], however, in other studies [Ψ13-14]Bn-pseudopeptide #5 had much lower affinity for mouse, guinea pig and rat GRP-receptors (60-430 nM)[64]. Our findings that D-amino substituted-somatostatin- or-substance P, and [D-Phe12]Bn-analogues have low affinity (>100 nM) for hGRP-receptors are consistent with other studies in cells containing hGRP-receptor[52] as well as with studies on mouse, rat and guinea pig GRP-receptors (Table-5). Lastly, our results with the peptoid analogue [#33, PD176252] showing it has low affinity (170-213 nM) for hGRP-receptors differ from a previous study where it is reported to have a high affinity (Ki=1 nM) in one study[1] and a moderately-high affinity (20-30 nM) in a second[42]. In contrast, our results with the other putative Bn peptoid antagonist [analogue #34, PD168368] showing it has a very-high affinity (IC50=0.25-0.5 nM) for hNMB-receptors are consistent with one study[1], but not with other studies on human cells which showed a much lower affinity (IC50=15 nM)[52] as well as studies showing a much lower affinity (30-502 nM) in cells containing rNMB-receptors[42,52]. These results demonstrate there are marked species differences in affinities of these putative antagonists and their affinity for human Bn-receptors are frequently not predictable from studies of nonhuman Bn-receptors.
Based on their binding affinities, the ten groups of putative antagonists, as well as members within each group, varied greatly in their selectivity for hGRP-receptor or hNMB-receptor. Of the 35 different compounds tested, 23 were hGRP-receptor-selective, 4 were hNMB-receptor-preferring and 8 nonselective. The hGRP-receptor selective-putative antagonists varied greatly in selectivity (105-to >80,000-fold) with the most selective (>10,000-fold) being various des-Met14Bn-esters [#17,18,20]; the D-Pro13, Ψ13-14pseudo-bond peptide Bn-analogue [#24]; followed by a number of highly selective analogues (1,000-9,999-fold) including various des-Met14Bn-alkylamides [#10-16], the des-Met14amide-Bn-analogue [#8]; and a few Ψ13-14-pseudopeptide-bond analogues [#5-7]. In contrast, for the hNMB-receptor-preferring antagonists (peptoids, D-amino acid-substituted-somatostatin analogues), only peptoids (#33,34) had high selectivity for hNMB-receptor (>1,000-fold), with the PD168368 peptoid being more selective than the PD176252 (7,000 vs 1,000-fold). Comparison of these results with the literature showed some similarities and differences. As a general finding we found many Bn-putative antagonists, which were GRP-receptor selective in nonhuman studies, showed higher selectivity for the hGRP-receptor. The putative antagonists we found to be the most highly selective (>10,000 fold) for the hGRP-receptor [#17,18,20,24] were all reported to be less selective for GRP-receptor in nonhuman cells[10,33,61](Table-5). For the hNMB-receptor, the peptoids [#33,34] had the highest selectivity for the hNMB-receptor (1,000-7,200-fold), whereas analogue #33 (PD176252) was reported to show only a 6-147-fold selectivity for hNMB-receptor in other studies[1,42]. Furthermore, with the rNMB-receptor, analogue #33 showed only a 24-fold selectivity over GRP-receptor [1]. Similarly, for the peptoid analogue #34 (PD168368) our results showing a 2,000-7,000-fold hNMB-receptor selectivity differed from the 40-fold in other studies of hNMB- or rNMB-receptors[52]. In contrast, the d-amino acid-substituted-somatostatin analogue [#30], which we found had a modest (9-fold) selectivity for hNMB-receptor, our results agree with other studies with hNMB- and rNMB-receptors[47,52].
To determine whether these putative antagonists were pure antagonists in human cells or had agonist activity, their abilities to stimulate/inhibit activation of hGRP/hNMB-receptors were analyzed by assessing changes in phospholipase C activity, which both receptors are coupled to, in all species studied[3,4,24]. A detailed study of their abilities to activate and/or inhibit human Bn-receptors is important, because previous studies in nonhuman cells show these different Bn-analogues had marked differences in ability to function as antagonists or agonists in different species (Table-5)[10,21,24,25,34,63]. To determine antagonist/agonist activity, it was important to include in the assessment, cells with receptor densities similar to that found in native cells, because various studies show Bn-receptor densities can affect the expression of agonist activity[24,59,67]. To address this issue we assessed hGRP-receptor responses in native hGRP-receptor in HuTu-80 cells, and with the hNMB-receptor, for which no native cells expressing sufficient receptors were available, we assessed activation in the lung cancer cell line, NCI-H1299, which was transfected with hNMB-receptor at the same low levels seen with native hGRP-receptors in HuTu-80 cells.
For the 24 hGRP-receptor-selective analogues, none activated phospholipase C at high concentrations. These results demonstrated the hGRP-receptor behaves more like the mouse and guinea pig GRP-receptor in the structural peptide requirements for initiating activation, than the rGRP-receptor, where the requirements for activation are less stringent and a number of these compounds showed full/partial-agonist activity[63,64]. Similarly, none of the 4 hNMB-receptor-selective analogues demonstrated agonist activity at hNMB-receptor. In contrast, 11 of 24 hGRP-receptor-preferring compounds showed agonist activity at the hNMB-receptor. Three (#2,7.5,17) showed some agonist activity at high concentrations only and eight (#6,10,11,12,14,15,18,20) were partial agonists (efficacies-34-61%). In particular, the highly selective hGRP-receptor analogues #18 and #20 were reported to show no agonist activity in rat or guinea pig[24,33,63,64], however all showed partial agonist activity with the hNMB-receptor. However, a number of the highly selective hGRP-receptor compounds [#7,8,22,23,24] had no agonist activity at the hNMB-receptor. Furthermore, the selectivity based on their potencies for inhibiting hGRP-receptor/hNMB-receptor activation (160-970-fold) demonstrating a number of this compounds are excellent candidates for studies investigating the role of hGRP-receptor in human responses. Lastly, the peptoids [#33,34], not only did not show agonist affinity for hNMB-receptor, but also not for the hGRP-receptor and they functioned as a potent antagonists for the hNMB-receptor, which is similar to the findings in rat and mouse GRP-receptors[52].
Although this initial study was not designed to provide detailed insights into the structure-function relations of agonist/antagonism at the hGRP- and/or hNMB-receptors, which would have required synthesis of a number of new, potentially novel Bn analogues related to those previously described and examined in this study, our results do prove some insights in this area. Previously, it was proposed from studies of constrained bombesin-analogues that there were marked differences in the active conformations of agonists for high affinity binding and/or activation for the NMB- and GRP-receptors[6,24]. Our results are consistent with this proposal, because different compounds varied markedly in their abilities to activate these two receptors and function as an agonist, partial agonist or antagonist for a given compound. Previous studies[7,24,61,63] showed the chemical nature of substitutions in the COOH terminal-amino acids of these compounds was critically important for either/or affinity/activity of NMB- and GRP-receptors. Some of our results are consistent with this conclusion, because Group V and VI analogues functioned as partial agonists (34-61%) at the hNMB-receptor, but were pure GRP-receptor antagonists. The chain-length of the alkyl group in Group V-analogues played an important role in determining the degree of partial agonism. These results suggest that increasing chain length of the alkyl-moiety either changes the peptide conformation or binds to additional receptor sites that are important in determining affinity for the GRP-receptor only, whereas with the NMB-receptor, they are important primarily in determining receptor activation and not changes in affinity. Previous studies of the affinity/activation of GRP-receptors have led to the proposal that NH2-terminus of agonists adopt a folded-β-pleated-sheet conformation, which is stabilized by hydrogen-bonding[7,24,61,63]. This model proposed the partial agonism/antagonism seen in various studies with synthetic Bn-related peptides[7,24,61,63], occurred because the COOH-terminal chemical alterations resulting in partial alterations of this conformation by promoting increased rotational-freedom/flexibility [7,24,61,63]. No such model exists for NMB-receptor, but our data suggest there are important difference in structural requirements for activation of the hGRP-receptor and hNMB-receptor that must be accounted for by any NMB-receptor model. The functional result of these differences is the peptide structural requirements for activation of the hNMB-receptor are less stringent that for the hGRP-receptor. These results, coupled with previous studies of the GRP-receptor showing a marked species variation in peptide structural requirements for the activation of the GRP-receptor[7,24,61,63], demonstrate that the Bn-family of receptors show great variability between both receptor subtypes and within a receptor subtype for different species, for peptide structural determinants of activation.
In conclusion, this study provides insights into the most useful potential receptor antagonists for studies involving Bn-receptors in humans. Our results show that analogue #24 ([(3-Ph-Pr6), His7, D-Ala11, D-Pro13, Ψ(13-14), Phe14]Bn(6-14)NH2) and analogue #7 [D-Phe6, Leu13, Ψ(CH2NH), Cpa14]Bn(6-14)], are the most selective, potent hGRP-receptor antagonists, and to a lesser degree analogue #7.5 ([D-Tpi6, Leu13, Ψ(CH2NH), Leu14]Bn(6-14) [RC-3095]), with each showing no agonist activity at either hBn-receptor. In the case of hNMB-receptor, the antagonist of choice is analogue #34 (peptoid-PD168368). A large number of other analogues have high selectivity for the hGRP-receptor, some of which have been used in vivo and in vitro studies in humans[19,57], however most have some agonist activity for hNMB-receptor at high concentrations, which could limit their general utility in various human studies.
Acknowledgments
This work was partially supported by intramural funds of NIDDK and NCI, NIH.
Abbreviations
- Bn
bombesin
- BRS-3
bombesin receptor subtype 3
- BSA
bovine serum albumin fraction V
- CNS
central nervous system
- DMEM
Dulbecco's minimum essential medium
- FBS
fetal bovine serum
- GI
gastrointestinal
- GPCR
G protein-coupled receptor
- GRP
gastrin-releasing peptide
- GRP-receptor
gastrin-releasing peptide receptor
- IP
inositol-phosphates
- NMB
neuromedin B
- NMB-receptor
neuromedin B receptor
- PBS
phosphate-buffered saline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ashwood V, Brownhill V, Higginbottom M, Horwell DC, Hughes J, Lewthwaite RA, et al. PD 176252 - The first high affinity non-peptide gastrin-releasing peptide (BB2) receptor antagonist. Bioorg Med Chem. 1998;8:2589–94. doi: 10.1016/s0960-894x(98)00462-4. [DOI] [PubMed] [Google Scholar]
- 2.Benya RV, Fathi Z, Pradhan T, Battey JF, Kusui T, Jensen RT. Gastrin-releasing peptide receptor-induced internalization, down-regulation, desensitization and growth: Possible role of cAMP. Mol Pharmacol. 1994;46(2):235–45. [PubMed] [Google Scholar]
- 3.Benya RV, Kusui T, Pradhan TK, Battey JF, Jensen RT. Expression and characterization of cloned human bombesin receptors. Mol Pharmacol. 1995;47:10–20. [PubMed] [Google Scholar]
- 4.Benya RV, Wada E, Battey JF, Fathi Z, Wang LH, Mantey SA, et al. Neuromedin B receptors retain functional expression when transfected into BALB 3T3 fibroblasts: analysis of binding, kinetics, stoichiometry, modulation by guanine nucleotide-binding proteins, and signal transduction and comparison with natively expressed receptors. Mol Pharmacol. 1992;42(6):1058–68. [PubMed] [Google Scholar]
- 5.Corjay MH, Dobrzanski DJ, Way JM, Viallet J, Shapira H, Worland P, et al. Two distinct bombesin receptor subtypes are expressed and functional in human lung carcinoma cells. J Biol Chem. 1991;266:18771–9. [PubMed] [Google Scholar]
- 6.Coy DH, Jiang NY, Kim SH, Moreau JP, Lin JT, Frucht H, et al. Covalently cyclized agonist and antagonist analogues of bombesin and related peptides. J Biol Chem. 1991;266(25):16441–7. [PubMed] [Google Scholar]
- 7.Coy DH, Jiang NY, Sasaki Y, Taylor J, Moreau JP, Wolfrey WT, et al. Probing peptide backbone function in bombesin. A reduced peptide bond analogue with potent and specific receptor antagonist activity. J Biol Chem. 1988;263(11):5056–60. [PubMed] [Google Scholar]
- 8.Coy DH, Mungan Z, Rossowski WJ, Cheng BL, Lin JT, Mrozinski JE, Jr et al. Development of a potent bombesin receptor antagonist with prolonged in vivo inhibitory activity on bombesin-stimulated amylase and protein release in the rat. Peptides. 1992;13:775–81. doi: 10.1016/0196-9781(92)90186-7. [DOI] [PubMed] [Google Scholar]
- 9.Coy DH, Taylor JE, Jiang NY, Kim SH, Wang LH, Huang SC, et al. Short-chain pseudopeptide bombesin receptor antagonists with enhanced binding affinities for pancreatic acinar and Swiss 3T3 cells display strong antimitotic activity. J Biol Chem. 1989;264:14691–7. [PubMed] [Google Scholar]
- 10.Coy DH, Wang LH, Jiang NZ, Jensen RT. Short chain bombesin pseudopeptides which are potent and more general bombesin receptor antagonists. Eur J Pharmacol. 1990;190(12):31–8. doi: 10.1016/0014-2999(90)94109-b. [DOI] [PubMed] [Google Scholar]
- 11.Cuttitta F, Carney DN, Mulshine J, Moody TW, Fedorko J, Fischler A, et al. Bombesin-like peptides can function as autocrine growth factors in human small-cell lung cancer cells. Nature. 1985;316:823–6. doi: 10.1038/316823a0. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson KE, Uemara N, Sekar MC, McDaniel HB, Anderson W, Coy DH, et al. Partial agonist activity of the bombesin-receptor antagonist [Leu14-psi-CH2-NH-Leu13]-bombesin in frog peptic cells. Biochem Biophys Res Commun. 1988;157:1154–8. doi: 10.1016/s0006-291x(88)80994-x. [DOI] [PubMed] [Google Scholar]
- 13.Eden JM, Hall MD, Higginbottom M, Horwell DC, Howson W, Hughes J, et al. PD 165929 - The first high affinity non-peptide neuromedin-B (NMB) receptor selective antagonist. Bioorg Med Chem Lett. 1996;6:2617–22. [Google Scholar]
- 14.Fathi Z, Corjay MH, Shapira H, Wada E, Benya R, Jensen R, et al. BRS-3: novel bombesin receptor subtype selectively expressed in testis and lung carcinoma cells. J Biol Chem. 1993;268(8):5979–84. [PubMed] [Google Scholar]
- 15.Frucht H, Gazdar AF, Park JA, Oie H, Jensen RT. Characterization of functional receptors for gastrointestinal hormones on human colon cancer cells. Cancer Res. 1992;52:1114–22. [PubMed] [Google Scholar]
- 16.Gonzalez N, Hocart SJ, Portal-Nunez S, Mantey SA, Nakagawa T, Zudaire E, et al. Molecular basis for agonist selectivity and activation of the orphan bombesin receptor subtype 3 receptor. J Pharmacol Exp Ther. 2008;324:463–74. doi: 10.1124/jpet.107.132332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez N, Moody TW, Igarashi H, Ito T, Jensen RT. Bombesin-related peptides and their receptors: recent advances in their role in physiology and disease states. Curr Opin Endocrinol Diabetes Obes. 2008;15:58–64. doi: 10.1097/MED.0b013e3282f3709b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinz-Erian P, Coy DH, Tamura M, Jones SW, Gardner JD, Jensen RT. [D-Phe12]bombesin analogues: a new class of bombesin receptor antagonists. Am J Physiol. 1987;252:G439–G442. doi: 10.1152/ajpgi.1987.252.3.G439. [DOI] [PubMed] [Google Scholar]
- 19.Hildebrand P, Lehmann FS, Ketterer S, Christ AD, Stingelin T, Beltinger J, et al. Regulation of gastric function by endogenous gastrin releasing peptide in humans: studies with a specific gastrin releasing peptide receptor antagonist. Gut. 2001;49:23–8. doi: 10.1136/gut.49.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horwell DC, Howson W, Rees DC. Peptoid design. Drug Des Discov. 1994;12:63–75. [PubMed] [Google Scholar]
- 21.Houben H, Denef C. Effect of the bombesin receptor blockers [Leu13, psi CH2NH-Leu14]bombesin and N-pivaloyl GRP(20-25) alkylamide (L 686,095-001C002) on basal and neuromedin C-stimulated PRL and GH release in pituitary cell aggregates. Peptides. 1991;12:371–4. doi: 10.1016/0196-9781(91)90028-n. [DOI] [PubMed] [Google Scholar]
- 22.Jensen JA, Carroll RE, Benya RV. The case for gastrin-releasing peptide acting as a morphogen when it and its receptor are aberrantly expressed in cancer. Peptides. 2001;22:689–99. doi: 10.1016/s0196-9781(01)00380-1. [DOI] [PubMed] [Google Scholar]
- 23.Jensen RT. Receptors on pancreatic acinar cells. In: Johnson LR, Jacobson ED, Christensen J, Alpers DH, Walsh JH, editors. Physiology of the Gastrointestinal Tract. Third. Vol. 2. New York: Raven Press; 1994. pp. 1377–446. [Google Scholar]
- 24.Jensen RT, Battey JF, Spindel ER, Benya RV. International Union of Pharmacology. LVIII. Mammalian Bombesin Receptors: Nomenclature, distribution, pharmacology, signaling and functions in normal and disease states. Pharmacol Rev. 2008;60:1–42. doi: 10.1124/pr.107.07108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen RT, Coy DH. Progress in the development of potent bombesin receptor antagonists. Trends Pharmacol Sci. 1991;12(1):13–9. doi: 10.1016/0165-6147(91)90483-9. [DOI] [PubMed] [Google Scholar]
- 26.Jensen RT, Heinz-Erian P, Mantey S, Jones SW, Gardner JD. Characterization of the ability of various substance P antagonists to inhibit action of bombesin. Am J Physiol. 1988;254:G883–G890. doi: 10.1152/ajpgi.1988.254.6.G883. [DOI] [PubMed] [Google Scholar]
- 27.Jensen RT, Jones SW, Folkers K, Gardner JD. A synthetic peptide that is a bombesin receptor antagonist. Nature. 1984;309:61–3. doi: 10.1038/309061a0. [DOI] [PubMed] [Google Scholar]
- 28.Jensen RT, Moody TW. Bombesin-related peptides and neurotensin: effects on cancer growth/proliferation and cellular signaling in cancer. In: Kastin AJ, editor. Handbook of Biologically active peptides. Amsterdam: Elsevier; 2006. pp. 429–34. [Google Scholar]
- 29.Kroog GS, Jensen RT, Battey JF. Mammalian bombesin receptors. Med Res Rev. 1995;15(5):389–417. doi: 10.1002/med.2610150502. [DOI] [PubMed] [Google Scholar]
- 30.Ladenheim EE, Jensen RT, Mantey SA, McHugh PR, Moran TH. Distinct distributions of bombesin receptor subtypes in the rat central nervous system. Brain Res. 1992;593:168–78. doi: 10.1016/0006-8993(92)91305-x. [DOI] [PubMed] [Google Scholar]
- 31.Leban JJ, Kull FC, Jr, Landavazo A, Stockstill B, McDermed JD. Development of potent gastrin-releasing peptide antagonists having a D-Pro-psi (CH2NH)-Phe-NH2 C terminus. Proc Natl Acad Sci U S A. 1993;90:1922–6. doi: 10.1073/pnas.90.5.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann FS, Beglinger C. Gastrin-releasing peptide. In: Kastin AJ, editor. Handbook of Biologically active peptides. Amsterdam: Elsevier; 2006. pp. 1047–55. [Google Scholar]
- 33.Lin JT, Coy DH, Mantey SA, Jensen RT. Peptide structural requirements for antagonism differ between the two mammalian bombesin receptor subtypes. J Pharmacol Exp Ther. 1995;275:285–95. [PubMed] [Google Scholar]
- 34.Maggi CA, Coy DH, Giuliani S. Effect of [D-Phe6] bombesin (6-13) methylester, a bombesin receptor antagonist, towards bombesin-induced contractions in the guinea-pig and rat isolated urinary bladder. J Auton Pharmacol. 1992;12:215–22. doi: 10.1111/j.1474-8673.1992.tb00335.x. [DOI] [PubMed] [Google Scholar]
- 35.Maina T, Nock BA, Zhang H, Nikolopoulou A, Waser B, Reubi JC, et al. Species differences of bombesin analog interactions with GRP-R define the choice of animal models in the development of GRP-R-targeting drugs. J Nucl Med. 2005;46:823–30. [PubMed] [Google Scholar]
- 36.Mantey S, Frucht H, Coy DH, Jensen RT. Characterization of bombesin receptors using a novel, potent, radiolabeled antagonist that distinguishes bombesin receptor subtypes. Mol Pharmacol. 1993;45:762–74. [PubMed] [Google Scholar]
- 37.Mantey SA, Coy DH, Entsuah LK, Jensen RT. Development of bombesin analogs with conformationally restricted amino acid substitutions with enhanced selectivity for the orphan receptor human bombesin receptor subtype 3. J Pharmacol Exp Ther. 2004;310:1161–70. doi: 10.1124/jpet.104.066761. [DOI] [PubMed] [Google Scholar]
- 38.Mantey SA, Coy DH, Pradhan TK, Igarashi H, Rizo IM, Shen L, et al. Rational design of a peptide agonist that interacts selectively with the orphan receptor, bombesin receptor subtype 3. J Biol Chem. 2001;276:9219–29. doi: 10.1074/jbc.M008737200. [DOI] [PubMed] [Google Scholar]
- 39.Mantey SA, Gonzalez N, Schumann M, Pradhan TK, Shen L, Coy DH, et al. Identification of bombesin receptor subtype-specific ligands: effect of N-methyl scanning, truncation, substitution, and evaluation of putative reported selective ligands. J Pharmacol Exp Ther. 2006;319:980–9. doi: 10.1124/jpet.106.107011. [DOI] [PubMed] [Google Scholar]
- 40.Mantey SA, Weber HC, Sainz E, Akeson M, Ryan RR, Pradhan TK, et al. Discovery of a high affinity radioligand for the human orphan receptor, bombesin receptor subtype 3, which demonstrates it has a unique pharmacology compared to other mammalian bombesin receptors. J Biol Chem. 1997;272(41):26062–71. doi: 10.1074/jbc.272.41.26062. [DOI] [PubMed] [Google Scholar]
- 41.Moody TW, Chan D, Fahrenkrug J, Jensen RT. Neuropeptides as autocrine growth factors in cancer cells. Curr Pharm Des. 2003;9:495–509. doi: 10.2174/1381612033391621. [DOI] [PubMed] [Google Scholar]
- 42.Moody TW, Jensen RT, Garcia L, Leyton J. Nonpeptide neuromedin B receptor antagonists inhibit the proliferation of C6 cells. Eur J Pharmacol. 2000;409:133–42. doi: 10.1016/s0014-2999(00)00828-1. [DOI] [PubMed] [Google Scholar]
- 43.Moody TW, Mantey SA, Pradhan TK, Schumann M, Nakagawa T, Martinez A, et al. Development of high affinity camptothecin-bombesin conjugates that have targeted cytotoxicity for bombesin receptor-containing tumor cells. J Biol Chem. 2004;279:23580–9. doi: 10.1074/jbc.M401938200. [DOI] [PubMed] [Google Scholar]
- 44.Moody TW, Merali Z. Bombesin-like peptides and associated receptors within the brain: distribution and behavioral implications. Peptides. 2004;25:511–20. doi: 10.1016/j.peptides.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Moody TW, Staley J, Zia F, Coy DH, Jensen RT. Neuromedin B binds with high affinity, elevates cytosolic calcium and stimulates the growth of small cell lung cancer cell lines. J Pharmacol Exp Ther. 1992;263:311–7. [PubMed] [Google Scholar]
- 46.Moody TW, Venugopal R, Hu V, Gozes Y, McDermed J, Leban JJ. BW 1023U90: a new GRP receptor antagonist for small-cell lung cancer cells. Peptides. 1996;17:1337–43. doi: 10.1016/s0196-9781(96)00195-7. [DOI] [PubMed] [Google Scholar]
- 47.Orbuch M, Taylor JE, Coy DH, Mrozinski JE, Jr, Mantey SA, Battey JF, et al. Discovery of a novel class of neuromedin B receptor antagonists: substituted somatostatin analogues. Mol Pharmacol. 1993;44(4):841–50. [PubMed] [Google Scholar]
- 48.Patel O, Shulkes A, Baldwin GS. Gastrin-releasing peptide and cancer. Biochim Biophys Acta. 2006;1766:23–41. doi: 10.1016/j.bbcan.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Pradhan TK, Katsuno T, Weber HC, Mantey SA, Coy DH, Jensen RT. Discovery of a universal ligand that interacts with high affinity with all four classes of bombesin (BN) receptors. Gastroenterology. 1997;112:A474. [Google Scholar]
- 50.Reubi JC. Peptide receptors as molecular targets for cancer diagnosis and therapy. Endocr Rev. 2003;24:389–427. doi: 10.1210/er.2002-0007. [DOI] [PubMed] [Google Scholar]
- 51.Reubi JC, Wenger S, Schumuckli-Maurer J, Schaer JC, Gugger M. Bombesin receptor subtypes in human cancers: detection with the universal radoligand (125)I-[D-TYR(6), beta-ALA(11),PHE(13), NLE(14)] bombesin(6-14) Clin Cancer Res. 2002;8:1139–46. [PubMed] [Google Scholar]
- 52.Ryan RR, Katsuno T, Mantey SA, Pradhan TP, Weber HC, Battey JF, et al. Comparative pharmacology of a nonpeptoid neuromedin B antagonist PD 168368. J Pharmacol Exp Ther. 1999;290:1202–11. [PubMed] [Google Scholar]
- 53.Ryan RR, Taylor JE, Daniel JL, Cowan A. Pharmacological profiles of two bombesin analogues in cells transfected with human neuromedin B receptors. Eur J Pharmacol. 1996;306(13):307–14. doi: 10.1016/0014-2999(96)00223-3. [DOI] [PubMed] [Google Scholar]
- 54.Ryan RR, Weber HC, Hou W, Sainz E, Mantey SA, Battey JF, et al. Ability of various bombesin receptor agonists and antagonists to alter intracellular signaling of the human orphan receptor BRS-3. J Biol Chem. 1998;273:13613–24. doi: 10.1074/jbc.273.22.13613. [DOI] [PubMed] [Google Scholar]
- 55.Ryan RR, Weber HC, Mantey SA, Hou W, Hilburger ME, Pradhan TK, et al. Pharmacology and intracellular signaling mechanisms of the native human orphan receptor BRS-3 in lung cancer cells. J Pharmacol Exp Ther. 1998;287:366–80. [PubMed] [Google Scholar]
- 56.Saeed ZA, Huang SC, Coy DH, Jiang NY, Heinz-Erian P, Mantey SA, et al. Effect of substitutions in position 12 of bombesin on antagonist activity. Peptides. 1989;10(3):597–603. doi: 10.1016/0196-9781(89)90149-6. [DOI] [PubMed] [Google Scholar]
- 57.Schwartsmann G, DiLeone LP, Horowitz M, Schunemann D, Cancella A, Pereira AS, et al. A phase I trial of the bombesin/gastrin-releasing peptide (BN/GRP) antagonist RC3095 in patients with advanced solid malignancies. Invest New Drugs. 2006;24:403–12. doi: 10.1007/s10637-006-6886-5. [DOI] [PubMed] [Google Scholar]
- 58.Tokita K, Hocart SJ, Katsuno T, Mantey SA, Coy DH, Jensen RT. Tyrosine 220 in the fifth transmembrane domain of the neuromedin B receptor is critical for the high selectivity of the peptoid antagonist PD168368. J Biol Chem. 2001;276:495–504. doi: 10.1074/jbc.M006059200. [DOI] [PubMed] [Google Scholar]
- 59.Tsuda T, Kusui T, Hou W, Benya RV, Akeson MA, Kroog GS, et al. Effect of gastrin-releasing peptide receptor number on receptor affinity, coupling, degradation and receptor modulation. Mol Pharmacol. 1997;51(5):721–32. doi: 10.1124/mol.51.5.721. [DOI] [PubMed] [Google Scholar]
- 60.von Schrenck T, Heinz-Erian P, Moran T, Mantey SA, Gardner JD, Jensen RT. Neuromedin B receptor in esophagus: evidence for subtypes of bombesin receptors. Am J Physiol. 1989;256:G747–G758. doi: 10.1152/ajpgi.1989.256.4.G747. [DOI] [PubMed] [Google Scholar]
- 61.von Schrenck T, Wang LH, Coy DH, Villanueva ML, Mantey S, Jensen RT. Potent bombesin receptor antagonists distinguish receptor subtypes. Am J Physiol. 1990;259:G468–G473. doi: 10.1152/ajpgi.1990.259.3.G468. [DOI] [PubMed] [Google Scholar]
- 62.Wang LH, Battey JF, Wada E, Lin JT, Mantey S, Coy DH, et al. Activation of neuromedin B-preferring bombesin receptors on rat glioblastoma C-6 cells increases cellular Ca2+ and phosphoinositides. Biochem J. 1992;286:641–8. doi: 10.1042/bj2860641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang LH, Coy DH, Taylor JE, Jiang NY, Kim SH, Moreau JP, et al. Desmethionine alkylamide bombesin analogues: a new class of bombesin receptor antagonists with a potent antisecretory activity in pancreatic acini and antimitotic activity in Swiss 3T3 cells. Biochemistry (Mosc) 1990;29(3):616–22. doi: 10.1021/bi00455a004. [DOI] [PubMed] [Google Scholar]
- 64.Wang LH, Coy DH, Taylor JE, Jiang NY, Moreau JP, Huang SC, et al. Des-Met carboxyl-terminally modified analogues of bombesin function as potent bombesin receptor antagonists, partial agonists, or agonists. J Biol Chem. 1990;265(26):15695–703. [PubMed] [Google Scholar]
- 65.Weber HC. Regulation and signaling of human bombesin receptors and their biological effects. Curr Opin Endocrinol Diabetes Obes. 2009;16:66–71. doi: 10.1097/med.0b013e32831cf5aa. [DOI] [PubMed] [Google Scholar]
- 66.Williams BY, Schonbrunn A. Bombesin receptors in a human duodenal tumor cell line: binding properties and function. Cancer Res. 1994;54(3):818–24. [PubMed] [Google Scholar]
- 67.Wu JM, Hoang DO, Feldman RI. Differential activation of human gastrin-releasing peptide receptor-mediated responses by bombesin analogs. Mol Pharmacol. 1995;47(4):871–81. [PubMed] [Google Scholar]