Abstract
Effective B cell–mediated immunity and antibody responses often require help from CD4+ T cells. It is thought that a distinct CD4+ effector T cell subset, called T follicular helper cells (TFH), provides this help; however, the molecular requirements for TFH differentiation are unknown. We found that expression of the transcription factor Bcl6 in CD4+ T cells is both necessary and sufficient for in vivo TFH differentiation and T cell help to B cells in mice. In contrast, the transcription factor Blimp-1, an antagonist of Bcl6, inhibits TFH differentiation and help, thereby preventing B cell germinal center and antibody responses. These findings demonstrate that TFH cells are required for proper B cell responses in vivo and that Bcl6 and Blimp-1 play central but opposing roles in TFH differentiation.
Each lineage of effector CD4+ T cells (TH1, TH2, TH17, and Treg) is defined and controlled by a unique master regulator transcription factor (T-bet, GATA3, RORγt, and Foxp3, respectively) (1). A proposed fifth effector subset, T follicular helper (TFH) cells, is thought to provide help for the generation of B cell–mediated immune responses, including class switch recombination, germinal center differentiation, and affinity maturation (2). Here, we identified Bcl6 as a TFH master regulator and found that germinal center formation does not occur in the absence of TFH cells.
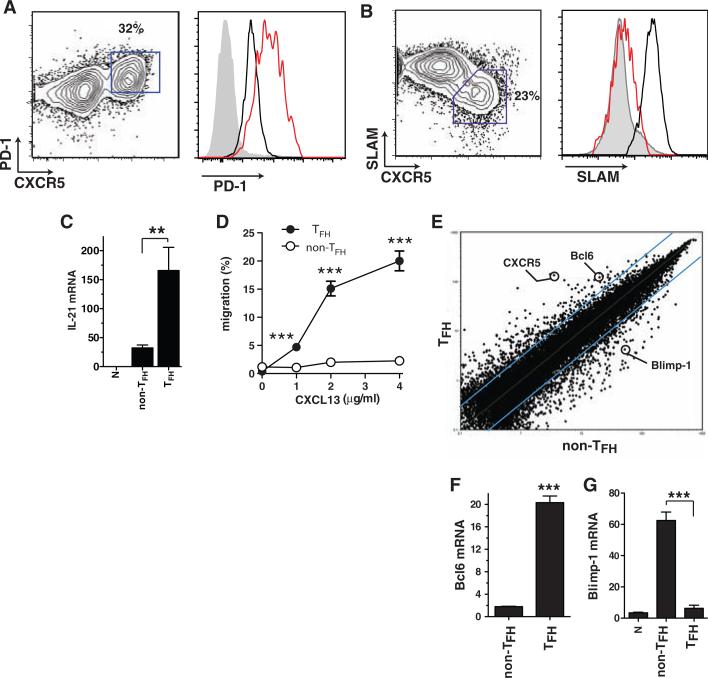
TFH cells are well described phenotypically in humans, and more recently in mice, as expressing high levels of the chemokine receptor CXCR5 and molecules such as ICOS, PD1, interleukin-21 (IL-21), and BTLA (2–9). Given that CD4+ T cells can up-regulate CXCR5 and/or ICOS after activation (2, 10), it is important to phenotypically distinguish TFH from highly activated CD4+ T cells. We identified TFH cells in mice in the context of acute infection with lymphocytic choriomeningitis virus (LCMV) by adoptively transferring T cell receptor (TCR) transgenic T cells specific for the LCMV epitope gp66-77 in the context of major histocompatibility complex (MHC) class II molecule I-Ab (SMtg). TFH cells were CXCR5high ICOShigh PD1high BTLAhigh CD200high SLAMlow (Fig. 1, A and B, and fig. S1) and capable of producing IL-21 (Fig. 1C). We confirmed these results for polyclonal LCMV-specific CD4+ T cell responses (fig. S2). CXCR5 is the receptor for the B cell follicle chemokine CXCL13 (11), and TFH cells were selectively able to migrate in response to CXCL13 in vitro (Fig. 1D), consistent with the importance of CXCR5 for TFH (6, 12).
Fig. 1.
Bcl6 is a TFH-specific transcription factor. Naïve SMtg CD4+ T cells were transferred into B6 mice. In all panels, splenocytes were analyzed 8 days after infection with LCMV. (A and B) SMtg expression of CXCR5 and PD-1 (A) or SLAM (CD150) (B). SMtg+ (CD45.1+) CD4+ gated cells are shown. CXCR5high TFH cells are boxed in fluorescence-activated cell sorter (FACS) plots. Histogram overlays depict TFH cells (red) as well as naïve CD4+ T cells (gray) and CXCR5low non-TFH SMtg cells (black). Data are representative of more than 10 independent experiments. (C) IL-21 mRNA in SMtg CD4+ T cells, normalized to the β-actin mRNA level (×10–4). **P = 0.008. (D) In vitro chemotaxis toward CXCL13 (BLC) by ex vivo SMtg CD4+ T cells. Results are expressed as percentages of SLAMlow TFH SMtg (solid circles) and SLAMhigh non-TFH SMtg (open circles) that migrated in a transwell assay. ***P ≤ 0.001. 1 μg, P = 0.001; 2 μg, P = 0.0006; 4 μg, P = 0.0006. Data are representative of three independent experiments; n = 2 per group. (E) Scatterplot of the average signal of biological replicates of TFH versus non-TFH SMtg gene expression microarray data. Blue lines indicate changes in gene expression by a factor of 3; 386 gene probes exhibited a factor of >3.0 increase in TFH. Data from one of two independent experiments are shown; n = 2 per group. (F and G) Quantitative reverse transcription PCR of Bcl6 (F) and Blimp-1 (G) mRNA expression, normalized to β-actin (×10–4). ***P < 0.0001. Data are representative of four independent experiments; n = 2 per group. Error bars in all graphs are SEM.
To understand how TFH differentiation is transcriptionally regulated, we performed gene expression microarray analysis of virus-specific TFH and non-TFH effector CD4+ T cells (Fig. 1E and figs. S3 and S4). Notably, the transcription factor B cell CLL/lymphoma 6 (Bcl6) was strongly up-regulated in TFH (Fig. 1E). This is in agreement with previous reports of elevated Bcl6 expression in murine and human TFH cells (3–5, 8). Furthermore, Blimp-1 (prdm1) was the most down-regulated transcription factor in TFH cells (fig. S4), consistent with a recent report (13). Bcl6 is essential for germinal center B cell differentiation (14–16), and Blimp-1 is well characterized as an antagonist of Bcl6 that can also be directly repressed by Bcl6 (16–20). Up-regulation of Bcl6 mRNA (Fig. 1F) and down-regulation of Blimp-1 mRNA (Fig. 1G) were confirmed by quantitative polymerase chain reaction (qPCR). Bcl6 protein expression was detected in germinal center CD4+ T cells (fig. S2), consistent with previous observations in human lymphoid tissue (4, 21).
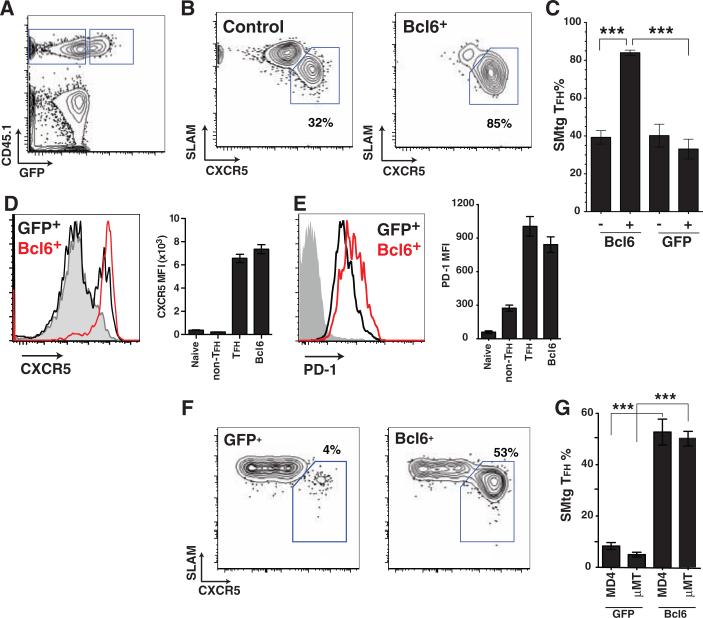
Although Bcl6 mRNA expression has been correlated with TFH, no experimental data supporting a specific role for Bcl6 in TFH differentiation have been reported. We expressed Bcl6 in SMtg CD4+ T cells via a retroviral vector (RV) with a bicistronic mRNA coexpressing green fluorescent protein (GFP) (fig. S5). Transduced Bcl6-RV+ SMtg and control untransduced SMtg CD4+ T cells were transferred into naïve C57BL/6 hosts, which were subsequently infected with LCMV, and TFH differentiation was examined (Fig. 2, A to E). Bcl6 expression drove nearly absolute TFH differentiation in vivo (80 to 90%; Fig. 2, B and C), in contrast to TFH differentiation in control untransduced (GFP–) SMtg cells in the same mice (Fig. 2B) or mice that received SMtg transduced with a control retrovirus expressing only GFP (GFP-RV+) and untransduced SMtg in equal proportions (Fig. 2C). Comparably striking results were seen in studies where only Bcl6-RV+ or GFP-RV+ SMtg CD4+ T cells were transferred into host mice (fig. S6). Bcl6 overexpression did not affect T cell expansion in vivo (fig. S5). Constitutive expression of Bcl6 drove up-regulation of CXCR5, PD-1, ICOS, CD200, and BTLA expression (Fig. 2, D and E, and fig. S6), as well as the inhibition of SLAM and Blimp-1 (fig. S6; see below). These results indicate that Bcl6 expression drives full TFH differentiation in vivo.
Fig. 2.
Bcl6 expression is sufficient for TFH differentiation in vivo. (A to E) Naïve SMtg CD4+ T cells were transduced with Bcl6-RV (Bcl6+) or left untransduced (control) and transferred into B6 mice subsequently infected with LCMV. (A) Gating of CD45.1+ untransduced SMtg (GFP–) and Bcl6-RV+ SMtg (GFP+) in the same host. CD4+ B220– gate is shown. (B) TFH (SLAMlow CXCR5high, boxed) and non-TFH (SLAMhigh CXCR5low) differentiation of untransduced SMtg (left) and Bcl6-RV+ SMtg (right). (C) Quantitation of SMtg TFH differentiation. Mice received Bcl6-RV+ SMtg and untransduced SMtg, or GFP-RV+ and untransduced SMtg. “–,” untransduced; “+,” transduced with indicated RV. ***P < 0.0001. Data are representative of three independent experiments; n = 4 per group. (D and E) CXCR5 expression (D) and PD-1 expression (E) on naïve CD4+ T cells (gray), GFP-RV+ SMtg (black), and Bcl6-RV+ SMtg (red). Bar graphs show mean fluorescence intensity (MFI) of naïve CD4+ T cells, GFP-RV+ SMtg non-TFH, GFP-RV+ SMtg TFH, and Bcl6-RV+ SMtg. For PD-1 MFI, non-TFH versus TFH or Bcl6-RV+, P < 0.05; TFH versus Bcl6-RV+, P > 0.05. Data are representative of three independent experiments; n = 4 to 6 per group. (F and G) GFP-RV+ or Bcl6-RV+ SMtg cells were adoptively transferred separately into B cell–deficient mice (μMT) or HEL-specific BCR transgenic mice (MD4) on a μMT background. Host mice were subsequently infected with LCMV. Each group is a composite of three experiments; n = 2 (GFP-RV+ μMT), 6 (Bcl6-RV+ MD4), 6 (Bcl6-RV+ μMT), or 8 (GFP-RV+ MD4) per group. (F) Differentiation of SMtg CD4+ T cells in MD4 BCR transgenic mice. TFH cells (SLAMlow CXCR5high) are boxed. CD4+ B220– CD45.1+ GFP+ gate is shown. (G) Quantitation of SMtg TFH differentiation. GFP-RV+ versus Bcl6-RV+ in MD4, ***P < 0.0001. GFP-RV+ versus Bcl6-RV+ in μMT, ***P < 0.0001.
TFH differentiation is known to require the presence of B cells and is thought to require the presence of antigen-specific B cells (6). We thus hypothesized that Bcl6 expression induced by interaction with antigen-specific B cells could be the event that commits a T cell to TFH differentiation. To test this, we examined whether Bcl6 expression in CD4+ T cells was sufficient to drive TFH differentiation in μMT B cell–deficient mice and in B cell receptor (BCR) transgenic mice of an irrelevant specificity (MD4, specific for hen egg lysozyme). GFP-RV+ SMtg CD4+ T cells failed to differentiate into TFH in μMT or MD4 mice infected with LCMV (Fig. 2, F and G), which demonstrates that TFH differentiation in the context of a viral infection is dependent on the presence of antigen-specific B cells. In contrast, Bcl6-RV+ SMtg cells differentiated into TFH in the absence of antigen-specific B cells or even in the total absence of B cells (Fig. 2, F and G). These results indicate that cognate T-B interactions induce Bcl6 expression in CD4+ T cells and that Bcl6 is sufficient to drive TFH differentiation, even in the absence of such interactions.
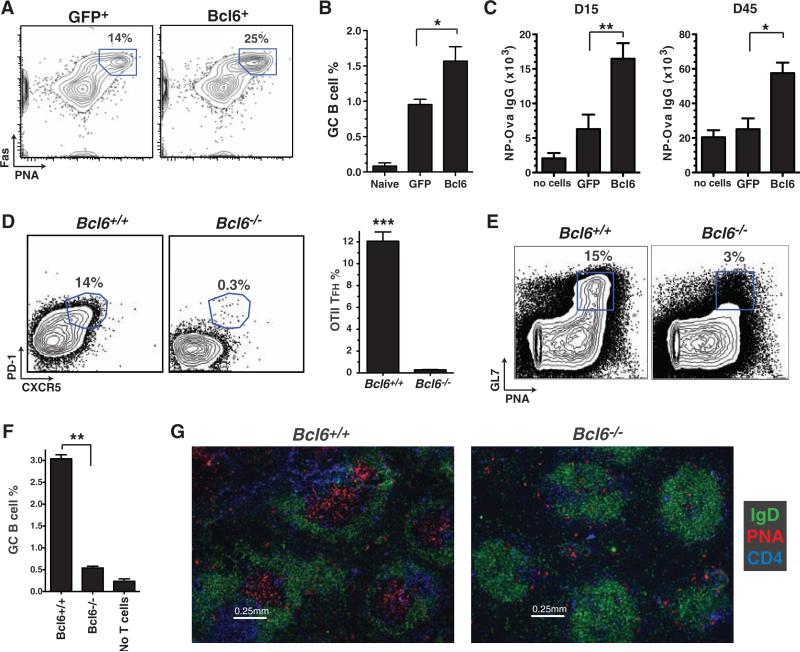
TFH cells are thought to provide B cell help in vivo (2, 22). We assessed the capacity of Bcl6-RV+ SMtg CD4+ T cells to help B cells in vivo by examining germinal center development in LCMV-infected mice. Overexpression of Bcl6 increased the already robust frequency of germinal center B cells after LCMV infection (Fig. 3, A and B). As an additional measure of B cell help, we also examined the role of Bcl6 in T cell–dependent antibody production. Constitutive expression of Bcl6 in OT-II CD4+ T cells enhanced NP-Ova serum immunoglobulin G (IgG) responses (Fig. 3C and fig. S7), which were sustained (Fig. 3C). Our results suggest that Bcl6 was specifically enhancing TFH differentiation and not skewing the TH1/TH2 profile of the CD4+ T cells, because all IgG isotypes were enhanced in the mice receiving Bcl6-expressing OT-II CD4+ T cells, with the strong IgG1 dominance maintained (fig. S7).
Fig. 3.
Bcl6 expression is necessary for inducing TFH B cell help in vivo. (A) Germinal center B cells (PNA+ Fas+, gated) in mice that received GFP-RV+ or Bcl6-RV+ SMtg CD4+ T cells and were subsequently infected with LCMV, analyzed at day 8 and gated on activated B cells (B220+ IgDlow). (B) Frequency of germinal center B cells of total splenocytes; n = 4 per group. Data are representative of three independent experiments. *P = 0.029. (C) GFP-RV+ or Bcl6-RV+ OT-II CD4+ T cells were transferred into B6 mice subsequently immunized with NP-Ova in alum. Control mice were immunized but received no OT-II cells. NP-Ova enzyme-linked immunosorbent assay (ELISA) was performed at day 15 and day 45; n = 6 per group. Data are representative of two independent experiments. Day 15 endpoint ELISA titers, **P = 0.008; day 45 endpoint ELISA titers, *P = 0.017.(D) Bcl6+/+ or Bcl6–/– OT-II CD4+ Tcells were transferred into congenically mismatched B6 mice subsequently immunized with Ova in alum. Splenocytes were analyzed 6 days after immunization; n = 4 per group. Data are representative of four independent experiments. OT-II+ CD44high gate is shown. Quantitation of OT-II TFH differentiation is also shown. ***P < 0.0001. (E to G) Bcl6+/+ or Bcl6–/– OT-II CD4+ T cells were cotransferred with B1-8 B cells into Icos–/– mice subsequently immunized with NP-Ova in alum; n = 2 per group. Data are representative of two independent experiments. (E) Germinal center B cells (PNA+ GL7+, boxed) 7 days after immunization. TCRβ– IgDlow gate is shown. (F) Quantitation of GC B cells as percent of spleen. **P = 0.0015. (G) Germinal center histology. Spleen sections were stained with IgD (green), PNA (red), and CD4 (blue).
The results of these experiments showed that Bcl6 expression was sufficient to drive the differentiation of functional TFH. To test whether Bcl6 was also necessary for TFH differentiation, we examined Bcl6–/– CD4+ T cells. Bcl6–/– mice have an abundance of highly activated CD4+ T cells (fig. S8) and succumb to early mortality (14, 15). To circumvent these issues, we transferred Bcl6+/+ or Bcl6–/– OT-II bone marrow into irradiated C57BL/6 recipients (fig. S8). Bcl6–/– OT-II CD4+ T cells obtained from chimeric mice did not exhibit lymphoproliferation or spontaneous activation upon transfer into C57BL/6 mice (fig. S8, D to F). Bcl6–/– or Bcl6+/+ OT-II recipient mice were subsequently immunized with Ova in alum. Strikingly, Bcl6–/– OT-II CD4+ T cells did not differentiate into TFH cells (Fig. 3D). We hypothesized that if TFH cells are necessary for B cell help in vivo, a cell-intrinsic CD4+ T cell block in TFH differentiation should result in a failure to generate antigen-specific B cell responses such as germinal center formation. To test this hypothesis, we transferred Bcl6–/– or Bcl6+/+ OT-II CD4+ T cells into Icos–/– mice, which have ineffective B cell help (2, 23). After NP-Ova immunization, Icos–/– mice that received Bcl6–/– OT-II CD4+ T cells were unable to form germinal centers, in contrast to mice that received wild-type OT-II CD4+ T cells (Fig. 3, E to G, and fig. S9). These data demonstrate that Bcl6 is necessary for TFH differentiation and that TFH cells are necessary for germinal center formation. Together, these results indicate that Bcl6 is a bona fide master regulator of TFH differentiation in vivo.
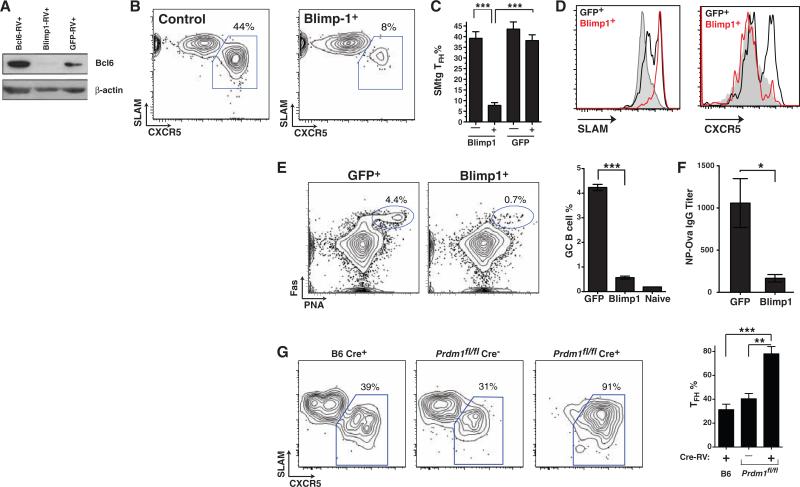
Blimp-1 is a known antagonist of Bcl6, capable of directly inhibiting Bcl6 expression in B and T cells (17, 18). Conversely, Blimp-1 expression can be inhibited by Bcl6 (16–18, 20). On the basis of our observations that Bcl6 drives TFH cell differentiation and function, and because Blimp-1 was the single most down-regulated transcription factor in TFH cells by gene expression array analysis (Fig. 1E and fig. S4) and qPCR (Fig. 1G), we hypothesized a role for Blimp-1 in blocking TFH differentiation in vivo. We constructed a Blimp-1 retroviral expression vector, Blimp1-RV (fig. S5), designed to express physiological levels of Blimp-1. Only CD4+ T cells expressing low levels of the GFP reporter were used for in vivo experiments (fig. S10A). Blimp-1 blocked Bcl6 protein expression in activated antigen-specific CD4+ T cells in vivo (Fig. 4A). To determine the effects of Blimp-1 on TFH differentiation, we mixed Blimp1-RV+ SMtg CD4+ T cells and untransduced control SMtg cells in equal proportions and transferred them into host mice subsequently infected with LCMV. We observed normal proliferation of Blimp-1–expressing SMtg CD4+ T cells (fig. S10); however, TFH differentiation was severely abrogated, with an 80% reduction in TFH frequency (Fig. 4, B and C). Blockade of TFH differentiation by Blimp-1 was also observed when mice separately received Blimp1-RV+ versus GFP-RV+ SMtg cells (fig. S10, F and G). Constitutive expression of Blimp-1 inhibited acquisition of the TFH phenotype: SLAM expression was increased (Fig. 4D), whereas CXCR5, ICOS, and PD-1 expression were all decreased (Fig. 4D and fig. S10). Inhibition of TFH differentiation by Blimp-1 was physiological and specific, because the expression levels of SLAM, ICOS, and PD-1 by Blimp1-RV+ SMtg CD4+ T cells were equivalent to the expression levels seen in wild-type activated non-TFH SMtg CD4+ T cells, and not naïve cells (fig. S10I). Blimp1-RV+ and wild-type non-TFH SMtg cells also expressed comparable amounts of the cytokines interferon-γ (IFN-γ) and IL-2 (fig. S11). High amounts of Blimp-1 expression can inhibit proliferation in B and T cells (17, 24, 25). The moderate level of Blimp-1 expression used in our experiments (fig. S10E) did not affect proliferation in vivo (fig. S10, C, D, and H), in agreement with previous in vitro studies (26) and our observation that non-TFH CD4+ T cells express 20 times as much Blimp-1 as do TFH cells and are still proliferative. Blimp-1 expression did not affect expression of the T helper lineage–specific transcription factors Foxp3, GATA3, and RORγt (fig. S11), which indicates that Blimp-1 did not induce differentiation into other helper lineages. Collectively, these data suggest that Blimp-1 acts specifically to repress Bcl6 and thus blocks TFH differentiation.
Fig. 4.
Blimp-1 and Bcl6 are antagonistic and reciprocal regulators of TFH differentiation. (A) Immunoblot of Bcl6 protein expression (and β-actin control) in transduced SMtg CD4+ T cells in vivo. (B) TFH (SLAMlow CXCR5high, boxed) differentiation of untransduced SMtg (left, “Control”) and Blimp1-RV+ SMtg (right, “Blimp-1+”) cells within a common host 8 days after LCMV infection. Gating is shown in fig. S10B. (C) Quantitation of SMtg TFH differentiation. “–,” untransduced; “+,” transduced with the indicated RV. ***P < 0.0001; n = 4 per group. Data are representative of two independent experiments. (D) SLAM and CXCR5 expression by naïve CD4+ T cells (gray), GFP-RV+ SMtg (black), and Blimp1-RV+ SMtg (red). (E and F) GFP-RV+ or Blimp1-RV+ OT-II CD4+ T cells were transferred into SAP-deficient mice subsequently immunized with NP-Ova in alum; n = 4 per group. Data are representative of three independent experiments. (E) Germinal center B cells (PNA+ Fas+, gated) in mice that received GFP-RV+ or Blimp1-RV+ OT-II CD4+ T cells. B220+ IgDlow gate is shown. Quantitation of germinal center B cells in the spleen is also shown. ***P < 0.0001. (F) NP-Ova IgG ELISA endpoint titers at day 10. *P = 0.016. (G) Purified naïve B6 and prdm1fl/fl CD45.2+ CD4+ T cells were transduced with SMtg-RV, with or without Cre-RV (Cre+ or Cre–), sorted, and transferred into CD45.1+ mice subsequently infected with LCMV. FACS plots depict TFH (CXCR5high SLAMlow, boxed) differentiation of control Cre+ SMtg+ B6 cells (left), Blimp-1–sufficient Cre– SMtg+ prdm1fl/fl cells (center), and Blimp-1–deficient Cre+ SMtg+ prdm1fl/fl cells (right). CD4+ CD45.1– CD44high 7AAD– gate is shown. Quantitation of TFH differentiation is also shown. Data are representative of two independent experiments. **P = 0.002, ***P = 0.0006; n = 4 to 5 per group.
Given that Blimp-1 is a physiological inhibitor of Bcl6 expression and TFH differentiation in vivo, we performed an additional test of the necessity of TFH for B cell help by transferring Blimp1-RV+ OT-II and GFP-RV+ OT-II CD4+ T cells into SAP-deficient (sh2d1a–/–) mice [SAP-deficient mice exhibit a CD4+ T cell–intrinsic defect in germinal center formation (27–29)] subsequently immunized with NP-Ova. We observed germinal centers and anti–NP-Ova serum IgG in GFP-RV+ OT-II CD4+ T cell recipient mice after immunization (Fig. 4, E and F). Strikingly, although OT-II cell numbers were normal in Blimp1-RV+ OT-II recipient mice (fig. S12), germinal centers were reduced by 90% (Fig. 4E). Constitutive Blimp-1 expression also inhibited the NP-Ova–specific IgG response, reducing the serum antibody concentration to only 16% of normal levels (Fig. 4F). All IgG isotypes were reduced (fig. S12), confirming that Blimp-1 was specifically inhibiting TFH differentiation. These results demonstrate both that Blimp-1 inhibits CD4+ T cell help to B cells and that TFH cells are required for B cell help in vivo.
To confirm the biological role of Blimp-1 in inhibiting TFH differentiation in vivo, we tested the ability of Blimp-1–deficient CD4+ T cells to differentiate into TFH. To avoid autoimmunity complications (30, 31), we deleted Blimp-1 (prdm1) in vitro in mature prdm1fl/fl CD4+ T cells (32) by means of a Cre-expressing RV. We transferred Cre+ SMtg+ prdm1fl/fl and control Cre– SMtg+ prdm1fl/fl CD4+ T cells into mice subsequently infected with LCMV. Deletion of prdm1 substantially enhanced TFH differentiation in vivo (Fig. 4G) without altering proliferation (fig. S13). These data indicate that Blimp-1 expression in vivo normally restricts Bcl6 expression and TFH differentiation. In sum, our results reveal that Bcl6 and Blimp-1 are reciprocal master regulators of TFH differentiation, with TFH differentiation in vivo requiring the presence of Bcl6 and the absence of Blimp-1.
There has been extensive speculation about a role for Bcl6 in TFH differentiation, based on gene expression data from human (2, 4) and murine TFH studies (3, 8, 9, 13). Our data directly show that Bcl6 specifically drives TFH differentiation and is a bona fide master regulator. The relationship between TFH and other CD4+ T cell lineages has been a long-standing problem. The predominant CD4+ T cell response to LCMV is TH1 (fig. S14), and it is notable that T-bet and IFN-γ were still expressed in the TFH in vivo, although at lower levels than in TH1/non-TFH LCMV-specific CD4+ T cells (fig. S14). These observations are consistent with a model in which TFH cells follow their own differentiation pathway but are not an isolated lineage and can exhibit partial characteristics of TH1/TH2 polarization depending on environmental conditions. This overlapping differentiation model would resolve the conundrum in the literature that neither TH1, TH2, nor TH17 are required for B cell help in vivo (8, 33, 34), but that cells with TH1, TH2, or TH17 phenotypes can provide B cell help in vivo (9, 35–39).
The capacity for B cell help is a central attribute of CD4+ T cells and is a cornerstone of protective immunity. It is well known that in B cells, Bcl6 and Blimp-1 are powerful antagonistic master regulators of germinal center B cell differentiation and plasma cell differentiation. Our findings that Bcl6 and Blimp-1 also control TFH differentiation illustrate the elegant use of the same antagonistic transcription factors to drive different functions in two lymphocyte populations differentiating in parallel: antigen-specific B cells and the TFH cells that provide their help. Manipulation of these signaling pathways in vivo may have substantial therapeutic benefit for enhancing vaccines or, conversely, blocking auto-antibody responses.
Supplementary Material
Footnotes
References and Notes
- 1.Zhu J, Paul WE. Blood. 2008;112:1557. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King C, Tangye S, Mackay C. Annu. Rev. Immunol. 2008;26:741. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- 3.Vinuesa CG, et al. Nature. 2005;435:452. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 4.Chtanova T, et al. J. Immunol. 2004;173:68. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 5.Rasheed AU, Rahn HP, Sallusto F, Lipp M, Müller G. Eur. J. Immunol. 2006;36:1892. doi: 10.1002/eji.200636136. [DOI] [PubMed] [Google Scholar]
- 6.Haynes NM, et al. J. Immunol. 2007;179:5099. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 7.Vogelzang A, et al. Immunity. 2008;29:127. doi: 10.1016/j.immuni.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Nurieva RI, et al. Immunity. 2008;29:138. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinhardt RL, Liang HE, Locksley RM. Nat. Immunol. 2009;10:385. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ansel KM, McHeyzer-Williams LJ, Ngo VN, McHeyzer-Williams MG, Cyster JG. J. Exp. Med. 1999;190:1123. doi: 10.1084/jem.190.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ansel KM, et al. Nature. 2000;406:309. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- 12.Hardtke S, Ohl L, Förster R. Blood. 2005;106:1924. doi: 10.1182/blood-2004-11-4494. [DOI] [PubMed] [Google Scholar]
- 13.Fazilleau N, McHeyzer-Williams LJ, Rosen H, McHeyzer-Williams MG. Nat. Immunol. 2009;10:375. doi: 10.1038/ni.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye BH, et al. Nat. Genet. 1997;16:161. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 15.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Science. 1997;276:589. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 16.Klein U, Dalla-Favera R. Nat. Rev. Immunol. 2008;8:22. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 17.Martins G, Calame K. Annu. Rev. Immunol. 2008;26:133. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 18.Cimmino L, et al. J. Immunol. 2008;181:2338. doi: 10.4049/jimmunol.181.4.2338. [DOI] [PubMed] [Google Scholar]
- 19.Shaffer AL, et al. Immunity. 2002;17:51. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 20.Shaffer AL, et al. Immunity. 2000;13:199. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 21.Cattoretti G, et al. Blood. 1995;86:45. [PubMed] [Google Scholar]
- 22.Vinuesa CG, Tangye S, Moser B, Mackay C. Nat. Rev. Immunol. 2005;5:853. doi: 10.1038/nri1714. [DOI] [PubMed] [Google Scholar]
- 23.Akiba H, et al. J. Immunol. 2005;175:2340. doi: 10.4049/jimmunol.175.4.2340. [DOI] [PubMed] [Google Scholar]
- 24.Reljic R, Wagner SD, Peakman LJ, Fearon DT. J. Exp. Med. 2000;192:1841. doi: 10.1084/jem.192.12.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K. J. Exp. Med. 2008;205:1959. doi: 10.1084/jem.20080526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong D, Malek TR. J. Immunol. 2007;178:242. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 27.Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R. Nature. 2003;421:282. doi: 10.1038/nature01318. [DOI] [PubMed] [Google Scholar]
- 28.Ma CS, Nichols KE, Tangye S. Annu. Rev. Immunol. 2007;25:337. doi: 10.1146/annurev.immunol.25.022106.141651. [DOI] [PubMed] [Google Scholar]
- 29.Schwartzberg PL, Mueller KL, Qi H, Cannons JL. Nat. Rev. Immunol. 2009;9:39. doi: 10.1038/nri2456. [DOI] [PubMed] [Google Scholar]
- 30.Kallies A, et al. Nat. Immunol. 2006;7:466. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 31.Martins GA, et al. Nat. Immunol. 2006;7:457. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 32.Shapiro-Shelef M, et al. Immunity. 2003;19:607. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 33.Tsiagbe VK, Thorbecke GJ. In: The Biology of Germinal Centers. Thorbecke GJ, Tsiagbe VK, editors. Springer-Verlag; Berlin: 1998. pp. 1–103. [Google Scholar]
- 34.Kopf M, Le Gros G, Coyle AJ, Kosco-Vilbois M, Brombacher F. Immunol. Rev. 1995;148:45. doi: 10.1111/j.1600-065x.1995.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 35.King IL, Mohrs M. J. Exp. Med. 2009;206:1001. doi: 10.1084/jem.20090313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaretsky AG, et al. J. Exp. Med. 2009;206:991. [Google Scholar]
- 37.Hsu HC, et al. Nat. Immunol. 2008;9:166. doi: 10.1038/ni1552. [DOI] [PubMed] [Google Scholar]
- 38.Smith KM, Brewer JM, Rush CM, Riley J, Garside P. J. Immunol. 2004;173:1640. doi: 10.4049/jimmunol.173.3.1640. [DOI] [PubMed] [Google Scholar]
- 39.Smith KM, et al. J. Immunol. 2000;165:3136. doi: 10.4049/jimmunol.165.6.3136. [DOI] [PubMed] [Google Scholar]
- 40.We thank R. Kageyama, L. Crickard, K. Hansen, C. Kim, and K. Van Gunst for technical assistance; S. Kaech for helpful discussions; A. Haberman and S. Kerfoot for technical help; and the NIH Tetramer Core for providing MHC class II tetramer reagents. Supported by LIAI institutional funds, a Pew Scholar Award, a Cancer Research Institute Award, and National Institute of Allergy and Infectious Diseases grants R01 072543 and NIAID R01 063107 (S.C.); Rheuminations Inc., the Arthritis Foundation, the Connecticut Chapter of the Lupus Foundation of America, and NIH grants AR40072, AR44076, and P30 AR053495 (J.C.); and fellowships from the UCSD/LIAI Immunology NIH Training Grant (I.Y. and R.J.J.). R.J.J. is a member of the UCSD Biomedical Sciences (BMS) graduate program. Microarray data have been deposited at the NCBI Gene Expression Omnibus (GSE16697). Author contributions are as follows: LCMV TFH identification and microarrays, I.Y. and S.C.; Bcl6-RV and Blimp1-RV experiments, R.J.J., D.D., B.B.; TFH migration, D.E.; Blimp-1 conditional knockout experiments, D.D.; Bcl6−/− mice, A.L.D.; Bcl6−/− experiments, A.C.P.; writing, S.C. (with intellectual and editorial contributions from the other authors); project conception and experimental design, S.C. and J.C.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.