Abstract
Proteins of the DUF199 family, present in all gram-positive bacteria and best characterized by the WhiA sporulation control factor in Streptomyces coelicolor, are thought to act as genetic regulators. The crystal structure of the DUF199/WhiA protein from Thermatoga maritima demonstrates that these proteins possess a bipartite structure, in which a degenerate N-terminal LAGLIDADG homing endonuclease (LHE) scaffold is tethered to a C-terminal helix-turn-helix (HTH) domain. The LHE domain has lost those residues critical for metal binding and catalysis, and also displays an extensively altered DNA-binding surface as compared to homing endonucleases. The HTH domain most closely resembles related regions of several bacterial sigma70 factors that bind the −35 regions of bacterial promoters. The structure illustrates how an invasive element might be transformed during evolution into a larger assemblage of protein folds that can participate in the regulation of a complex biological pathway.
Modern protein folds are believed to have arisen from the expansion of a much smaller complement of proteins that were found in the last common ancestor of prokarya, archaea and eukarya (Caetano-Anolles et al., 2007; Orengo and Thornton, 2005). Evidence for the ability of early proteins to diversify into increasingly elaborate structures, and to acquire novel functions, includes the observations that (i) many modern families of protein folds (such as the TIM barrels) encompass a wide variety biological activities (Nagano et al., 2002); (ii) individual proteins can exhibit multiple unrelated activities (a property termed 'moonlighting') (Jeffery, 2003); and (iii) certain protein sequences can adopt multiple folded states that each display unique functional properties (Tuinstra et al., 2008).
Among the many protein folds that have been diversified over the course of evolution, those that are associated with DNA cleavage and modification stand out. For example, the retroviral RNAse H and integrase folds are utilized by many transposases, nucleotidyl transferases, resolvases and nucleases (Nowotny, 2009). Similarly, the folds found within homing endonucleases (proteins that promote the genetic mobility of microbial introns and inteins) are also found in an impressive array of enzymes, including those involved in phage restriction, DNA replication, DNA repair, and recombination (Stoddard, 2005). These proteins are constructed around catalytic core folds containing HNH, PD…(D/E)xK and GIY-YIG active site motifs.
Homing endonucleases have also been domesticated and employed by their biological hosts for more disparate biological purposes. For example, many homing endonucleases have been adapted by their hosts to assist in RNA folding and splicing (Ho et al., 1997), while others (such as the yeast HO endonuclease) have been recast for the purpose of initiating nuclear gene conversion events (Koufopanou and Burt, 2005). In most of these cases, the new host-specific function still involves the ability of these enzymes to catalyze phosphotransfer reactions or to promote the rearrangement of nucleic acid substrates.
However, at least three instances have been documented where the biological function of homing endonuclease scaffolds have been more dramatically altered--with the protein in each case finding new employment as a regulator of a more complex biological pathway. In the first example, the DNA-binding domains found in 'Smad' proteins (eukaryotic transcription factors involved in TGF-β signaling) were found to be comprised of a 'ββα-metal' endonuclease fold (resembling the I-PpoI endonuclease (Flick et al., 1998)) that has lost its catalytic activity (Grishin, 2001). In the second case, the DNA binding domain of the AP2/ERF family of plant transcription regulators contain a recognizable HNH endonuclease domain (a structure common to many phage-derived homing endonucleases) (Magnani et al., 2004). Finally, proteins of the DUF199 superfamily, found throughout most if not all gram-positive bacteria, are postulated to contain a degenerated LAGLIDADG homing endonuclease domain (Knizewski and Ginalski, 2007). The most well-characterized member of this family, the WhiA protein from the soil bacterium Streptomyces coelicolor (WhiASc), is required for sporulation and regulates the expression of multiple sporulation-specific 'Whi' genes, including its own reading frame. (Ainsa et al., 2000). It is unknown whether this regulation occurs through direct or indirect interaction with DNA promoter elements or other proteins. Regardless, it is likely that WhiA homologues in other gram-positive bacteria function in a similar manner, since those microbes all contain similar Whi operons including a single recognizable DUF199/WhiA protein (Ainsa et al., 2000).
In order to visualize the structural basis for the creation of a transcriptional regulatory protein from a protein fold typically associated with a mobile endonuclease, we have determined the crystal structure of the DUF199/WhiA protein from T. maritima (WhiATm), and examined similarities and differences of its structure and primary sequence relative to its closest bacterial homologues and also to more distantly related LAGLIDADG homing endonucleases. The structure illustrates how the unique evolutionary pressures that are placed upon a genetic regulator, versus those placed on an invasive endonuclease, might produce individually tailored structural and biochemical features that are appropriate for each function. Studies of these proteins also indicate a likely scenario by which an invasive element could be converted into a genetic regulator. This chain of events would involve progression from a simple role as an autoregulator of its own expression, to the subsequent acquisition of novel domains and properties resulting in a more complex protein assemblage that can participate in highly coordinated transcriptional regulation.
RESULTS
We expressed an untagged, full-length DUF199/WhiA construct originally encoded in Thermotoga maritima (WhiATm) as a soluble protein in E. coli (Supplementary Figure 1A). Proteolytic digest experiments with trypsin revealed that the full-length protein could be digested into two stable domains, consistent with a bipartite structural organization (Supplementary Figure 1B). We were initially able to obtain crystals of the protein's isolated LAGLIDADG domain, that diffracted to 2.6 Å resolution. Similar crystals were grown using selenomethionyl-derivatized protein that allowed us to calculate phases to 3.0 Å resolution by SAD phasing. The structure was determined and ultimately refined to 2.6 Å resolution, revealing a pair of LAGLIDADG domains in the asymmetric unit (ASU, Supplementary Figure 2). The resulting model of these LAGLIDADG domains was refined to final values of Rwork and Rfree of 19.0% and 26.1%, respectively (Table 1). The α-carbons of the two individual molecules in the ASU superimpose with an RMSD of 0.75 Å.
Table 1.
Crystallographic Statistics
| Data Collection | |||
|---|---|---|---|
| Protein | LAGLIDADG Se-Met | LAGLIDADG | Full length |
| Beamline (Å) | ALS 5.0.2 | Home source | Home source |
| Wavelength | 0.98 | 1.54 | 1.54 |
| Space group | P212121 | P212121 | P212121 |
| Unit cell (Å) | a=50.9, b=79.7, c=115.3 | a=50.5, b=79.9, c=115.4 | a=51.65, b=61.30, c=97.22 |
| Resolution (Å)a | 50–2.9 (3.0–2.9) | 50–2.6 (2.69–2.60) | 50–2.34 (2.42–2.34) |
| Rmerge (%) | 10.8 (37.6) | 4.3 (18.5) | 5.3 (34.7) |
| I/σI | 13.5 (4.07) | 18.1 (6.35) | 27.0 (5.0) |
| Redundancy | 9.2 (9.2) | 2.5 (2.4) | 4.7 (4.6) |
| Completeness (%) | 97.7 (98.0) | 93.8 (89.0) | 98.6 (98.3) |
| Unique Reflections | 10776 | 14107 | 13400 |
| Refinement Statistics | |||
| Rwork (%) | - | 19.8 | 22.7 |
| Rfree (%)b | - | 26.0 | 27.6 |
| Number of atoms | - | 3069 | 2265 |
| Protein | - | 2997 | 2190 |
| Water | - | 57 | 62 |
| Est. Coor. Error (Å) | - | 0.24 | 0.21 |
| Geometry | |||
| RMSD Bonds Å | - | 0.012 | 0.005 |
| RMSD angles (°) | - | 1.456 | 0.868 |
| RMSD chiral (Å3) | - | 0.104 | 0.05 |
| Average B (Å2) | - | 45.7 | 42.1 |
| Protein monomer B factors (Å2) | - | 45.2 (mol A), 47.1 (mol B) | 42.2 |
| Water B factors (Å2) | - | 19.4 | 40.4 |
| Ramachandran Distribution | |||
| Most favored (%) | - | 89.5 | 94.7 |
| Additionally allowed (%) | - | 10.2 | 4.9 |
| Generously allowed (%) | - | 0.3 | 0.0 |
| Disallowed (%) | - | 0.0 | 0.4 |
| PDB Accession Code | - | ||
Numbers in parentheses correspond to the highest resolution shells
Calculated using a test set corresponding to 5% of the data
Subsequently, we obtained crystals of the full-length WhiATm protein that diffracted to 2.35 Å resolution under different crystallization conditions. This structure (Figure 1) was solved via molecular replacement using the refined coordinates of a single LAGLIDADG domain described above as a search model, and refined to final values of Rwork and Rfree of 22.7% and 27.6%, respectively (Table 1). The full-length protein contained a single WhiA molecule in the ASU, which was comprised of an N-terminal LAGLIDADG (LHE) domain, a linker region and a C-terminal helix-turn-helix (HTH) domain. The crystallographic protein-protein contacts in the structure of the full-length protein are completely different from those in crystals of the isolated LAGLIDADG domain, indicating that the contacts between LAGLIDADG domains observed in the two structures represent lattice contacts that differ between different crystal packing arrangements. Overall, the three independent crystallographic views of the LAGLIDADG domain obtained in this study correspond very closely to one other (pairwise α-carbon RMSD values of 0.75 Å to 1.50 Å). Except where noted below, the remainder of this manuscript describes the structure of the full length WhiATm protein.
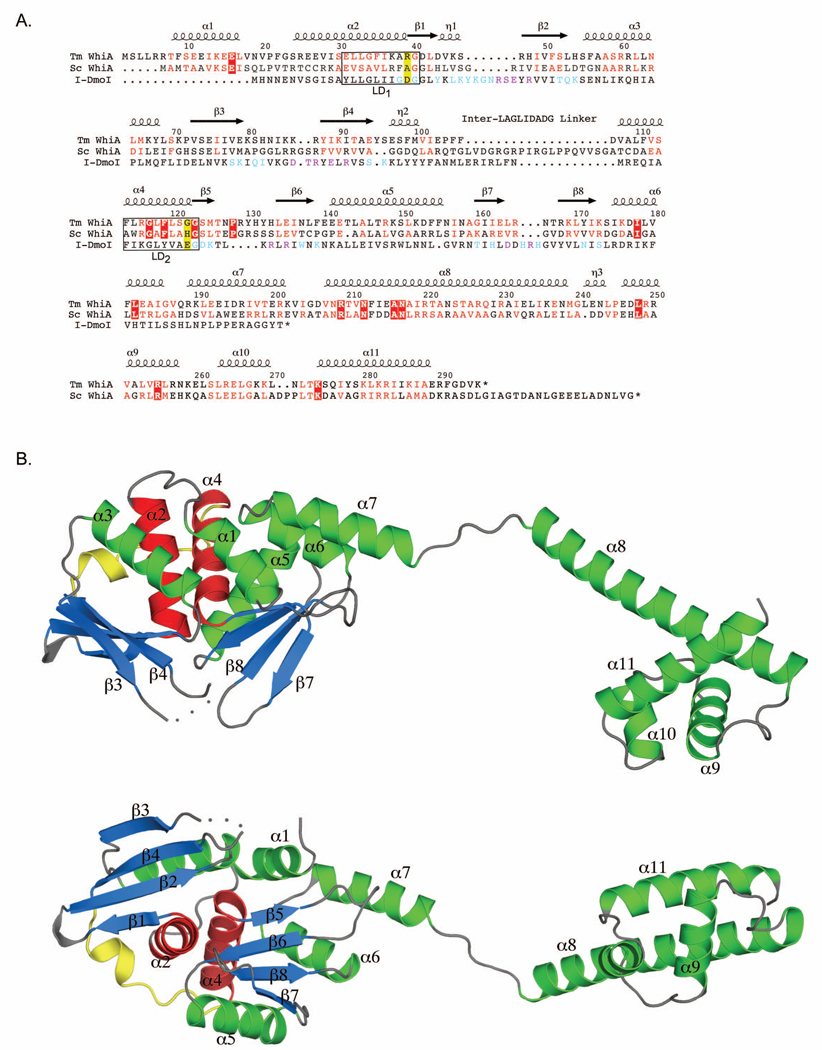
Figure 1. Structure and sequence conservation of Duf199 / WhiA.
A. A multiple sequence alignment of 14 different WhiA homologues (the same subset described by Knizewski et al. (Knizewski and Ginalski, 2007)) was generated using MUSCLE (Edgar, 2004). From this group alignment we show the alignment of WhiATm and WhiASc. We also performed a structure-based alignment of I-DmoI (PDB code 2VS7) compared to WhiATm. The residue numbering and location of α helices and β sheets shown above the alignment correspond to WhiATm and were generated with ESPript (Gouet et al., 1999). Residues blocked in red are identical amongst the WhiA family members used in the alignment; residues highlighted in red indicate conservative substitutions. I-DmoI residues highlighted in cyan contact the DNA backbone; residues highlighted in purple make base contacts. The yellow-highlighted residues indicate the position of acidic residues present in all LAGLIDADG homing endonucleases that are required for DNA cleavage and which are absent in the WhiA family. Boxes indicate the two LAGLIDADG motifs (LD1 and LD2).
B. Structure of full-length WhiATm shown in two different orientations. The β strands of the LAGLIDADG domain are highlighted in blue; the LAGLIDADG helices (α2 and α4) are red; the interdomain linker is yellow. The disordered loop connecting β-strands 3 and 4 is represented by dots. Labeled secondary structural elements correspond to the alignment in A.
The structure of WhiATm reveals that the linker region between its LAGLIDADG and HTH domains consists of two separate α-helices (α7 and α8) connected by a less structured series of residues (201–206) that may act as a flexible hinge (Figure 1). The overall dimensions of the protein are 105 Å by 30 Å by 25 Å; the distance between the center of the two independent domains is approximately 70 Å. One of the most conserved regions of the WhiA protein corresponds to the N-terminal end of the α8 helix, which contains four invariant residues (209R, 212N, 216A, and 217N) and numerous conservative substitutions amongst WhiA sequences obtained from 14 divergent bacterial organisms (Figure 1A; Supplementary Figure 4). This degree of conservation suggests either an essential function (such as interacting with a nucleic acid target region or with an additional protein factor) or a structural role, such as providing rigidity to the long α8 helix. The putative hinge region likely allows both domains a relatively large range of motion relative to each other; we therefore discuss the structural features of each domain independently.
The LAGLIDADG Domain
The N-terminal region of WhiATm contains the same protein fold topology that is observed in monomeric LAGLIDADG homing endonculeases. This region is comprised of two structurally similar domains, each containing an αββαββ core fold, that are connected by a short peptide linker (Figure 1 and Figure 2). The closest structural homologue of this domain, identified using the DALI webserver (Holm et al., 2008), is the I-DmoI homing endonuclease (an archaeal enzyme encoded within a mobile group I intron (Silva et al., 1999)). Despite overall limited sequence homology (13% identity; Figure 1A) both structures superimpose closely, with an a-carbon RMSD across all aligned residues (Figure 1A) of 2.4Å (Figure 2A). The most conserved elements within this region are those residues that comprise the two LAGLIDADG helices (α2 and α4 in WhiA) that form the core of the domain interface (Figure 2B). These helices are closely superimposable, including intimate packing between backbone atoms in the helices that is facilitated by the presence of small side chains at positions located near the helical interface (Figure 2B)
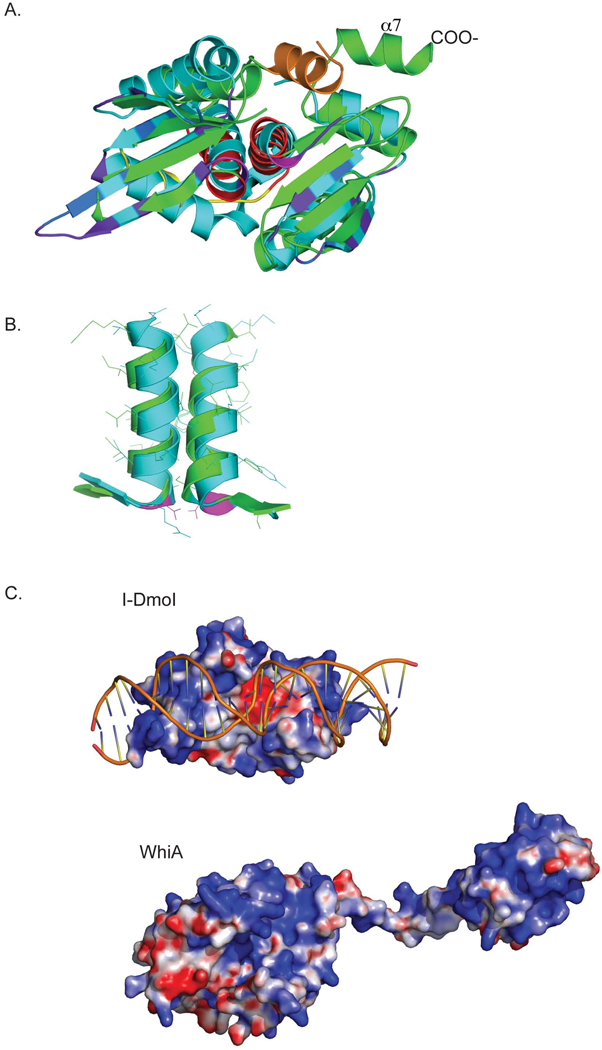
Figure 2. Structural comparison of the WhiATm LAGLIDADG domain with its closest structural homolog, the I-DmoI homing endonuclease.
A. Superposition of the LAGLIDADG domain of WhiATm (green) with I-DmoI (cyan, PDB code 2VS7), with the DNA binding domain of I-DmoI oriented towards the viewer. In WhiATm,, the N-terminal α1 helix present in the WhiA family but absent in I-DmoI is orange; the LAGLIDADG helices are red; and the interdomain linker is yellow. In I-DmoI, the conserved catalytic acidic residues at the base of the LAGLIDADG helices are magenta; residues that contact DNA bases are purple, and residues that contact the DNA backbone are dark blue.
B. Superposition of the LAGLIDADG helices of WhiATm (green; α2 left, α4 right) with I-DmoI (cyan).
C. Surface electrostatic charge potential (red: negative charge; blue: positive charge) of I-DmoI bound to its cognate DNA (top); and of full-length WhiATm (bottom; the LAGLIDADG domain is to the left, and the HTH domain to the right). The LAGLIDADG domains from the endonuclease and from WhiA are shown in the same relative orientation, looking into the β-sheet surfaces of each. The electrostatic potentials were calculated using program APBS (Baker et al., 2001). The large negative charge in I-DmoI corresponds to the pair of acidic residues (D21, E117) in the active site. The metal ions that these residues normally coordinate (and thus neutralize the active site residues) are not present in the electrostatic calculation.
A critical difference between WhiA family members and LAGLIDADG homing endonucleases is that the WhiA proteins lack acidic residues at the base of the LAGLIDADG helices that are strongly conserved in homing endonucleases (in I-DmoI these residues are D20 and E117). In the endonucleases, these residues coordinate divalent cations and are required for DNA cleavage. In WhiATm the corresponding residues are R39 and G123; in the independently determined structures of its LAGLIDADG domain described above divalent cations are clearly absent. In addition, homing endonuclease active sites contain conserved basic residues that are involved in transition-state stabilization (such as K43 and K120 in I-DmoI). These positions are occupied by a histidine and methionine (H54 and M125, respectively) in the WhiATm structure, and similarly nonconserved in its closest homologues. Therefore, WhiA family members are almost certainly not endonucleases, a conclusion supported by DNA digest experiments in our lab with WhiATm and its homologue from Streptomyces coelicolor, WhiASc (data not shown).
The crystal structure of the WhiATm protein also indicates that the mechanism of DNA recognition and binding by its LAGLIDADG domains might differ significantly from that displayed by the same domains in homing endonucleases. Enzymes such as I-DmoI utilize a pair of antiparallel β sheets and associated loops that make extensive contacts with their DNA substrates, via interactions with the DNA backbone and with individual nucleotide base-pairs across the entire DNA target. Each LAGLIDADG domain is responsible for recognition of a single DNA half-site, and their DNA-contact surfaces are uniformly positively charged--a feature interrupted only by the presence of conserved acid residues in the active sites at the center of the domain interface (Figure 2C).
In contrast, a substantial region of the same surface of WhiATm, corresponding to the the N-terminal LAGLIDADG domain, displays significant negative surface charge (Figure 2C). Furthermore, the C-terminal LAGLIDADG domain displays a positively charged surface that extends well beyond its β-sheet region. It therefore seems likely that the DUF199/WhiA protein family interacts with its DNA target in a manner unique from the mode of DNA binding exhibited by LAGLIDADG homing endonucleases such as I-DmoI.
There are several additional differences between the LAGLIDADG folds found in WhiA proteins versus homing endonucleases. First, the WhiA family contains an additional N-terminal α helix (α1, Figure 2A) that is not present in homing endonucleases. The function of this helix is not clear, although it makes extensive contacts with both LAGLIDADG helices and with helix α7 in the linker region. In addition, the length, sequence and structure of the peptide that connects the two LAGLIDADG domains in monomeric homing endonucleases and in the WhiA family (Figure 1B,2A) is highly variable, ranging from 12 residues in WhiATm to 30 residues in the orthologous WhiA protein from S. coelicolor. In I-DmoI this region contains an α-helix spanning almost three full turns, whereas in WhiATm this region consists of largely random coil architecture.
Finally, WhiATm and its most closely related homologues are unique in containing an additional five residues at the N-terminus that are not present in other WhiA members. In the structure of the isolated LAGLIDADG domains (Supplementary Figure 2), these additional residues form an interchain β strand interaction with β3 of its crystallographic dimeric partner, but in the full-length structure (a monomer in the asymmetric unit) these residues are disordered.
The helix-turn-helix domain
The C-terminal region of WhiATm forms a canonical three-helical bundle, termed a 'helix-turn-helix' (HTH) domain, comprised of the α9, α10 and α11 helices of the full-length protein (Figure 1 and Figure 3). Although WhiATm does not display any additional elaborations upon this core fold, other WhiA homologues contain additional C-terminal residues; for example, WhiA from S. coelicolor contains 23 additional residues not present in WhiATm (Figure 3A). The closest structural homologues of the WhiATm HTH domain, identified by a three-dimensional similarity search using the DALI webserver (Holm et al., 2008), are similar HTH domains comprising 'domain 4' of the bacterial sigma 70 protein family (Figure 3A, B). The most similar structure, domain 4 from the E. coli SigmaE protein, superposes on the Tm WhiA HTH domain with an α-carbon RMSD of 1.96Å over 65 residues.
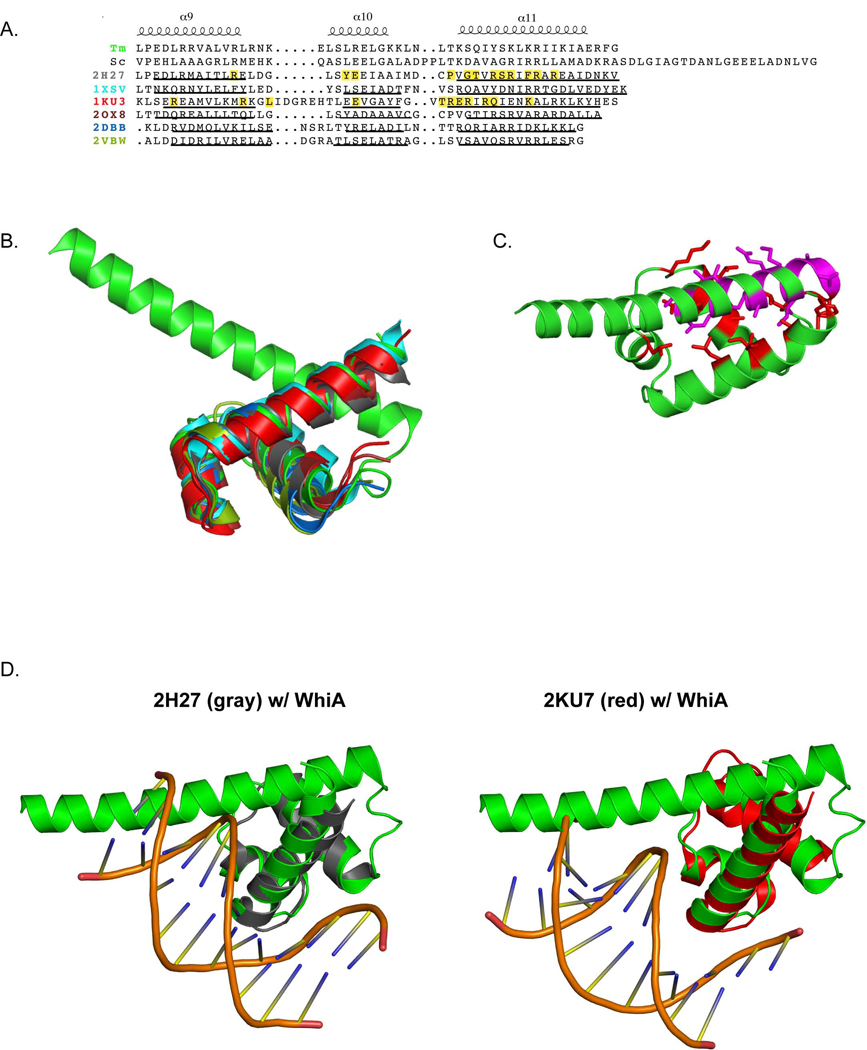
Figure 3. The HTH domain of WhiATm is structurally related to domain 4 of bacterial σ factors.
A. Structure-based alignment of WhiATm (top), and its closest structural homologues identified using the DALI server. The secondary structure elements shown above the alignment correspond to WhiATm. Residues highlighted yellow in 2H27 and 1KU3 correspond to amino acids that contact DNA. Underlined regions indicate α helices. PDB codes correspond to the following structures: 2H27: domain 4 of E. coli SigmaE (1.96 Å α-carbon RMSD over 65 equivalent positions); 1XSV: S. aureus hypothetical UPF0122 protein SAV123 (2.59 Å over 67 equivalent positions); 1KU3: T. aquaticus RNA Polymerase Sigma subunit, domain 4 (2.44 Å over 57 equivalent positions); 2OX8: M. tuberculosis SigC −35 element promoter recognition domain (2.57 Å over 69 equivalent positions); 2DBB: P. horikoshii HTH-type transcriptional regulator (4.09 Å over 119 equivalent positions); 2VBW: M. tuberculosis Rv3291c, transcriptional regulator (2.31 Å over 96 equivalent positions).
B. Superposition of the structures (color-coded as in panel A). The superposition was created by FATCAT (Ye and Godzik, 2003).
C. The α8 “linker” helix binds to a cleft formed between the WhiATm α9 and α11 helices. Residues on the α8 helix that make contacts with the cleft are in magenta; residues of the HTH domain that contact α8 are colored red.
D. Structures of 2H27 (left, gray) and 2KU7 (right, red) bound to DNA. WhiATm (green) is superposed over the structures, demonstrating a potential clash of α8 with the minor groove.
The HTH domains from bacterial sigma70 factors typically bind the −35 region of bacterial promoters, and the structure of two of these factors (E. coli SigmaE, PDB code 2H27; T. aquaticus RNA Polymerase Sigma subunit, PDB code 1KU3) have been solved bound to DNA. Superposition of the WhiATm HTH domain onto these structures indicates that the third (α11) helix of the WhiATm HTH domain might make significant DNA contacts (Figure 3D), which would be consistent with the principal mode of DNA binding most commonly displayed by HTH domains (Aravind, 2005). In addition, the electrostatic potential of the HTH domain would be compatible with DNA binding (Figure 2C). However, in this orientation, the α8 linker helix of WhiATm would closely approach the minor groove of DNA (Figure 3C). Therefore, if the HTH domain of WhiATm does bind DNA, it may do so in a manner requiring distortion of its DNA target, and might use additional residues from this region to make further contacts with the DNA backbone, as has been observed for other HTH-containing proteins (Khare et al., 2004).
An additional important structural feature of the HTH domain is a cleft formed between the α9 and α11 helices, into which the C-terminal end of the α8 helix is docked (Figure 3C). A combination of hypdrophobic interactions and hydrogen bonds stabilize this interaction, which includes nearly half (14 out of 32 residues) of the α8 helix. A cleft in this configuration is typical of HTH domains and is often utilized to pack additional stabilizing structural elements. It is noteworthy that the N-terminal half of the long α8 helix is well conserved throughout bacterial species, while the C-terminal half seems to be less constrained (Supplementary Figure 4). Two possible explanation are either that (i) WhiATm’’s function requires the helical conformation of N-terminal end of α8 to be stable in the absence of additional packing interactions (which may require a conserved sequence of amino acids), or (ii) that this region is involved in additional molecular interactions in order to support the biological function of WhiA.
DISCUSSION
Evolution of transcription regulatory activity from a homing endonuclease?
There is no direct experimental evidence demonstrating either DNA-binding or direct transcriptional activation functions for the WhiA proteins. However, previous biochemical and genetic studies of WhiA from S. coelicolor (Knizewski and Ginalski, 2007) and its homologue from S. ansochromogenes (originally termed sawC in that organism) (Xie et al., 2007) indicate that those particular proteins are involved in septation and sporulation, and effect the expression of several genes, including their own, that are involved in those processes. Combined with the observation that these proteins contain two separate domains known for their DNA-binding activity, it is reasonable to hypothesize that the WhiA proteins might function as transcriptional regulators.
After invasion of a genomic target by a homing endonuclease, there is little selective pressure imposed by the host for the maintenance of a functional enzyme, leading to the gradual accumulation of mutations that reduce its activity. This evolutionary degradation eventually leads to loss of the endonuclease gene and its associated intervening sequence (Burt and Koufopanou, 2004). Homing endonucleases often avoid this fate by acquiring novel activities that are beneficial to their host during evolution, a situation that places them under selective pressure to maintain a well-behaved protein fold (Stoddard, 2005). Several examples of domesticated LAGLIDADG-containing proteins have been documented (such as the HO endonuclease and maturase intron splicing factors), however the WhiA family is particularly noteworthy because it is the first example of a putative transcription factor containing a LAGLIDADG fold, and (if this functional annotation is found to be true) would be the third known example of a domesticated transcription factor derived from any homing endonuclease (Grishin, 2001; Knizewski and Ginalski, 2007).
In a manner analogous to the hypothesized recruitment of the LAGLIDADG protein scaffold for transcriptional regulation by the DUF199/WhiA proteins in bacteria, a variety of additional protein folds that are primarily associated with enzymatic activity have also been co-opted and employed as genetic regulators. For example, the eukaryotic Gal80, TAFII150, and Cdc68/Spt16 transcription factors are derived from (or share common ancestors with) oxidoreductase, aminopeptidase N and aminopeptidase P enzyme families, respectively (Aravind and Koonin, 1998). During this evolutionary transformation, these proteins are often observed to sacrifice their catalytic activity and adopt novel transcriptional regulatory functions.
Assuming that the WhiA family of transcription regulators was derived from mobile endonuclease ancestors, several key events would have occurred during the evolutionary creation and expansion of this protein family. First, an ancestor to modern day bacteria would have acquired an active LAGLIDADG homing endonuclease. At some point subsequent to this initial genetic transfer the homing endonuclease gained an additional HTH protein domain, lost its ability to cleave DNA, and became a completely domesticated transcription factor.
A key question regarding such an evolutionary scenario is whether a bifunctional intermediate might have existed during this process, in which the endonuclease activity and the ability to act as a transcriptional regulator were shared by a single protein scaffold (a relationship commonly termed 'moonlighting'). Many modern homing endonucleases require relatively tight regulation of their own expression, primarily to avoid toxicity that might be associated with its own overexpression. At least one such endonuclease (the phage-derived I-TevI enzyme) also serves as its own transcriptional autorepressor (Edgell et al., 2004). A scenario in which a homing endonuclease first adopted a very simple form of transcriptional regulatory activity to regulate its own expression, followed by subsequent incorporation into more complex forms of gene regulation and loss of its original endonuclease activity, seems attractive.
WhiA proteins and transcriptional regulation
Genetic studies in S. coelicolor suggest that WhiASc, an essential sporulation factor, functions as a transcriptional activator by regulating the expression of numerous genes, including its own (Ainsa et al., 2000). WhiASc contains two promoters, a low level upstream promoter that is expressed independently of WhiASc, and a sporulation-specific promoter more proximal to the WhiA transcriptional start site that requires WhiASc for expression (Ainsa et al., 2000). Bacterial transcriptional activators often function by binding on or near the −35 region of promoters and recruiting the bacterial RNA polymerase holoenzyme to the promoter. It is therefore noteworthy that the closest structural homologues of the WhiATm HTH domain are HTH domains (i.e. domain 4) of bacterial sigma70 factors, which bind to −35 promoter elements.
Given the data summarized above, it seems likely that the bacterial Duf199/WhiA proteins might be involved in interactions both with a DNA target and also with additional protein factors within the transcriptional apparatus. The structure of WhiA from T. maritima reveals a striking combination of two individual domains that are each well known for their abilities to facilitate both DNA recognition and protein-protein association. LAGLIDADG endonucleases typically recognize long (twenty or more basepair) targets with variable fidelity, while HTH domains recognize shorter (six to eight) basepair targets. However, these domains are also capable of facilitating packing interactions with additional structural protein domains: LAGLIDADG endonucleases are often fused to protein splicing domains (termed 'inteins'), and HTH domains can facilitate a variety of protein binding and dimerization interactions, primarily in eukaryotes (Aravind et al., 2005). Both the LAGLIDADG and HTH domains would therefore seem to be well-suited to adopting novel functions as part of a transcription factor complex, as they are both often found in multi-domain architectures and can apparently facilitate a wide variety of macromolecular interactions.
Given the broad spectrum of environmental niches of organisms that contain WhiA, it is likely that during evolution WhiA has been utilized to regulate diverse biological pathways. For example, while WhiA from S. coelicolor has a well- established role in sporulation, in other non-sporulating bacteria WhiA homologues likely regulate distinct pathways. In the course of adopting novel functions in various organisms, WhiA would have also likely developed novel protein interaction partners. A possible WhiA-binding candidate from S. coelicolor is the iron-sulfur protein WhiB, which shares a similar genetic phenotype as WhiA in sporulation. However, this interaction would only occur in a subset of organisms that contain WhiA since the WhiB family is only present in Actinomycetes.
EXPERIMENTAL METHODS
Protein production
The WhiATm gene was amplified from T. maritima genomic DNA (ATCC) using PCR, cloned into the pET24 expression vector (Novagen) and expressed in BL21(DE3)RIL bacteria (Novagen) in LB media supplemented with 1% glucose and antibiotics (kanamycin and chloramphenicol). A starter culture was grown overnight at 37°C, and diluted 1:50 the next morning into media with antibiotics. WhiA expression was induced with the addition of 1 mM IPTG when the OD600 reached 0.6–0.8, and incubated for an additional 3 hrs at 37°C. Cells were then centrifuged and stored at −20°C. Pellets were thawed and lysed by sonication on ice in 300 mM NaCl, 50 mM Tris pH 8.0, 1 mM PMSF. After centrifuging for 30 minutes in a SS34 rotor (Sorvall) at 43,000 g, the supernatant was incubated in a 70°C water bath for 15 minutes followed by a 60 minute spin at 43,000 g in an SS34 rotor. The cleared lysate was then loaded onto a 1 mL Heparin HiTrap column (GE Healthcare) at room temperature (using a Biorad peristaltic pump), and eluted on a Pharmacia AktaPrime FPLC with a 300 mM to 1 M NaCl gradient in 25 mM Tris, pH 8.0 buffer over 30 column volumes. The peak fraction typically eluted at ~600–700 mM NaCl. Peak fractions were pooled, precipitated with 25% (w/v) ammonium sulfate and centrifuged in 2 mL eppendorf tubes at 4°C, 13,000 rpm in an Eppendorf tabletop centrifuge. The pellet was resuspended in 25 mM Tris, pH 7.5 to ~5 mg/mL and dialyzed against 25 mM Tris pH 7.5, 150 mM (NH4)2SO4. Protein concentration was estimated using optical absorbance at 280 nm with a calculated molar extinction coefficient of 10430 M−1cm−1.
For production of selenomethionyl-containing protein we used BL21(DE3)-RIL bacteria and followed the method of (Doublie, 1997) in which the methionine biosynthesis pathway was inhibited prior to induction by the addition of Ile, Lys and Thr, and supplemented with selenomethionine (Fisher, Acros). Briefly, an O/N culture of BL21(DE3)-RIL bacteria transformed with pET24_WhiATm was grown in a 10 ml overnight culture in LB/1% glucose, kanamycin, chloramphenicol. The next morning the starter culture was pelleted and resuspended in 10 mL of minimal media, diluted into 1 L of minimal media with antibiotics, and grown to OD600 of 0.6. Amino acids that shut down cellular methionine production and selenomethionine were added, the culture incubated for 15 minutes, and then induced with 1 mM IPTG for 3 hrs. The remainder of the protein purification was performed as described above.
Crystallization
Crystals of the isolated LAGLIDADG were grown from a solution of the full-length protein that had been subjected to in situ proteolysis with trypsin (Dong et al., 2007). Trypsin (Sigma/Aldrich) was added to the full-length protein solution on ice in a range of ratios (1:1000 to 1:10,000 w/w) just before setting up crystallization trials. Drops were set using the hanging drop vapor diffusion method (1 µl of protein/protease plus 1 µl of mother liquor) and incubated at 18°C. Trypsin-treated WhiATm crystallized in 20% ethanol, 0.1 M Tris, pH 9.0 and 200 mM NaCl. Selenomethionine-containing crystals (also treated with trypsin) were grown in 20% ethanol, 0.1 M Tris pH 9.3 and 0.2 M KCl. For cryopreservation, crystals were transferred to mother liquor containing 25% glycerol and flash frozen in liquid nitrogen.
Crystals of the full-length WhiATm protein were generated by the hanging drop method in 0.1 M Tris, 8.0, 0.2 M NaCl and 10% PEG8000 at 18°C. Crystals typically grew in clusters after about two weeks, and had to be separated for cryopreservation (as described above).
Data collection
X-ray data sets on native crystals were collected using an in-house rotating anode HF-007 x-ray generator equipped with a RAXIS IV++ imaging plate area detector (both instruments from Rigaku, Inc.). A single wavelength dataset at the peak energy (12.661 KEV) with inverse-beam geometry was collected for a SeMet containing crystal (trypsin form only) at the Advanced Light Source synchrotron facility (Berkeley, CA), beamline 5.0.2. Data were indexed and scaled using HKL2000 software (Otwinowski and Minor, 1997). Phases for the crystal containing the isolated LAGLIDADG domain were solved using SOLVE (Terwilliger and Berendzen, 1999) and solvent flattened using RESOLVE (Terwilliger and Berendzen, 1999). The model was built using COOT (Emsley and Cowtan, 2004) and was refined using TLS restrained refinement in Refmac5 (Murshudov et al., 1997) while monitoring Rfree (Kleywegt and Brunger, 1996), and also monitoring the overall geometric quality of the model using PROCHECK (Laskowski et al., 1993). TLS parameters were defined using the TLS online server http://skuld.bmsc.washington.edu/~tlsmd/ (Painter and Merritt, 2006). The resulting models of the WhiA LAGLIDADG domain (of which there were two in the asymmetric unit) were then used as search models in molecular replacement to solve the phases of the full-length crystals using PHASER (McCoy et al., 2007). The density for the HTH domain of WhiATm was initially built using ARP/WARP (Perrakis et al., 2001) and unaccounted density was manually built in COOT (Emsley and Cowtan, 2004). The CCP4i suite of programs (1994) was extensively used throughout the structure solving process to implement programs and adjust files.
The coordinates for the refined models of the isolated LAGLIDADG domain and the full length WhiATm protein have been deposited in the RCSB protein structure database (PDB ID codes 3HYI and 3HYJ).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the staff of the Advanced Light Source (ALS) beamline 5.0.2 and members of the FHCRC structural biology program for technical assistance, advice and discussion. Funding provided by the NIH (GM49857 and CA133833) and the FHCRC Division of Basic Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
REFERENCES
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Ainsa JA, Ryding NJ, Hartley N, Findlay KC, Bruton CJ, Chater KF. WhiA, a protein of unknown function conserved among gram-positive bacteria, is essential for sporulation in Streptomyces coelicolor A3(2) J Bacteriol. 2000;182:5470–5478. doi: 10.1128/jb.182.19.5470-5478.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Eukaryotic transcription regulators derive from ancient enzymatic domains. Curr Biol. 1998;8:R111–R113. doi: 10.1016/s0960-9822(98)70982-0. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr Opin Genet Dev. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Caetano-Anolles G, Kim HS, Mittenthal JE. The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc Natl Acad Sci U S A. 2007;104:9358–9363. doi: 10.1073/pnas.0701214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A, Xu X, Edwards AM, Chang C, Chruszcz M, Cuff M, Cymborowski M, Di Leo R, Egorova O, Evdokimova E, et al. In situ proteolysis for protein crystallization and structure determination. Nat Methods. 2007;4:1019–1021. doi: 10.1038/nmeth1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgell DR, Derbyshire V, Van Roey P, LaBonne S, Stanger MJ, Li Z, Boyd TM, Shub DA, Belfort M. Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor. Nat Struct Mol Biol. 2004;11:936–944. doi: 10.1038/nsmb823. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Flick KE, Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature. 1998;394:96–101. doi: 10.1038/27952. [DOI] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Grishin NV. Mh1 domain of Smad is a degraded homing endonuclease. J Mol Biol. 2001;307:31–37. doi: 10.1006/jmbi.2000.4486. [DOI] [PubMed] [Google Scholar]
- Ho Y, Kim SJ, Waring RB. A protein encoded by a group I intron in Aspergillus nidulans directly assists RNA splicing and is a DNA endonuclease. Proc Natl Acad Sci U S A. 1997;94:8994–8999. doi: 10.1073/pnas.94.17.8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery CJ. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 2003;19:415–417. doi: 10.1016/S0168-9525(03)00167-7. [DOI] [PubMed] [Google Scholar]
- Khare D, Ziegelin G, Lanka E, Heinemann U. Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat Struct Mol Biol. 2004;11:656–663. doi: 10.1038/nsmb773. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Brunger AT. Checking your imagination: applications of the free R value. Structure. 1996;4:897–904. doi: 10.1016/s0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- Knizewski L, Ginalski K. Bacterial DUF199/COG1481 proteins including sporulation regulator WhiA are distant homologs of LAGLIDADG homing endonucleases that retained only DNA binding. Cell Cycle. 2007;6:1666–1670. doi: 10.4161/cc.6.13.4471. [DOI] [PubMed] [Google Scholar]
- Koufopanou V, Burt A. Degeneration and domestication of a selfish gene in yeast: molecular evolution versus site-directed mutagenesis. Mol Biol Evol. 2005;22:1535–1538. doi: 10.1093/molbev/msi149. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Magnani E, Sjolander K, Hake S. From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell. 2004;16:2265–2277. doi: 10.1105/tpc.104.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology 276: Macromolecular Crystallography, Part A. 1997 doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- Silva GH, Dalgaard JZ, Belfort M, Van Roey P. Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI. J Mol Biol. 1999;286:1123–1136. doi: 10.1006/jmbi.1998.2519. [DOI] [PubMed] [Google Scholar]
- Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc Natl Acad Sci U S A. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Li W, Tian Y, Liu G, Tan H. Identification and characterization of sawC, a whiA-like gene, essential for sporulation in Streptomyces ansochromogenes. Arch Microbiol. 2007;188:575–582. doi: 10.1007/s00203-007-0278-x. [DOI] [PubMed] [Google Scholar]
- Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19 Suppl 2:ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.