Abstract
Cloning and sequencing of the upstream region of the gene of the CC chemokine HCC-1 led to the discovery of an adjacent gene coding for a CC chemokine that was named “HCC-2.” The two genes are separated by 12-kbp and reside in a head-to-tail orientation on chromosome 17. At variance with the genes for HCC-1 and other human CC chemokines, which have a three-exon-two-intron structure, the HCC-2 gene consists of four exons and three introns. Expression of HCC-2 and HCC-1 as studied by Northern analysis revealed, in addition to the regular, monocistronic mRNAs, a common, bicistronic transcript. In contrast to HCC-1, which is expressed constitutively in numerous human tissues, HCC-2 is expressed only in the gut and the liver. HCC-2 shares significant sequence homology with CKβ8 and the murine chemokines C10, CCF18/MRP-2, and macrophage inflammatory protein 1γ, which all contain six instead of four conserved cysteines. The two additional cysteines of HCC-2 form a third disulfide bond, which anchors the COOH-terminal domain to the core of the molecule. Highly purified recombinant HCC-2 was tested on neutrophils, eosinophils, monocytes, and lymphocytes and was found to exhibit marked functional similarities to macrophage inflammatory protein 1α. It is a potent chemoattractant and inducer of enzyme release in monocytes and a moderately active attractant for eosinophils. Desensitization studies indicate that HCC-2 acts mainly via CC chemokine receptor CCR1.
Keywords: bicistronic mRNA, cDNA, chemotaxis, cytosolic free calcium
We recently have isolated and characterized a new human CC chemokine, HCC-1 (1), which is structurally similar to macrophage inflammatory protein (MIP)-1α (46% amino acid identity) and occurs in high concentrations in human plasma like MIP-1γ in murine blood. Northern blot analysis revealed that, in contrast to other chemokines, HCC-1 is expressed constitutively at high levels in numerous human tissues. We further showed that HCC-1 interacts with receptors that also recognize MIP-1α and RANTES and stimulates the proliferation of CD34+ myeloid progenitor cells in vitro. In the present paper, we report the discovery and characterization of a CC chemokine, HCC-2, that arises from a gene that is arranged in tandem with the gene of HCC-1 on chromosome 17. Mono- as well as bicistronic transcripts of the two genes were detected. Recombinant HCC-2 was expressed and shown to act mainly on monocytes and eosinophils in a similar manner as MIP-1α.
MATERIALS AND METHODS
Cell Culture.
Cell lines (T84, HUH-7) were purchased from the American Type Culture Collection and maintained in DMEM supplemented with 10% fetal calf serum.
Oligonucleotides.
The oligo-deoxyribonucleotides used were synthesized chemically (Perkin—Elmer) and are listed in Table 1.
Table 1.
Oligonucleotides used
| Primer | Sequence* |
|---|---|
| CC-1/L1 | TGACTTGCCCTCTCCGATGTC |
| CC-1/L2 | GTTCCCTCCTCTGTAAATGAG |
| CC-1/X1 | AGCCTCTGAAGCTCCCACCAGG |
| INT-1 | AGGGAAGCTCCAAGAGGGTGAC |
| CCPF-7 | TCTCACAAAGCATCCCGTG |
| CCPF-19 | TTTAAGTATTTCAGACCAAG |
| CC-3AS | TAAGGTCCCCCCACCAACT |
| CC-3S | GACTGAATCCTCCTCACAA |
| CC-34b | TGGGAGCTTCAGAGGCTCAG |
| IgU-6 | ACCTCAGCTGCAGCTCGAGGGIIGGGIIGGGII |
| CCPF-2 | AGCTTTTTCATGCAATCCTGAACTC |
| UNIP-6 | ACCTCAGCTGCAGCTCGAG |
| CCPF-4 | ATGACACCTGGCTTGGAGC |
| EXCC-2A | ACGGTACCCCAGGAAGCAGTGAGCCC |
| EXCC-2B | TTTTTTTTTTTTTTTGAAG |
| EXCC-2C | TCTCGAGTCTAGATTTTTTTTTGAAGAGCATG |
| CCPF-1 | GTGGGTGGCTGGCCTCTTTTGTCTC |
| CC-2/XbaI | GACTCTAGAGCCCTGCCTGCCCACCAG |
| CC2/1 SstI | TAGAGCTCCTGGGTCACTCAGTTCTC |
| CC-1/PE | GTCTTGGTCCCTAGGGCGATGGTG |
| CC-1/T1 | AAATATTTGGCCTCAATGATC |
| CC-1/T2 | CCGCAGGAGCCTCTGAAG |
| EX1 | CTTCTCGAGAAAAGACAGTTCACAAATGATGCAGAGAC |
| EX2 | CGGAATTCTTATATTGAGTAGGGCTTCAG |
*Sequences of the oligonucleotides are listed in 5′ to 3′ orientation.
Gene Cloning and Characterization.
A human genomic library in λ phage (Stratagene) was screened as described (2) by using a partial 269-bp, HCC-1-specific cDNA fragment (1) as a probe. Three independent clones were isolated, and the SstI restriction fragments of one of them were subcloned into pBSK+. Both strands of the HCC-1 and HCC-2 genes were sequenced by the method of Sanger (3) by using an automatic fluorescence sequencer (ABI 373A, Perkin–Elmer) (4) and the primer walking strategy. Sequence comparisons and analysis of the promoter regions were performed with the macmolly program package (Soft Gene, Berlin) and matinspector, Ver. 1.0 (5).
Chromosomal Localization.
Chromosomal localization of the HCC-1 and HCC-2 genes was determined by PCR analysis (30 cycles) with DNA from the three murine/human DNA hybrid cell lines CPS 324, 811, and 909 (Bios, New Haven, CT) as recommended by the supplier. The HCC-1-specific primer combination CC-1/L1 and CC-1/L2 was used. The amplified product was sequenced as described above.
RT-PCR.
Total RNA was extracted from 107 cells (T84, HUH-7) or 10–30 mg of tissue (liver, tonsils, bone marrow) by using the RNeasy kit (Qiagen, Chatsworth, CA). RNA (5 μg) was reverse transcribed by using oligo(dT) primers, and the integrity of the cDNA first strand was tested by the amplification of a β-tubulin fragment. For analytical RT-PCR, the first cDNA strand was amplified for 38 cycles by using the primer combinations CC-1/X1/INT-1, CCPF-7/CCPF-19, CCPF-7/INT-1, CCPF-7/CC-3AS, and CC-3S/INT-1. 5′ rapid amplification of cDNA ends PCR for extension of the 5′ terminus of the HCC-1 cDNA into the HCC-2 ORF was performed by reverse transcription of mRNA from 5 μg of total RNA of T 84 cells by using the oligonucleotide CC-34b followed by dC-tailing of the cDNA first strands (6). Three successive amplification steps were performed (first step: 35 cycles, oligonucleotides CC-34b/IgU-6; second step: 25 cycles, oligonucleotides CCPF-2 and UNIP-6; third step: 35 cycles, oligonucleotides CCPF-4 and UNIP-6). The full length bicistronic HCC-2/HCC-1 cDNA including the 3′ terminus of the HCC-1 cDNA was amplified in two steps, 36 cycles each, from human liver first strand cDNA (first step: oligonucleotides EXCC-2A/EXCC-2B; second step: oligonucleotides EXCC-2A/EXCC-2C) and subsequently cloned into the KpnI/XbaI sites of the pcDNA3 cloning vector (Invitrogen).
Northern Blot Analysis.
Commercially available tissue mRNA blots were used for Northern analysis (CLONTECH). The blots were hybridized at 60°C with radiolabeled PCR fragments specific for HCC-2 (oligonucleotides CCPF-1 and CC-2/XbaI) and HCC-1 (CC-1/X1 and CC2/1 SstI) according to the manufacturer’s instructions followed by autoradiography.
Detection of the Transcriptional Start Points of the HCC-1 Gene.
Primer extension was performed (7) by using 80 μg of total RNA from human liver, tonsils, bone marrow, and HUH-7 cells with the oligonucleotide CC-1/PE. Alternative transcription start points were investigated by RT-PCR (33 cycles) by using three different primers (CC-1/T1, CC-1/T2, and CC-1/X1) in combination with the primer INT-1.
Expression and Purification of HCC-2.
Recombinant HCC-2 was expressed in Pichia pastoris by using the pPIC9 vector (Invitrogen). A fragment encoding the amino acid residues 21–113 of the HCC-2 precursor was generated by PCR by using the primers EX1 and EX2 and was inserted after cleavage with XhoI and EcoRI beneath the coding region for the α-factor and a KEX recognition site. The resulting vector pPIC-CC2 was propagated into yeast spheroblasts after linearization with BglII. Recombinant clones were screened by immunodot blotting by using a polyclonal rabbit anti-HCC-2 serum. HCC-2-secreting yeast cells were cultured in shaking flasks (250 ml) at 30°C with different media. The main secreted form of HCC-2 was purified by RP- and cation exchange HPLC, checked by capillary zone electrophoresis and microbore RP-HPLC, and analyzed by amino acid sequencing and MS analysis (matrix-assisted laser desorption ionization–MS, LC-MS coupling). MS analysis also was carried out after carboxamidomethylation of the purified rHCC-2 to determine free sulfhydryl groups. Enzymatic degradation of the recombinant molecule was carried out in two subsequent steps by using trypsin and chymotrypsin. HPLC-separated fragments were characterized further by sequence analysis and electrospray ionization–MS to localize the disulfide bonds.
Biological Assays.
Monocytes (8), lymphocytes (9), and neutrophils (10) were isolated according to established methods. The release of N-acetyl-β-d-glucosaminidase from monocytes (8) and the release of elastase from neutrophils were tested as described (10). Chemotaxis was assessed in 48-well chambers (Neuroprobe, Cabin John, MD) by using polyvinylpyrrolidone-free polycarbonate membranes (Nucleopore, Neuroprobe) with 5-μm pores for 5 × 104 monocytes, eosinophils, and neutrophils and 3-μm pores for 105 lymphocytes (11). All assays were carried out in triplicate, and the migrated cells were counted in five randomly selected fields at 1,000-fold magnification after migration for 30 min (polymorph nuclear neutrophils) or 1 h (monocytes, eosinophils, and lymphocytes). Changes in the cytosolic free Ca2+ concentration ([Ca2+]i) were measured in monocytes, eosinophils, neutrophils, and lymphocytes loaded with Fura-2 (12). Receptor desensitization was tested in monocytes, eosinophils, neutrophils, and lymphocytes by monitoring [Ca2+]i changes upon repeated chemokine stimulation at 90-s intervals (8).
RESULTS
HCC-1 and HCC-2 Genes.
Analyzing the 5′-flanking region of the gene encoding HCC-1 (1), we identified an adjacent gene encoding a CC chemokine, HCC-2. 5′-rapid amplification of cDNA ends–PCR of the HCC-1 cDNA and analysis of the 20-kbp insertion in the λ phage clone that was isolated during the genomic screening for HCC-1 showed that an additional ORF of 339 bp encoding HCC-2 (113 amino acids, calculated Mr 12, 238, pI 7.9) was located upstream of the HCC-1 ORF. The two genes are separated by 12 kbp and reside in a head-to-tail orientation (Fig. 1) on chromosome 17 (data not shown), which is in agreement with the YAC contig-based mapping of human CC chemokine genes on chromosome 17q11.2 (13).
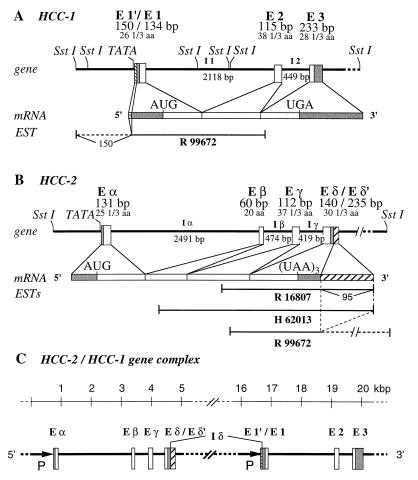
Figure 1.
Schematic representation of the genes and mRNAs for the human HCC-1 (A) and HCC-2 (B), and the HCC-2/HCC-1 gene complex (C). The open and filled boxes represent coding and untranslated regions, respectively. Hatched boxes correspond to the nontranslated regions for monocistronic messages of the tandem gene. The sizes (bp) of exons (E) and introns (I), start (AUG) and stop (UGA or UAA) codons, the numbers of amino acids (aa), and the expressed sequence tag (EST) clones (R99672, R16807, H62013) are indicated. Promoters (P) are marked by arrows. The entire gene complex consists of six SstI restriction fragments (16 kbp, 2.2 kbp, 1.813 kbp, 0.542 kbp, 196 bp, and 16 bp). TATA-box motifs are located upstream of the transcription start in both genes. Exon E δ′ and E 1′ are only present in the monocistronic cDNAs.
Like other human CC chemokine genes, the HCC-1 gene consists of three exons and two introns (Fig. 1A). The promoter was identified by primer extension and RT-PCR analysis by using total RNA from human liver, tonsils, bone marrow, and HUH-7 hepatoma cells. No sequence homology to promoters of other chemokine genes was found. The region contains putative binding sites for the transcription factors Myc-Max (−332 to −319) (14), E47 (−684 to −699) (15), and AP-2 (−243 to −236 and −750 to −757) (16). In contrast, the HCC-2 gene consists of four exons and three introns (Fig. 1B), and the promoter contains putative binding sites for the transcription factors IRF-2 (−357 to −345) (17), ELK-1 (−46 to −61) (18), and YY-1 (−738 to −756) (19).
Transcripts of the HCC-2/HCC-1 Gene Complex.
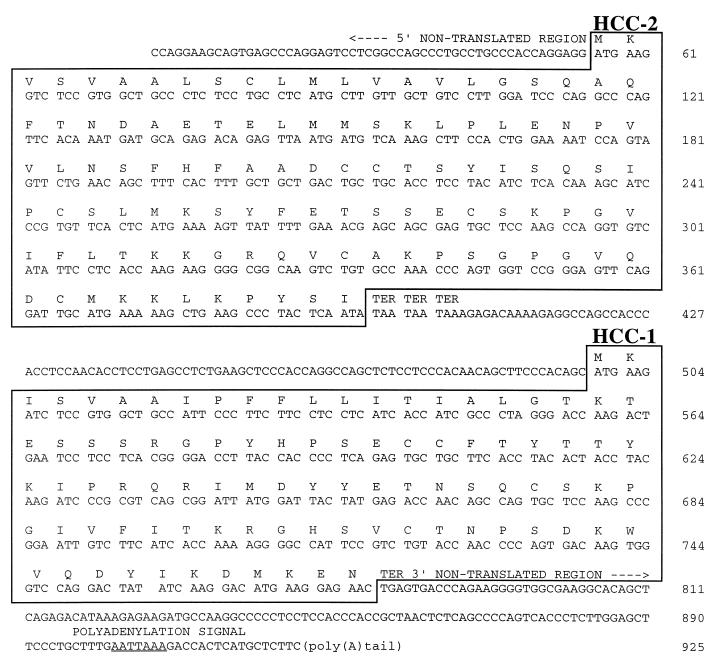
Recently, an HCC-1-specific expressed sequence tag (EST) clone was described (GenBank accession no. R99672) that lacks the first 16 nucleotides of exon E1′ of the HCC-1 gene but has a 5′-terminal extension of 150 nucleotides (Fig. 1A). As determined by sequence comparison, this region is represented within the HCC-2 gene. From the arrangement of the genes encoding HCC-2 and HCC-1 on chromosome 17 and the nucleotide sequence of the EST clone R99672, the existence of a mature bicistronic mRNA is obvious (Fig. 1). To ascertain that the bicistronic HCC-2/HCC-1 transcript is generated in normal cells, we amplified the full length cDNA from adult liver cDNA. The complete transcript of 925 nucleotides contains the HCC-2 and the HCC-1 ORF and has the same 3′-nontranslated region as the monocistronic HCC-1 mRNA. As shown in Fig. 2, the two ORFs are separated by 104 nucleotides.
Figure 2.
Nucleotide and amino acid sequence of bicistronic HCC-2/HCC-1 cDNA. The 5′-nontranslated region consists of 55 bp, the HCC-2 coding region of 339 bp, the intergene sequence of 104 bp, the HCC-1 coding region of 279 bp, and the 3′-nontranslated region of 148 bp. The ORFs are highlighted by boxes and the polyadenylation signal is underlined. TER indicates the translation stop. The sequence has been deposited in the EMBL database (accession no. Z70292).
In addition to the EST clone R99672, which represents part of the bicistronic HCC-2/HCC-1 cDNA, we found two HCC-2 cDNA fragments within the EST database (GenBank accession numbers R16807 and H62013), which are indicative of monocistronic HCC-2 mRNA (Fig. 1B). The 3′-nontranslated region of these clones matches the first 95 bases of the intron δ (which is the intergenic sequence in monocistronic expression), suggesting that the corresponding mRNA is derived from the same gene. Not present in the exon δ of the bicistronic transcripts (Fig. 1C), this mRNA contains an AAUAAA motif 72 nucleotides downstream of the exon δ/intron δ-splice junction serving as a signal for cleavage and polyadenylation of the transcript. The fourth exon of the monocistronic HCC-2 transcript is designated as exon δ’ (Fig. 1B).
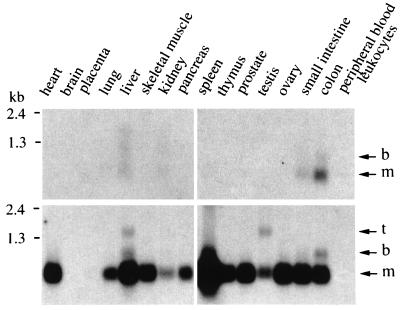
The expression of the bicistronic and monocistronic transcripts was assessed by Northern blot analysis in several tissues (Fig. 3). Monocistronic HCC-1 mRNA was highly expressed in all tissues examined except brain, placenta, and leukocytes, whereas monocistronic HCC-2 mRNA was only detectable in the colon and, at low levels, in the small intestine and liver. The bicistronic HCC-2/HCC-1 transcript occurred in colon and liver. A third transcript hybridizing with the HCC-1-specific cDNA probe was detected in the testis and liver.
Figure 3.
HCC-2 and HCC-1 expression in tissues. The blots were sequentially hybridized with 32P-labeled cDNA probes specific for HCC-2 (Upper) and HCC-1 (Lower). The position of the RNA markers are indicated. Arrows mark the positions of the monocistronic (m) and bicistronic (b) HCC-2 and HCC-1 transcripts and an additional, third (t) HCC-1 transcript.
Expression and Analysis of HCC-2.
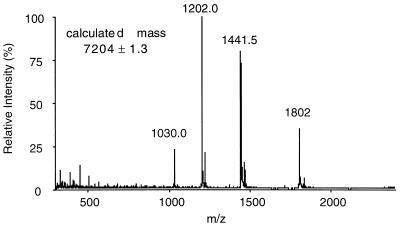
The main form of HCC-2 secreted by the recombinant yeast strain used for expression was a protein of 66 amino acids. At more than 99% purity, HCC-2 had a mass of 7,204 ± 1.3 Da (Fig. 4). The mass value remained unchanged after carboxamidomethylation, indicating the absence of free sulfhydryl groups. Analysis of proteolytic fragments clearly showed that the two additional cysteines (Fig. 5, Cys c and f) form a disulfide bond (data not shown). In Fig. 5, HCC-2 is aligned with several related human CC chemokines. It shares 68% identical amino acids with CKβ8 (20), a recently identified chemokine with six cysteines at identical positions. The amino acid sequence identity of HCC-2 with MIP-1α, HCC-1, and I-309 was 56%, 39%, and 33%, respectively.
Figure 4.
Electrospray MS of purified recombinant HCC-2. The molecular mass was calculated from the four multiple charged ions and did not change after carboxamidomethylation, indicating the presence of the three disulfide bonds.
Figure 5.
Amino acid sequence of HCC-2 aligned with CKβ8, MIP-1α, HCC-1, and I-309. The cysteines are highlighted by boxes, and identical amino acids are marked by asterisks.
Chemotaxis.
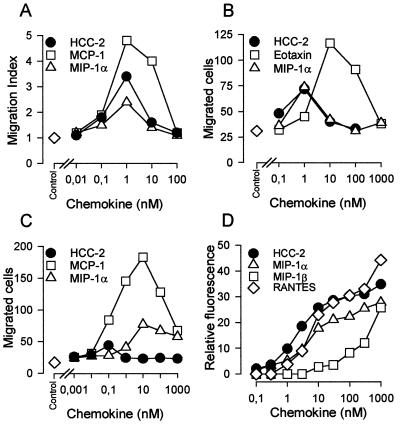
HCC-2 was tested as a potential chemoattractant for neutrophils, eosinophils, monocytes, and lymphocytes. As shown in Fig. 6 A-C, HCC-2 induced migration of monocytes, eosinophils, and lymphocytes with a typical bimodal concentration dependence. No activity, however, was found on neutrophils. In monocytes, HCC-2 was as potent as MIP-1α and monocyte chemoattractant protein (MCP)-1, as indicated by the concentration (1 nM) at which maximum migration was observed and had an intermediate efficacy (numbers of migrated cells). On eosinophils, HCC-2 and MIP-1α had similar chemotactic activity. They were both more potent, but less efficacious, than eotaxin. HCC-2 also induced the migration of interleukin 2-conditioned T lymphocytes but was considerably less effective than MCP-1 and MIP-1α. In three independent experiments, the chemotactic index (mean ± SD) was 7.2 ± 2.4 for MCP-1, 4.0 ± 0.8 for MIP-1α, and 2.9 ± 0.5 for HCC-2.
Figure 6.
Chemotaxis of human leukocytes and enzyme release from monocytes induced by recombinant HCC-2 and other CC chemokines. Chemotactic responses of human monocytes (A), eosinophils (B), and lymphocytes (C) are shown. For monocytes, the migration index is given rather than mean numbers of migrated cells per five high power fields in triplicate wells. One experiment representative for three performed with cells from different donors is shown. (D) Release of N-acetyl-β-d-glucosaminidase from cytochalasin B-treated human blood monocytes in response to increasing concentrations of HCC-2 and related CC chemokines. Enzyme activity is expressed as relative fluorescence, and each symbol represents mean values from duplicate determinations. One experiment representative for three performed with cells from different donors is shown.
Enzyme Release.
In view of the chemotactic activity observed, HCC-2 was tested as inducer of N-acetyl-β-d-glucosaminidase release in monocytes. As shown in Fig. 6D, HCC-2 was as effective as MIP-1α and RANTES and was clearly superior to MIP-1β. In agreement with the lack of chemotaxis, HCC-2 did not induce elastase release in neutrophils up to a concentration of 1,000 nM (data not shown).
[Ca2+]i Changes and Receptor Usage.
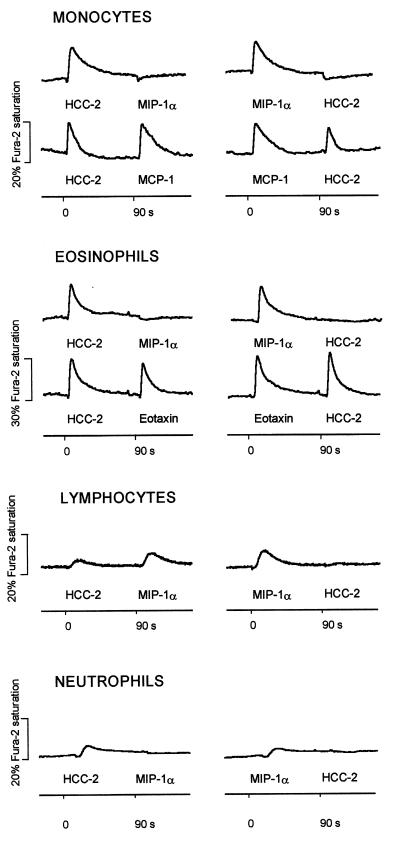
HCC-2 elicited a marked, transient rise in [Ca2+]i in monocytes and eosinophils. Weak responses were observed in lymphocytes and, interesting to note, in neutrophils (Fig. 7). [Ca2+]i changes after repeated stimulation were monitored to assess receptor usage. Complete cross-desensitization between HCC-2 and MIP-1α was observed in monocytes, eosinophils, and neutrophils, indicating that both chemokines fully share receptors on these cells. In monocytes, cross-desensitization also was observed between HCC-2 and RANTES or MIP-1β (data not shown). On the other hand, stimulation with HCC-2 did not affect the response of monocytes to MCP-1, indicating that CCR2, the MCP-1 receptor, does not recognize HCC-2. In eosinophils, no cross-desensitization was observed between HCC-2 and eotaxin, indicating that HCC-2 does not act with high affinity on CCR3. Lymphocytes express receptors that recognize MIP-1α and HCC-2. The response to HCC-2 was abrogated by pretreatment with MIP-1α, whereas the response to MIP-1α was only slightly attenuated by pretreatment of HCC-2. These observations suggest that lymphocytes may express receptors that recognize MIP-1α but not HCC-2. We can, however, not exclude the point that MIP-1α and HCC-2 bind to the same receptors, with MIP-1α having a higher affinity.
Figure 7.
[Ca2+]i changes and cross-desensitization. Fura-2-loaded cells were stimulated sequentially at 90-s intervals with 50 nM HCC-2 and other CC chemokines, and [Ca2+]i-dependent fluorescence changes were recorded. The tracings are representative for three separate experiments performed under identical conditions with cells from different donors.
DISCUSSION
HCC-2 is a chemokine whose gene is located on chromosome 17 in close apposition to the gene of HCC-1. Different transcripts for both were found, including a bicistronic mRNA. Because the two genes are separated by a 12-kbp region, the bicistronic mRNA is most likely generated from a 20-kbp pre-mRNA whose synthesis starts upstream of the HCC-2 gene and ends downstream of the HCC-1 gene. Based on the splice consensus rule (21), the junction sequences (exon δ/GTGAGT-12 kbp-AG/exon 1) are recognized as splice sites, and therefore the 12-kbp intergene sequence (intron, I δ) is removed resulting in the mature bicistronic mRNA. Bicistronic mRNAs are known to occur in bacteria and viruses but are very rare in mammals. It has been reported that mammalian mRNAs contain short ORFs located upstream of the main coding region (for review see ref. 22), but their importance is unclear. An mRNA with two large ORFs has been described for the human and the murine growth/differentiation factor 1 genes (23). Because growth/differentiation factor 1 is encoded by the downstream ORF and was detected in tissues exclusively generating the bicistronic mRNA, the existence of an internal ribosome entry site within this mRNA was proposed (24). Similarly, both ORFs within the HCC-2/HCC-1 bicistronic mRNA, which is generated in colon and liver, code for functional proteins. In view of the close proximity of the two genes, however, it is interesting that monocistronic HCC-1 mRNA is expressed at high levels in many different tissues whereas HCC-2 mRNA only was found in intestine and liver. A third transcript of 1.5 kb was detected in liver and testis by using a HCC-1-specific probe. The nature of this mRNA is still unknown. The occurrence of bi- or even polycistronic mRNA may represent a mechanism for the fine-tuning of chemokine gene expression.
Unlike other human CC chemokine genes, the HCC-2 gene consists of four exons and three introns, a gene arrangement that was reported for the murine CC chemokine C10 (25). Exon 2 codes for a 20-amino acid stretch in the NH2-terminal region of HCC-2 (16 amino acids for C10), which does not occur in other CC chemokines. It is possible that CKβ8, which contains six cysteines and is structurally closely related to HCC-2, exhibits the same four-exon-three-intron gene structure.
HCC-1 and HCC-2 belong to the CC chemokines that, on the basis of sequence similarity, can be subdivided into two subgroups, one including the MCPs and eotaxin and the other including RANTES, MIP-1α, MIP-1β, and CKβ8 (26). HCC-1 and HCC-2 clearly belong to the second group. Like the human CKβ8 and the murine C10 (25), CCF18/MRP-2 (27), and MIP-1γ (28), HCC-2 has six instead of four conserved cysteines. As shown by the present data, the two additional cysteines of HCC-2 form a third disulfide bond. Because the positions of the additional cysteines are conserved, it can be assumed that the third disulfide bond also occurs in CKβ8 and the murine homologues. The additional disulfide fixes the COOH-terminal region to the core of the molecule and is likely to restrict its mobility to a great extent. Ongoing NMR studies will clarify the three-dimensional structure of this type of chemokine in comparison to CC chemokines with four cysteines where the distal COOH-terminal region is disordered conformationally (29, 30).
The biological activities of HCC-2 were tested on peripheral blood leukocytes. HCC-2 shows striking functional similarity to MIP-1α. HCC-2 is a chemoattractant with high potency for monocytes and eosinophils, like MIP-1α (31). It mainly interacts with the receptors for MIP-1α as shown by cross-desensitization experiments. Full cross-desensitization between HCC-2 and MIP-1α on eosinophils and neutrophils clearly suggests that HCC-2 acts via the RANTES and MIP-1α receptor CCR1. It has been shown previously that CCR1 (32) is expressed at low levels in eosinophils and neutrophils (33). The present desensitization experiments showed a complete lack of cross-desensitization between HCC-2 and MCP-1 on monocytes and HCC-2 and eotaxin on eosinophils, indicating that HCC-2 does not interact with high affinity with the corresponding receptors, CCR2 and CCR3. The lack of functional responses on neutrophils, where only minor [Ca2+]i changes were observed, is not surprising because similar results were obtained previously with MIP-1α (34). In lymphocytes, HCC-2 induced only weak calcium fluxes and chemotaxis. The fact that MIP-1α was significantly more effective suggests the involvement of receptors that are recognized by MIP-1α but not by HCC-2. Except for this difference, our results indicate that HCC-2 is biologically more related to MIP-1α than to any other known chemokine.
Note.
Meanwhile, two additional papers on a chemokine identical to HCC-2 have been published. In these papers, the CC chemokine is designated as MIP-5 (35) and leukotactin-1 (36), respectively. The data reported are mainly in agreement with the data of our group. However, the organization of the HCC-2/HCC-1 gene complex is not reported within these papers.
Acknowledgments
We thank Dr. Ian Clark-Lewis and Dr. Knut Adermann for samples of chemically synthesized chemokines and Andrea Blaser and Peter Pietrzyk for expert technical assistance. This study was supported in part by a grant from the Swiss National Science Foundation (31–039744.93) and the “Stiftung zur Förderung der wissenschaftlichen Forschung” of the University of Bern. Human donor blood buffy coats were provided by the Swiss Central Laboratory Blood Transfusion Service, SRK.
ABBREVIATIONS
- MIP
macrophage inflammatory protein
- MCP
monocyte chemoattractant protein
- [Ca2+]i
cytosolic free calcium concentration
- CCR
CC chemokine receptor
- I
inosine nucleotide
- r
recombinant
- EST
expressed sequence tag
- CC
chemokine type CC
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Z70292).
References
- 1.Schulz-Knappe P, Mägert H J, Dewald B, Meyer M, Cetin Y, Kubbies M, Tomeczkowski J, Kirchhoff K, Raida M, Adermann K, et al. J Exp Med. 1996;183:295–299. doi: 10.1084/jem.183.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill O, Kuhn M, Zucht H D, Cetin Y, Kulaksiz H, Adermann K, Klock G, Rechkemmer G, Forssmann W G, Mägert H J. Proc Natl Acad Sci USA. 1995;92:2046–2050. doi: 10.1073/pnas.92.6.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith L M, Sanders J Z, Kaiser R J, Hughes P, Dodd C, Connell C R, Heiner C, Kent S B H, Hood L E. Nature (London) 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 5.Quandt K, Frech C, Karas H, Wingender E, Werner T. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill O, Cetin Y, Cieslak A, Mägert H J, Forssmann W G. Biochem Biophys Acta. 1995;1253:146–149. doi: 10.1016/0167-4838(95)00204-4. [DOI] [PubMed] [Google Scholar]
- 7.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 1–3. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 8.Uguccioni M, D’Apuzzo M, Loetscher M, Dewald B, Baggiolini M. Eur J Immunol. 1995;25:64–68. doi: 10.1002/eji.1830250113. [DOI] [PubMed] [Google Scholar]
- 9.Loetscher P, Seitz M, Baggiolini M, Moser B. J Exp Med. 1996;184:569–577. doi: 10.1084/jem.184.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peveri P, Walz A, Dewald B, Baggiolini M. J Exp Med. 1988;167:1547–1559. doi: 10.1084/jem.167.5.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uguccioni M, Loetscher P, Forssmann U, Dewald B, Li H, Lima S H, Li Y, Kreider B, Garotta G, Thelen M, et al. J Exp Med. 1996;183:2379–2384. doi: 10.1084/jem.183.5.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Tscharner V, Prod’hom B, Baggiolini M, Reuter H. Nature (London) 1986;324:369–372. doi: 10.1038/324369a0. [DOI] [PubMed] [Google Scholar]
- 13.Naruse K, Ueno M, Satoh T, Nomiyama H, Tei H, Takeda M, Ledbetter D H, van Coillie E, Opdenakker G, Gunge N, et al. Genomics. 1996;34:236–240. doi: 10.1006/geno.1996.0274. [DOI] [PubMed] [Google Scholar]
- 14.Blackwood E M, Eisenman R N. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 15.Voronova A, Baltimore D. Proc Natl Acad Sci USA. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell P J, Wang C, Tjian R. Cell. 1987;50:847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- 17.Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 18.Hipskind R A, Rao V N, Mueller C G F, Reddy E S P, Nordheim A. Nature (London) 1991;54:531–534. doi: 10.1038/354531a0. [DOI] [PubMed] [Google Scholar]
- 19.Shi Y, Seto E, Chang L S, Shenk T. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 20.Forssmann U, Delgado M B, Uguccioni M, Loetscher P, Garotta G, Baggiolini M. FEBS Lett. 1997;408:211–216. doi: 10.1016/s0014-5793(97)00408-0. [DOI] [PubMed] [Google Scholar]
- 21.Padgett R A, Grabowski P J, Konarska M M, Seiler S, Sharp P A. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. Cell. 1986;47:481–483. doi: 10.1016/0092-8674(86)90609-4. [DOI] [PubMed] [Google Scholar]
- 23.Lee S J. Proc Natl Acad Sci USA. 1991;88:4250–4254. doi: 10.1073/pnas.88.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mountford P S, Smith A G. Trends Genet. 1995;11:179–184. doi: 10.1016/S0168-9525(00)89040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger M S, Kozak C A, Gabriel A, Prystowsky M B. DNA Cell Biol. 1993;12:839–847. doi: 10.1089/dna.1993.12.839. [DOI] [PubMed] [Google Scholar]
- 26.Baggiolini M, Dewald B, Moser B. Adv Immunol. 1994;55:97–179. [PubMed] [Google Scholar]
- 27.Youn B S, Jang I K, Broxmeyer H E, Cooper S, Jenkins N A, Gilbert D J, Copeland N G, Elick T A, Fraser M J, Kwon B S. J Immunol. 1995;155:2661–2667. [PubMed] [Google Scholar]
- 28.Poltorak A N, Bazzoni F, Smirnova I I, Alejos E, Thompson P, Luheshi G, Rothwell N, Beutler B. J Inflam. 1995;45:207–219. [PubMed] [Google Scholar]
- 29.Clark-Lewis I, Kim K S, Rajarathnam K, Gong J H, Dewald B, Moser B, Baggiolini M, Sykes B D. J Leukocyte Biol. 1995;57:703–711. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- 30.Clore G M, Gronenborn A M. FASEB J. 1995;9:57–62. doi: 10.1096/fasebj.9.1.7821760. [DOI] [PubMed] [Google Scholar]
- 31.Rot A, Krieger M, Brunner T, Bischoff S C, Schall T J, Dahinden C A. J Exp Med. 1992;176:1489–1495. doi: 10.1084/jem.176.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neote K, DiGregorio D, Mak J Y, Horuk R, Schall T J. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- 33.Combadiere C, Ahuja S K, Murphy P M. DNA Cell Biol. 1995;14:673. doi: 10.1089/dna.1995.14.673. [DOI] [PubMed] [Google Scholar]
- 34.McColl S R, Hachicha M, Levasseur S, Neote K, Schall T J. J Immunol. 1993;150:4550–4560. [PubMed] [Google Scholar]
- 35.Coulin F, Power C A, Alouani S, Peitsch M C, Schroeder J M, Moshizuki M, Clark-Lewis I, Wells T N. Eur J Biochem. 1997;248:507–515. doi: 10.1111/j.1432-1033.1997.00507.x. [DOI] [PubMed] [Google Scholar]
- 36.Youn B S, Zhang S M, Lee E K, Park D H, Broxmeyer H E, Murphy P M, Locati M, Pease J E, Kim K K, Antol K, et al. J Immunol. 1997;159:5201–5205. [PubMed] [Google Scholar]