Abstract
As genetic model systems, fish have played a key role in our understanding of a wide range of biological processes, including vertebrate pigmentation. In this review, we focus on one aspect of pigmentation, skin pigmentation, which has been of momentous importance in human history. Two fish models, medaka and zebrafish, played important roles in demystifying skin color and, by extension, the concept of “race.” Related thinking has the potential to make two additional contributions to human welfare. Fish can be used to validate gene candidates from genome-wide association studies (GWAS) in what has been called “Systems Genetics.” Because fish are familiar vertebrates, and share genetic mechanisms of skin color with humans, they also have outstanding potential as an educational tool—to “demystify” race, to increase public understanding of the role of model systems and evolution in science, and to enhance appreciation of both genetic and environmental factors that impact human health and society.
Introduction
This special issue of Zebrafish is dedicated to pigmentation, a subject associated with a fascinating diversity of interesting scientific questions. This paper focuses on one of those questions—what is the origin of variation in human skin color?—and implications of this work for research using fish, for science, and for society. We begin by reviewing the key role that medaka and zebrafish played in the discovery of two genes that make the largest known contributions to the lighter skin color of Europeans. The first is melanoma-associated transporter protein (MATP)1 (also known as associated in melanoma 1 [AIM1] and solute carrier protein 45A2 [SLC45A2]). The second gene is SLC24A52 (also known as golden and NCKX5–potassium-dependent sodium–calcium exchanger). We discuss some of the important contributions that these fish models have made to research in these proteins, and the contributions that fish functional genomics can make to genome-wide association studies in humans. We will close by using basic fish crosses to illustrate how a better public understanding of genetics and environment can make positive contributions to societal understanding of research, model systems, and human genetics and ancestry.
Medaka, Zebrafish, Mice, and Skin Color
Dozens of genes affecting pigmentation have been found in zebrafish3,4 and medaka.5 The first fish pigmentation gene to be linked to normal variation in human skin color is the first gene to be positionally cloned in medaka, the B gene, known in humans as melanoma-associated transporter protein (MATP).1 In concurrent work, this gene was also linked to the mouse pigmentation defect, underwhite, which the investigators recognized to be involved in human oculocutaneous albinism type 4, or OCA4.6 Polymorphisms in this gene have been linked to European skin color. In particular, single-nucleotide polymorphism (SNP), rs16891982, is a C-to-A trans-version that changes the ancestral leucine at amino acid 374 to phenylalanine (Leu374Phe). This polymorphism is associated with a significant contribution to the light color of European skin,7 and is predominant in Northern Europeans, with decreasing frequency as one proceeds to the southern parts of Europe and the Middle East.8 An impressive characterization of a centuries-old collection of medaka pigmentation mutants9,10 and a set of radiation- and ethylnitrosourea (ENU)-induced mutations in the B gene contributed to the original finding in medaka.11 Of those mutants, a number of spontaneous medaka mutants, including allele, b, have light skin, but normally colored eyes. Recent work with this mutant has revealed the presence of alternative transcripts, the shorter of which is eye specific and expressed in the presence of the b mutation, and the longer of which is missing in b, and appears to be required for expression of slc45a2 in the skin. The b mutation was found in upstream noncoding sequence.12 This work will allow the study of cis/trans regulatory mechanisms underlying the ocular and cutaneous expression of slc45a2, which may in turn help to elucidate the relative roles of SLC45A2 in human skin, eye, and hair color.7
In contrast to the story for MATP, SLC24A5 has not been associated with albinism in humans or other mammals. The gene was discovered in the context of the positional cloning of the first mutation studied in zebrafish—the light-skinned variant, golden.13 The first hint that this mutation might relate to humans was the ultrastructural phenotype of golden, as compared with wild-type melanophores, the fish equivalent of human melanocytes. The golden phenotype was associated with a diminution of number, size, and pigment density of the melanin-containing organelle of melanophores/melanocytes: melanosomes—changes shared with those of human light skin. The golden b1 mutation was a nonsense mutation in a potassium-dependent sodium–calcium exchanger, whose function is conserved from zebrafish to humans, as demonstrated by complementation of the golden melanophore pigmentation defect by the human gene. Work with human SNP data at hapmap.org showed that the human ortholog SLC24A5 contains a single coding SNP, rs1426654, changing the ancestral guanine to adenine, corresponding to a change from alanine (conserved in all vertebrate orthologs sequenced to date) to threonine at amino acid position 111 (Ala111Thr). This polymorphism diminishes sodium–calcium exchange14 and contributes to 25–38% of the skin color difference between Africans and Europeans.2 It was particularly striking to find that the region around SLC24A5 shows the strongest signature of evolutionary selection in the entire genome—exclusively for the CEU Northwestern European population of the HapMap (www.hapmap.org). The size of the accompanying region of diminished variation (about 150 kb around SLC24A5) is the largest in the entire genome in that population. This compares with about 22 kb for MATP. Due to the necessity for UV light to generate adequate Vitamin D in the skin, it appears that the adaptive mutation resulting in the Ala111Thr polymorphism was necessary for Europeans to live at high latitudes.15
One of the questions raised is why the gene was not recognized as a pigment gene in mice. Mutations in Slc24a5 were not found in any of the hundreds of pigment variants of mice because, as we now know, knocking out this gene does not cause a visible hair color change.16 Because skin lightening was noted in the skin of the ears and nose of these knockouts, however, mouse mutant alleles for this gene may potentially be found in mutant screens for lighter skin in mice had they been done; they were not found in a screen for darker skin in mice.17 Close inspection revealed defects in ocular pigmentation in Slc24a5 knockout mice. A defect in the retinal pigmentation in these mice, together with the presence of SLC24A5 in a region associated with human age-related macular degeneration in a genome-wide association study (GWAS),18 is consistent with a possible role for SLC24A5 in this most common form of acquired blindness in humans. In contrast to the mouse phenotype, the original zebrafish golden b1 mutants, which lack slc24a5, show a more obvious skin color phenotype. Fish melanocytes are more human in their epidermal location than mouse body melanocytes, though no transfer of melanosomes to keratinocytes occurs in zebrafish, as it does in humans.19 These findings support the contention that work on non-mammalian model systems such as fish yields information that cannot be as readily obtained from mice.
Systems Genetics
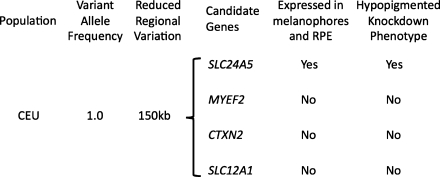
So what role might fish play in determining which genes contain phenotypically important variations in humans? The approach that takes advantage of the power of zebrafish reverse genetics is systems genetics, in which model systems are used in conjunction with GWAS to verify the involvement of specific genes and polymorphisms that are associated with disease by GWAS. An avalanche of GWAS for a host of human disease phenotypes has been completed.20 The systems genetics approach provides opportunities for both validation and the study of disease mechanism using fish (Fig. 1). The inquiries typically used for validation include the determination of expression pattern, which in zebrafish is whole-mount in situ hybridization21 and antisense morpholino knockdown phenotypes.22 So what is an ideal class of phenotypes that might be used to test this idea? Given that phenotypes such as high blood pressure and diabetes have strong environmental factors that can be difficult to control for, it will be beneficial to test the idea of systems genetics using a phenotype that can be measured quantitatively, and has a minimal environmental contribution. Human skin pigmentation can be quantitatively measured from areas of skin that do not receive much light (inner aspect of the upper arm), using instruments that measure reflected light.23 Skin color therefore represents an ideal model for testing the potential power of systems genetics. Indeed, morpholino analysis in zebrafish has been shown to be useful as a functional genomics tool for analysis of pigmentation.24 As illustrated in Fig. 1, the combination of in situ and morphant analysis for zebrafish slc24a5, in combination with GWAS mapping of skin color in South Asia to SLC24A5,25 validates this systems genetics approach. If existing methods are unable to figure out how pigmentation works, how likely is it that we can understand more complicated phenotypes?
FIG. 1.
Concept of Systems Genetics. Work with SLC24A52,25 taken in reverse, provides a paradigm for systems genetics. A genome-wide association study (GWAS) performed for light skin color using South Asians25 revealed a non-ancestral allele of SLC24A5 that played a role in skin color in that population. Our complementary studies with zebrafish and HapMap populations2 showed near-exclusivity of this allele within the CEU population, within a region of diminished variation that is not present in a control population (YRI). Testing each of four candidate genes within this region for tissue patterns of gene expression and function in zebrafish using whole-mount in situ hybridization and morpholino knockdowns, respectively, pointed to SLC24A5 as a skin color gene.
There appear to be some 1500 proteins in human melanosomes.26 Some of these proteins may have been contaminants of the melanosome preparation. Among the actual melanosome-specific proteins, there might be some that will not affect pigmentation if they are missing. The number of known polymorphisms is highly variable. Most of these will be phenotypically inconsequential. A small proportion of the polymorphisms will correlate with phenotypic variations. These would be the target for GWAS to identify, for example, the genes involved in causing the light skin of East Asians and Amerindians. It will be interesting to see a decade from now how much zebrafish and medaka will have contributed to validation of disease markers found in GWAS.
Potential Educational Roles for Fish Genetics in Science, Education, and Society
I assert that a basic understanding of genetics is important to society and easy to explain in the context of pigment genetics. People are generally aware of the range of human skin color, and in light of the 2008 American Presidential campaign, readily motivated by the issues surrounding it. The visual nature of pigmentation makes it easier for people to envision and understand. Fish are a good introductory animal for people who prefer to see relevance to vertebrates, and yet prefer not to consider mammals in experiments. Fish also provide compelling examples of the important role model systems have played in the ongoing revolution in the biological sciences.
“School of Fish”: Fish pigmentation thought experiments for education
Fish can be readily used to demystify skin color and to convey a more balanced appreciation of the important roles of both environmental and genetic aspects of human phenotypes. The concepts underlying these thought experiments may be most useful to educators at the high school and undergraduate levels, and for the public. While these concepts are obvious to most geneticists, they were clearly underappreciated by proponents of Eugenics (see article by Farber in this issue). Given today's gene-centric view of the biological world, society can benefit from increased appreciation of these issues among members of the lay public, and members of the scientific, medical, and pharmaceutical communities.
Thinking about different genotypic arrangements of fish living in different tanks can clarify the difference between the effects of genotype and environment. Let us start with an ancestrally dark fish that is homozygous wild-type (+/+), and a homozygous mutant (m/m) fish for a codominant pigmentation mutation in which heterozygotes are of an intermediate color. A cross between the two original strains yields all m/+ heterozygotes. So what do scientists call these individuals? We define their skin color phenotype as intermediate color, know that they are heterozygotes by genotype, and also know that half of their genomes are from the wild-type background and half from the mutant background.
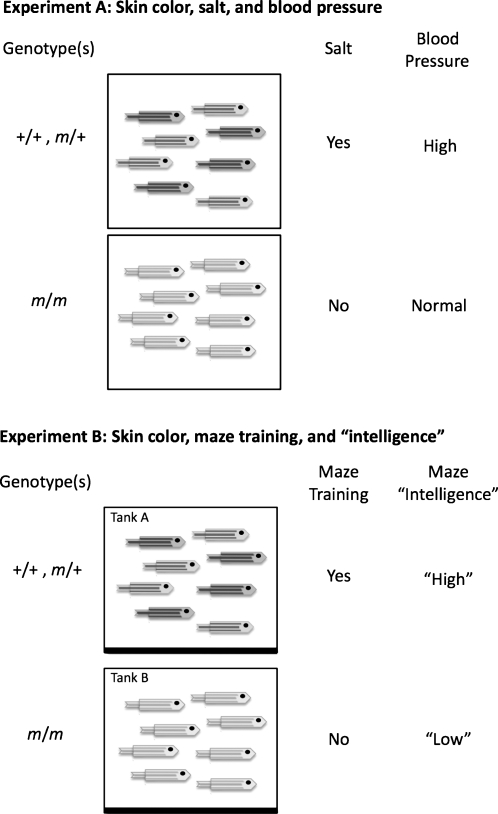
Let us now consider some simple experiments with these populations that demonstrate the importance of environment (Fig. 2). In the first experiment, we put the wild types and heterozygotes in tank A and the m homozygotes in B, and feed the A population food that is more salty, causing high blood pressure (Fig. 2A). It would be important to distinguish the A and B populations from each other for the sake of dealing with the disease. In this case, we just change to a lower salt diet (environmental) to normalize blood pressure. A color association between darker pigmentation and blood pressure shown by GWAS would be a red herring, because reversing the diet would cause the B population to have high blood pressure. This experiment indicates how genetic factors can mislead us into under-appreciating environmental factors in disease.
FIG. 2.
A “School of Fish” for teaching the importance of both environment and genetics in human phenotypes. (A) In the first thought experiment, a GWAS for high blood pressure reveals a linkage to dark skin color. However, salt fed to fish in that tank was responsible for the blood pressure elevation, not skin color. (B) In the second thought experiment, a GWAS for a specific measure of “intelligence” reveals a linkage to dark skin color. However, special maze education, not the skin color, was responsible for the difference observed. Other environmental influences can be substituted to show how misleading genetics is when environmental influences are not adequately considered.
So what are the human implications of this experiment? Most people of varying proportions of African/European ancestry self-identify as black or African-American. This designation is medically useful when this self-identification is associated with cultural factors such as salt intake that are important in disease. If a person of higher-risk ancestry lives in a healthier environment, the cultural environment, whether defined by skin color or nationality, or language, will matter less. If a person with a lower-risk ancestry lives in an unhealthy environment, their ancestry may be of no help. The growing cultural diversity of African-American populations is diminishing the medical relavance of pigmentation phenotypes.
In our second experiment, let us provide the residents of tank A (the darker ones) with maze training, which offers a learning opportunity to get more food, but not allow fish in tank A to have that training. Let us consciously label maze ability as “intelligence” and only test that. The fish in tank A will be measured as more “intelligent” than those in tank B (Fig. 2B). Does this mean that dark fish are more intelligent? No. Now let us reverse the training, which would cause fish in tank B to be more intelligent. Is this due to genotype? Of course not. It is again due to the environment. It is this very issue that renders invalid attempts to link intelligence to human skin color.
To study the genetic link between color polymorphisms and fractional ancestry, let us do a third experiment in which we look at the progeny of crosses between heterozygotes (m/+). As you have learned from Mendelian genetics, such crosses yield ¼ +/+, ½ m/+, and ¼ m/m (the classic 1:2:1 ratio). On average, for all of these fish, ½ of their genomes will come from each of the original two populations. Due to re-assortment and recombination during meiosis, the color does not necessarily correlate with the average fraction of the original wild-type genome. This experiment illustrates the scientific invalidity of the one-drop rule, which labels people with “any” African ancestry as “black.”27
Let us now use a fourth fish experiment in which we allow many generations of interbreeding. A wide range of proportionality of genomes of each of the original strains will be generated. Let us say that polymorphism x is present originally only in the light-skinned m population, is unlinked to the m chromosome, and causes high blood pressure. Due to lack of linkage, if a descendant happens to have light skin color (m/m), the presence of the x polymorphism is not assured. The overall probability of having the x polymorphism is proportional to the fraction of their genomes being of the genetic background of the original m population, but on an individual basis, there is no assurance of a correspondence between skin color and high blood pressure. This experiment clarifies the issue of gene segregation, and explains why testing at defined loci is far superior to using race or skin color as a proxy for the real susceptibility. These arguments obviously apply to the first racial drug, BiDil.28
Using these thought experiments, we illustrated how the frame of thinking associated with “race” can mislead us into thinking that phenotypes are caused exclusively by genotype, and causes us to ignore the responsible environmental factors. Ancestry, as determined by haplotype, is interesting to be aware of. We can today assign proportions of particular ancestries contributing to an individual's genome29 if we need to. As illustrated above, on an individual basis, and especially for individuals of multiple ancestries, it is most desirable to focus on the specific polymorphisms in specific genes that are associated with specific responses to drugs. In particular, pigmentation-based proxies for disease susceptibility genotypes that can segregate from the relevant locus can be misleading to the individual. But is genotyping for specific disease polymorphisms the end-all? No, because genetic background often has a significant effect on phenotype. It will take education for the broader society to acquire a balanced understanding of both the need for genotypic analysis and the importance of environment in their health.
Skin pigmentation and “race”
Skin pigmentation is of such great human relevance because it is the most notable factor involved in the complicated and contentious concept of race. Due to the obvious nature of differences in skin color, it has been the most obvious focus of attempts of scientists to classify humans. Linnaeus started with four groups: white, red, yellow, and black.30 Notably, different (and progressively less desirable) personality traits were assigned exclusively to each. Further, of these four classifications, the “red” Indians found by Christopher Columbus in the Caribbean we now know to have been derived from the “yellow” East Asians—but painted red.31 Blumenbach,32 the founder of anthropology, increased the number of human categories, and declared the Caucasian skull the most beautiful. Given these unscientific classifications of people, the term “race” over the years has not surprisingly focused on differences, in the context of discussions in which superiority of people with white skin over people with darker colors was assumed. Human skin color was a primary mechanism of facilitating government-sponsored discrimination, mass sterilization, and an extension of tribalism, genocide. An alarming history of two wild distortions of Darwinism-Eugenics and Social Darwinism (reviewed by Steven Farber in this issue and by others33–35) illustrates why it is important for scientists to speak against abuses of science. Geneticists such as Dobzhansky have spoken out cogently and eloquently against racist thinking.34 These discussions among scientists are important, since they point out to the uninformed that scientists today do not believe in the social Darwinism that contaminated evolutionary teaching in the early 20th century. Whatever damage may be caused to scientific education and progress, anti-evolutionists must be first acknowledged for pointing out that many early teachings of evolution were racist, and that early anti-evolutionist legislators such as William Jennings Bryan were motivated by a sense of justice. What needs to be clarified through writings such as this is that we as scientists clearly dissociate ourselves from that past, and that we wish to share an acknowledgment of the truth of the long history of our common ancestry.
The problems with race cause scientists who work on skin color to avoid the topic, even if it is the most immediate thing people will ask about.36 It would seem responsible at this time to work toward elimination of the use of such a scientifically imprecise and misused term, and to use the available and more precise term, “ancestry.” It is equally important to recognize, when applicable, environmental factors (such as diet and toxin exposure), ethnicity, nationality, language, and culture. Sharing our understanding of pigmentation genetics with the public will serve to demystify skin color and race. Effective and appropriate communication of this understanding to the public has the potential of diminishing racist thinking and improving human health. A substantial part of this work will be to improve what has been a poor appreciation for the impacts of environment on human welfare, health, and disease. Out of our excitement for genetics in the last century, undue credit has at times been given to genetics, and important environmental factors that affect society and health, ignored. If we have learned anything from past, future education will present a more balanced view of nature and nurture.
To accomplish these educational goals, scientists—in particular, geneticists—can facilitate public understanding of genetics through participation in local, regional, and national programs such as the National Science Foundation's Geneticist-Educator Network of Alliances project (GENA). In this effort, we should take the extra trouble to separate genetic from environmental principles in the classroom. This will yield a more educated public that understands the process of science, uses critical scientific evidence to make decisions, and does not succumb to old, illogical prejudices. Meeting this challenge will result in greater scientific progress, equal opportunity, and medical advancement in our genomic era—in short, a better society.
Summary
Fish have great utility in the analysis of a range of biological processes associated with pigmentation, and have contributed fundamentally to our understanding of human skin color. Published work on MATP and SLC24A5 in zebrafish, medaka, and humans together suggests that zebrafish and medaka have the potential to be used to validate function inferred from human GWAS. Due to their prominent role in human skin color genetics, fish have great educational power to increase understanding of skin color genetics, to increase public awareness of the power of model systems, and to improve balance in scientific and public thinking about the roles of genetics and the environment in biology and human disease. The similarity between human and zebrafish pigment genes, a growing collection of fish pigmentation mutants, and accessibility of fish pigmented cells to study make it likely that fish will continue to play a significant role in elucidating the biology of pigmentation in the foreseeable future.
Acknowledgments
This work was supported by 1RO1 AR 052535 from NIAMS, the Jake Gittlen Cancer Research Foundation, and Pennsylvania Tobacco Settlement Funds. I also thank Mary Burns for insightful suggestions.
Disclosure Statement
No competing financial interests exist.
References
- 1.Fukamachi S. Shimada A. Shima A. Mutations in the gene encoding B, a novel transporter protein, reduce melanin content in medaka. Nat Genet. 2001;28:381–385. doi: 10.1038/ng584. [DOI] [PubMed] [Google Scholar]
- 2.Lamason RL. Mohideen MP. Mest JR. Wong AC. Norton HL. Aros MC, et al. SLC24A5 affects pigmentation in zebrafish and man. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 3.Kelsh RN. Brand M. Jiang YJ. Heisenberg CP. Lin S. Haffter P, et al. Zebrafish pigmentation mutations and the processes of neural crest development. Development. 1996;123:369–389. doi: 10.1242/dev.123.1.369. [DOI] [PubMed] [Google Scholar]
- 4.Parichy D. Evolution of Danio pigment pattern development. Heredity. 2006;97:200–210. doi: 10.1038/sj.hdy.6800867. [DOI] [PubMed] [Google Scholar]
- 5.Kelsh RN. Inoue C. Momoi A. Kondoh H. Furutani-Seiki M. Ozato K, et al. The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech Dev. 2004;127:841–859. doi: 10.1016/j.mod.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Newton JM. Cohen-Barak O. Hagiwara N. Gardner JM. Davisson MT. King RA, et al. Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet. 2001;69:981–988. doi: 10.1086/324340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graf J. Hodgson R. van Daal A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat. 2005;25:278–284. doi: 10.1002/humu.20143. [DOI] [PubMed] [Google Scholar]
- 8.Norton HL. Kittles RA. Parra E. McKeigue P. Maio X. Cheng KC, et al. The evolution of human skin pigmentation variation: a candidate gene and admixture mapping approach reveals the independent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 2007;24:710–722. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- 9.Packer A. Medaka on the move. Nat Genet. 2001;28:302. doi: 10.1038/91042. [DOI] [PubMed] [Google Scholar]
- 10.Wittbrodt J. Shima A. Schartl M. Medaka—a model organism from the far East. Nat Rev Genet. 2002;3:53–64. doi: 10.1038/nrg704. [DOI] [PubMed] [Google Scholar]
- 11.Shimada A. Fukamachi S. Wakamatsu Y. Ozato K. Shima A. Induction and characterization of mutations at the b locus of the medaka, Oryzias latipes. Zoolog Sci. 2002;19:411–417. doi: 10.2108/zsj.19.411. [DOI] [PubMed] [Google Scholar]
- 12.Fukamachi S. Kinoshita M. Tsujimura T. Shimada A. Oda S. Shima A, et al. Rescue from Oculocutaneous Albinism Type 4 using medaka slc45a2 cDNA driven by its own promoter. Genetics. 2008;178:761–769. doi: 10.1534/genetics.107.073387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Streisinger G. Walker C. Dower N. Knauber D. Singer F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio) Nature. 1981;291:293–296. doi: 10.1038/291293a0. [DOI] [PubMed] [Google Scholar]
- 14.Ginger RS. Askew SE. Ogborne RM. Wilson S. Ferdinando D. Dadd T, et al. SLC24A5 Encodes a trans-Golgi network protein with potassium-dependent sodium-calcium exchange activity that regulates human epidermal melanogenesis. J Biol Chem. 2008;283:5486–5495. doi: 10.1074/jbc.M707521200. [DOI] [PubMed] [Google Scholar]
- 15.Jablonski NG. Skin: A Natural History. Berkeley, CA: University of California Press; 2006. [Google Scholar]
- 16.Vogel P. Read RW. Vance RB. Platt KA. Troughton K. Rice DS. Ocular albinism and hypopigmentation defects in Slc24a5-/- Mice. Vet Pathol. 2008;45:264–279. doi: 10.1354/vp.45-2-264. [DOI] [PubMed] [Google Scholar]
- 17.Fitch KR. McGowan KA. van Raamsdonk CD. Fuchs H. Lee D. Puech A, et al. Genetics of dark skin in mice. Genes Dev. 2003;17:214–228. doi: 10.1101/gad.1023703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyengar SK. Song D. Klein BE. Klein R. Schick JH. Humphrey J, et al. Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration. Am J Hum Genet. 2004;74:20–39. doi: 10.1086/380912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beckwith LG. Moore JL. Tsao-Wu GS. Harshbarger JC. Cheng KC. Ethylnitrosourea induces neoplasms in zebrafish (Danio rerio) Lab Invest. 2000;80:379–385. doi: 10.1038/labinvest.3780042. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy MI. Abecasis GR. Cardon LR. Goldsten DB. Little J. Ioannidis JA. Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 21.Thisse C. Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 22.Nasevicius A. Ekker SC. Effective targeted gene “knockdown” in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 23.Stamatas GN. Zmudzka BZ. Kollias N. Beer JZ. Noninvasive measurements of skin pigmentation in situ. Pigm Cell Res. 2004;17:618–626. doi: 10.1111/j.1600-0749.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 24.Pickart MA. Sivasubbu S. Nielsen Al. Shriram S. King RA. Ekker SC. Functional genomics tools for the analysis of zebrafish pigment. Pigm Cell Res. 2004;17:461–470. doi: 10.1111/j.1600-0749.2004.00189.x. [DOI] [PubMed] [Google Scholar]
- 25.Stokowski RP. Krishna Pant PV. Dadd T. Fereday A. Hinds DA. Jarman C, et al. A genomewide association study of skin pigmentation in a South Asian population. Am J Hum Genet. 2007;81:1119–1132. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi A. Valencia JC. Hu ZZ. Watabe H. Yamaguchi H. Mangini NM, et al. Proteomic and bioinformatic characterization of the biogenesis and function of melanosomes. J Proteome Res. 2006;5:3135–3144. doi: 10.1021/pr060363j. [DOI] [PubMed] [Google Scholar]
- 27.Davis FJ. University Park: Pennsylvania State University Press; 1991. Who Is Black? One Nation's Definition. [Google Scholar]
- 28.Schwartz RS. Editorial: Racial profiling in medical research. N Engl J Med. 2001;344:1392–1393. doi: 10.1056/NEJM200105033441810. [DOI] [PubMed] [Google Scholar]
- 29.Li JZ. Absher DM. Tang H. Southwick AM. Casto AM. Ramachandran S, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 30.Linne C. 13th. Vindobonae [Vienna]: Ioannis Thomae; 1763. Systema Naturae. [Google Scholar]
- 31.Oppenheimer S. Out of Eden. London, England: Constable & Robinson; 2004. [Google Scholar]
- 32.Blumenbach JF. The Anthropological Treateses of Johann Friedrich Blumenbach and the Inaugural Dissertation of John Hunter, M.D. on the Varieties of Man. In: Bendyshe T, editor. London: Longman, Green, Longman, Roberts & Gates; 1865. [Google Scholar]
- 33.Black E. New York: Four Walls Eight Windows; 2003. War Against the Weak: Eugenics and America's Campaign to Create a Master Race. [Google Scholar]
- 34.Dunn LC. Dobzhansky T. Heredity, Race and Society. New York: New American Library; 1952. [Google Scholar]
- 35.Reed SC. A short history of human genetics in the USA. Am J Med Genet. 1978;3:282–295. doi: 10.1002/ajmg.1320030308. [DOI] [PubMed] [Google Scholar]
- 36.Cheng KC. Demystifying skin color and race. In: Hall RE, editor. Racism in the 21st Century. New York: Springer; 2008. pp. 3–23. [Google Scholar]