Although Cholesteryl Ester Transfer Protein (CETP) mediates the transfer of cholesteryl esters and triglycerides between lipoprotein particles and thus plays a crucial role in reverse cholesterol transport, the association of variations in the CETP gene with acute myocardial infarction (MI) remains unclear. In this study we examined whether common genetic variation in the CETP gene is related to early-onset non-fatal MI risk in a population-based case-control study from western Washington State.
Genotyping for the CETP -2708 G/A, -971 A/G, -629 A/C, Intron-I TaqI G/A and exon-14 A/G (I405V) SNPs was performed in 578 cases with first acute non-fatal MI and in 666 demographically similar controls, free of clinical cardiovascular disease, identified randomly from the community. In-person interviews and non-fasting blood specimens provided data on coronary heart disease risk factors.
In men, there was little evidence for an association between single SNPs and MI risk, but in women the age- and race-adjusted OR was found to be significant in 4 out of the 5 CETP single variants.
Haplotype analysis revealed two haplotypes associated with MI risk among men. As compared to men homozygous for the most common haplotype D (-2708 G, -971 G, -629 C, TaqI G and exon-14 A), the fully-adjusted multiplicative model identified haplotype G (-2708 G, -971 A, -629 A, TaqI G and exon-14 G) was associated with a 4.0-6.0-fold increased risk of MI for each additional copy; [95%CI 2.4-14.8] and haplotype B (-2708 G, -971 G, -629 A, TaqI A and exon-14 A) showed a significant decreased risk for early onset MI [OR=0.18; 95%CI 0.04 - 0.75]. An evolutionary-based haplotype analysis indicated that the two haplotypes associated with the MI risk are most evolutionarily divergent from the other haplotypes.
Variation at the CETP gene locus is associated with the risk of early-onset non-fatal MI. This association was found to be independent of HDL-C levels. These data and the sex-specific findings require confirmation in other populations
Introduction
Cholesteryl Ester Transfer Protein (CETP) mediates the transfer of neutral lipids such as cholesteryl esters and triglycerides between lipoprotein particles and thus plays a crucial role in reverse cholesterol transport from peripheral tissues to the liver (1). Its unique role in exchanging cholesterol esters and triglycerides from high-density lipoprotein cholesterol (HDL-C) to low density cholesterol (LDL-C) and to very low density lipoprotein cholesterol (VLDL-C) make it an appealing candidate for treatment modulation. Raising HDL-C is a relatively novel strategy in the efforts to treat atherosclerosis and CETP inhibitors have been recently developed to accomplish this (2,3). However, while on one hand lowering CETP activity may lead to increased HDL-C levels, yet on the other hand it may adversely affect reverse cholesterol transport which is considered an athero-protective mechanism by which myocardial infarction (MI) is prevented (4,5).
MI is a complex disorder with an estimated heritability that ranges between 25% and 60% (6). While numerous studies have shown that family history is a significant risk factor for atherosclerosis, its contribution cannot be fully accounted for by traditional cardiovascular risk factors, such as dyslipidemia, hypertension, smoking, and obesity (7, 8). Integration of genetic testing for determination of cardiovascular at-risk patients is an attractive idea that is widely studied. One possible setback in studying the association of polymorphisms and specific phenotypes such as early-onset MI may stem from the uncertainties as to the nature of the gene to be investigated. CETP in its unique physiological role serves as an interesting candidate gene for such studies. The gene encoding CETP was cloned (9) and mapped to chromosome 16q21, and encompasses 22 Kb consisting of 16 exons. A multitude of CETP genetic variants have been so far identified through re-sequencing efforts and some have been exhaustively analyzed and characterized for associations with CETP mass/activity and HDL-C (10).
In addition to numerous SNPs associated with amino acid changes, SNPs located within the promoter area are significantly associated with CETP and HDL-C concentrations (11). Rare mutations and natural genetic variation at the CETP locus that are associated with decreased transfer of cholesteryl ester from the core of HDL particles to apoB containing lipoproteins in exchange for triglycerides, are associated with higher concentrations of HDL-C. Yet, the association between these CETP genotypes and MI risk is less clear (12). The main objective of the present study was to examine the association of the common genetic variation in the CETP gene, at the level of either SNPs or haplotypes, with the risk of acute MI in a population-based case-control study.
Methods
The basic design of the case-control study has been described in detail (13). Briefly, the subjects were drawn from a study of incident MI among women 18 to 59 years of age and men 18-49 years of age, residing in three contiguous counties of western Washington State. Eligible case patients were subjects diagnosed with a first fatal or nonfatal MI between July 1998 and December 2000 (women) and July 1998 and March 2000 (men). Cases were identified through the review of hospital discharge diagnoses provided by all hospitals within the study region. Criteria for MI were adapted from the Cardiovascular Health Study (14) and were defined by evidence of symptoms, elevated enzymes, and electrocardiographic changes. Using these criteria, we identified 1,192 MI patients (508 women and 684 men) of whom 1,181 were living at the time that we initiated recruitment. Nine hundred sixty three were contacted and among these, we were able to determine eligibility for 670 through a telephone interview and 578 cases (257 women and 321 men) participated in an in-person interview, for an overall case response rate of 54.9%. We used random-digit telephone dialing to identify a control group of women 18 to 59 years of age and men 18 to 49 years of age living in the same area during the time period of the study (13). We recruited 666 such controls frequency matched on age. The estimated response rate, incorporating both the household screening and interview participation rates, was 54.7%.
Participating cases and controls were interviewed in person by trained interviewers who used a structured questionnaire that elicited information about known and suspected cardiovascular risk factors. Information was collected on age, race, education, weight and height, family history of cardiovascular disease, physician-diagnosed diabetes, hypertension, or hypercholesterolemia, cigarette smoking and alcohol consumption, physical activity and reproductive and menstrual characteristics. The structured interview elicited information for the time period preceding the MI or an equivalent date for controls. Non-fasting plasma lipid and lipoprotein variables were measured by enzymatic procedures (13). Case blood samples were obtained at least 6 months after the event.
We used data from the “Pharmacogenetics and Risk of Cardiovascular Disease” resequencing project (http://droog.gs.washington.edu/parc/data/cetp/welcome.html) and a linkage disequilibrium-based single nucleotide polymorphism selection algorithm, LDSelect (available at http://pga.gs.washington.edu/)(15) to choose a set of tagging SNPs to represent all variants in the 5′ end of the CETP gene. We limited our assessment of SNPs to those with a minor allele frequency ≥ 0.05 and formed bins of highly correlated SNPs based on R2 ≥ 0.64). It has been shown that using an r2 threshold of 0.64 permits the identification of the major haplotypes (∼80%) that can be exhaustively,yet efficiently used for association studies. (15).
In addition, the common SNPs located in Intron 1 (TaqI) and in exon 14 (amino acid change - I405V) that have been extensively analyzed in the past for association with plasma lipid phenotypes, were also chosen for genotyping. Of the ten LDs between the 5 tagSNPs, four r2 were lower than 0.1, two r2 ranged from 0.1 to 0.29, two r2 ranged from 0.3 to 0.4, and the LD between TaqI and -629 was somewhat higher (r2 = 0.68).
Genotyping for the CETP -2708 A/G (rs12149545), -971 A/G (rs4783961), -629 A/C (rs1800775), Intron-I TaqI (rs708272) and Exon-14 (I405V) (rs5882) polymorphisms was performed by fluorescence-based minisequencing using the ABI SNaPShot kit (Applied Biosystems, Foster City, CA, U.S.A). The primer sequences used for amplification of the PCR fragments of the selected CETP SNPs and the sequences of the SNaPShot extension primers are outlined in tables S1 and S2 in Supplementary material. The PCR products were pooled (2 μL each), and all primers, dNTPs, enzymes and buffers were then removed from the mix using High Pure™ PCR product purification kit (Applied Biosystems, Foster City, CA, U.S.A) incubating at 37°C for one hour and then at 75°C for 15 minutes. Primers with 5′ end terminating one base form the SNP corresponding to the various SNPs were mixed 2 μM each. Then 5 μL of SNapShot Multiplex ready reaction mix™ (containing Taq polymerase and the labeled dideoxy-NTPs), 3 μL of pooled PCR product, 1 μL of pooled primers and 1 μL DDW were mixed. The mix was subjected to 25 thermal cycles, each one as follows: 96°C for 10′, 50°C for 5′ and 60°C for 30′. The SNaPShot reaction products were loaded onto the ABI Prism 310 genetic sequencer for analysis. The results were analyzed using Gene-Scan analysis software version 3.1 (Applied Biosystems, Foster City, CA, U.S.A).
Statistical Methods
Standard descriptive methods were used for comparing cases and controls on frequency distributions for categorical variables and measures of central tendency (e.g., means, medians) for continuous variables. Departures of allele frequencies from the Hardy-Weinberg equilibrium were tested among controls to identify potential bias in the genotypic distributions. Crude initial analyses included comparisons of genotype and allele frequencies between cases and controls based on gene counting.
We used the HaploScore program to 1) infer haplotypes in study participants and 2) within a general linear model framework, use an efficient score statistic to test the association between the haplotypes and the risk of MI (16). HaploScore assigns the probability for each haplotype pair in each individual and then directly models an individual's phenotype as a function of each inferred haplotype pair, weighted by their estimated probability, to account for haplotype ambiguity. In addition, a Bayesian approach, implemented through the software package PHASE (17) was used to infer the most likely haplotypes for each individual and these haplotypes were then introduced into an unconditional logistic regression model in order to estimate the odds ratio (OR), after controlling for characteristics related to the outcome to increase precision. Ninety-five percent confidence intervals on the ORs were calculated as measures of precision of the estimates, using standard errors of the coefficients from the logistic model. Because of potential gender differences and differences in the age cut-off for early MI event, all analyses were conducted separately for males and females, and results were compared across gender.
We also performed a cladistic-based association analysis to identify specified contrasts of association among the haplotypes and MI risk (18). We used the program EHAP (19) which permits analyses of the information contained in the evolutionary relationships among haplotypes and in the study population using generalized linear models. After a cladogram of the haplotypes considered in the analyses was constructed using the principle of parsimony, rather than perform an omnibus test to determine whether the effects if the haplotypes in the cladogram differ significantly, a sequential series of nested tests were performed. At each step of the algorithm, a “full model” was compared with a series of reduced models (1 degree-of-freedom tests) aimed to group together evolutionary closely related haplotypes. The full model changed between steps contingent on the results of previous step. Haplotypes that fall into the external nodes of the cladogram (zero-step clades) were initially considered. Subsequently, one-step clades (produced by moving backward one mutational step from the zero-step clades toward internal nodes), two-step clades and so forth, were examined.
Results
The characteristics of the MI patients and control subjects are summarized in Table 1. The study subjects were predominantly white and MI cases were, on average, 1-2 years older than the controls.
Table 1. Characteristics of Myocardial Infarction Cases and Controls.
| Men | Women | |||
|---|---|---|---|---|
| Characteristic a | Case Patients
(n = 321) |
Control Subjects
(n = 308) |
Case Patients
(n = 256) |
Control Subjects
(n = 351) |
| Age, y | 44 (5.2) | 42.2 (5.3) | 50.5 (6.6) | 49.5 (6.8) |
| Caucasian, % | 84.7 | 87.7 | 86.7 | 89.3 |
| Education ≤ high school, % | 42.4 | 28.3 | 55.6 | 29.6 |
| Diabetes, % | 10.8 | 2.0 | 22.0 | 4.8 |
| Hypertension, % | 37.5 | 17.2 | 45.0 | 23.9 |
| Hypertension Medications, % | 70.0 | 6.2 | 81.1 | 17.1 |
| Hypercholesterolemia, % | 44.9 | 25.0 | 42.0 | 28.2 |
| Hypercholesterolemia Medications, % | 65.9 | 3.9 | 56.8 | 8.6 |
| Current smoking, % | 46.8 | 20.3 | 53.7 | 12.8 |
| Vigorous physical activity % | 33.8 | 59.4 | 23.6 | 41.3 |
| Family history of early onset of CHD % | 37.8 | 25.7 | 42.1 | 34.5 |
| Family history of early onset of MI % | 48.1 | 25.3 | 57.0 | 36.0 |
| Frequency of alcohol intake (drinks/wk) b | 7.4 (2.2-18.0) | 6.6 (2.9-11.0) | 4.1 (0.5-8.2) | 4.2 (1.3-9.0) |
| Premenopausal % | - | - | 30.9 | 40.7 |
| Body mass index, (kg/m2) | 29 (5.0) | 26.6 (4.2) | 29.7 (8.1) | 26.9 (6.1) |
| Triglyceridec, (mg/dl) | 247.4 (338.3) | 197.5 (153.7) | 217.9 (185.6) | 160.5 (118.5) |
| HDL-Cc, (mg/dl) | 38.2 (10.5) | 43.3 (13.4) | 47.4 (15.1) | 59.4 (16.7) |
| Lp(a)c, (nmol/L) | 79.5 (87.0) | 52.9 (72.0) | 83.0 (98.3) | 50.4 (66.2) |
Values are expressed as percent or as mean (SD) unless otherwise indicated.
Values are expressed as median (Inter-quartile range)
Non-fasting values
Genotype frequencies for the five CETP SNPs are shown in Table 2. Among the controls, except for CETP -2708 in women, genotype distributions did not deviate from Hardy–Weinberg proportions (P-values 0.12-0.86). When adjusted for multiple comparisons, the disequilibrium for CETP -2708 in women did not remain significant (P-value = 0.22). In men, the frequencies of CETP-971G, -629 and TaqI A alleles were somewhat lower in cases than in controls. In women, there was evidence that the CETP polymorphisms were distributed differently in cases and in controls.
Table 2. Distribution of CETP Genotypes and Minor Allele Frequencies in Cases and Controls.
| SNP | Genotype and Minor Allele | Men | Women | ||
|---|---|---|---|---|---|
| Case | Control | Case | Control | ||
| -2708 | AA | 20 (6.4) | 16 (5.4) | 13 (5.3) | 26 (7.8) |
| AG | 134 (42.6) | 126 (42.9) | 101 (40.9) | 165 (49.2) | |
| GG | 160 (51.0) | 152 (51.7) | 133 (53.84) | 144 (43.0) | |
| Freq A (95% CI) | 0.277 (0.23-0.33) | 0.269 (0.23-0.30) | 0.257 (0.22-0.31) | 0.324 (0.29-0.36) | |
| -971 | AA | 89 (28.5) | 70 (23.7) | 59 (24.1) | 112 (33.5) |
| AG | 148 (47.4) | 140 (47.5) | 108 (44.1) | 157 (47.0) | |
| GG | 75 (24.1) | 85 (28.8) | 78 (31.8) | 65 (19.5) | |
| Freq G (95% CI) | 0.478 (0.44-0.52) | 0.525 (0.49-0.57) | 0.539 (0.48-0.60) | 0.430 (0.39-0.47) | |
| -629 | AA | 80 (25.6) | 70 (23.7) | 55 (22.5) | 104 (31.2) |
| AC | 170 (54.5) | 155 (51.6) | 131 (53.7) | 166 (49.9) | |
| CC | 62 (19.9) | 70 (23.7) | 58 (23.8) | 63 (18.9) | |
| Freq C (95% CI) | 0.471 (0.42-0.53) | 0.500 (0.46-0.54) | 0.506 (0.44-0.57) | 0.438 (0.40-0.48) | |
| TaqI | AA | 51 (16.7) | 54 (18.8) | 44 (18.0) | 80 (24.0) |
| GA | 155 (50.8) | 148 (51.6) | 127 (51.8) | 172 (51.7) | |
| GG | 99 (32.5) | 85 (29.6) | 74 (30.2) | 81 (24.3) | |
| Freq A (95% CI) | 0.421 (0.38-0.46) | 0.446 (0.41-0.49) | 0.439 (0.39-0.48) | 0.498 (0.46-0.54) | |
| Ex14 | AA | 134 (42.7) | 130 (44.1) | 112 (45.3) | 147 (43.9) |
| AG | 145 (46.2) | 126 (42.7) | 102 (41.3) | 144 (43.0) | |
| GG | 36 (11.1) | 39 (13.2) | 33 (13.4) | 44 (13.1) | |
| Freq G (95% CI) | 0.342 (0.29-0.39) | 0.346 (0.31-0.38) | 0.309 (0.25-0.38) | 0.346 (0.31-0.38) | |
Next, we used the logistic model to examine the association between CETP SNPs and MI risk in both gender groups after taking into consideration the effects of age, race and other characteristics. In men, the age- and race-adjusted association of the CETP genotypes with MI risk indicated that under a multiplicative model, there was no association with each of the CETP SNPs (Table 3). These associations did not change considerably when, in addition to age and race, the multivariate models included the adjustment for education, smoking, body mass index and HDL-C. Only for CETP -971 SNP did the multivariate results indicate that the minor allele was associated with the risk of MI (OR=0.77, 95%CI=0.60-0.98).
Table 3. Risk of MI associated with CETP genotypes, by sex.
| Men | Women | |||||||
|---|---|---|---|---|---|---|---|---|
| Frequencya (%) | Minimally Adjusted Model | Fully Adjusted Model | Frequencya (%) | Minimally Adjusted Model | Fully Adjusted Model | |||
| SNP | Cases
(2N =624) |
Controls
(2N = 590) |
ORb,c 95% CIb |
ORb,d 95% CIb |
Cases
(2N =490) |
Controls
(2N = 668) |
ORb,d 95% CIb |
ORb,d 95% CIb |
| -2708 A | 28 | 27 | 1.10 | 1.20 | 26 | 32 | 0.69** | 0.82 |
| 0.84-1.44 | 0.89-1.62 | 0.52-0.91 | 0.59-1.13 | |||||
| -971 G | 48 | 52 | 0.82 | 0.77* | 54 | 43 | 1.50** | 1.27 |
| 0.65-1.02 | 0.60-0.98 | 1.19-1.89 | 0.97-1.66 | |||||
| -629 C | 47 | 50 | 0.90 | 0.87 | 51 | 44 | 1.33* | 1.26 |
| 0.71-1.14 | 0.67-1.13 | 1.05-1.70 | 0.95-1.68 | |||||
| TaqI A | 42 | 45 | 0.88 | 0.95 | 44 | 50 | 0.78* | 0.83 |
| 0.69-1.12 | 0.73-2.15 | 0.62-1.00 | 0.63-1.15 | |||||
| Ex14 G | 34 | 35 | 0.99 | 1.02 | 31 | 35 | 0.98 | 1.02 |
| 0.78-1.25 | 0.79-1.34 | 0.77-1.24 | 0.77-1.35 | |||||
Frequency of the minor allele.
Odds ratios (OR) and 95% confidence interval (95% CI) are for each additional copy of the minor allele, relative to subjects homozygous for the more common allele.
OR estimates are adjusted for age and race.
OR estimates are adjusted for age and race, education, smoking, BMI and HDL-C.
p≤ 0.05
p≤ 0.01
In women, the univariate associations with single CETP variants was more pronounced, yet, upon the additional adjustment for the set of co-variables, none of the CETP SNPs was associated with MI risk. In multivariate analyses that included all five CETP SNPs simultaneously, in men only, the TaqI was the only SNP associated with MI risk (Data not shown).
The association between CETP gene variants and MI was also examined at the level of haplotypes. Seventy five percent of the haplotype were inferred with greater than 99.5 percent probability, and in an additional 15 percent of the inferred haplotypes, the probability of certainty ranged between 71% and 99%. Similar rates of certainty were observed in women (mean=93.6%) and men (mean= 93.3%). Haplotype inference from the five SNPs resolved 11 common haplotypes (frequency ≥ 2%) out of which 6 had a frequency that exceeded 5 percent in the control population (Table 4). The 11 CETP haplotypes accounted for 93% of male chromosomes and 91% of female chromosomes, in the control group. The six most common haplotypes (Hap D, Hap L, Hap K, Hap J, Hap A, and Hap C) accounted for 80-81% of the men and women chromosomes. None of the single variants tagged only one common haplotype.
Table 4. Risk of MI Associated with CETP Haplotypes.
| CETP SNP | Men | Women | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | -2708 | -971 | -629 | TaqI | Ex 14 | Haplotype Frequencies | ORa (95% CI) | Haplotype Frequencies | ORa (95% CI) | ||
| Alleles | Case | Control | Case | Control | |||||||
| Hap A | G | G | A | A | G | 0.086 | 0.080 | 1.36 (0.83-2.20) | 0.082 | 0.073 | 0.94 (0.59-1.49) |
| 1.32 (0.76-2.27) | 0.82 (0.49-1.39) | ||||||||||
| Hap B | G | G | A | A | A | 0.009 | 0.029 | 0.37 (0.11-1.21)* | 0.030 | 0.024 | 1.17 (0.47-2.91) |
| 0.18 (0.04-0.75)** | 1.02 (0.33-3.08) | ||||||||||
| Hap C | G | G | C | G | G | 0.091 | 0.082 | 1.15 (0.66-1.99) | 0.089 | 0.063 | 1.03 (0.61-1.74) |
| 1.23 (0.65-2.33) | 1.14 (0.62-2.12) | ||||||||||
| Hap D | G | G | C | G | A | 0.268 | 0.293 | Reference group | 0.302 | 0.257 | Reference group |
| Hap E | G | A | A | A | G | 0.016 | 0.032 | 0.76 (0.39-1.49) | 0.020 | 0.032 | 0.67 (0.34-1.29) |
| 0.91 (0.43-1.93) | 0.86 (0.40-1.84) | ||||||||||
| Hap F | G | A | A | A | A | 0.049 | 0.028 | 1.56 (0.79-3.04) | 0.035 | 0.050 | 0.60 (0.31-1.16) |
| 1.35 (0.61-2.97) | 0.59 (0.27-1.29) | ||||||||||
| Hap G | G | A | A | G | G | 0.046 | 0.018 | 4.05 (1.84-8.91)** | 0.032 | 0.026 | 0.79 (0.31-1.58) |
| 5.98 (2.40-14.78)** | 0.96 (0.43-2.17) | ||||||||||
| Hap H | G | A | A | G | A | 0.026 | 0.020 | 1.32 (0.55-3.15) | 0.014 | 0.0276 | 0.54 (0.22-1.34) |
| 1.02 (0.39-2.62) | 0.46 (0.16-1.35) | ||||||||||
| Hap J | G | A | C | G | A | 0.101 | 0.086 | 1.47 (0.94-2.30) | 0.098 | 0.107 | 0.79 (0.51-1.22) |
| 1.48 (0.89-2.47) | 0.85 (0.51-1.42) | ||||||||||
| Hap K | A | A | A | A | G | 0.070 | 0.093 | 0.89 (0.56-1.43) | 0.095 | 0.131 | 0.65 (0.43-1.00) |
| 0.94 (0.55-1.61) | 0.82 (0.50-1.34) | ||||||||||
| Hap L | A | A | A | A | A | 0.181 | 0.169 | 1.16 (0.80-1.67) | 0.158 | 0.181 | 0.67 (0.45-0.99) |
| 1.31 (0.87-1.97) | 0.81 (0.51-1.28) | ||||||||||
|
| |||||||||||
| Global Scoreb& P value | 19.07 (0.0599) | 13.98 (0.234) | |||||||||
| 24.86 (0.0009) | 9.83 (0.545) | ||||||||||
Odds ratios (OR) and 95% confidence interval (95% CI) are for each additional copy of the haplotype allele, relative to subjects homozygous for the reference haplotype group. Upper estimates are adjusted for age and race and lower estimates are adjusted for age and race, education, smoking, BMI and HDL-C.
Score and p-value in parenthesis from a global association test between CETP haplotypes and MI risk
p≤ 0.05 from haplotype specific score test
p ≤ 0.01 from haplotype specific score test
Next we used logistic models for univariate associations between haplotypes and MI risk (Data not shown). In men, the multiplicative age- and race-adjusted model (1 degree of freedom), showed a significant decreased risk associated with the haplotype B (OR=0.30 for each additional copy of haplotype B) when compared with the reference group, i.e. subjects carrying no haplotype B. Similar results were obtained from the fully-adjusted model (OR=0.14 for each additional copy of haplotype B). The logistic model showed that haplotype D was also associated with a decreased risk of MI (OR=0.72; 95 percent CI: 0.54–0.99). Haplotype G was associated with a 3.8-5.7-fold increased risk of MI for each additional copy; this association was highly statistically significant (95 percent CI: 1.8-8.1 and 2.4-13.9 from the minimally-adjusted, and from the fully-adjusted model, respectively). The remaining haplotypes were not significantly associated with MI. In women, the univariate logistic models have shown no association of CETP haplotypes with MI.
In men, the multiplicative fully-adjusted model showed similar results; a significant decreased risk associated with haplotype B (OR=0.18) and a significant increased risk (OR=5.98) associated with haplotype G, when compared with the reference group, i.e. subjects homozygotes for the common haplotype D (Table 4). In women, all CETP haplotypes were not significantly associated with MI.
In men, the age- and race-adjusted global score statistic from a multivariate model which include all haplotypes was 19.1 and, with 11 df, the P-value from the χ2 distribution was 0.0599; this P value was identical to the empirical P value based on 1,000 simulation repetitions (Table 4). For the full model the global score statistic was 24.9 and the χ2P-value was 0.0009. An advantage of the HaploScore method is demonstrated by the haplotype-specific scores, which allow the evaluation of which haplotypes have the strongest association with the risk of MI after adjustments for other co-variables. In men, as judged by the specific scores, haplotypes B and G were significantly associated with MI risk. The score for haplotype B was negative (-2.3 and -3.6 for the minimally and fully adjusted models, respectively), i.e. the haplotype B is associated with decreased MI risk, and the score for haplotype G was positive (2.6 and 2.9 for the minimally and fully adjusted models, respectively), which indicates that carriers of haplotype G have significantly higher MI risk.
In women, the age- and race-adjusted global score statistic was 14.0, 11 df, P- value 0.234. For the full model the score statistic was even lower, 9.8 and the P- value was 0.545. The haplotype-specific scores indicated that age- and race-adjusted haplotype D was associated with increased MI risk; yet, this haplotype showed little association with MI risk when the model was fully adjusted for other co-variables.
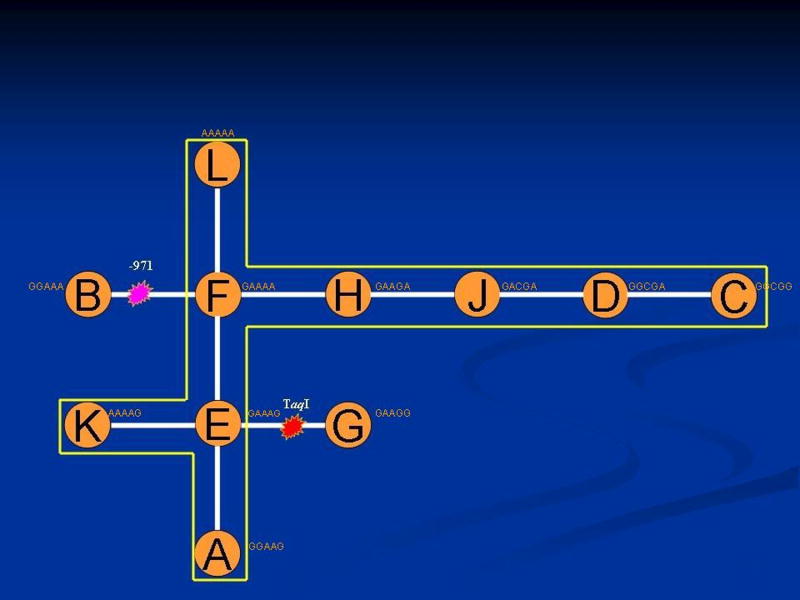
Figure 1 presents the most parsimonious cladogram of the 11 haplotypes (A through L) defined by the 5 SNP markers of the CETP gene. This network of haplotypes represents the minimum number of evolutionary steps (as estimated from nucleotide sequence similarity) that interrelates the 5 closely linked markers. This cladogram characterizes the haplotypes by degree of genetic relatedness. Haplotypes that share marker types are grouped together into clades and haplotypes that are dissimilar for marker types are positioned farthest apart. These relationships were then used to select comparisons among groups of haplotypes that may be informative for the presence of a polymorphism with an effect on MI risk.
Figure 1. Cladogram estimated by Templeton et al. algorithm relating the CETP haplotypes presented in Table 4.
Asterisks indicate the localization of significant changes in MI risk within the cladogram.
Table S3 (Supplementary material) presents the cladistic analysis in males that is defined by the nesting of the 11 CETP haplotypes into 0-, 1- and 2-step. There are 10 pairwise comparisons (6 of 0-step clades, 2 within step-1 and 2 within step-2 clades). This nested analysis suggested that four of these phenotypic comparisons are statistically significant. As a result, haplotypes C, D, J, H, L, F, E, K, and A are clustered together in one clade. As can be seen from Table S4, a significant contrast is detected for MI risk that is localized to the transition between haplotype E and haplotype G (p=0.005 after correction for multiple testing). This suggests that these haplotypes may differ for a functional mutation which is tagged by the TaqI site within the CETP gene that influences MI risk. In addition, a borderline significant contrast (p=0.04 after correction for multiple testing) is localized to the transition between haplotype F and haplotype B, which is associated with the -971 site. In women similar analysis showed that no null hypotheses was rejected and all haplotypes were collapsed into a single clade (Results not shown).
Discussion
In this population-based case-control study, we found two CETP haplotypes that were associated with the occurrence of early-onset non-fatal MI in men. When compared to subjects who were homozygous for the most common haplotype (D), those who harbored haplotype G were at a 4.0 to 6.0-fold increased MI risk. The analyses also showed that subjects who carried haplotype B tended to be at a significantly lower risk for developing early-onset MI. The risk of MI associated with these CETP haplotypes was found to be strongly modified by gender since similar associations were not observed in women.
CETP has a central role in the metabolism of HDL by transferring lipids between plasma lipoproteins and may therefore alter the susceptibility to atherosclerotic vascular disease. The exact mechanism for the contribution of CETP to the atherosclerotic process is unclear. On one hand, increased CETP levels associated with lower HDL-C have been positively associated with intima media thickening (20) and with increased risk of fatal and non-fatal coronary artery disease (CAD) in subjects with high serum TG levels (21). Alternatively, CETP is claimed to have a protective role against atherosclerosis since cholesteryl ester transfer process provides a potentially beneficial pathway for delivery of HDL-derived cholesteryl esters to the liver (22). Indeed, several studies have shown that individuals with reduced CETP activity in conjunction with reduced hepatic lipase function were at increased risk of CAD, independent of their high HDL-C levels (23, 24). Some of the SNPs included in this study were previously evaluated as single SNP association-studies with cardiovascular risk (12, 25-28). In our study the associations between single CETP variants and the risk MI were not consistent across gender groups and dependent on the set of covariables included in the models.
A number of studies have demonstrated associations between CETP haplotypes and change in CETP mass/activity and HDL concentrations. While some of these reported associations were similar to a single SNP analysis (28-34), other studies showed stronger associations between CETP haplotypes and change in the HDL-C levels than that observed in the SNP analysis (32, 35). This is consistent with previously suggested benefits of haplotype analysis for examining SNPs in LD (36).
Our study is one of the few which have investigated the association between CETP haplotypes and the risk of CHD (34, 37-38). Haplotype G (GAAGG), which was associated with a significantly higher MI risk among men, differs from the most common Haplotype D (GGCGA) at the -971A, -629A and Ex14G loci. This association was independent of the adjustment for additional co-variables, including HDL-C. From common haplotype D which is most likely to be the oldest (19), we identified several haplotypes arising from single SNP changes that were not associated with MI risk. Only a more recent clade marked by a reversion back to allele G on TaqI and a transition from haplotype E to haplotype G was associated with a profound association with MI risk.
A number of studies have suggested that the C-629A polymorphism is functional, with the –629A allele altering Sp1/Sp3 binding and associated with reduced promoter activity, decreased CETP mass and increased HDL levels (31, 39) and with reduced risk of MI (28, 34). However, a recent cohort study found an increased risk of coronary disease in carriers of the -629A allele compared to CC homozygotes (40). The Taq1 SNP has been found to be in strong linkage disequilibrium with the -629 SNP. However, it is not yet clear whether the -629 SNP is entirely responsible for the functional effects or whether other changes linked to these SNPs may also be involved (12, 27, 34, 39). For example, a VNTR located 1,946 bp upstream of the transcription start site of the CETP gene was found to be linked to TaqI SNP and appears to have an independent effect on CETP and HDL-C levels (12, 41). Yet, in our study, the associations between the TaqI SNP and haplotypes with MI risk were independent of HDL-C levels.
A number of studies have demonstrated weak or no associations between CETP haplotypes and CHD risk (34, 37, 38). Our results suggested that associations between the CETP haplotypes and MI risk are sex specific. Relatively young women are intrinsically less susceptible to MI, and therefore we would anticipate that women with early-onset MI would be expected to carry a heavier burden of predisposing genes. Nonetheless, both in our study and in the Framingham study the associations with CETP polymorphisms were limited to men (42). Animal models also support such sex differences: transgenic rats heterozygous for human CETP were shown to have increased atherogenic lipid profile and coronary plaques with decreased survival outcome in male transgenic rats compared with females CETP transgenics (43).
A number of limitations are inherent in the present study. We included a moderate number of MI survivors, and therefore we cannot exclude the possibility that associations seen in our study are due to chance or in part due to differential early case fatality among the MI patients related to these polymorphisms. We computed the power of the present study for detecting MI risk associated with the putative high-risk genotypes, using the PS program (44). The results demonstrate that the proposed study has a reasonable good statistical power to detect medium OR levels for common alleles/haplotypes and for detecting relatively high OR for rare haplotypes. For example, assuming an α = 0.05 and a common allele/haplotype prevalence of 30%, our sample size of men provides 65% power to detect an OR=1.5 and 80% power to detect an OR=1.62. For common allele/haplotype prevalence of 40%, the power estimates are 70% and 0.83%, respectively. For a rare haplotype prevalence of 2.5%, our sample size of men provides 95% power to detect an OR=4.0. The sample size of women provides higher power estimates. Additional studies that involve larger numbers of subjects, especially women and fatal events, are required to confirm the present study's associations. Over 85% percent of our study sample was Caucasians and thus spurious associations due to population stratification are not likely to occur. Moreover, when restricting our analyses to Caucasians only, similar results were obtained. In case-control studies, selection and recall biases are potential problems. We do not think that appreciable bias has resulted due to non response, as 1) it is not likely that a person's decision not to participate is related to his/her CETP genotypes, 2) the distribution of “traditional” cardiovascular risk factors between recruited cases and controls exhibited well-established differences, and 3) the CETP SNPs frequencies among our controls are similar to those reported in other populations of European descent individuals (12, 25, 31, 34, 37, 38, 42). Finally, a large number of CETP SNPs are not covered in our analyses and thus part of the CETP variation was not captured. Yet, a multitude of the unstudied SNPs are rare with a limited usefulness in describing the major genetic determinants of MI risk in the general population.
In conclusion, this study of relatively young men and women suggests an association between the CETP gene variation and MI risk in men. Further elucidation of the pathways through which variation in this gene exerts its effect on MI risk is warranted.
Supplementary Material
Acknowledgments
Grants and contracts: This work was supported by grants P30ES07033 and R01HL056931 from the National Institutes of Health.
Footnotes
Conflict of Interest: No conflict exists for all authors.
References
- 1.Bruce C, Tall AR. Cholesteryl ester transfer protein, reverse cholesterol transport and atherosclerosis. Curr Opin Lipidol. 1995;6:306–11. doi: 10.1097/00041433-199510000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Kothari HV, Poirier KJ, Lee WH, Satoh Y. Inhibition of cholesterol ester transfer protein CGS 25159 and changes in lipoproteins in hamsters. Atherosclerosis. 1997;128:59–66. doi: 10.1016/s0021-9150(96)05981-3. [DOI] [PubMed] [Google Scholar]
- 3.Okamoto H, Yonemori F, Wakitani K, Minowa T, Maeda K, Shinkai H. A cholesteryl ester transfer protein inhibitor attenuates atherosclerosis in rabbits. Nature. 2000;406:203–7. doi: 10.1038/35018119. [DOI] [PubMed] [Google Scholar]
- 4.Fielding CG, Havel RJ. Cholesterol ester transfer protein: friend or foe? J Clin Invest. 1996;97:2687–8. doi: 10.1172/JCI118719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevenson CG. Cholesterol ester transfer protein: a molecule with three faces? Crit Rev Clin Lab Sci. 1998;35:517–46. doi: 10.1080/10408369891234264. [DOI] [PubMed] [Google Scholar]
- 6.Lusis AJ, Mar R, Pajukanta P. Genetics of atherosclerosis. Annu Rev Genomics Hum Genet. 2004;5:189–218. doi: 10.1146/annurev.genom.5.061903.175930. [DOI] [PubMed] [Google Scholar]
- 7.Snowden CB, McNamara PM, Garrison RJ, Feinleib M, Kannel WB, Epstein FH. Predicting coronary heart disease in siblings--a multivariate assessment: the Framingham Heart Study. Am J Epidemiol. 1982;115:217–22. doi: 10.1093/oxfordjournals.aje.a113293. [DOI] [PubMed] [Google Scholar]
- 8.Friedlander Y, Arbogast P, Schwartz SM, Marcovina SM, Austin MA, Rosendaal FR, Reiner AP, Psaty BM, Siscovick DS. Family history as a risk factor for early myocardial infarction in women. Atherosclerosis. 2001;156:201–7. doi: 10.1016/s0021-9150(00)00635-3. [DOI] [PubMed] [Google Scholar]
- 9.Drayna D, Jarnagin AS, McLean J, Henzel W, Kohr W, Fielding C, et al. Cloning and sequencing of human cholesteryl ester transfer protein cDNA. Nature. 1987;327:632–4. doi: 10.1038/327632a0. [DOI] [PubMed] [Google Scholar]
- 10.Boekholdt SM, Thompson JF. Natural genetic variation as a tool in understanding the role of CETP in lipid levels and disease. J Lipid Res. 2003;44:1080–93. doi: 10.1194/jlr.R200018-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Hinds DA, Seymour AB, Durham LK, et al. Genomic scan of 71 candidate regions spanning 17.1 Mb for association with HDL cholesterol levels. Hum Genomics. 2004;1:421–34. doi: 10.1186/1479-7364-1-6-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson JF, Durham LK, Lira MF, Shear C, Milos PM. CETP polymorphisms associated with HDL cholesterol may differ from those associated with cardiovascular disease. Atherosclerosis. 2005;181:45–53. doi: 10.1016/j.atherosclerosis.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Friedlander Y, Schwartz SM, Durst R, Meiner V, Robertson AS, Leitersdorf E, Siscovick DS. SREBP-2 and SCAP Isoforms and Risk of Early Onset Myocardial Infarction. Atherosclerosis. 2008;196:896–904. doi: 10.1016/j.atherosclerosis.2007.02.006. Mar 23 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Ives DG, Fitzpatrick AL, Bild DE, Psaty BM, Kuller LH, Crowley PM, et al. Surveillance and ascertainment of cardiovascular events. The Cardiovascular Health Study. Ann Epidemiol. 1995;5:275–85. doi: 10.1016/1047-2797(94)00093-9. [DOI] [PubMed] [Google Scholar]
- 15.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a Maximally Informative Set of Single-Nucleotide Polymorphisms for Association Analyses Using Linkage Disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Templeton A, Boerwinkle E, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of alcohol dehydrogenase activity in drosophila. Genetics. 1987;117:343–51. doi: 10.1093/genetics/117.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seltman H, Roeder K, Devlin B. Evolutionary-based association analysis using haplotype data. Genet Epidemiol. 2003;25:48–58. doi: 10.1002/gepi.10246. [DOI] [PubMed] [Google Scholar]
- 20.de Vries R, Perton FG, Dallinga-Thie GM, van Roon AM, Wolffenbuttel BHR, van Tol A, Dullaart RPF. Plasma Cholesteryl Ester Transfer Is a Determinant of Intima-Media Thickness in Type 2 Diabetic and Nondiabetic Subjects. Diabetes. 2005;54:3554–9. doi: 10.2337/diabetes.54.12.3554. [DOI] [PubMed] [Google Scholar]
- 21.Boekholdt SM, Kuivenhoven JA, Wareham NJ, Peters RJG, Jukema W, Luben R, Bingham SA, Day NE, Kastelein JJP, Khaw KT. Plasma Levels of Cholesteryl Ester Transfer Protein and the Risk of Future Coronary Artery Disease in Apparently Healthy Men and Women. Circulation. 2004;110:1418–23. doi: 10.1161/01.CIR.0000141730.65972.95. [DOI] [PubMed] [Google Scholar]
- 22.de Grooth GJ, Klerkx AH, Stroes ES, Stalenhoef AF, Kastelein JJ, Kuivenhoven JA. A review of CETP and its relation to atherosclerosis. J Lipid Res. 2004;45:1967–74. doi: 10.1194/jlr.R400007-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Hirano KS, Yamashita S, Kuga Y, Sakai N, Nozaki S, Kihara S, Arai T, Yanagi K, Takami S, Menju M. Atherosclerotic disease in marked hyperalphalipoproteinemia. Combined reduction of cholesteryl ester transfer protein and hepatic triglyceride lipase. Arterioscler Thromb Vasc Biol. 1995;15:1849–56. doi: 10.1161/01.atv.15.11.1849. [DOI] [PubMed] [Google Scholar]
- 24.Zhong S, Sharp DS, Grove JS, Bruce C, Yano K, Curb JD, Tall AR. Increased coronary heart disease in Japanese-American men with mutation in the cholesteryl ester transfer protein gene despite increased HDL levels. J Clin Invest. 1996;97:2917–23. doi: 10.1172/JCI118751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corbex M, Poirier O, Fumeron F, et al. Extensive association analysis between the CETP gene and coronary heart disease phenotypes reveals several putative functional polymorphisms and gene-environment interaction. Genet Epidemiol. 2000;19:64–80. doi: 10.1002/1098-2272(200007)19:1<64::AID-GEPI5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 26.Agerholm-Larsen B, Tybjaerg-Hansen A, Schnohr P, et al. Common cholesteryl ester transfer protein mutations, decreased HDL cholesterol, and possible decreased risk of ischemic heart disease: The Copenhagen City Heart Study. Circulation. 2000;102:2197–203. doi: 10.1161/01.cir.102.18.2197. [DOI] [PubMed] [Google Scholar]
- 27.Kakko S, Tamminen M, Paivansalo M, et al. Variation at the cholesteryl ester transfer protein gene in relation to plasma high density lipoproteins cholesterol levels and carotid intima-media thickness. Eur J Clin Invest. 2001;31:593–602. doi: 10.1046/j.1365-2362.2001.00859.x. [DOI] [PubMed] [Google Scholar]
- 28.Blankenberg S, Rupprecht HJ, Bickel C, Jiang XC, Poirier O, Lackner KJ, Meyer J, Cambien F, Tiret L. Common genetic variation of the cholesteryl ester transfer protein gene strongly predicts future cardiovascular death in patients with coronary artery disease. J Am Coll Cardiol. 2003;41:1983–9. doi: 10.1016/s0735-1097(03)00408-x. [DOI] [PubMed] [Google Scholar]
- 29.Bauerfeind A, Knoblauch H, Schuster H, Luft FC, Reich JG. Single nucleotide polymorphism haplotypes in the cholesteryl-ester transfer protein (CETP) gene and lipid phenotypes. Hum Hered. 2002;54:166–73. doi: 10.1159/000070662. [DOI] [PubMed] [Google Scholar]
- 30.Gudnason V, Kakko S, Nicaud V, Savolainen MJ, Kesaniemi YA, Tahvanainen E, Humphries S. Cholesteryl ester transfer protein gene effect on CETP activity and plasma high-density lipoprotein in European populations. The EARS Group. Eur J Clin Invest. 1999;29:116–28. doi: 10.1046/j.1365-2362.1999.00412.x. [DOI] [PubMed] [Google Scholar]
- 31.Klerkx AH, Tanck MW, Kastelein JJ, Molhuizen HO, Jukema JW, Zwinderman AH, Kuivenhoven JA. Haplotype analysis of the CETP gene: not TaqIB, but the closely linked -629C ≥ A polymorphism and a novel promoter variant are independently associated with CETP concentration. Hum Mol Genet. 2003;12:111–23. doi: 10.1093/hmg/ddg013. [DOI] [PubMed] [Google Scholar]
- 32.Knoblauch H, Bauerfeind A, Krahenbuhl C, Daury A, Rohde K, Bejanin S, Essioux L, Schuster H, Luft FC, Reich JG. Common haplotypes in five genes influence genetic variance of LDL and HDL cholesterol in the general population. Hum Mol Genet. 2002;11:1477–85. doi: 10.1093/hmg/11.12.1477. [DOI] [PubMed] [Google Scholar]
- 33.Lu H, Inazu A, Moriyama Y, Higashikata T, Kawashiri MA, Yu W, Huang Z, Okamura T, Mabuchi H. Haplotype analyses of cholesteryl ester transfer protein gene promoter: a clue to an unsolved mystery of TaqIB polymorphism. J Mol Med. 2003;81:246–55. doi: 10.1007/s00109-002-0414-7. [DOI] [PubMed] [Google Scholar]
- 34.Tobin MD, Braund PS, Burton PR, Thompson JR, Steeds R, Channer K, Cheng S, Lindpaintner K, Samani NJ. Genotypes and haplotypes predisposing to myocardial infarction: a multilocus case-control study. Eur Heart J. 2004;25:459–67. doi: 10.1016/j.ehj.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 35.Winkelmann BR, Hoffmann MM, Nauck M, Kumar AM, Nandabalan K, Judson RS, Boehm BO, Tall AR, Ruano G, Marz W. Haplotypes of the cholesteryl ester transfer protein gene predict lipid-modifying response to statin therapy. Pharmacogenom J. 2003;3:284–96. doi: 10.1038/sj.tpj.6500195. [DOI] [PubMed] [Google Scholar]
- 36.Akey J, Jin L, Xiong M. Haplotypes vs single marker linkage disequilibrium tests: what do we gain? Eur J Hum Genet. 2001;9:291–300. doi: 10.1038/sj.ejhg.5200619. [DOI] [PubMed] [Google Scholar]
- 37.Freeman DJ, Samani NJ, Wilson V, McMahon AD, Braund PS, Cheng S, Caslake MJ, Packard CJ, Gaffney D. A polymorphism of the cholesteryl ester transfer protein gene predicts cardiovascular events in non-smokers in the West of Scotland Coronary Prevention Study. Eur Heart J. 2003;24:1833–42. doi: 10.1016/j.ehj.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 38.McCaskie PA, Beilby JP, Chapman CML, Hung J, McQuillan BM, Thompson PL, Palmer LJ. Cholesteryl ester transfer protein gene haplotypes, plasma high-density lipoprotein levels and the risk of coronary heart disease. Hum Genet. 2007;121:401–11. doi: 10.1007/s00439-007-0326-2. [DOI] [PubMed] [Google Scholar]
- 39.Dachet C, Poirier O, Cambien F, et al. New functional promoter polymorphism, CETP/-629, in cholesteryl ester transfer protein (CETP) gene related to CETP mass and high density lipoprotein cholesterol levels: role of Sp1/Sp3 in transcriptional regulation. Arterioscler Thromb Vasc Biol. 2000;20:507–15. doi: 10.1161/01.atv.20.2.507. [DOI] [PubMed] [Google Scholar]
- 40.Borggreve SE, Hillege HL, Wolffenbuttel BHR, de Jong PE, Zuurman MW, van der Steege G, van Tol A, Dullaart RPF. An Increased Coronary Risk Is Paradoxically Associated with Common Cholesteryl Ester Transfer Protein Gene Variations That Relate to Higher High-Density Lipoprotein Cholesterol: A Population-Based Study. Journal Clin Endo Metab. 2006;91:3382–8. doi: 10.1210/jc.2005-2322. [DOI] [PubMed] [Google Scholar]
- 41.Hayes WSL, Elsenboss L, Williams A, Andre C, Abramson R, Thompson JF, Milos PM. Sequencing of the cholesteryl ester transfer protein 5′ regulatory region using artificial transposons. Gene. 1997;197:101–7. doi: 10.1016/s0378-1119(97)00247-3. [DOI] [PubMed] [Google Scholar]
- 42.Ordovas JM, Cupples LA, Corella D, Otvos JD, Osgood D, Martinez A, Lahoz C, Coltell O, Wilson PWF, Schaefer EJ. Association of Cholesteryl Ester Transfer Protein–TaqIB Polymorphism With Variations in Lipoprotein Subclasses and Coronary Heart Disease Risk The Framingham Study. Arterioscler Thromb Vasc Biol. 2000;20:1323–9. doi: 10.1161/01.atv.20.5.1323. [DOI] [PubMed] [Google Scholar]
- 43.Herrera VL, Tsikoudakis A, Didishvili T, Ponce LR, Bagamasbad P, Gantz D, Herscovitz H, Van Tol A, Ruiz-Opazo N. Analysis of gender-specific atherosclerosis susceptibility in transgenic [hCETP]25DS rat model. Atherosclerosis. 2004;177:9–18. doi: 10.1016/j.atherosclerosis.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 44.Dupont WD, Plummer WD., Jr Power and sample size calculations for studies involving linear regression. Control Clin Trials. 1998;19:589–601. doi: 10.1016/s0197-2456(98)00037-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.