Abstract
Multiple lines of evidence from molecular studies indicate that individual taste qualities are encoded by distinct taste receptor cells. In contrast, many physiological studies have found that a significant proportion of taste cells respond to multiple taste qualities. To reconcile this apparent discrepancy and to identify taste cells that underlie each taste quality, we investigated taste responses of individual mouse fungiform taste cells that express gustducin or GAD67, markers for specific types of taste cells. Type II taste cells respond to sweet, bitter or umami tastants, express taste receptors, gustducin and other transduction components. Type III cells possess putative sour taste receptors, and have well elaborated conventional synapses. Consistent with these findings we found that gustducin-expressing Type II taste cells responded best to sweet (25/49), bitter (20/49) or umami (4/49) stimuli, while all GAD67 (Type III) taste cells examined (44/44) responded to sour stimuli and a portion of them showed multiple taste sensitivities, suggesting discrimination of each taste quality among taste bud cells. These results were largely consistent with those previously reported with circumvallate papillae taste cells. Bitter-best taste cells responded to multiple bitter compounds such as quinine, denatonium and cyclohexamide. Three sour compounds, HCl, acetic acid and citric acid, elicited responses in sour-best taste cells. These results suggest that taste cells may be capable of recognizing multiple taste compounds that elicit similar taste sensation. We did not find any NaCl-best cells among the gustducin and GAD67 taste cells, raising the possibility that salt sensitive taste cells comprise a different population.
Sweet, salty, sour, bitter and umami are thought to represent the five basic taste qualities. Among them, sweet, bitter and umami tastes are detected by different G-protein coupled receptors (GPCRs) and transduced by a common signalling pathway involving gustducin, PLCβ2, IP3R3 and TRPM5 (Lindemann, 2001; Chandrashekar et al. 2006). Bitter (T2rs), sweet (T1r2/T1r3) and umami (T1r1/T1r3) receptors are expressed in different sets of taste cells (Adler et al. 2000; Nelson et al. 2001), suggesting that these qualities may be discriminated at the receptor cell level. In contrast, sour and salty tastes are thought to be mediated by channel type receptors, such as ASIC, HCN, PKD1L3–PKD2L1, V1R variant and amiloride sensitive Na+ channel (Lindemann, 2001; Chandrashekar et al. 2006). The putative sour receptors PKD1L3 and PKD2L1 are not co-expressed with T1r3, T2rs, TRPM5 and IP3R3 (Ishimaru et al. 2006; Huang et al. 2006; Kataoka et al. 2008), suggesting that sour-responding taste cells may comprise a different population from the sweet-, bitter- and umami-responding taste cells. Thus, molecular studies imply that individual taste modalities are encoded by different taste receptor cells.
Previous physiological studies with mouse circumvallate (posterior tongue) and fungiform papillae (anterior tongue) have demonstrated that a significant proportion of taste cells respond to multiple taste stimuli (Caicedo et al. 2002; Yoshida et al. 2006a; Tomchik et al. 2007). In addition, some gustatory nerve fibres and gustatory neurons in geniculate ganglion also responded to multiple taste stimuli (Ninomiya et al. 1982, 1984; Breza et al. 2006), and taste cells and gustatory nerve fibres were shown to share similar response characteristics including breadth of responsiveness (Yoshida et al. 2006a,b;). These data suggest some taste cells may have multiple sensitivities to basic taste qualities and transmit their information to gustatory nerve fibres. However, in the above studies the majority of taste cells were narrowly tuned to single taste qualities. These specialized taste cells may have great impact on the discrimination of basic taste qualities.
Taste buds contain a variety of morphologically and functionally distinct types of taste cells (Type I∼IV cells). Type I (dark) cells may have a supportive role similar to glial cells (Lawton et al. 2000) and Type IV (basal) cells are assumed to be progenitor cells (Murray, 1973). Type II (light) cells express GPCRs and transduction components for sweet, bitter and umami (Yang et al. 2000a; Clapp et al. 2004, DeFazio et al. 2006), suggesting that Type II cells may mediate these tastes. Type III cells form conventional synapses with gustatory nerve fibres (Royer & Kinnamon, 1988) and express synapse related genes such as SNAP25 (Yang et al. 2000b), and the putative sour receptor PKD2L1 (Kataoka et al. 2008), suggesting that Type III cells may be responsible for sour taste. Thus, molecular data suggest that cell types may be closely related to response properties of taste cells. A recent study in mouse circumvallate papillae demonstrated that ‘Receptor’ (Type II) cells elicited Ca2+ responses to bitter, sweet and umami stimuli and ‘Presynaptic’ (Type III) cells showed Ca2+ responses to all taste qualities (Tomchik et al. 2007). However, regional subpopulations of taste cells differ in certain response properties such as amiloride sensitivity (Ninomiya & Funakoshi, 1988; Ninomiya et al. 1991; Ninomiya, 1998) and in expression of gustducin and taste receptors (Kim et al. 2003; Shigemura et al. 2008), raising the possibility that Type II and III cells in fungiform papillae may differ from those in circumvallate papillae.
In this study, we focused on mouse fungiform taste cells expressing either α-gustducin (a G protein α-subunit and Type II cell marker) or GAD67 (a Type III cell marker). To identify Type II and III cells we used transgenic mice expressing green fluorescent protein (GFP) under control of the gustducin or GAD67 promoters and single cell RT-PCR. To clarify response profiles of Type II and III cells we examined responses to five basic taste stimuli (NaCl, saccharin, HCl, quinine and monosodium glutamate). We also investigated responses to multiple taste stimuli that elicited similar taste sensation in bitter and sour sensitive taste cells. Our results indicate that sweet, bitter, umami and sour taste qualities might be discriminated at the level of the taste receptor cell.
Methods
Recording responses of taste cells
All experimental procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the committee for Laboratory Animal Care and Use at Kyushu University, Japan. The procedures for recording of taste cell responses were similar to those used previously (Yoshida et al. 2005, 2006a). Subjects were >8 weeks old C57BL/6N mice (n= 19) and transgenic mice expressing GFP under control of the gustducin (Wong et al. 1999) or GAD67 (GAD-GFP (Δneo) mice, Tamamaki et al. 2003) promoter (n= 20 and 31, respectively). Animals were anaesthetized with ether and killed by cervical dislocation. The anterior part of the tongue was removed and injected with 100 μl of Tyrode solution containing 0.2∼1 mg ml−1 elastase (Elastin Products, Owensville, MO, USA). After incubation for 10∼15 min at room temperature, the lingual epithelium was peeled and pinned out in a Sylgard coated culture dish with the mucosal side down and washed several times with Tyrode solution. Individual fungiform taste buds with a piece of epithelium were excised from this sheet and transferred to a recording chamber. The residual sheet was stored at 4°C for another series of experiments.
The recording chamber containing excised taste buds was mounted on the stage of a laser scanning confocal microscope (FV-1000 and Fluoview; Olympus, Tokyo, Japan). The mucosal side of an excised epithelium with single taste bud was drawn into the orifice of the stimulating pipette. Tyrode solution was always perfused inside the stimulating pipette except during the period of recording. Tyrode solution was continuously flowed into the recording chamber with a peristaltic pump at approximately 2 ml min−1. The receptor membrane was rinsed with distilled water (DW) at least 30 s before and after taste stimulation.
The electrical responses of taste cells in isolated taste buds were recorded extracellularly from the basolateral side at room temperature (25°C). Taste bud cells containing GFP were identified under confocal laser scanning microscopy (excitation 488 nm, emission 500∼600 nm) and were approached by a recording electrode (inner diameter 1∼3 μm, pipette resistances 1.5∼3.5 MΩ, see Figs 1, 7). Seal resistances were typically 3∼10 times the pipette resistances. Electrical signals from taste bud cells were recorded by a high-impedance patch-clamp amplifier (Axopatch 200B; Axon Instruments, Union City, CA, USA) interfaced to a computer (Windows XP) by an analog-to-digital board (Digidata 1320A; Axon Instruments). Signals were filtered at 1 kHz, sampled at 5∼10 kHz and stored on the hard-disk drive of a computer using pCLAMP software (Gap-Free mode; Axon Instruments) for later analysis. After recording responses from taste cells, Ca2+–Mg2+ free Tyrode solution was introduced into the recording chamber to loosen connections between taste bud cells. Several minutes after incubation, the recorded taste cell was drawn out from a taste bud using the recording electrode. The cell was transferred into a PCR tube containing 0.5 μl RNase inhibitor (RNase OUT: Invitrogen, Carlsbad, CA, USA) by breaking the tip of the electrode in a PCR tube. Collected samples were immediately frozen with liquid nitrogen and stored at –80°C for later single cell RT-PCR analysis.
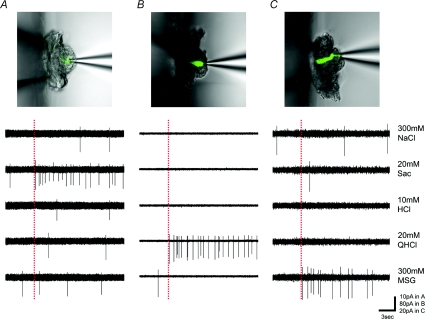
Figure 1. Sample recordings from gustducin-GFP taste cells.
Upper panels show pictures of gustducin-GFP taste cells from which taste responses were recorded. Lower panels show taste responses to 300 mm NaCl, 20 mm saccharin (Sac), 10 mm HCl, 20 mm quinine-HCl (QHCl), and 300 mm monosodium glutamate (MSG). Dotted lines show the onset of taste stimulation.
Figure 7. Sample recordings from GAD67-GFP taste cells.
Upper panels show pictures of GAD67-GFP taste cells from which taste responses were recorded. Lower panels show taste responses to five basic taste stimuli (NaCl, saccharin, HCl, quinine, MSG). Dotted lines show the onset of taste stimulation.
Solutions
Tyrode solution contained (in mm): NaCl, 140; KCl, 5; CaCl2, 1; MgCl2, 1; Hepes, 10; glucose, 10; sodium pyruvate, 10; pH adjusted to 7.4 with NaOH. Ca2+–Mg2+ free Tyrode solution contained (in mm): NaCl, 140; KCl, 5; EDTA, 2; Hepes, 10; glucose, 10; sodium pyruvate, 10; pH adjusted to 7.4 with NaOH. Taste stimuli were the following (mm): 300 NaCl, 1∼10 HCl, 20 saccharin sodium (Sac), 20 quinine-HCl (QHCl), 300 monosodium glutamate (MSG), 20 denatnium benzoate (Den), 0.1 cyclohexamide (CX), 10 caffeine, 0.5 sucrose octaacetate (SOA), 1∼10 citric acid (CA), 3∼30 acetic acid (AA). Chemicals were dissolved in distilled water (DW) and used at room temperature (25°C). All chemicals were purchased from Wako Pure Chemical Industries (Osaka, Japan).
Data analysis
To analyse data, the number of spikes per unit time was counted throughout the recording. The mean spontaneous impulse discharge for each unit was calculated by averaging the number of spikes over the 10 s period that distilled water flowed over the taste pore prior to each stimulation. The final criteria for the occurrence of a response were the following: (1) number of spikes was larger than the mean plus 2 standard deviations of the spontaneous discharge in two repeated trials; (2) more than three spikes were evoked by taste stimulation. The magnitude of response to a particular stimulus was obtained by counting the total number of impulses for the first 10 s after the onset of stimulus application and subtracting the spontaneous impulse discharge.
Breadth of responsiveness of the taste cells was quantified using the following entropy equation (Smith & Travers, 1979; Travers & Smith, 1979).
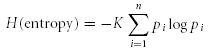 |
where H is the breadth of responsiveness, K is a scaling constant (2.096 for three stimuli, 1.431 for five stimuli), pi is the proportional response to each taste stimulus, and logarithms of pi are taken to the base 10. This entropy value varies continuously from 0.0 for a cell that responds exclusively to one stimulus to 1.0 for a cell that responds equivalently to all of the taste stimuli (Smith & Travers, 1979). All data in the text are means ±s.e.m.
Single cell RT-PCR
The protocol for the multiplex single cell RT-PCR was as used previously (Yoshida et al. 2005, 2009). Reverse transcription (RT) and first round amplification took place in the same tube using OneStep RT-PCR kit (Qiagen, Ratingen, Germany) according to the manufacturer's instructions. A 50 μl reaction mixture contained the following: 10 μl Qiagen OneStep RT-PCR buffer (×5), 2 μl Qiagen OneStep RT-PCR enzyme mix, 0.4 mm of each dNTP, 1 μl RNase inhibitor, 0.2∼0.6 mm of each outside primers (Table 1) and the sample (containing 0.5 μl of RNase inhibitor). After the RT reaction at 50°C for 30 min, the first round of PCR was subsequently performed in the same tube with a 15 min preincubation at 95°C followed by 30 cycles of denaturation (94°C for 30 s), annealing (53°C for 60 s), and amplification (72°C for 90 s) in a thermal cycler (TaKaRa PCR thermal cycler: Takara, Tokyo, Japan). Subsequently, the first round PCR products were re-amplified for 40 cycles (94°C for 30 s, 58°C for 30 s, 72°C for 60 s) in separate reactions using the internal primer pairs for each template. Each 10 μl second round reaction mix contained the following: 0.25 units of Taq DNA polymerase (TaKaRa Ex Taq™ HS: Takara), 1 μl of 10× PCR buffer containing 20 mm Mg2+, 0.2 mm of each dNTP, 0.6 mm of each internal primer pair (Table 1) and 0.2 μl of first round PCR products. After second round amplification, reaction solutions were subjected to 2% agarose gel electrophoresis with ethidium bromide. Positive control reaction with mRNA purified from a single taste bud and negative control reaction with 0.5 μl of electrode solution (Tyrode solution) were run in parallel from the RT-PCR. β-Actin was used as the internal control. All primer sets were designed to span exon–intron boundaries to distinguish PCR products derived from genomic DNA and mRNA.
Table 1.
Nucleotide sequences of primers used in single cell RT-PCR experiments
| Gene | Accession no. | Forward | Reverse | Product size |
|---|---|---|---|---|
| SNAP25 | NM_011428 | AAGGGATGGACCAAATCAAT | CAATGGGGGTGACTACTCTG | 601 bp |
| AAAAAGCCTGGGGCAATAAT | AGCATCTTTGTTGCACGTTG | 304 bp | ||
| Gustducin | NM_001081143 | ACGAGATGCAAGAACTGTGA | TATCTGTCACGGCATCAAAC | 941 bp |
| TGCTTTGAAGGAGTGACGTG | GTAGCGCAGGTCATGTGAGA | 341 bp | ||
| T1r3 | NM_031872 | TGCCTGAATTTTCCCATTAT | AGGACACTGAGGCAGAAGAG | 889 bp |
| CTACCCTGGCAGCTCCTGGA | CAGGTGAAGTCATCTGGATGCTT | 343 bp | ||
| β-Actin | NM_007393 | CCTGAAGTACCCCATTGAAC | GTAACAGTCCGCCTAGAAGC | 943 bp |
| GGTTCCGATGCCCTGAGGCTC | ACTTGCGGTGCACGATGGAGG | 370 bp |
Upper: outside primers; lower: inside primers
Results
Taste cells expressing GFP generate action potentials
Previous studies suggested that information derived from taste cells generating action potentials provide the major component of taste information that is transmitted to gustatory nerve fibres (Yoshida et al. 2006a,b;). Therefore, we first tested whether taste cells expressing GFP under control of the gustducin promoter (gustducin-GFP taste cells) or GAD67 promoter (GAD67-GFP taste cells) generate action potentials. Both gustducin-GFP taste cells and GAD67-GFP taste cells were readily visible in our isolated taste bud preparation under confocal laser scanning microscopy (Figs 1 and 7). We were able to attach a recording electrode to individual GFP-expressing taste cells. Taste cells expressing GFP in both types of transgenic mice generated action potentials (Figs 1 and 7).
Response properties of gustducin taste cells
Gustducin is a G-protein that contributes to sweet, bitter and umami taste (Wong et al. 1996; He et al. 2004) and is a Type II cell marker (Yang et al. 2000a), suggesting that taste cells expressing gustducin (Type II cells) may respond to these taste stimuli. We examined taste responses of gustducin-GFP taste cells to five basic taste stimuli applied in a restricted fashion to the apical side of taste cells. Stimuli used were NaCl (salty), saccharin (sweet), HCl (sour), quinine (bitter) and MSG (umami). As expected, all gustducin-GFP taste cells responded best to sweet, bitter or umami taste stimuli. Figure 1 shows sample recordings from three different gustducin-GFP taste cells: sweet best, bitter best and umami best cells are shown. The sweet best cell responded to 20 mm saccharin but not to the other four taste stimuli (Fig. 1A). The bitter best cell responded specifically to 20 mm quinine (Fig. 1B) and the umami best cell responded specifically to 300 mm MSG (Fig. 1C). We recorded taste responses from 29 gustducin-GFP taste cells. Among them, 10 cells responded best to saccharin, 17 cells responded best to quinine and two cells responded best to MSG. No gustducin-GFP taste cell responded best to NaCl or HCl.
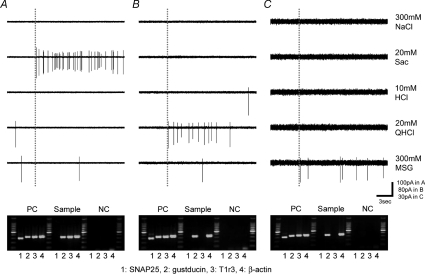
We next examined responses of subtypes of taste cells from non-transgenic wild-type mice (C57BL/6N) using a combination of recording of taste cell responses and single cell RT-PCR (Yoshida et al. 2005). We first recorded taste responses to five basic taste stimuli (NaCl, saccharin, HCl, quinine and MSG) from taste cells chosen at random and then harvested the taste cell. Expression of gustducin (Type II cells) or SNAP25 (Type III cells) was examined by multiplex single cell RT-PCR. We tested 40 cells in this way and detected expression of gustducin mRNA in 20 cells and expression of SNAP25 mRNA in three cells. We failed to detect both gustducin and SNAP25 mRNA in 17 cells although these were detected in all positive control samples (purified mRNA from a taste bud). This may be due to the difficulty of detection of mRNAs in single cell RT-PCR after recording of taste responses and may indicate that false negative results could be obtained in these experiments. Despite these technical limitations of our single cell RT-PCR experiments, we found no gustducin-positive taste cells expressed SNAP25. These results were consistent with those in previous studies (DeFazio et al. 2006; Clapp et al. 2006). Figure 2 shows three examples of gustducin mRNA positive cells, a sweet best cell (Fig. 2A), a bitter best cell (Fig. 2B), and a umami best cell (Fig. 2C). Fifteen of 20 gustducin mRNA-positive taste cells responded best to saccharin, three responded best to quinine, and two responded best to MSG. Similarly to the results with gustducin-GFP taste cells, no gustducin mRNA-positive taste cells responded best to NaCl or HCl. In contrast, three SNAP25 mRNA positive cells responded best to HCl. We also examined sweet-sensitive cells for expression of T1r3, a component of sweet and umami receptors (Bachmanov et al. 2001; Kitagawa et al. 2001; Max et al. 2001; Montmayeur et al. 2001; Nelson et al. 2001; Sainz et al. 2001). We detected the expression of T1r3 mRNA in 8 of 15 sweet best gustducin mRNA-positive taste cells. Expression of T1r3 mRNA was not found in two MSG best cells and three bitter best cells. Degradation of mRNA during long-time recording of taste responses, false-negative results and T1r3 independent receptor systems for sweet taste may explain the T1r3-negative sweet sensitive cells in our single cell RT-PCR experiments.
Figure 2. Sample recordings from taste cells in which expression of gustducin was detected by single cell RT-PCR.
Upper panels show taste responses to five basic taste stimuli. Dotted lines show the onset of taste stimulation. Lower panels show the gel electrophoretic detection of expression of SNAP25, gustducin, T1r3 and β-actin by multiplex single cell RT-PCR. Purified mRNA from a single taste bud and electrode solution without sample were used as positive control (PC) and negative control (NC), respectively. The predicted sizes of PCR products were the following: SNAP25 (304 bp), gustducin (341 bp), T1r3 (343 bp), β-actin (370 bp).
We pooled the data from gustducin-GFP cells and gustducin mRNA-positive taste cells as ‘gustducin-positive’ taste cells. The response properties of all 49 gustducin-positive taste cells are shown in Fig. 3. In total, 25 cells responded best to saccharin, 20 cells responded best to quinine, and four cells responded best to MSG. Thirty-eight of 49 (78%) taste cells responded to only one, 10 of 49 (20%) responded to two, and 1 of 49 (2%) responded to three of five basic taste stimuli. The mean entropy value for the breadth of responsiveness of 49 gustducin-positive taste cells was 0.087 ± 0.024 (mean ±s.e.m.). This value is very similar to that of receptor cells in mouse circumvallate papillae (0.07 ± 0.02, Tomchik et al. 2007). This small mean entropy value indicates that gustducin-positive taste cells are likely to be specifically tuned to particular basic taste stimuli.
Figure 3. Response profiles of 49 gustducin-positive taste cells.
Taste responses are shown of each taste cell to 300 mm NaCl (NaCl, blue), 20 mm saccharin (Sac, red), 20 mm quinine-HCl (QHCl, purple), 300 mm monosodium glutamate (MSG, yellow), and 10 mm HCl (HCl, green). Gustducin-positive taste cells are arranged according to the best stimulus (sweet best: 1∼25, bitter best: 26∼45, umami best: 46∼49) and response magnitudes (impulses per 10 s).
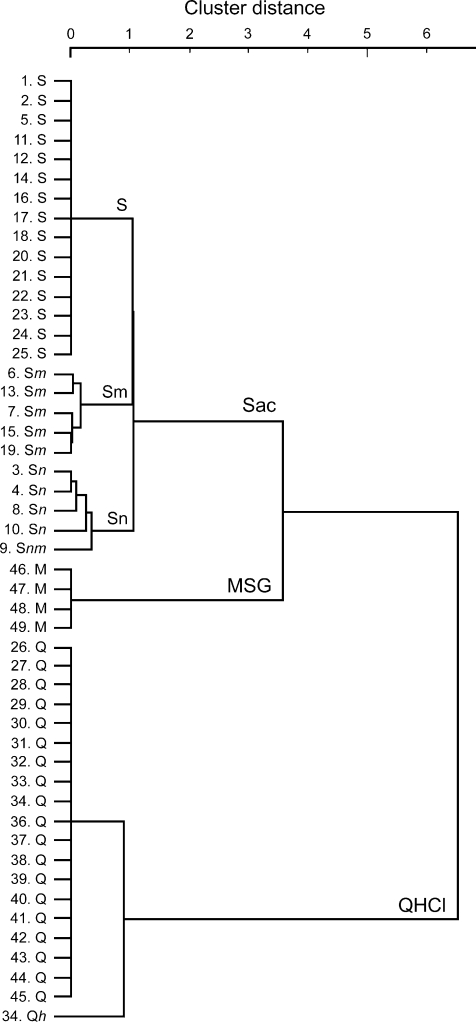
Using hierarchical cluster analysis we classified gustducin-positive taste cells into several groups according to the response profile to five basic taste stimuli (Fig. 4). Gustducin-positive taste cells were classified into three groups, sweet best (labelled Sac), bitter best (QHCl), and umami best (MSG). The sweet best group was further classified into three groups: sweet specific (labelled S), sweet + umami (Sm), and sweet + salty (Sn). The mean entropy value for sweet best cells (0.153 ± 0.039, n= 25) was significantly greater than that for bitter best cells (0.021 ± 0.021, n= 20, P < 0.01, t-test), suggesting that bitter sensitive cells may be more narrowly tuned to particular taste stimuli than are sweet sensitive cells.
Figure 4. Cluster dendrogram showing the relationships among response profiles of gustducin-expressing taste cells.
The cell number and response profile of each cell is indicated on the left. Capital letters indicate the stimulus producing the maximum response (shown first) and all others with responses ≥50% of maximum. Lower-case letters indicate responses <50% of the maximum. The order of the letters indicates the relative magnitude of the response to each stimulus (S or s: Sac; N or n: NaCl; H or h: HCl, Q or q: quinine, M or m: MSG). The three clusters are labelled Sac, MSG and quinine according to the best stimulus. The Sac cluster is further divided into 3 groups, sweet specific (labelled S), sweet and salty (labelled Sn), sweet and umami (Sm).
Response properties of bitter sensitive taste cells
A previous study with mouse circumvallate papillae (Caicedo & Roper, 2001) demonstrated that most bitter sensitive taste cells were activated by only one of five bitter compounds tested (CX, QHCl, Den, SOA and phenylthiocarbamide). In contrast, molecular studies demonstrated that individual taste receptor cells express a large repertoire of bitter-responsive T2r taste receptors, suggesting that each cell may be capable of recognizing multiple tastants (Adler et al. 2000). We examined quinine-sensitive gustducin-GFP taste cells in mouse fungiform papillae to determine if five different bitter compounds (20 mm QHCl, 0.1 mm CX, 20 mm Den, 0.5 mm SOA and 10 mm Caffeine) could elicit responses from these cells. We tested 14 quinine sensitive cells and found that most of them responded to three bitter compounds: QHCl, Den and CX (Fig. 5). These cells did not respond to SOA or caffeine. By hierarchical cluster analysis, bitter sensitive taste cells were classified into two groups according to the breadth of responsiveness to three bitter compounds (Fig. 6). The mean entropy of the breadth of responsiveness for the narrower and broader clusters was 0.577 ± 0.1 (n= 5, s.e.m.) and 0.907 ± 0.053 (n= 9), respectively, which differ significantly (P < 0.01, t-test). These results suggest that most bitter sensitive cells expressing gustducin in mouse fungiform papillae may be capable of recognizing multiple bitter compounds.
Figure 5. Bitter taste response profiles of 14 quinine sensitive taste cells.
Taste responses are shown of each taste cell to 20 mm quinine-HCl (QHCl), 20 mm denatnium (Den) and 0.1 mm cyclohexamide (CX). Bitter taste cells are arranged according to the magnitude of response to quinine. No cells responded to 0.5 mm sucrose octaacetate or 10 mm caffeine.
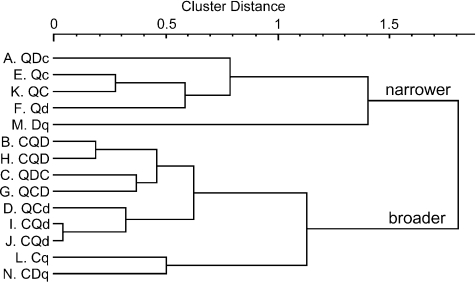
Figure 6. Cluster dendrogram showing the relationships among response profiles of bitter sensitive taste cells.
The cell number and response profile of each cell is indicated on the left (Q or q: quinine, D or d: denatnium, C or c: cyclohexamide). The two clusters are labelled narrower and broader according to the breadth of responsiveness to three bitter compounds. The mean entropy of the breadth of responsiveness for the narrower and broader cluster is 0.577 ± 0.1 (n= 5) and 0.907 ± 0.053 (n= 9), respectively, which differ significantly (P < 0.01, t-test).
Response properties of GAD67 taste cells
As noted above we found that gustducin-positive (Type II) taste cells responded to sweet, bitter or umami taste stimuli. Sour and salty taste may be mediated by other types of taste bud cells. In taste buds, GAD67 is expressed in a subset of type III cells (DeFazio et al. 2006; Nakamura et al. 2007, Tomchik et al. 2007). We used GAD67-GFP mice to examine taste response properties of type III cells. Figure 7 shows sample recordings from two GAD67-GFP taste cells: one specifically responded to 10 mm HCl (Fig. 7A) and the other responded to multiple taste stimuli (Fig. 7B). We recorded taste responses from 44 GAD67-GFP taste cells and found that all GAD67-GFP cells responded to 10 mm HCl (Fig. 8). Eleven of 44 GAD67-GFP taste cells responded to multiple taste stimuli, and the rest (33 cells) responded specifically to HCl. The mean entropy value for the breadth of responsiveness of 44 GAD67-GFP taste cells was 0.123 ± 0.034 (mean ±s.e.m.). This value is significantly smaller than the mean entropy value observed with ‘presynaptic cells’ (Type III cells) in mouse circumvallate papillae (0.47 ± 0.04, Tomchik et al. 2007), suggesting that type III cells in fungiform papillae may have different response properties or functions from those in circumvallate papillae.
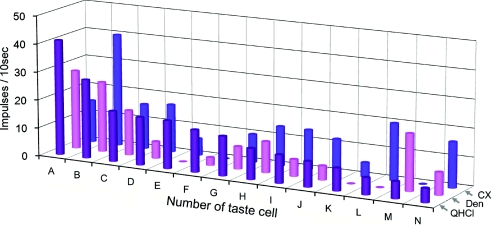
Figure 8. Response profiles of 44 GAD67-expressing taste cells.
Taste responses are shown of each taste cell to 300 mm NaCl (NaCl, blue columns), 20 mm saccharin (Sac, red columns), 20 mm quinine-HCl (QHCl, purple columns), 300 mm monosodium glutamate (MSG, yellow columns), and 10 mm HCl (HCl, green columns). All taste cells responded best to HCl. Taste cells are arranged according to sensitivity to five basic taste stimuli (broad sensitive: 1∼11, sour specific: 12∼44), and magnitude of response to HCl.
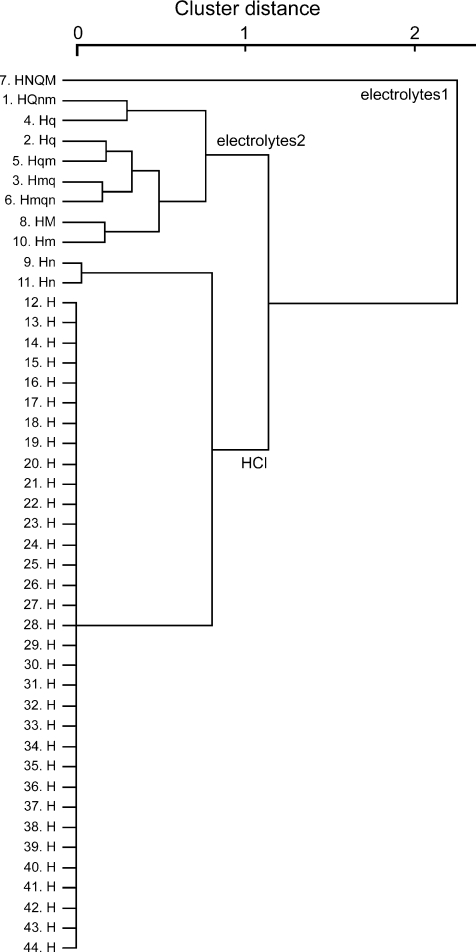
By hierarchical cluster analysis, GAD67-GFP taste cells are classified into three groups according to the response profile of each cell to five basic taste stimuli (Fig. 9). Two groups (electrolytes1 and electrolytes2) responded to multiple tastants and one group showed specific responses to HCl except for two taste cells that responded to HCl and NaCl (Fig. 9, cells 9 and 11). The mean entropy value for taste cells in the electrolyte2 group (0.473 ± 0.053, n= 8) was very close to the mean entropy value of presynaptic cells (Type III cells) in mouse circumvallate papillae (0.47 ± 0.04, Tomchik et al. 2007), suggesting that those cells may have similar function in both fungiform and circumvallate papillae.
Figure 9. Cluster dendrogram showing the relationships among response profiles of GAD67 taste cells.
The cell number and response profile of each cell is indicated on the left (S or s: Sac; N or n: NaCl; H or h: HCl, Q or q: quinine, M or m: MSG). The three clusters are labelled electrolytes1, electrolytes2 and HCl. Taste cells in the electrolytes1 and electrolytes2 clusters showed multiple responses to electrolytes; taste cells in the HCl cluster showed specific response to HCl, except 2 cells (Nos 9 and 11).
Response properties of sour sensitive taste cells
Because all mouse fungiform papillae GAD67-GFP cells tested responded to HCl (Fig. 8) we investigated their responses to three sour tastants: 10 mm acetic acid (pH 3.4), 10 mm citric acid (pH 2.6) and 10 mm HCl (pH 2.0). We tested 14 HCl sensitive cells and found that most of them responded to all three sour compounds (Fig. 10). The mean entropy for the breadth of responsiveness was 0.894 ± 0.026 (n= 14), suggesting broad tuning to sour stimuli in HCl sensitive taste cells. By hierarchical cluster analysis, sour sensitive taste cells were classified into three groups labelled ACH, CH and H (Fig. 11). Taste cells in the ACH group showed relatively equal responses to all three sour compounds. Taste cells in the CH group showed smaller response to acetic acid, and taste cells in the H group showed smaller responses to acetic acid and citric acid.
Figure 10. Sour taste response profiles of 14 HCl sensitive taste cells.
Taste responses to 10 mm acetic acid (AA), 10 mm citric acid (CA) and 10 mm HCl (HCl) in each taste cell are shown. Sour sensitive taste cells are arranged according to the magnitude of response to HCl.
Figure 11. Cluster dendrogram showing the relationships among response profiles of sour sensitive taste cells.
The cell number and response profile of each cell are indicated on the left (A or a: acetic acid, C or c: citric acid, H or h: HCl). The three clusters are labelled ACH, CH and H according to the sour stimuli that elicited ≥50% of maximum response.
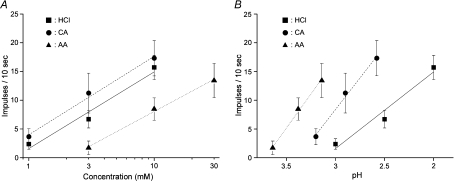
We examined concentration and pH dependency of taste responses to these sour compounds (Fig. 12). The magnitude of sour taste responses depended on the concentration of sour tastants applied (Fig. 12A). At the same concentration, citric acid and HCl elicited similar responses (range = 1∼10 mm, repeated ANOVA, F(1,66)= 1.13, P > 0.1) and acetic acid elicited smaller responses than citric acid (range = 3∼10 mm, F(1,45)= 9.65, P < 0.01) and HCl (range = 3∼10 mm, F(1,48)= 9.00, P < 0.01). We converted acid concentration into pH and examined pH dependency of sour taste response (Fig. 12B). At the same pH level, acetic acid elicited the largest and HCl the smallest responses. These properties were similar to those reported previously (Beidler, 1967; Lyall et al. 2001, Huang et al. 2008).
Figure 12. Concentration–response and pH–response relationships for HCl, citric acid and acetic acid.
A, concentration–response relationships for HCl (squares), citric acid (CA: circles) and acetic acid (AA: triangles) (n= 6∼21). B, pH–response relationships for HCl, citric acid and acetic acid (n= 6∼21). Values indicated are means ±s.e.m.
Discussion
The present study examined response properties of individual mouse fungiform taste cells identified by expression of two specific molecules, gustducin and GAD67. Gustducin and GAD67 are well known markers for Type II and Type III taste cells, respectively. Our results demonstrated that gustducin-expressing taste cells responded best to sweet, bitter or umami stimuli, and that all GAD67-expressing taste cells responded to sour stimuli (Figs 3 and 8). These response properties were fully consistent with known properties of Type II and III cells, respectively, indicating that our molecular markers did identify the appropriate subtypes of taste cells. In addition, our results were mostly consistent with previous findings obtained with mouse circumvallate taste cells using a calcium imaging technique (Tomchik et al. 2007; Huang et al. 2008). Thirty-eight of 49 (78%) gustducin-positive taste cells and 33 of 44 (75%) GAD67-GFP taste cells showed specific responses to only one of five basic taste qualities, suggesting that the majority of gustducin-positive and GAD-positive cells in mouse fungiform papillae are specifically tuned to a single taste quality. Both Type II and III taste cells generated action potentials in response to taste stimuli, indicating that these cells may transmit taste information to the gustatory nerve fibres (Yoshida et al. 2006a,b;). Thus, taste information from these specialized taste cells may be transmitted directly to gustatory nerve fibres and then to higher order neurons, and may be important to evoke specific taste sensation for each taste quality.
Gustducin is a transduction component for sweet, bitter and umami taste. α-Gustducin knockout mice have greatly diminished, but not entirely abolished, behavioural and nerve responses to sweet, bitter and umami compounds (Wong et al. 1996, He et al. 2004), indicating that gustducin plays a key role in the transduction of these tastes and that a gustducin-independent pathway for these tastes also exists. In the present study, we found that gustducin-positive taste cells responded best to sweet, bitter or umami taste stimuli, but not best to HCl or NaCl (Figs 1–3) and the breadth of responsiveness of these cells was narrow (mean entropy = 0.087 ± 0.024, n= 49), indicating that each gustducin-expressing taste cell may be specifically tuned to sweet, bitter or umami taste. In α-gustducin knockout mice, the reduction of taste information produced from these cells may lead to greatly diminished neural and behavioural responses to these tastes. The residual responses in α-gustducin knockout mice may come from: (1) the gustducin lineage of taste cells that lack α-gustducin, or (2) taste cells other than the gustducin lineage.
In mouse circumvallate taste buds, Defazio et al. (2006) demonstrated that GAD1 (GAD67) was coexpressed with SNAP25, NCAM and AADC but not with TRPM5 and PLCβ2. Clapp et al. (2006) demonstrated that SNAP25 was expressed in a separate population of mouse taste cells from those expressing T1r3 or TRPM5. In addition, Tomchik et al. (2007) demonstrated that almost all GAD-GFP mouse taste cells in circumvallate papillae expressed serotonin or SNAP25 but not PLCβ2. Our preliminary single cell RT-PCR data using mouse fungiform taste bud cells are consistent with these data (R. Yoshida, A. Miyauchi & Y. Ninomiya, unpublished observation). All these data indicate that in mice GAD67-expressing taste cells are synaptic (Type III) cells but not receptor (Type II) cells. Other laboratories using rats demonstrated that a subpopulation of SNAP25-expressing cells also expressed gustducin and PLCβ2 (Oike et al. 2006; Ueda et al. 2006), and that some GAD-expressing cells were gustducin immunoreactive (Cao et al. 2009), indicating that SNAP25 and GAD may not be a specific marker for Type III cells in rats. These differences are most likely to be due to mouse vs. rat species differences in expression patterns of marker molecules. In mouse circumvallate papillae, expression of GAD is likely to be restricted in the Type III cell population. Expression patterns of GAD and other markers are not confirmed in fungiform taste buds. However, if expression patterns of GAD-expressing cells were not significantly different between fungiform and circumvallate papillae, GAD67-GFP cells in this study might be Type III cells. Type III cells express the putative sour taste receptor PKD2L1 (Kataoka et al. 2008), and have been shown to sense sour taste (Huang et al. 2008). Consistently with these studies we found that all GAD67 taste cells examined responded to sour taste stimuli (Figs 7, 8). It is known that at the same pH organic acids such as acetic acid are more intensely sour than mineral acids such as HCl (Harvey, 1920). Our results demonstrated that sour sensitive Type III cells respond to a variety of acids (acetic acid, citric acid and HCl) and that at the same pH organic acids elicited greater responses than did mineral acids (Figs 10, 11), suggesting that these cells may be sour taste receptor cells. We observed the expression of PKD2L1 in some of the GAD67 taste cells that responded to sour taste stimuli (R. Yoshida & Y. Ninomiya, unpublished observation). Genetic ablation of these cells may result in the loss of taste responses to sour stimuli in mice lacking PKD2L1-expressing taste cells (Huang et al. 2006).
In both the present and previous studies (Yoshida et al. 2006a) we found broadly tuned and narrowly tuned taste cells. The number of broadly tuned cells in this study did appear lower than in our previous study. The most likely explanation for this is that the source of taste bud cells differed between these two studies. In our previous study we recorded taste responses from taste bud cells chosen at random. In contrast, in the present study we recorded taste responses from only two identified types of taste cells (gustducin-expressing cells and GAD-expressing cells). Some of the taste bud cells other than the gustducin-positive and GAD-positive cells may show multiple sensitivities to taste stimuli. For example, SNAP25-positive, GAD-negative cells may respond to multiple taste stimuli by means of cell–cell communication.
Our findings provide independent and important validation of previous results gained in mouse circumvallate taste cells using the calcium imaging technique (Tomchik et al. 2007; Huang et al. 2008). However, there may be some differences arising from technical and/or regional differences in preparation. In mouse circumvallate papillae, Type II (‘receptor’) cells were shown to be narrowly tuned to bitter, sweet or umami taste stimuli (Tomchik et al. 2007). The mean entropy value of receptor cells in mouse circumvallate papillae (0.07 ± 0.02) was very similar to gustducin taste cells in fungiform papillae (0.087 ± 0.024). Taken together, Type II cells may be devoted to detection and transmission of sweet, bitter and umami taste information. However, coexpression patterns of gustducin and taste GPCRs showed regional differences (Kim et al. 2003; Shigemura et al. 2008), suggesting that gustducin-expressing taste cells in fungiform vs. circumvallate papillae may have different response properties, such as gurmarin sensitivity (Ninomiya & Imoto, 1995; Ninomiya et al. 1997). These differences have not yet been revealed by physiological studies in taste cells. Huang et al. (2008) reported that Type III (‘presynaptic’) cells in circumvallate papillae specifically respond to acid taste stimulation and release serotonin. Consistent with these findings, GAD67 taste cells in fungiform papillae responded to sour taste stimuli (Figs 7, 8). In addition, Tomchik et al. (2007) demonstrated that Type III cells in circumvallate taste cells respond to multiple taste qualities. In fungiform taste buds, a portion of Type III cells responded to multiple taste qualities and the breadth of responsiveness of these cells (0.473 ± 0.053) was very similar to those in circumvallate taste cells (0.47 ± 0.04). Therefore, Type III cells may be divided into two groups, one for sour specific responses and the other for multiple taste responses. However, fungiform Type III cells tend to respond more specifically to sour taste stimuli than do circumvallate Type III cells. This may be due to different numbers of Type III cells between fungiform and circumvallate taste buds. The number of Type III cells in mouse circumvallate taste buds may be 3–5 times larger than that in fungiform taste cells (R. Yoshida & Y. Ninomiya, unpublished observation). This may imply a functional difference between Type III cells from fungiform vs. circumvallate papillae. Another possibility is the existence of Type III cells that do not express GAD67. Most GAD-expressing cells coexpressed SNAP25 or serotonin but 20∼30% of SNAP25-expressing cells and serotonin-expressing cells did not possess GAD67 (DeFazio et al. 2006; Tomchik et al. 2007). In addition, high K+ depolarization was used to identify Type III cells in circumvallate taste buds (Tomchik et al. 2007) but not in this study. These facts may imply that GAD-GFP cells used in this study represent only a subpopulation of Type III cells. Therefore, we might miss recording from Type III cells that have multiple taste sensitivities. These multiple taste sensitivities may be caused by cell–cell communication (Roper, 2006) or multiple chemosensory receptor(s). Future studies may shed light on this.
In addition, bitter sensitivities may differ between fungiform and circumvallate taste cells. Bitter taste is mediated by a family of ∼30 GPCRs (T2rs, Adler et al. 2000; Matsunami et al. 2000). Gene expression analyses in the rat gustatory system demonstrated that a single taste receptor cell expresses a large repertoire of T2rs (Adler et al. 2000), suggesting that each bitter sensitive taste cell may be capable of detecting multiple bitter compounds. Gene expression analyses of mouse and human gustatory tissues showed a more limited coexpression of T2rs (Matsunami et al. 2000, Behrens et al. 2007), suggesting heterogeneous populations of bitter taste cells. Previous physiological experiments demonstrate that most bitter taste cells in rat circumvallate papillae respond to one or two of five bitter stimuli (QHCl, CX, Den, SOA and phenylthiocarbamide; Caicedo & Roper, 2001). In contrast, our data on bitter sensitive cells in fungiform papillae showed that most taste cells responded to multiple bitter compounds (quinine, cyclohexamide and denatonium: Fig. 5) but not to two other bitter compounds (0.5 mm SOA and 10 mm caffeine). One possible explanation for this discrepancy may be different expression patterns of T2rs between fungiform and circumvallate taste cells. There are little data on the expression patterns of T2rs in mouse fungiform taste buds, therefore, further studies are needed. In any event, these differences may explain different sensitivities to bitter compounds between the chorda tympani (CT) and the glossopharyngeal (IXth) nerve (Danilova & Hellekant, 2003; Damak et al. 2006). Denatnium and cyclohexamide evoke a large response in the IXth nerve but only a very slight response in the CT nerve. Caffeine and SOA evoke almost no response in the CT nerve but a slight response in the IXth nerve. Quinine-HCl evokes a large response in both the CT and the IXth nerve. In circumvallate papillae, a large population of bitter sensitive cells responded specifically to cyclohexamide or denatonium (Caicedo & Roper, 2001), suggesting that these cells may contribute to a large response to these compounds in the IXth nerve. Twenty-three of 69 circumvallate bitter sensitive cells showed multiple sensitivities to bitter compounds (Caicedo & Roper, 2001). These cells may be comparable to fungiform bitter sensitive cells expressing gustducin. In the CT nerve, quinine evokes a larger response than does cyclohexamide or denatonium; however, all these compounds evoked large responses in bitter sensitive cells expressing gustducin (Fig. 5). α-Gustducin knockout mice showed large residual CT nerve responses to quinine (Wong et al. 1996). Taken together, there is the possibility that quinine sensitive taste cells that do not express gustducin may exist in fungiform taste buds and these cells may be more specifically tuned to quinine.
Based on this study, sweet, bitter, umami and sour taste may be detected by gustducin-expressing (Type II) or GAD67-expressing (Type III) taste cells and these tastes may be discriminated at the receptor cell level. However, we have not found any salt sensitive taste cells amongst the gustducin-expressing and GAD67-expressing taste cells we examined, indicating that taste responsive cells are not restricted to gustducin-positive and GAD-positive cells. Which type of taste cell is responsible for salt taste detection? NaCl sensitive taste cells are classified into two groups according to amiloride sensitivities, amiloride sensitive (AS) and insensitive (AI) cells (Ninomiya & Funakoshi, 1988; Ninomiya, 1996; Yoshida et al. 2009). AS cells are narrowly responsive to NaCl, whereas AI cells are broadly responsive to NaCl, KCl and HCl. Response properties of AI cells are similar to those GAD67-expressing taste cells which showed taste sensitivities to multiple taste modalities, suggesting that AI cells may be Type III cells. AS cells may be included in the cell population that does not express gustducin or GAD67. Type I cells may be one of the candidates because amiloride sensitive channels were expressed in Type I fungiform taste cells (Vandenbeuch et al. 2008). The other candidate is Type II or III cells that do not possess gustducin or GAD67. In any case, salt specific taste cells exist in mouse fungifrom taste cells. These data indicate the existence of specific coding channels from taste cells to the central nervous system for each of five basic taste qualities.
Acknowledgments
This work was supported by KAKENHI 20019010 (Y.Y.), 18109013, 18077004 (Y.N.) and 21791808 (R.Y.).
Glossary
Abbreviations
- AA
acetic acid
- ASIC
amiloride sensitive cation channel
- CA
citric acid
- CT
corda tympani
- CX
cyclohexamide
- Den
denatnium benzoate
- DW
distilled water
- GAD67
glutamate decarboxylase 67
- GFP
green fluorescent protein
- GPCR
G-protein coupled receptor
- HCN
hyperpolarization activated cyclic nucleotide gated potassium channel
- IP3R3
inositol 1,4,5-trisphosphate receptor 3
- IXth
glossopharyngeal
- MSG
monosodium glutamate
- PKD1L3
polycystic kidney disease 1 like 3
- PKD2L1
polycystic kidney disease 2 like 1
- PLCβ2
phosphlipase Cβ2
- QHCl
quinine-HCl
- Sac
saccharin sodium
- SNAP25
synaptosomal associated protein 25
- SOA
sucrose octaacetate
- T1r
taste receptor type 1
- T2r
taste receptor type 2
- TRPM5
transient receptor potential cation channel subfamily M member 5
- V1R
transient receptor potential cation channel subfamily V member 1.
Author contributions
R.Y. and Y.N. designed and performed the research and wrote the initial draft. A.M., T.Y., M.J. and Y.M. performed recordings of taste responses. K.Y. and N.S. designed the research and analysed data. Y.Y., K.O., H.U. and R.F.M. provided transgenic mice and critically revised the final article. All authors read and approved the final version. All experiments were done in the Graduate School of Dental Sciences, Kyushu University.
Authors' present addresses
Y. Murata: Department of Physiology, Kochi Medical School, Nankoku, Kochi 783-8505, Japan.
K. Yasumatsu: Department of Oral Physiology, Asahi University School of Dentistry, Gifu, 501-0296, Japan.
References
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJP, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Bachmanov AA, Li X, Reed DR, Ohmen JD, Li S, Chen Z, Tordoff MG, de Jong PJ, Wu C, West DB, Chatterjee A, Ross DA, Beauchamp GK. Positional cloning of the mouse saccharin preference (Sac) locus. Chem Senses. 2001;26:925–933. doi: 10.1093/chemse/26.7.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M, Foerster S, Staehler F, Raguse JD, Meyerhof W. Gustatory expression pattern of the human TAS2R bitter receptor gene family reveals a heterogenous population of bitter responsive taste receptor cells. J Neurosci. 2007;27:12630–12640. doi: 10.1523/JNEUROSCI.1168-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beidler LM. Anion influences on taste receptor response. In: Hayashi T, editor. Olfaction and Taste II. Vol. 8. Oxford, UK: 1967. pp. 509–534. [Google Scholar]
- Breza JM, Curtis KS, Contreras RJ. Temperature modulates taste responsiveness and stimulates gustatory neurons in the rat geniculate ganglion. J Neurophysiol. 2006;95:674–685. doi: 10.1152/jn.00793.2005. [DOI] [PubMed] [Google Scholar]
- Caicedo A, Roper SD. Taste receptor cells that discriminate between bitter stimuli. Science. 2001;291:1557–1560. doi: 10.1126/science.291.5508.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo A, Kim K, Roper SD. Individual mouse taste cells respond to multiple chemical stimuli. J Physiol. 2002;544:501–509. doi: 10.1113/jphysiol.2002.027862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Zhao FL, Kolli T, Hivley R, Herness S. GABA expression in the mammalian taste bud functions as a route of inhibitory cell-to-cell communication. Proc Natl Acad Sci U S A. 2009;106:4006–4011. doi: 10.1073/pnas.0808672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- Clapp TR, Yang R, Stoick CL, Kinnamon SC, Kinnamon JC. Morphologic characterization of rat taste receptor cells that express components of the phospholipase C signalling pathway. J Comp Neurol. 2004;468:311–321. doi: 10.1002/cne.10963. [DOI] [PubMed] [Google Scholar]
- Clapp TR, Medler KF, Damak S, Margolskee RF, Kinnamon SC. Mouse taste cells with G protein-coupled taste receptors lack voltage-gated calcium channels and SNAP-25. BMC Biol. 2006;4:7. doi: 10.1186/1741-7007-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damak S, Rong M, Yasumatsu K, Kokrashvili Z, Perez CA, Shigemura N, Yoshida R, Mosinger B, Glendinning JI, Ninomiya Y, Margolskee RF. Trpm5 null mice respond to bitter, sweet, and umami compounds. Chem Senses. 2006;31:253–264. doi: 10.1093/chemse/bjj027. [DOI] [PubMed] [Google Scholar]
- Danilova V, Hellekant G. Comparison of the responses of the chorda tympani and glossopharyngeal nerves to taste stimuli in C57BL/6J mice. BMC Neurosci. 2003;4:5. doi: 10.1186/1471-2202-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFazio RA, Dvoryanchikov G, Maruyama Y, Kim JW, Pereira E, Roper SD, Chaudhari N. Separate populations of receptor cells and presynaptic cells in mouse taste buds. J Neurosci. 2006;26:3971–3980. doi: 10.1523/JNEUROSCI.0515-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RB. The relation between the total acidity, the concentration of the hydrogen ion, and the taste of acid solutions. J Am Chem Soc. 1920;42:712–714. [Google Scholar]
- He W, Yasumatsu K, Varadarajan V, Yamada A, Lem J, Ninomiya Y, Margolskee RF, Damak S. Umami taste responses are mediated by α-transducin and α-gustducin. J Neurosci. 2004;24:7674–7680. doi: 10.1523/JNEUROSCI.2441-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AL, Chen X, Hoon MA, Chandrashekar J, Guo W, Trankner D, Ryba NJ, Zuker CS. The cells and logic for mammalian sour taste detection. Nature. 2006;442:934–938. doi: 10.1038/nature05084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YA, Maruyama Y, Stimac R, Roper SD. Presynaptic (Type III) cells in mouse taste buds sense sour (acid) taste. J Physiol. 2008;586:2903–2912. doi: 10.1113/jphysiol.2008.151233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Inada H, Kubota M, Zhuang H, Tominaga M, Matsunami H. Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc Natl Acad Sci U S A. 2006;103:12569–12574. doi: 10.1073/pnas.0602702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka S, Yang R, Ishimaru Y, Matsunami H, Sevigny J, Kinnamon JC, Finger TE. The candidate sour taste receptor, PKD2L1, is expressed by type III taste cells in the mouse. Chem Senses. 2008;33:243–254. doi: 10.1093/chemse/bjm083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MR, Kusakabe Y, Miura H, Shindo Y, Ninomiya Y, Hino A. Regional expression patterns of taste receptors and gustducin in the mouse tongue. Biochem Biophys Res Commun. 2003;312:500–506. doi: 10.1016/j.bbrc.2003.10.137. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Kusakabe Y, Miura H, Ninomiya Y, Hino A. Molecular genetic identification of a candidate receptor gene for sweet taste. Biochem Biophys Res Commun. 2001;283:236–242. doi: 10.1006/bbrc.2001.4760. [DOI] [PubMed] [Google Scholar]
- Lawton DM, Furness DN, Lindemann B, Hackney CM. Localization of the glutamate-aspartate transporter, GLAST, in rat taste buds. Eur J Neurosci. 2000;12:3163–3171. doi: 10.1046/j.1460-9568.2000.00207.x. [DOI] [PubMed] [Google Scholar]
- Lindemann B. Receptors and transduction in taste. Nature. 2001;413:219–225. doi: 10.1038/35093032. [DOI] [PubMed] [Google Scholar]
- Lyall V, Alam RI, Phan DQ, Ereso GL, Phan TH, Malik SA, Montrose MH, Chu S, Heck GL, Feldman GM, DeSimone JA. Decrease in rat taste receptor cell intracellular pH is the proximate stimulus in sour taste transduction. Am J Physiol Cell Physiol. 2001;281:C1005–1013. doi: 10.1152/ajpcell.2001.281.3.C1005. [DOI] [PubMed] [Google Scholar]
- Matsunami H, Montmayeur JP, Buck LB. A candidate of taste receptors in human and mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- Max M, Shanker YG, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee RF. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- Montmayeur JP, Liberles SD, Matsunami H, Buck LB. A candidate taste receptor gene near a sweet taste locus. Nat Neurosci. 2001;4:492–498. doi: 10.1038/87440. [DOI] [PubMed] [Google Scholar]
- Murray R. The ultrastructure of taste buds. In: Friedemann I, editor. The Ultrastructure of Sensory Organs. Amsterdam: North Holland; 1973. pp. 1–81. [Google Scholar]
- Nakamura Y, Yanagawa Y, Obata K, Watanabe M, Ueno H. GABA is produced in taste bud. Chem Senses. 2007;32:J19. [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJP, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y. Salt taste responses of mouse chorda tympani neurons: Evidence for existence of two different amiloride-sensitive receptor components for NaCl with different temperature dependencies. J Neurophysiol. 1996;76:3550–3554. doi: 10.1152/jn.1996.76.5.3550. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y. Reinnervation of cross-regenerated gustatory nerve fibres into amiloride-sensitive and amiloride-insensitive taste receptor cells. Proc Natl Acad Sci U S A. 1998;95:5347–5350. doi: 10.1073/pnas.95.9.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya Y, Funakoshi M. Amiloride inhibition of responses of rat single chorda tympani nerve fibres to chemical and electronical tongue stimuli. Brain Res. 1988;451:319–325. doi: 10.1016/0006-8993(88)90777-9. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Imoto T. Gurmarin inhibition of sweet taste responses in mice. Am J Physiol Regul Integr Comp Physiol. 1995;268:R1029–1025. doi: 10.1152/ajpregu.1995.268.4.R1019. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Tonosaki K, Funakoshi M. Gustatory neural response in the mouse. Brain Res. 1982;244:370–373. doi: 10.1016/0006-8993(82)90100-7. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Mizukoshi T, Higashi T, Katsukawa H, Funakoshi M. Gustatory neural responses in three different strains of mice. Brain Res. 1984;302:305–314. doi: 10.1016/0006-8993(84)90244-0. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Tanimukai T, Yoshida S, Funakoshi M. Gustatory nerve responses in preweanling mice. Phisiol Behav. 1991;49:927–934. doi: 10.1016/0031-9384(91)90203-z. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Inoue M, Imoto T, Nakashima K. Lack of gurmarin sensitivity of sweet taste receptors innervated by the glossopharyngeal nerve in C57BL mice. Am J Physiol Regul Integr Comp Physiol. 1997;272:R1002–1006. doi: 10.1152/ajpregu.1997.272.3.R1002. [DOI] [PubMed] [Google Scholar]
- Oike H, Matsumoto I, Abe K. Group IIA phospholipase A2 is coexpressed with SNAP-25 in mature taste receptor cells of rat circumvallate papillae. J Comp Neurol. 2006;494:876–886. doi: 10.1002/cne.20848. [DOI] [PubMed] [Google Scholar]
- Roper SD. Cell communication in taste bud. Cell Mol Life Sci. 2006;63:1494–1500. doi: 10.1007/s00018-006-6112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer SM, Kinnamon JC. Ultrastructure of mouse foliate taste buds: synaptic and nonsynaptic interactions between taste cells and nerve fibres. J Comp Neurol. 1988;270:11–24. doi: 10.1002/cne.902700103. [DOI] [PubMed] [Google Scholar]
- Sainz E, Korley JN, Battey JF, Sullivan SL. Identification of a novel member of the T1R family of putative taste receptors. J Neurochem. 2001;77:896–903. doi: 10.1046/j.1471-4159.2001.00292.x. [DOI] [PubMed] [Google Scholar]
- Shigemura N, Nakao K, Yasuo T, Murata Y, Yasumatsu K, Nakashima A, Katsukawa H, Sako N, Ninomiya Y. Gurmarin sensitivity of sweet taste responses is associated with co-expression patterns of T1r2, T1r3, and gustducin. Biochem Biophys Res Commun. 2008;367:356–363. doi: 10.1016/j.bbrc.2007.12.146. [DOI] [PubMed] [Google Scholar]
- Smith DV, Travers JB. A metric for the breadth of tuning of gustatory neurons. Chem Senses Flav. 1979;4:215–229. [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- Tomchik SM, Berg S, Kim JW, Chaudhari N, Roper SD. Breadth of tuning and taste coding in mammalian taste buds. J Neurosci. 2007;27:10840–10848. doi: 10.1523/JNEUROSCI.1863-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers JB, Smith DV. Gustatory sensitivities in neurons of the hamster nucleus tractus solitarius. Sens Processes. 1979;3:1–6. [PubMed] [Google Scholar]
- Ueda K, Ichimori Y, Okada H, Honma S, Wakisaka S. Immunolocalization of SNARE proteins in both type II and type III cells of rat taste buds. Arch Histol Cytol. 2006;69:289–296. doi: 10.1679/aohc.69.289. [DOI] [PubMed] [Google Scholar]
- Vandenbeuch A, Clapp TR, Kinnamon SC. Amiloride-sensitive channels in type I fungiform taste cells in mouse. BMC Neurosci. 2008;9:1. doi: 10.1186/1471-2202-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GT, Gannon KS, Margolskee RF. Transduction of bitter and sweet taste by gustducin. Nature. 1996;381:796–800. doi: 10.1038/381796a0. [DOI] [PubMed] [Google Scholar]
- Wong GT, Ruiz-Avila L, Margolskee RF. Directing gene expression to gustducin-positive taste receptor cells. J Neurosci. 1999;19:5802–5809. doi: 10.1523/JNEUROSCI.19-14-05802.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Tabata S, Crowley HH, Margolskee RF, Kinnamon JC. Ultrastructural localization of gustducin immunoreactivity in microvilli of type II taste cells in the rat. J Comp Neurol. 2000a;425:139–151. doi: 10.1002/1096-9861(20000911)425:1<139::aid-cne12>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Yang R, Crowley HH, Rock ME, Kinnamon JC. Taste cells with synapses in rat circumvallate papillae display SNAP-25-like immunoreactivity. J Comp Neurol. 2000b;424:205–215. doi: 10.1002/1096-9861(20000821)424:2<205::aid-cne2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Sanematsu K, Shigemura N, Yasumatsu K, Ninomiya Y. Taste receptor cells responding with action potentials to taste stimuli and their molecular expression of taste related genes. Chem Senses. 2005;30:i19–i20. doi: 10.1093/chemse/bjh092. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Shigemura N, Sanematsu K, Yasumatsu K, Ishizuka S, Ninomiya Y. Taste responsiveness of fungiform taste cells with action potentials. J Neurophysiol. 2006a;96:3088–3095. doi: 10.1152/jn.00409.2006. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Yasumatsu K, Shigemura N, Ninomiya Y. Coding channels for taste perception: information transmission from taste cells to gustatory nerve fibres. Arch Histol Cytol. 2006b;69:233–242. doi: 10.1679/aohc.69.233. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Horio N, Murata Y, Yasumatsu K, Shigemura N, Ninomiya Y. NaCl responsive taste cells in the mouse fungiform taste buds. Neuroscience. 2009;159:795–803. doi: 10.1016/j.neuroscience.2008.12.052. [DOI] [PubMed] [Google Scholar]