Abstract
In spite of the many recent developments in the field of vector sialomics, the salivary glands of larval mosquitoes have been largely unexplored. We used whole-transcriptome microarray analysis to create a gene-expression profile of the salivary gland tissue of fourth-instar Anopheles gambiae larvae, and compare it to the gene-expression profile of a matching group of whole larvae. We identified a total of 221 probes with expression values that were (a) significantly enriched in the salivary glands, and (b) sufficiently annotated as to allow the prediction of the presence/absence of signal peptides in their corresponding gene products. Based on available annotation of the protein sequences associated with these probes, we propose that the main roles of larval salivary secretions include: (a) immune response, (b) mouthpart lubrication, (c) nutrient metabolism, and (d) xenobiotic detoxification. Other highlights of the study include the cloning of a transcript encoding a previously unknown salivary defensin (AgDef5), the confirmation of mucus secretion by the larval salivary glands, and the first report of salivary lipocalins in the Culicidae.
Keywords: Anopheles gambiae, salivary gland, Diptera, gene expression, salivary defensin, transcriptome, salivary lipocalin
1. Introduction
The study of the salivary secretions of arthropod vectors of disease (a field known as ‘vector sialomics’) has gained considerable momentum since Ribeiro (1987) first proposed that certain components of the salivary secretions of hematophagous arthropods play a key role in regulating the host’s hemostatic and immune reactions. Currently, the list of vector species for which a published salivary transcriptome and/or proteome exists include Anopheles gambiae (Arca et al.,1999; Lanfrancotti et al.,2002; Arca et al.,2005, Calvo et al.,2006a), An. funestus (Calvo et al.,2007), Aedes aegypti (Ribeiro et al.,2007), Ae. albopictus (Arca et al.,2007), Triatoma brasiliensis (Santos et al.,2007), Triatoma infestans (Assumpcao et al.,2008) Glossina morsitans (Van Den Abbeele et al.,2007), Culicoides nubeculosus (Langner et al.,2007) and Xenopsylla cheopis (Andersen et al.,2007), among others. Within the mosquitoes (Diptera: Culicidae), most research efforts have been aimed at characterizing the salivary glands (SG) of adult stages, leaving the larval SG largely unexplored.
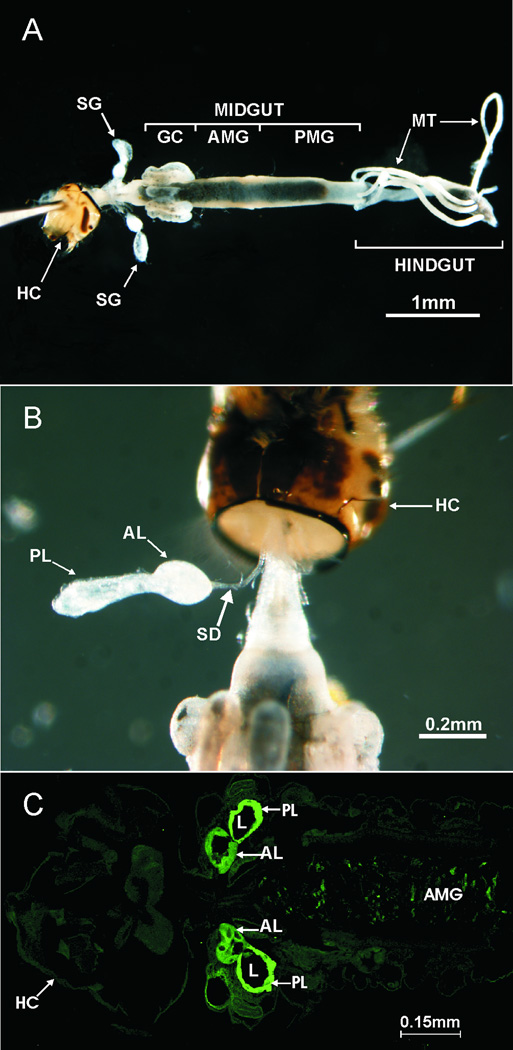
In mosquito larvae, the SG are two well developed structures located in the thorax, on each side of the digestive tract (fig. 1A). In Anopheles species, each gland is divided into two lobes (fig. 1B and 1C) and is connected to the digestive tract by a salivary duct that opens directly into the mouth cavity. A ring of imaginal cells, which will develop into the adult SG during pupation, is located in the most anterior part of each gland (Jensen and Jones, 1957; Christophers, 1960).
Fig. 1.
Location and structure of the salivary glands in the fourth-instar larva of An. gambiae. A) Micrograph showing the location of the salivary glands in relation to the digestive tract. Exoskeleton, fat body and other tissues were previously removed. B) External structure of the salivary gland’s lobes and the salivary duct (only one gland shown). C) Internal structure of the salivary glands. The lumen of both lobes can be visualized in this laser confocal micrograph of a paraffin section labeled with FITC-conjugated Erythrina cristagalli lectin, which displays high binding affinity for the salivary gland tissue (shown in bright green). AL, anterior lobe. AMG, anterior midgut. GC, gastric caeca. HC, head capsule. L, lumen. MT, Malpighian tubes. PL, posterior lobe. PMG, posterior midgut. SD, salivary duct. SG, salivary gland.
Information regarding the nature and purpose of the mosquitoes’ larval salivary secretions is extremely scarce. In contrast with other Diptera, mosquito larvae do not need to secrete glue proteins to attach their pupae to a substrate (as Drosophila larvae do), nor do they bind particles together to form protective structures (as some Chironomus larvae do). In fact, the only published reference we are aware of regarding the function of the larval SG in mosquitoes, states that they have “no such special function to serve” (Christophers, 1960). It seems unlikely, however, that such well developed structures as the larval SG would have evolved in the Culicidae if they had no physiological role to fulfill. In other insect species, larval salivary secretions have been associated with digestion (Verma and Baylan, 1972; Agarwal, 1976; Verma et al, 1977), immunity (Turillazzi et al.,2004; Korayem et al.,2004, Candido-Silva et al.,2007; Liu, 2004), larval molting (Gelman et al, 1991; Zheng et al, 2003) and even social interaction (Cummings et al, 1999; Hunt, 1991).
In this work, we aimed at identifying the gene products most abundantly synthesized by the salivary glands of An. gambiae larvae, and therefore throw some light on the nature and function of their secretions. We performed whole-genome microarray analysis of gene expression, a technique that has been successfully used in the past to create transcriptional profiles of different tissues of An. gambiae larvae (Neira Oviedo et al.,2008) and adults (Marinotti et al.,2005, 2006; Warr et al.,2007). Based on available annotation of the selected gene products, we discuss the potential physiological roles of the larval salivary glands and their secretions.
2. Materials and methods
2.1. Mosquito rearing and dissection
An. gambiae (G3 strain) eggs were obtained from the Centers for Disease Control and Prevention (CDC; Atlanta, Georgia). Mosquito larvae were reared in distilled water at 28°C, 12h: 12h light: darkness cycle, and provided with ground Tetramin® flakes (Tetra Werke) as food.
For each biological replicate of the experiment, 50 fourth-instar larvae were anesthetized on ice and dissected in nuclease-free 70% ETOH. Their salivary glands were extracted and stored at −20°C in nuclease-free 70% ETOH until RNA extraction. Three biological replicates of the experiment were performed.
2.2. RNA sample preparation, processing and hybridization to Affymetrix GeneChip® Arrays
Total RNA was extracted using the RNAeasy-Mini Kit™ (Qiagen) following the manufacturer’s recommendations. RNA processing and hybridization were performed at the University of Florida’s microarray core facility. Briefly, RNA concentration was determined on a NanoDrop Spectrophotometer (NanoDrop Technologies) and sample quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies) as per the manufacturers guidelines. cDNA was synthesized from 5ug of total RNA using the One-Cycle Eukaryotic Target Labeling reagents (Affymetrix, Inc.). The cDNA was used as a template for in vitro transcription (IVT) in the presence of T7 RNA Polymerase and a biotinytlated nucleotide/ribonucleotide mix for cRNA amplification and biotin labeling using the IVT labeling kit (Affymetrix, Inc). The biotin-labeled cRNA was fragmented and hybridized for 16 hours to the Affymetrix GeneChip® Plasmodium/Anopheles Genome Array according to the manufactures’ protocols.
The arrays were then washed and stained with streptavidin-phycoerythrin on an Affymetrix Fluidics Station 450, and scanned on a GeneChip Scanner 3000 (Affymetrix, Inc) using default values to generate signal intensities. Quality control of hybridized chips was performed following Affymetrix recommendations (Affymetrix, 2004a).
As of December 2008, the microarray design used in this study probes for ~11,200 An. gambiae transcripts, as annotated in the VectorBase database (http://www.vectorbase.org/Genome/MartView/).
2.3. Statistical analysis
The GeneSifter™ (VizX Labs) software package was used to perform all statistical analyses. Transcripts were considered as present in a group (whole larvae or SG) if they were scored as such in all three biological repeats by Affymetrix built-in detection call (Affymetrix, 2004a). Expression values were background-adjusted, normalized, and log2-transformed by applying the ‘robust multiarray average’ (RMA) algorithm (Irizarry et al.,2003). Using a t-test, we compared the average signal intensity of each transcript in the SG to that found in a matching group of whole 4th instar larvae processed identically as part of a recently published study by our laboratory (Neira Oviedo et al.,2008). False discovery rate was adjusted to 1% using a Benjamini & Hochberg correction (Benjamini and Hochberg, 1995). Transcripts showing an intensity value two-fold or higher (P < 0.01) in the SG as compared to whole larvae were considered to be significantly enriched in the SG and therefore selected for further analysis.
2.4. Bioinformatics
The predicted amino acid sequences of transcripts encoding a methionine within the first 50 residues were submitted to the SignalP server (Bendtsen et al.,2004) to identify potentially secreted gene products. Functional annotation of selected transcripts was obtained from the June 2007 version of AnoXcel (Ribeiro et al.,2004), a comprehensive annotation repository that compiles information from several relevant sources, including databases such as Gene Ontology (Asburner et al.,2001), SWISSPROT (Bairoch and Boeckmann, 1992), KOG (Tatusov et al.,2003), PFAM (Bateman et al.,2000), SMART (Letunic et al.,2002), among others. This database can be downloaded from http://exon.niaid.nih.gov/transcriptome/A_gambiae/AnoXcel-Nov-2007-Web.zip; the stand alone Affymetrix annotated data can be downloaded from http://exon.niaid.nih.gov/transcriptome/A_gambiae/AffyXcel-Nov-2007.tar.gz.
Based on the results of the aforementioned analyses, transcripts were classified as putative secreted, putative non-secreted, or unknown. Assignment to sub-categories within these groups was based on available functional annotation of the known or predicted gene products. All microarray data discussed in this publication have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE9642. This data will also be available to the public through the expression data repository at VectorBase. (http://www.vectorbase.org).
Sequence alignments were performed using ClustalW (Thompson et al.,1994) available through the BioEdit software package (Hall, 1999) using default parameters. No manual adjustments of the alignment were necessary. Phylogenetic and molecular evolutionary analyses were conducted in the MEGA (version 4) software package (Kumar et al.,2004), using the maximum parsimony (MP) method and neighbor joining (NJ) algorithms. Specifically, we run NJ analysis with 100,000 bootstrap replicates and pair-wise comparisons. The putative annotation of disulfide bridges shown in figure 6a was performed using the crystal structure of sapecin from the flesh fly Sarcophaga peregrina (GenBank accession number P18313) as a model.
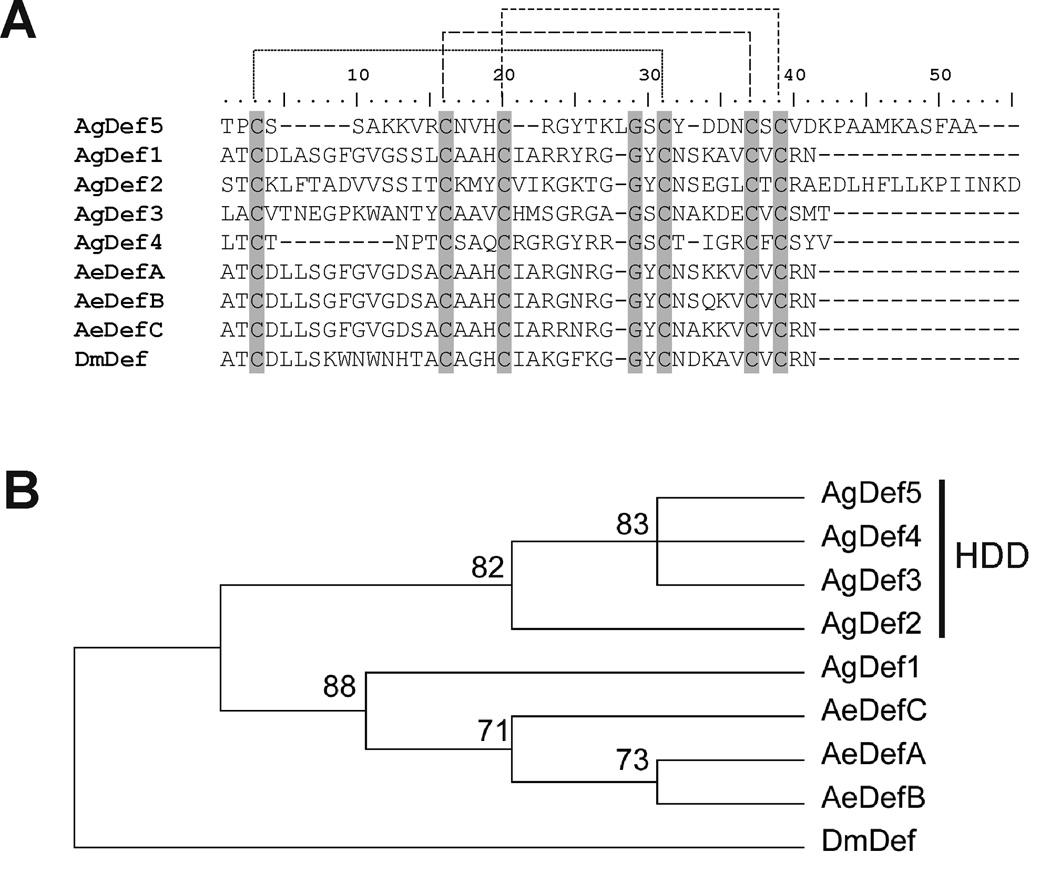
Fig. 6.
Comparison of the amino acid sequence of AgDef5 with other known dipteran defensins. A) The predicted mature peptide sequence of Anopheles gambiae AgDef5 is aligned with those of Anopheles gambiae AgDef1-4 (ENSEMBL accessions AGAP011294-PA, AGAP004632-PA, AGAP007199-PA, AGAP005416-PA, respectively), Aedes aegypti AeDefA-C (ENSEMBL accessions AAEL003841-PA, AAEL003857-PA, AAEL003832-PA, respectively), and Drosophila melanogaster DmDef (ENSEMBL accession FBgn0010385). Conserved residues are shown in grey background; the peptide sequence of AgDef5 contains the six conserved cystein residues characteristic of all insect defensins. Corresponding cysteins for disulfide bridge formation are connected with dashed lines. Homology annotation of these residues was performed using the known structure of sapecin from the flesh fly Sarcophaga peregrina (GenBank accession number P18313) as a model. B) Neighbor-joining phylogenetic analysis of dipteran defensin sequences using 100,000 bootstrap replicates and pairwise comparison. AgDef5 is placed in the cluster of highly divergent defensins (HDD) formed by AgDef3, AgDef4 and AgDef2 (Christophides et al., 2002). Numbers in the phylogram nodes indicate the percent bootstrap support. Overall tree topology was further confirmed using maximum parsimony analysis (data not shown).
2.5. RT-PCR
Total RNA was extracted from dissected SG using the RNAeasy-Mini Kit™ (Qiagen) following the manufacturer’s recommendations and using the kit’s on-column DNAse treatment. cDNA synthesis was performed using the Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen) using random hexamers as primers and following the manufacturer’s recommendations.
Amplification reactions were assembled using 22µl PCR Supermix HiFidelity (Invitrogen), 1µl (≥100ng) cDNA, 1µl forward primer (10µM), and 1µl reverse primer (10µM). Primer sequences are listed in the supplementary materials, table 1. PCR conditions were: 94°C for 3 min; 35 cycles of [94°C for 30 sec, 50°C for 30 sec, 72°C for 1 min]; 72°C for 3 min; hold at 4°C. PCR products were visualized on a 1% agarose/EtBr gel.
2.6. Quantitative real-time PCR
RNA extraction and cDNA construction were performed as described for RT-PCR analysis. Quantitative real-time PCR (qRT-PCR) reactions were prepared using the SYBR Green PCR Master Mix (Applied Biosystems), following the manufacturer’s recommendations. Reactions were prepared in triplicate and run in an ABI PRISM® 7000 Sequence Detection System (Applied Biosystems). Relative abundance levels were calculated by the ΔΔCT method (Applied Biosystems, 2004), using the ribosomal protein S7 transcript as endogenous control (Warr et al.,2007). All primer sequences used for qRT-PCR are listed in the supplementary materials, table 1.
2.7. An. gambiae defensin 5 (AgDef5) cDNA cloning and sequencing
An. gambiae defensin 5 PCR product was excised from the agarose gel and extracted using the MinElute gel extraction kit (Qiagen). Purified PCR product was cloned and used to transform TOP10® Escherichia coli cells using the TOPO TA Cloning® Kit for Sequencing (Invitrogen), following the manufacturer’s instructions. Colonies producing a PCR product of the appropriate size were cultured overnight at 37°C in 5ml of LB medium containing 50µg/ml carbenicilin. Plasmid vectors were isolated from these bacterial cultures using the Qiaprep Spin Miniprep Kit (Qiagen), following the manufacturer’s recommendations.
Sequencing was performed by using the ABI Prism® BigDye™ Terminator Cycle Sequencing Reaction (Applied Biosystems), following the manufacturer’s guidelines. Output sequence files were edited using SeqMan software (DNASTAR).
2.8. Two-dimensional (2D) gel electrophoresis and protein identification by mass spectrometry (MS)
Approximately 50µg of sample proteins (200 pairs of larval SG) were used to perform 2D gel electrophoresis using ZOOM IPGRunner System (Invitrogen) under manufacturer’s recommended running conditions. Protein identification of 2D gel-separated proteins was performed on reduced and alkylated trypsin-digested samples prepared by standard mass spectrometry protocols. Tryptic digests were analyzed by coupling the Nanomate (Advion BioSciences) – an automated chip-based nano-electrospray interface source – to a quadrupole time-of-flight mass spectrometer, QStarXL MS/MS System (Applied Biosystems/Sciex). Computer-controlled, data-dependent automated switching to MS/MS provided peptide sequence information. AnalystQS software (Applied Biosystems/Sciex) was used for data acquisition. Data processing and databank searching were performed with Mascot software (Matrix Science). The NR protein database from the NCBI, National Library of Medicine, NIH, was used for the search analysis.
2.9. Microscopy and histochemistry
For images showing the gross morphology of the larval alimentary canal and SG, early 4th instar larvae were dissected in 70% ethanol and photographed using an Olympus SZX12 stereoscope (Olympus Life Sciences) connected to a Nikon DS-L1 digital camera (Nikon Corporation).
For histochemical staining, longitudinal paraffin sections of whole 4th instar larvae were prepared as previously described (Smith et al.,2008). Rehydrated sections were incubated for 1 hr at 37°C with a 1/500 dilution of either FITC-conjugated Erythrina cristagalli lectin or FITC-conjugated wheat-germ agglutinin (Vector laboratories) in pre-incubation buffer (TBS, 1% bovine serum albumin, 1% normal goat serum, 0.1% TritonX100). Sections were then washed three times in TBS, mounted in 60% glycerol in TBS, and analyzed using a Leica LSCM SP2 laser scanning confocal microscope (Leica Microsystems).
3. Results and discussion
The gene expression platform used in this work (GeneChip® Plasmodium/ Anopheles array, Affymetrix Inc.) assigns both a qualitative value (present/absent call) and a quantitative ‘expression level’ value to each one of its probes. By comparing the expression profile of the fourth-instar larval SG (fig. 1) to that of whole larvae, we identified a total of 4,719 probes that were scored as ‘present’ in the SG, 747 of which were unique to this tissue (i.e. were scored as either ‘absent’ or ‘undetermined’ in one or more repeats of the whole larva sample) (supplementary materials, table 2). Among these, 221 probes (associated with 292 genes, encoding 318 transcripts) were selected for further analysis because they were (a) significantly (P<0.01) enriched at least twofold in the salivary glands, and (b) sufficiently annotated as to allow the prediction of the presence or absence of a signal peptide in their associated proteins.
The mismatch between the number of probes (221), transcripts (318) and genes (292) we list as significantly enriched in the larval SG can be explained by the constant evolution of our knowledge of the An. gambiae genome: the design of the Affymetrix microarray chip used in our study was based on the genomic information available in 2003 (Affymetrix, 2004b), and attempted to provide a probe for each transcript known at the time. However, many novel genes and transcripts have been identified since the commercial release of this design, and these can often be associated with existing probes. As a result, more than one transcript (sometimes from different genes) can be associated with several of the chip’s probes.
The 221 probes selected for this study, as well as their corresponding transcripts and relevant functional annotation, are listed in the supplementary materials, table 2. Scatter and volcano plots of the transcriptomic comparison between larval SG and whole larvae are available in the supplementary materials, fig. 1.
In order to check for any potential contamination of our SG samples with tissues from other regions of the digestive tract, we compared the enrichment levels of the aforementioned 221 selected probes in the SG, gastric caeca, anterior midgut, posterior midgut and hindgut. For this purpose, we used whole-transcriptome expression profiles of each gut section previously generated by our laboratory (Neira Oviedo et al., 2008) (accession number GSE7149 in NCBI’s gene expression omnibus, available at http://www.ncbi.nlm.nih.gov/geo/). The clear enrichment that virtually all the selected probes showed in the SG (supplementary materials, figure 2) suggests that contamination of our tissue samples is rather unlikely.
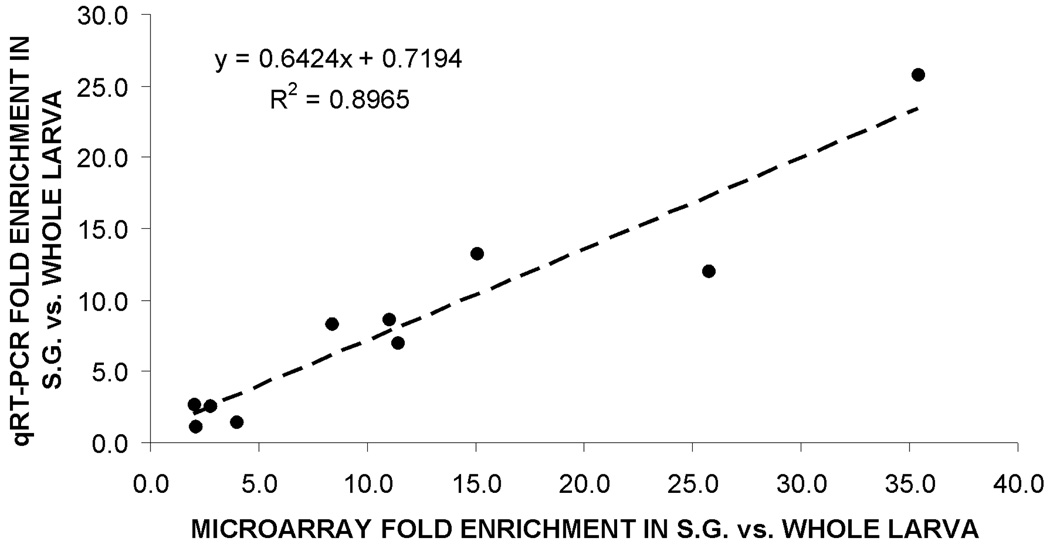
Ten transcripts found to be significantly enriched in the SG by our microarray analysis were randomly selected to cover the entire spectrum of transcript enrichment, and used to perform qRT-PCR. A high degree of correlation (R2=0.89) was observed between the expression values obtained by these two methods (fig. 2), validating the accuracy of our microarray-generated gene expression data.
Fig. 2.
Validation of microarray data. A linear regression model (equation and dashed line) revealed a high degree of correlation (R2>0.89) between results obtained by microarray analysis (x-axis) and qRT-PCR analysis (y-axis).
3.1. Functional annotation analysis of SG-enriched transcripts
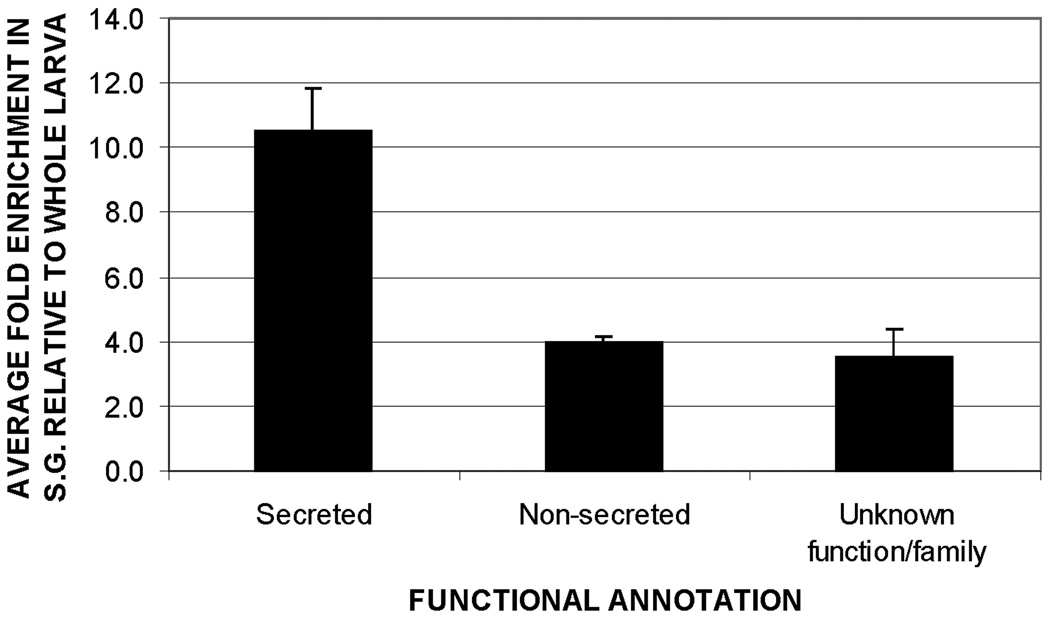
Based on available annotation for the known or predicted protein sequences encoded by the selected transcripts, 48 were identified as encoding putative components of the salivary secretions, 267 as corresponding to putative non-secreted proteins, and three as encoding proteins of unknown function. In accordance with the secretory nature of the SG, the average enrichment of probes associated with putative secreted proteins (11.1 fold vs. whole larva) was 2.8 times that of probes associated with putative non-secreted proteins (4 fold vs. whole larvae) (fig. 3).
Fig. 3.
Relative abundance of transcripts encoding putative secreted, non-secreted and unknown proteins in the larval SG of An. gambiae, as suggested by microarray analysis. Transcripts encoding secreted proteins were 2.8 times more abundant than those encoding non-secreted proteins. Bar height represents the mean enrichment of transcripts within each functional group. Error bars show the standard error of the mean.
3.2. Transcripts encoding putative secreted products
Probes associated with putative secreted gene products enriched in the SG were classified based on their available functional annotation. Among those with a known or predicted function, immune-related transcripts were found to be the most diverse and enriched (fig. 4 and table 1). To confirm that this enrichment of immune transcripts is a constitutive characteristic of the larval SG (and not the result of a microbial assault suffered by the particular cohort of insects used to obtain our SG samples) we measured, by qRT-PCR, the expression of five of these immune transcripts in the SG and whole body of a separate group of larvae reared specifically for this purpose. The enrichment in SG of all five transcripts observed in these independent samples (shown in supplementary fig. 3) supports the notion that immune-related gene products are constitutively enriched in the larval salivary secretions.
Fig. 4.
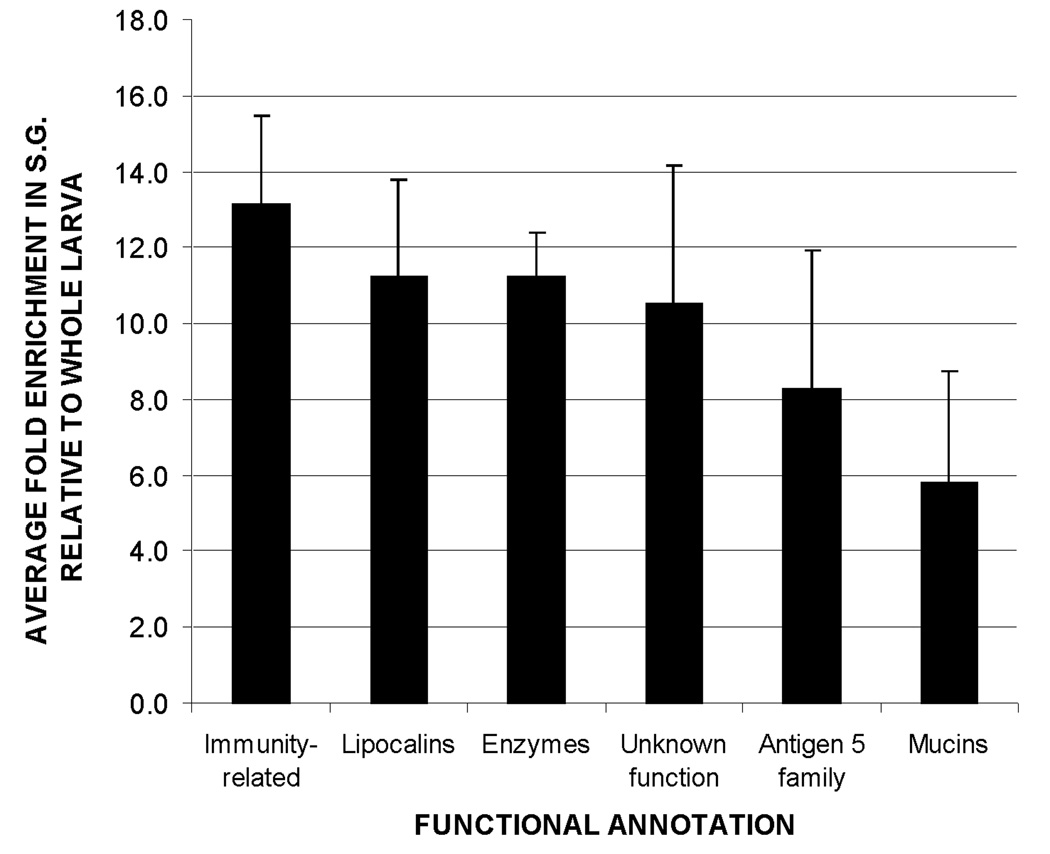
Relative abundance of transcripts associated with putative secreted proteins, grouped by functional annotation. Transcripts associated with immune functions were the most abundant, followed by lipocalins and digestive/detoxifying enzymes. Bar height represents the mean enrichment of transcripts within each functional group. Error bars show the standard error of the mean.
Table 1.
Diversity of secreted gene products found to be enriched in the larval SG, grouped by functional annotation.
| Functional category | No. of probes | No. of transcripts |
|---|---|---|
| Immune-related gene products | ||
| Antimicrobial factors | ||
| Defensins | 2 | 3 |
| Lysozyme | 1 | 1 |
| TIL-domain containing proteins | 4 | 4 |
| Pattern-recognition factors | ||
| Fibrinogen-related proteins | 1 | 3 |
| Immune-related serine proteases | 4 | 5 |
| Total- immune related gene products | 12 | 16 |
| Mucins | 3 | 3 |
| Lipocalins | 2 | 2 |
| AG5 Family | 3 | 3 |
| Digestive and detoxifying enzymes | ||
| Lipid digestion | 2 | 2 |
| Carbohydrate digestion | 4 | 4 |
| Protein digestion | 1 | 1 |
| Nucleotide digestion | 1 | 1 |
| Xenobiotic detoxification | 1 | 3 |
| Total- digestive and detoxifying enzymes | 9 | 11 |
| Gene products of unknown function | ||
| Conserved family | 3 | 3 |
| Other | 9 | 10 |
| Total – gene products of unknown function | 12 | 13 |
Other significantly enriched functional groups include digestive and detoxifying enzymes, members of the antigen 5 (AG5) family of genes, mucins and lipocalins (fig. 4 and table 1). Probes for 13 additional transcripts encoding putatively secreted proteins without any associated functional annotation were also found to be significantly enriched in the SG.
3.2.1. Immunity-related transcripts
3.2.1.1. Anti-microbial factors
Our analysis revealed enrichment in the SG of three functional groups associated with anti-microbial functions: (a) defensins, (b) lysozymes, and (c) proteins containing trypsin inhibitor-like (TIL) domains. Each one of these groups will be discussed separately. Additionally, salivary proteins assigned to other functional groups (such as the mucins and chitinases, discussed in sections 3.2.2 and 3.2.5, respectively) have the potential to significantly contribute to the anti-microbial effect of the saliva.
Defensins are small anti-microbial peptides (usually 36–46 residues long), present in a wide diversity of plant, vertebrate and invertebrate species. They are mainly active against Gram-positive bacteria, but also exhibit activity against fungi, yeast and protozoa (Bulet et al.,1999; Zhou et al.,2007). Four defensin genes (AgDef1 to 4) have been described to date in the An. gambiae genome (Christophides et al.,2002).
Our analysis identified the expression in larval SG of defensins AgDef2 and AgDef3, which have been previously reported to be present in the adult male and female SG, respectively (Arca et al.,2005; Calvo et al.,2006a). Additionally, we identified the expression of a previously undescribed defensin (henceforth referred to as AgDef5) which is encoded by the ENSEMBL gene AGAP007200, transcript AGAP007200-RA. We successfully cloned and sequenced the transcript encoding this novel defensin. The sequence of this transcript has been deposited in GenBank, with accession number EU273600.
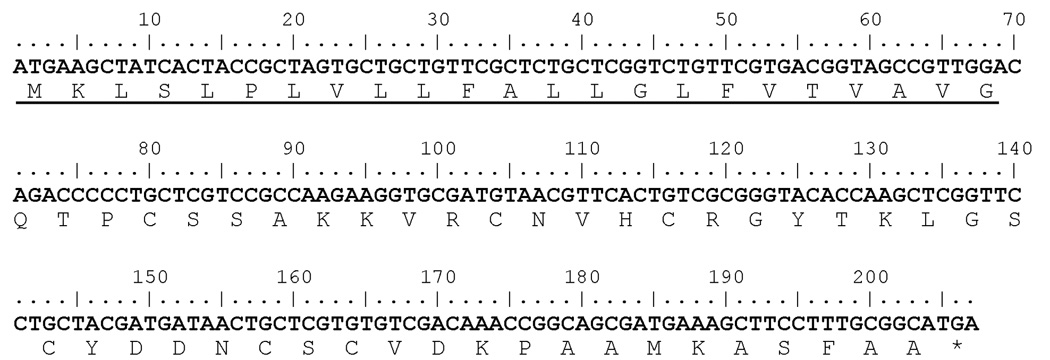
The AgDef5 gene contains a single exon and is located on the reverse strand of chromosome 2L (starting at position 44,248,847), immediately adjacent to the AgDef3 gene. cDNA corresponding to the AgDef5 transcript presents a 207 bp open reading frame that translates into a 68 amino-acid peptide (fig. 5). The first 23 residues of this peptide encode a signal sequence, indicating that it is a secreted protein. The mature 44 amino acid-long peptide contains the six conserved cystein residues characteristic of all insect defensins (fig. 6a; Bulet et al.,1999). A phylogenetic analysis (fig. 6b) places AgDef5 closest to AgDef3, AgDef4 and AgDef2, which have been previously classified as a highly divergent group of defensins (Christophides et al.,2002). The exact role of this new defensin in the larval mosquito’s immune system remains to be determined.
Fig. 5.
Nucleotide and deduced amino acid sequence of AgDef5. The first 23 amino acids (underlined) form a signal sequence, suggesting a secreted protein. The asterisk indicates the position of the stop codon.
Lysozymes are traditionally associated with antibacterial activity, which they exert either by catalytic activity (hydrolyzing glycosidic bonds in the peptidoglycan layer of the bacterial cell walls), or by non-catalytic mechanisms such as membrane perturbation and the stimulation of autolysin activity (During et al.,1999; Ibrahim et al.,2001). Additionally, it has been recently shown that the immune role of lysozyme in mosquitoes extends beyond its antibacterial activity, playing a role in the regulation of melanotic encapsulation of foreign bodies (Li and Paskewitz, 2006).
Eight different lysozyme-encoding genes (Lys c-1 to Lys c-8) have been reported in the genome of An. gambiae (Li et al.,2005). Our analysis revealed one lysozyme, encoded by transcript AGAP007386-RA, to be significantly enriched in the larval SG. This transcript is encoded by the gene Lys c-7, and it has been reported to be expressed in all developmental stages, and in virtually all tissues of the adult female, with the exception of the ovaries (Li et al.,2005). Interestingly, the peptide encoded by this transcript contains the residues necessary to bind carbohydrates, but lacks two amino acids thought to be essential for catalytic function (Li et al.,2005). Therefore, this lysozyme is likely to exert its antibacterial activity via the aforementioned non-enzymatic functions.
Further support for the immune role of Lys c-7 comes from a study recently published by our laboratory, in which we generated whole-genome expression data for each compartment of the larval digestive tract (Neira Oviedo et al.,2008). Analysis of this data set (available at http://www.ncbi.nlm.nih.gov/geo/, series number GSE7149), reveals abundant expression of Lys c-7 in two regions of the digestive tract: the gastric caeca and hindgut / Malpighian tubules; both of these areas have been identified as being heavily involved in immune responses in larval Diptera (Neira Oviedo et al.,2008, McGettigan et al.,2005).
Four transcripts corresponding to proteins containing a trypsin inhibitor-like (TIL) domain were detected in the larval SG (transcripts AGAP006813-RA, AGAP002636-RA, AGAP002445-RA, and AGAP002450-RA). Proteins containing TIL domains fall within the larger category of serine-protease inhibitors, which have been associated with several functions, including the regulation of blood and hemolymph coagulation, activation of phenoloxidase and cytokines, inactivation of microbial proteases, regulation of endogenous proteases and inhibition of bacterial growth (Friedrich, 1993; Stubbs, 1997; Kanost, 1999; Fogaca, 2006). Several reports exist regarding the presence of serine protease inhibitors in the salivary glands of vertebrate and invertebrate organisms, including humans (Hochstrasser et al.,1993; Kolho et al.,2005), adult mosquitoes (Calvo et al.,2006a; Ribeiro et al.,2007; Arca et al.,2007) and hematophagous hemipterans (Santos et al.,2007; Assumpcao et al.,2008). Serine-protease inhibitors present in the salivary secretions of adult female mosquitoes have been shown to facilitate blood feeding by interfering with the host’s inflammatory response and coagulation cascade (Stark and James, 1995; Lanfranciotti et al.,2002; Ribeiro et al.,2007). However, the presence of these transcripts in the non-hematophagous larvae suggests they play a role as part of the immune system, rather than acting as anticoagulant factors. A similar function has been proposed for serine-protease inhibitors found in adult male SG of An. gambiae (Calvo et al.,2006a).
3.2.1.2. Pattern recognition factors
Our results revealed significant enrichment in the SG of one probe (Ag.UNKN.836.0_CDS_s_at) associated with three transcripts (AGAP004998-RA, AGAP004999-RA, AGAP012651-RA) that encode proteins similar to the fibrinogen-related protein ficolin. Among these, transcripts AGAP004999-RA and AGAP012651-RA have been previously found to be expressed in the adult female SG of An. gambiae (Ribeiro, unpublished data).
Fibrinogen-related proteins (FREPs) are found in both vertebrates and invertebrates and are mainly associated with immune functions, although structural roles have also been reported (Wang et al.,2005). FREPs associated with immune reactions (such as ficolin) contain fibrinogen-like domains capable of recognizing and binding carbohydrates characteristic of the microbial surface (such as lipoteichoic acid, peptidoglycan and lipopolysaccharides). Once bound, they can act as opsonins, or trigger the complement cascade (Endo et al.,2007; Wang et al, 2004).
The presence of transcripts encoding ficolin-like products in the larval SG of mosquitoes is consistent with previous studies which have reported the expression of immune-related FREPs in different tissues of dipteran insects, including immature stages (Wang et al.,2005, 2004). Interestingly, it has been suggested that mosquitoes present a large expansion of FREP-encoding genes (when compared to other dipterans, such as Drosophila), probably reflecting the particularly diverse microbial challenges faced throughout their life cycle (Loker et al.,2004). Ribeiro et al. (2007) presented evidence of ficolins in the salivary secretions of adult female mosquitoes, and suggested they play a role related to blood-feeding. In the case of the larval salivary FREPs, however, the absence of blood-feeding implies that their role is likely to be either immune or structural.
3.2.1.3. Immune-related serine proteases
Several transcripts annotated as encoding immune-related serine proteases were found to be enriched in the larval SG. Transcripts AGAP011908-RA, AGAP011909-RA, AGAP011913-RA, AGAP011914-RA encode CUB -domain serine proteases, while transcript AGAP003057-RA encodes the clip-domain serine protease CLIPB8. With the exception of AGAP011909-RA, all these transcripts have been reported from the adult female SG of An. gambiae (Ribeiro, unpublished data).
The CUB domain consists of ~110 residues and is found almost exclusively in extracellular proteins. It was first discovered in the complement subcomponents C1s/C1r, and it has been reported to be present in several developmentally-regulated proteins (Bork and Beckmann, 1993), immune-related salivary proteins (Ligtenberg et al.,2007) and proteins involved in receptor-mediated endocytosis (Chrisrensen and Gburek, 2004), among others. Although the specific roles of this domain are not well understood, it has been suggested that it is involved in substrate recognition and oligomerization (Blanc et al.,2007). It is widely accepted that CUB domains present in complement-associated serine proteases mediate the interaction of these enzymes with non-self recognition proteins (such as ficolins), triggering the complement’s characteristic cascade of enzymatic activations. A role in substrate-recognition has been suggested for enzymes of this kind present in the saliva of adult female mosquitoes (Ribeiro et al.,2007).
Clip-domain serine proteases are major regulators of developmental and immune-related signaling processes such as the Toll signaling pathway, and the activation of phenoloxidase (one of the principal components of the melanization immune response) (Jang et al.,2008; Barillas-Mury, 2007). The ‘clip’ domain receives its name from its unusual ‘paper clip’ configuration, resulting from unique disulfide bonds (Muta et al.,1990). The particular clip-domain serine protease found in our study (CLIPB8) has been reported to promote melanization in the G3 strain of Anopheles gambiae (Barillas-Mury, 2007; Paskevitz et al.,2006). Melanization-related enzymes have been reported to exist in the salivary secretions of larval insects, although their functional roles are unknown (Satoh et al.,1999).
3.2.2. Mucins
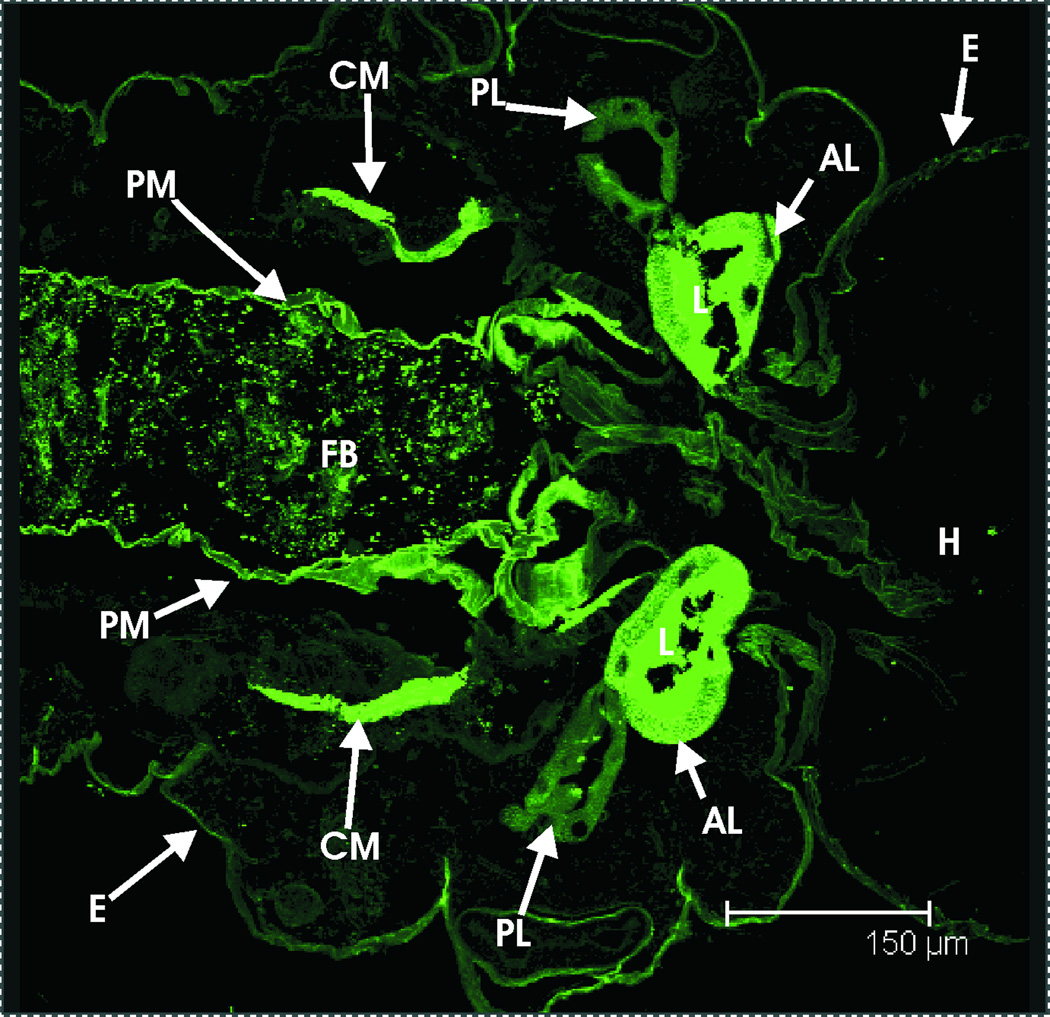
Mucins are extensively glycosilated proteins previously reported to be abundant in the salivary secretions of a wide diversity of animals, including adult male and female mosquitoes (Calvo et al.,2006a; Arca et al.,2005). Three mucin-encoding transcripts (AGAP006067-RA, AGAP010727-RA and AGAP008177-RA) were identified in our analysis as being significantly enriched in the larval SG. Interestingly, one of these (transcript AGAP010727-RA) has also been reported to exist in the adult salivary glands (Ribeiro, unpublished data), suggesting that it plays a role in aiding the digestive process throughout the mosquito’s life cycle. Further support for the presence of mucins in the larval salivary secretions comes from the staining of the SG lumen contents with fluorescently-labeled wheat-germ agglutinin (fig. 7), a plant lectin that selectively binds sialic acid and N-acetylglucosamine, and can be used to identify mucin-type glycoproteins (Furukawa et al.,1986).
Fig. 7.
Histochemical detection of mucins in the larval SG. Confocal micrograph showing a section of the thorax of a 4th instar An. gambiae larva stained with fluorescently-labeled wheat-germ agglutinin. Clear staining can be seen in the anterior lobe of both salivary glands (lumen), as well as in the food bolus, caecal membranes and perithtophic membrane. AL, anterior lobe of salivary gland. CM, caecal membrane. E, exosqueleton. FB, food bolus. H, head. L, lumen of salivary gland’s anterior lobe. PL, posterior lobe of salivary gland. PM, peritrophic matrix.
Traditionally, mucins have been exclusively associated with the formation of coats that protect and lubricate the epithelia where they are produced. However, recent studies have suggested that these proteins are also involved in a variety of complex physiological processes such as development, epithelial renewal, carcinogenesis and immunity (Corfield et al.,2000; Korayem et al.,2004; Moal et al.,2006; Wei and Bobek, 2005; Habte et al.,2006). In vertebrates, salivary mucins have shown potent antimicrobial activity, being able to inhibit viral replication and fungal growth (Wei and Bobek, 2005; Habte et al.,2006; Ogasawara et al.,2007). In Drosophila, Korayem et al. (2004) reported mucins to be co-expressed in larval SG and immune tissues, and suggested that they help form clots that immobilize potentially pathogenic microorganisms, preventing their dissemination and rendering them more susceptible to the insect’s immune system.
Although previous studies have reported the presence of mucus in the lumen of the foregut (including the pharynx) and midgut of larval mosquitoes, they have failed to identify the glands responsible for its secretion (Fry, 1996; Dahl et al.,1990). Dahl et al. (1990) stated that the SG “would be an obvious structure” for the secretion of this mucus, but were unable to provide histochemical evidence in support of this hypothesis. On the other hand, our results provide both transcriptomic and histochemical evidence supporting the secretion of mucin-type proteins by the larval SG. We believe this contradiction can be attributed to the technical limitations of the Alcian Blue / periodic acid Schiff’s reagent staining methods used by Dahl et al.,which (as the authors of that study acknowledge) are very sensitive to pH conditions, and can be difficult to interpret.
In light of our results, it seems plausible for the larval SG to be the site of origin of at least some of the mucus reportedly present in the digestive tract of mosquito larvae. The roles of this mucus are likely to include, in addition to food bolus lubrication, the immobilization and neutralization of ingested microorganisms.
3.2.3. Lipocalins
Two transcripts (AGAP004799-RA and AGAP009281-RA) encoding proteins annotated as lipocalins were identified as significantly enriched in the larval SG; one of these (transcript AGAP004799-RA) has also been reported to be particularly abundant in the adult male antennae of An. gambiae, where its function is unknown (Justice et al.,2003).
Lipocalins are a large group of small, generally extracellular proteins characterized by their ability to bind small hydrophobic molecules. Recent studies have revealed diverse roles for lipocalins, including transport, metabolism, immune regulation, smell perception, tissue development and behaviour modification (Akerstrom et al.,2000). Lipocalins have been reported to be the most abundant proteins in the salivary secretions of blood-feeding hemipterans and ticks, where they act as anticoagulants, anti-complement, and vasodilators (Assumpcao et al.,2008; Santos et al.,2007; Mans et al, 2008); however, no evidence of their existence in the saliva of mosquitoes has been presented to date (Assumpcao et al.,2008), making this the first report of salivary lipocalins in the Culicidae. Although their exact function in the saliva of the non-blood feeding mosquito larvae is yet to be determined, their potential role in sensory perception (Akerstrom et al.,2000) and their ability to diffuse in aqueous environments makes it tempting to speculate that salivary lipocalins can act as pheromones, providing chemical cues to other individuals about factors such as larval density. Alternatively, they might serve to capture lipidic nutrients from the diet.
3.2.4. AG5 family
Three transcripts annotated as encoding proteins belonging to the AG5 family were found to be significantly enriched in the larval SG (transcripts AGAP007584-RA, AGAP006443-RA and AGAP006419-RA). All of these transcripts have also been reported to be expressed in the SG of adult An. gambiae (Arca et al.,2005; Calvo et al.,2006a; Ribeiro, unpublished data) (supplementary table 2), suggesting that their functions are not related to a particular feeding strategy.
The AG5 family of proteins is related to venom allergens found in social hymenopterans (Hoffman, 1993) and to antifungal proteins found in plants (Szyperski et al.,1998). Although members of the AG5 family have been reported to exist in the salivary secretions of several hematophagous insect species (Arca et al.,2005; Ribeiro et al, 2007; Santos et al.,2007), their exact function remains unknown. Megraw et al. (1998) reported the presence of AG5 proteins in the digestive tract of Drosophila larvae, where the authors suggested they function as protease inhibitors, either regulating the activity of digestive proteases, or exerting an antimicrobial effect.
3.2.5. Digestive and detoxifying enzymes
The list of digestive and detoxifying enzymes found to be enriched in the larval SG is presented in the supplementary materials, table 2; it includes enzymes involved in the metabolism of all major groups of nutrients (lipids, proteins and carbohydrates), as well as one apyrase-like nucleotidase (encoded by transcript AGAP011026-RA). Although the presence of digestive enzymes in the larval saliva is not surprising, the presence of an apyrase is rather puzzling: apyrases are a type of 5’ nucleotidases traditionally associated with the saliva of adult female mosquitoes, where they facilitate blood feeding by inhibiting platelet aggregation in the host (Ribeiro, 1987). In the non-blood feeding larva, the role of such an enzyme is unclear, and is probably related to the terminal digestion of nucleic acids ingested as part of the insect’s normal diet (i.e. algal and microbial DNA). Interestingly, Lombardo et al. (2000) reported a complex developmental pattern of expression for transcript AGAP011026-RA (named AgApyL1 by the authors of that study), being found in larvae, pupae, and both male and female adults. In agreement with our results, these findings support the notion that this enzyme plays a role unrelated to blood feeding.
Three transcripts annotated as putatively secreted chitinases (AGAP005339-RA, AGAP005634-RA and AGAP000789-RA) were found to be significantly enriched in the larval SG. Of these, transcript AGAP005339-RA has been previously detected in the SG of adult female An. gambiae (Ribeiro, unpublished data). Additionally, a chitinase-like protein has been previously reported from the saliva of adult Anopheles mosquitoes (Owhashi et al.,2008); unfortunately, neither its exact molecular identity nor its biological roles have been conclusively defined.
In vertebrates, salivary chitinases have been associated with protection against pathogenic fungi (Van Steijn et al.,1999). Shi and Paskewitz (2004) demonstrated that bacterial challenges triggered the production of chitinase-like proteins in the haemolymph of adult An. gambiae, suggesting a putative immune-related role. Although none of the chitinases identified in our study show a significant degree of identity with those reported by Shi and Paskewitz (table 3 and suplementary materials, fig. 4), the secretion of salivary chitinases as a line of defense against environmental microbes seems logical when we consider that chitin-containing structures (such as fungi) are likely to constitute a large fraction of the larval mosquito’s regular diet (Clements, 1992). Further studies are required in order to confirm the enzymatic activity of the salivary chitinases identified by our study.
Table 3.
Percent amino acid identity between putative salivary chitinases found in our study (AGAP000789-PA, AGAP005339-PA, AGAP005634-PA) and putatively immune chitinases previously reported from An. gambiae (AgBR1 and AgBR2).
| AgBR1 | AgBR2 | AGAP000789-PA | AGAP005339-PA | AGAP005634-PA | |
|---|---|---|---|---|---|
| AgBR1 | - | 61.9% | 5.8% | 20.5% | 17.9% |
| AgBR2 | - | 6% | 20.9% | 21.6% | |
| AGAP000789-PA | - | 5.1% | 4.4% | ||
| AGAP005339-PA | - | 25.6% | |||
| AGAP005634-PA | - |
One probe (Ag.2L.1123.0_CDS_a_at) associated with three transcripts encoding flavin-containing monooxygenases (FMOs) was found to be significantly enriched in the larval SG. These enzymes have an important role in the metabolism of toxic compounds, often acting in parallel with (and on the same substrates as) cytochromes P450, another major group of detoxifying enzymes that are highly abundant in the digestive tract of An. gambiae larvae (Neira Oviedo et al.,2008, Testa and Kramer, 2007; Strode et al.,2006). The general metabolic function of FMOs consists in transforming toxic lipophilic substrates into more hydrophilic products, which are then easily transported and excreted with the urine (Testa and Kramer, 2007). The presence of this kind of enzymes in the saliva of larval An. gambiae might be related to the abundance of potentially toxic compounds (such as bacterial, algal and fungal toxins, as well as phenolic products of plant degradation) in their natural diet (Strode et al.,2006).
3.3. Transcripts encoding putative non-secreted and unknown proteins
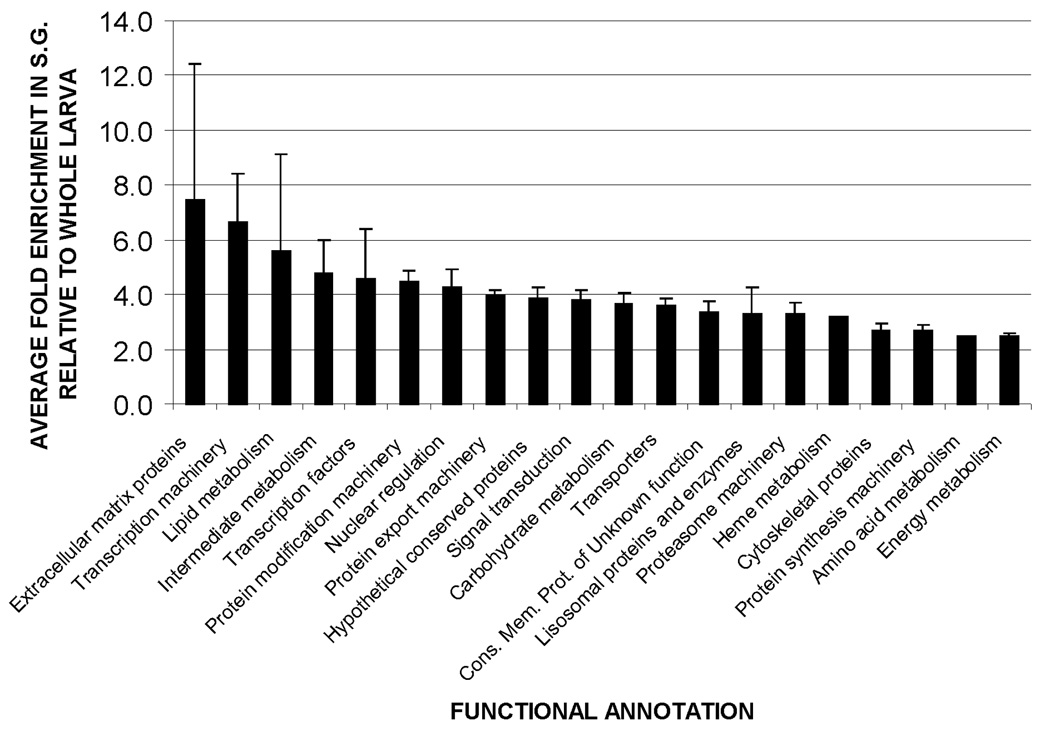
The 186 probes (corresponding to 267 transcripts) associated with putative non-secreted proteins found to be significantly enriched in the larval SG are listed in the supplementary materials, table 2. Based on available functional annotation, these transcripts can be catalogued into 20 functional categories (table 2). Transcripts annotated as encoding hypothetical conserved proteins of unknown function are the most diverse, followed by transcripts associated with protein export machinery, and protein modification machinery. As shown in fig. 8, probes associated with extracellular matrix proteins show the highest average relative abundance (7.4 fold increase vs. whole larvae), followed by those associated with transcription machinery (6.6 fold increase vs. whole larvae) and lipid metabolism (5.6 fold increase vs. whole larvae).
Table 2.
Diversity of housekeeping gene products found to be enriched in the larval SG, grouped by functional annotation.
| Functional category | No. of probes | No. of transcripts |
|---|---|---|
| Hypothetical conserved proteins of unknown function | 36 | 43 |
| Protein export machinery | 35 | 37 |
| Protein modification machinery | 31 | 36 |
| Signal transduction | 28 | 35 |
| Transporters | 19 | 20 |
| Conserved membrane proteins of unknown function | 17 | 20 |
| Carbohydrate metabolism | 12 | 14 |
| Proteasome machinery | 10 | 10 |
| Protein synthesis machinery | 9 | 9 |
| Transcription machinery | 5 | 5 |
| Lipid metabolism | 5 | 6 |
| Cytoskeletal proteins | 5 | 8 |
| Transcription factors | 4 | 5 |
| Nuclear regulation | 4 | 4 |
| Intermediate metabolism | 3 | 3 |
| Lisosomal proteins and enzymes | 3 | 5 |
| Energy metabolism | 3 | 3 |
| Extracellular matrix proteins | 2 | 2 |
| Heme metabolism | 1 | 1 |
| Amino acid metabolism | 1 | 2 |
Fig. 8.
Relative abundance of transcripts associated with putative non-secreted proteins, grouped by functional annotation. Bar height represents the mean enrichment of transcripts within each functional group. Error bars show the standard error of the mean.
Additionally, three transcripts encoding proteins not associated with any known function (AGAP007064-RA, AGAP003812-RA and AGAP008178-RA) were found to be significantly enriched in the SG. Interestingly, the protein encoded by transcript AGAP003812-RA has partial homology to a protein predicted to be a transposase in NCBI’s KOG database (accession NP_566626). Transposable elements have been detected in the salivary transcriptome of adult mosquitoes, where they have been proposed to either be markers of active transposition, or to act as regulators of transposable elements recently included in the genome (Ribeiro et al.,2007).
3.4. Protein identification by 2D gel electrophoresis / MS
Nine proteins could be identified by 2D gel electrophoresis followed by MS of the most intense bands (fig. 9). One of these proteins corresponds to a putatively secreted trypsin (protein AGAP001246-PA), with the remaining nine corresponding to putative non-secreted products (i.e. products lacking a signal peptide). This virtual absence of secreted products detectable by 2D gel electrophoresis / MS in larval SG contrasts with the wider diversity of secreted proteins detected by similar methods in the salivary glands of adult female mosquitoes (Ribeiro et al.,2007). This discrepancy is most likely due to differences in the salivation strategies used by the different developmental stages: larval mosquitoes are constantly feeding and therefore require a permanent flow of saliva, which precludes the accumulation of secreted proteins in the gland’s lumen at any point. In contrast, adult mosquitoes only discharge saliva while feeding (an event that normally occurs once every several hours or even days), allowing the glands to accumulate relatively large amounts of salivary secretions between discharges.
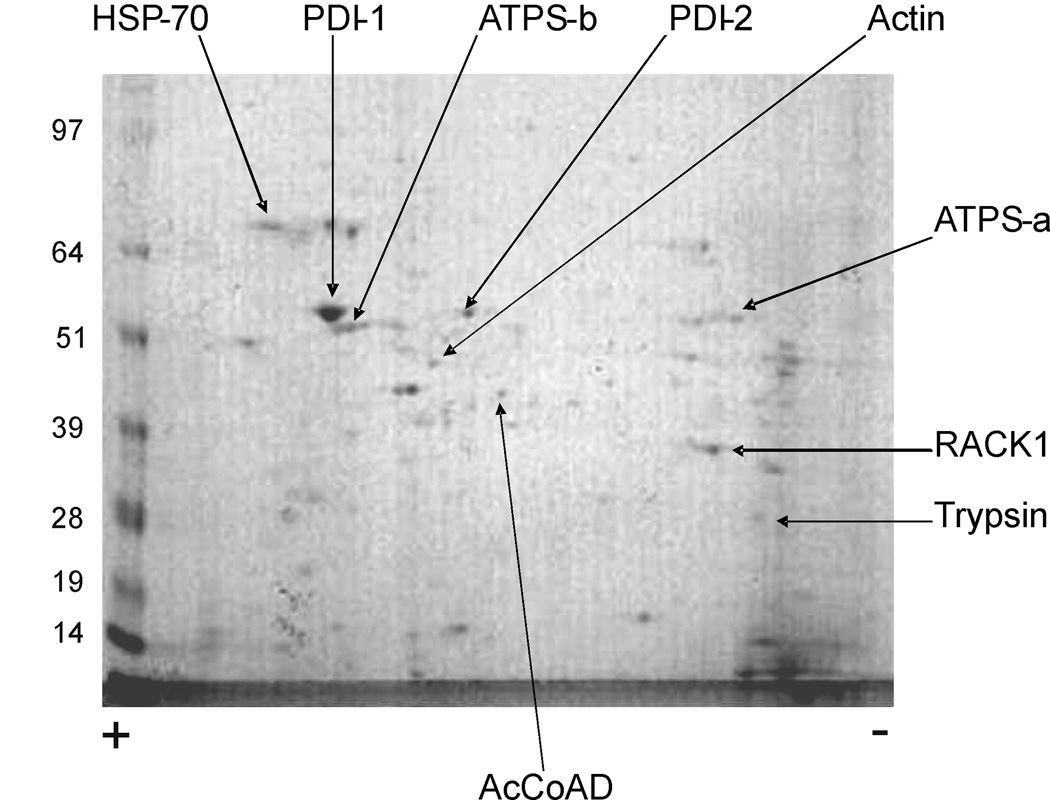
Fig. 9.
Two-dimensional gel electrophoresis of larval SG. Bands identified to a protein by triptic digestion / MS are shown. Numbers on the left correspond to molecular weight marker bands shown in the gel. The positive (+) and negative (−) signs below the gel mark the polarity of the isoelectrofocusing dimension. AcCoAD, acyl-coenzyme A dehydrogenase. ATPS-a, ATP synthase alpha subunit. ATPS-b, ATP synthase beta subunit. HSP-70, heat shock protein 70. PDI-1, protein disulfide isomerase 1. PDI-2, protein disulfide isomerase 2. RACK1, receptor for activated protein kinase c. ENSEMBL protein identifiers associated with each band are listed in the supplementary materials, table 3.
Transcripts corresponding to three putative non-secreted proteins detected by MS (protein disulfide isomerases 1 and 2, and heat shock protein 70), were identified in our analysis as significantly enriched in the salivary glands (transcripts AGAP012407-RA, AGAP007393-RA and AGAP004192-RA, respectively). Heat shock protein 70 (Hsp70) and at least one protein disulfide isomerase have also been reported in 2D gel studies in adult female Aedes aegypti mosquitoes (Ribeiro et al.,2007), suggesting a role common to larval and adult stages.
The sequence of Hsp70 lacks a signal peptide and is therefore considered to be intracellular in our study; however, recent studies have reported its ability to be secreted via alternative mechanisms involving lipid rafts and/or exosomes (Lancaster and Febbraio, 2005) and have provided evidence linking salivary Hsp70 to diverse immune functions including complement activation, antigen-display, and the prevention of bacterial adhesion to mucosal surfaces (See Fabian et al.,2007 for an extensive review). Considering the abundance of this protein revealed by our 2D gel analysis, it might represent an important component of the mosquito’s salivary immune arsenal.
Protein AGAP001246-PA (the aforementioned secreted trypsin) is unfortunately not associated with any probe in the commercial microarray design used in this experiment, and therefore its transcript could not be detected. Other putative non-secreted proteins detectable by MS whose transcripts were not found to be enriched in the SG (actin, acyl-coenzyme A dehydrogenase, ATP synthase sub-units A and B, and the receptor for activated protein kinase c) are at least equally abundant in other tissues of the whole larvae as they are in the SG, which explains why our experimental design did not detected them as significantly enriched in the SG tissue.
3.5. Comparison to adult sialomes
In clear contrast with the diet of adult mosquitoes (which consists of blood and/or sugary plant secretions), the diet of mosquito larvae consists mainly of microorganisms and various organic detritus collected from their aquatic environment (Clements, 1992). This radical difference in food sources is reflected in the different physiological roles played by the salivary secretions of the two life stages: while the saliva of adult female mosquitoes is used as a potent chemical cocktail aimed at countering the hemostatic and immune responses of their hosts (Ribeiro, 1987), the saliva of larval mosquitoes is unlikely to possess any particular anti-hemostatic activity, and is instead likely to be rich in elements that facilitate the ingestion and digestion of microbes.
In accordance with this, a comparative analysis of the larval salivary transcripts found in our study with that of adult An. gambiae available from previous studies (Arca et al.,2005; Calvo et al.,2006a, Ribeiro, unpublished data) (Supplementary materials, table 2) reveals some interesting differences and similarities. Notably, proteins of the D7 family (which are among the most abundant in the saliva of adult female mosquitoes, and are believed to act as anti-hemostatic factors that facilitate blood feeding) (Valenzuela et al.,2002; Arca et al.,2005; Calvo et al.,2006 a and b) were not detected in the larval SG. Similarly, proteins of the SG1 family were not detected in the larval saliva. SG1-type proteins are abundant in the saliva of adult female anopheline mosquitoes, where they play a yet undetermined role (Arca et al.,2005). The absence of this type of proteins in the non blood-feeding adult males (Calvo et al.,2006a) and larvae (this study) is strongly suggestive of a role related to the blood-feeding process.
On the other hand, our study suggests that the saliva of An. gambiae larvae is particularly rich in proteins with antimicrobial activity, such as chitinases, defensins and lysozyme, as well as other components that can help immobilize microorganisms (mucins), make them recognizable by the immune system (FREPs), or detoxify their potentially dangerous byproducts (FMOs). Additionally, other proteins for which an immune function is suspected (such as Hsp70 and members of the AG5 family) could increase the antimicrobial potency of the larval mosquito saliva. As previously mentioned, it is not surprising to find constitutive expression of antimicrobial factors in the saliva of an organism that feeds mostly on microorganisms. Larval mosquito saliva is discharged directly in the mouth (Clements, 1992), allowing for its components to start interacting with (and possibly neutralizing) any microbes immediately upon ingestion, and before they reach the metabolically-active midgut tissue. The fact that some of these antimicrobial factors are also present in the salivary secretions of adult mosquitoes (Supplementary materials, table 2) highlights the need to control microbes inevitably ingested with the regular diet of all developmental stages.
Our data supporting the presence of lipocalins in the larval saliva of An. gambiae is quite interesting, since proteins of this kind are reportedly absent in the salivary transcriptome of adult mosquitoes (Assumpcao et al.,2008), but are highly abundant the saliva of other blood-sucking arthropods (Montfort et al.,2000). It remains to be established why evolutionary forces have restricted their expression to the larval stages in An. gambiae, and what the physiological role of these proteins is.
Finally, we found an extensive set of transcripts encoding putative non-secreted proteins that are present in both the larval and adult SG (supplementary materials, table 2), suggesting that both developmental stages use, at least to some extent, similar molecular mechanisms for the production and secretion of salivary proteins.
4. Conclusions
Our study represents, to the best of our knowledge, the first report of the larval salivary transcriptome of any species in the Culicidae. By identifying the most abundant transcripts and proteins in the larval SG of An. gambiae, and analyzing their available functional annotation, we are able to provide an insight into the functional roles of the salivary secretions of mosquito larvae. Based on our results, we propose that these roles include: (a) immune response, (b) food bolus lubrication, (c) nutrient metabolism, and (d) xenobiotic detoxification.
In agreement with the feeding habits of the An. gambiae larvae, a clear abundance and diversity of immune-related gene products was observed, highlighting the crucial role of saliva as the first line of defense against the ingestion of potentially pathogenic microorganisms. In addition, some of the most abundant proteins in the salivary transcriptome of adult female An. gambiae (such as those of the D7 and SG1 families) were not detected by our study, supporting the notion that these proteins’ functions are mainly related to the blood feeding process.
Finally, the abundance of salivary transcripts that have no associated functional annotation (or for which a clear functional role in the saliva cannot be discerned) is a strong reminder of the large gaps that still exist in our understanding of the basic biology of one of the most important vectors of human disease.
Supplementary Material
Scatter and volcano plots of the transcriptomic comparison between larval salivary glands and whole larvae in An. gambiae.
Heat map showing enrichment of selected probes across all major compartments of the digestive tract.
qRT-PCR evaluation of immune-related transcript enrichment in salivary glands.
Comparison of the amino-acid sequences of putative salivary and putative immune chitinases of An. gambiae.
Sequence of PCR and qRT-PCR primers used in the study.
Probes present and significantly enriched in the larval salivary glands, and their associated functional annotation.
ENSEMBL identifiers associated with proteins detected by 2D gel electrophoresis / MS.
Acknowledgements
The authors would like to thank Dr. Mick Popp, Dr. Marina Thelonis-Scott and Dr. Gigi Ostrow from the University of Florida ICBR microarray core for their invaluable help in microarray processing. Dr. George Dimopoulos and Ms. Lindsey Garver for providing the sequence of the ribosomal protein S7 primers. We are also grateful to the NIAID Core facility led by Dr. Robert Hohman for their support, and to Dr. Robert MacCallum for sharing his expertise in the use of the VectorBase data repository. This project was supported by the National Institutes of Health grant R01-AI045098 to P.J.L. This work was also supported in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract NO1 CO 12400, and by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the government of the United States of America.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Affymetrix, Inc. GeneChip expression analysis - data analysis fundamentals. 2004a https://www.affymetrix.com/support/downloads/manuals/data_analysis_fundamentals_manual.pdf.
- Affymetrix, Inc. GeneChip Plasmodium/Anopheles genome array. 2004b http://www.affymetrix.com/support/technical/datasheets/plasmodium_datasheet.pdf.
- Agarwal AK. Digestive enzymes of sugarcane pink borer, Sesamia inferens Walker (Lepidoptera) J. Res. Lepidoptera. 1976;15:153–162. [Google Scholar]
- Akerstrom B, Flower DR, Salier JP. Lipocalins: unity in diversity. Biochimica Et Biophysica Acta-Protein Structure and Molecular Enzymology. 2000;1482:1–8. doi: 10.1016/s0167-4838(00)00137-0. [DOI] [PubMed] [Google Scholar]
- Andersen JF, Hinnebusch BJ, Lucas DA, Conrads TP, Veenstra TD, Pham VM, Ribeiro JMC. An insight into the sialome of the oriental rat flea, Xenopsylla cheopis (Rots) BMC Genomics. 2007;8:102. doi: 10.1186/1471-2164-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applied Biosystems. Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR. 2004 http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_042380.pdf.
- Arca B, Lombardo F, Capurro MD, della Torre A, Dimopoulos G, James AA, Coluzzi M. Trapping cDNAs encoding secreted proteins from the salivary glands of the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1516–1521. doi: 10.1073/pnas.96.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Francischetti IMB, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JMC. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem. Mol. Biol. 2007;37:107–127. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Valenzuela JG, Francischetti IMB, Marinotti O, Coluzzi M, Ribeiro JAC. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J. Exp. Biol. 2005;208:3971–3986. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assumpcao TCF, Francischetti IMB, Andersen JF, Schwarz A, Santana JM, Ribeiro JMC. An insight into the sialome of the blood-sucking bug Triatoma infestans, a vector of Chagas' disease. Insect Biochem. Mol. Biol. 2008;38:213–232. doi: 10.1016/j.ibmb.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Boeckmann B. The Swiss-Prot Protein-Sequence Data-Bank. Nucleic Acids Res. 1992;20:2019–2022. doi: 10.1093/nar/20.suppl.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillas-Mury C. CLIP proteases and Plasmodium melanization in Anopheles gambiae. Trends in Parasitology. 2007;23:297–299. doi: 10.1016/j.pt.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer ELL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Blanc G, Font B, Eichenberger D, Moreau C, Ricard-Blum S, Hulmes DJS, Moali C. Insights into how CUB domains can exert specific functions while sharing a common fold - Conserved and specific features of the CUB1 domain contribute to the molecular basis of procollagen C-proteinase enhancer-1 activity. J. Biol. Chem. 2007;282:16924–16933. doi: 10.1074/jbc.M701610200. [DOI] [PubMed] [Google Scholar]
- Bork P, Beckmann G. The Cub Domain - a widespread module in developmentally-regulated proteins. J. Mol. Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- Bulet P, Hetru C, Dimarcq JL, Hoffmann D. Antimicrobial peptides in insects: structure and function. Dev. Comp. Immunol. 1999;23:329–344. doi: 10.1016/s0145-305x(99)00015-4. [DOI] [PubMed] [Google Scholar]
- Calvo E, Dao A, Pham VM, Ribeiro JMC. An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families. Insect Biochem. Mol. Biol. 2007;37:164–175. doi: 10.1016/j.ibmb.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Pham VM, Lombardo F, Arca B, Ribeiro JMC. The sialotranscriptome of adult male Anopheles gambiae mosquitoes. Insect Biochem. Mol. Biol. 2006a;36:570–575. doi: 10.1016/j.ibmb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Andersen JF, Ribeiro JMC. Function and evolution of a mosquito salivary protein family. J. Biol. Chem. 2006b;281:1935–1942. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Candido-Silva JA, Zanarotti GM, Giallina AP, de Almeida JC. Developmental regulation of BhSGAMP-1, a gene encoding an antimicrobial peptide in the salivary glands of Bradysia hygida (Diptera, Sciaridae) Genesis. 2007;45:630–638. doi: 10.1002/dvg.20337. [DOI] [PubMed] [Google Scholar]
- Christensen EI, Gburek J. Protein reabsorption in renal proximal tubule-function and dysfunction in kidney pathophysiology. Pediatr. Nephrol. 2004;19:714–721. doi: 10.1007/s00467-004-1494-0. [DOI] [PubMed] [Google Scholar]
- Christophers SR. Aedes aegypti (L.) The yellow fever mosquito - its life history, bionomics and structure. London: Cambridge university press; 1960. [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa N, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Muller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu JN, Zheng LB, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Clements AN. Biology of Mosquitoes: Development, Nutrition and Reproduction. London: Chapman & Hall; 1992. [Google Scholar]
- Corfield AP, Myerscough N, Longman R, Sylvester P, Arul S, Pignatelli M. Mucins and mucosal protection in the gastrointestinal tract: new prospects for mucins in the pathology of gastrointestinal disease. Gut. 2000;47:589–594. doi: 10.1136/gut.47.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings DLD, Gamboa GJ, Harding BJ. Lateral vibrations by social wasps signal larvae to withhold salivary secretions (Polistes fuscatus, Hymenoptera : Vespidae) Journal of Insect Behavior. 1999;12:465–473. [Google Scholar]
- Dahl C, Craig DA, Merritt RW. Sites of possible mucus-producing glands in the feeding system of mosquito larvae (Diptera, Culicidae) Ann. Entomol. Soc. Am. 1990;83:827–833. [Google Scholar]
- During K, Porsch P, Mahn A, Brinkmann O, Gieffers W. The non-enzymatic microbicidal activity of lysozymes. FEBS Lett. 1999;449:93–100. doi: 10.1016/s0014-5793(99)00405-6. [DOI] [PubMed] [Google Scholar]
- Endo Y, Matsushita M, Fujita T. Role of ficolin in innate immunity and its molecular basis. Immunobiology. 2007;212:371–379. doi: 10.1016/j.imbio.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Fabian TK, Fejerdy P, Nguyen MT, Soti C, Csermely P. Potential immunological functions of salivary Hsp70 in mucosal and periodontal defense mechanisms. Arch. Immunol. Ther. Exp. (Warsz) 2007;55:91–98. doi: 10.1007/s00005-007-0012-z. [DOI] [PubMed] [Google Scholar]
- Fogaca AC, Almeida IC, Eberlin MN, Tanaka AS, Bulet P, Daffre S. Ixodidin, a novel antimicrobial peptide from the hemocytes of the cattle tick Boophilus microplus with inhibitory activity against serine proteinases. Peptides. 2006;27:667–674. doi: 10.1016/j.peptides.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Friedrich T, Kroger B, Bialojan S, Lemaire HG, Hoffken HW, Reuschenbach P, Otte M, Dodt J. A Kazal-type inhibitor with thrombin specificity from Rhodnius prolixus. J. Biol. Chem. 1993;268:16216–16222. [PubMed] [Google Scholar]
- Fry KM. Endogenous glycoconjugates are not associated with filter feeding in mosquito larvae (Diptera: Culicidae) Can. J. Zool.-Rev. Can. Zool. 1996;74:413–422. [Google Scholar]
- Furukawa K, Minor JE, Hegarty JD, Bhavanandan VP. Interaction of sialoglycoproteins with wheat-germ agglutinin-sepharose of varying ratio of lectin to sepharose - use for the purification of mucin glycoproteins from membrane extracts. J. Biol. Chem. 1986;261:7755–7761. [PubMed] [Google Scholar]
- Gelman DB, Demilo AB, Thyagaraja BS, Kelly TJ, Masler EP, Bell RA, Borkovec AB. 3-oxoecdysteroid 3-beta-reductase in various organs of the European corn-borer, Ostrinia nubilalis (Hubner) Arch. Insect Biochem. Physiol. 1991;17:93–106. [Google Scholar]
- Habte HH, Mall AS, de Beer C, Lotz ZE, Kahn D. The role of crude human saliva and purified salivary MUC5B and MUC7 mucins in the inhibition of Human Immunodeficiency Virus type 1 in an inhibition assay. Virology Journal. 2006;3 doi: 10.1186/1743-422X-3-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hochstrasser K, Wachter E, Reisinger PWM, Greim M, Albrecht GJ, Gebhard W. Amino-acid-sequences of mammalian Kazal-type proteinase-inhibitors from salivary-glands. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology. 1993;106:103–108. doi: 10.1016/0305-0491(93)90014-v. [DOI] [PubMed] [Google Scholar]
- Hoffman DR. Allergens in Hymenoptera Venom .25. The amino-acid-sequences of antigen 5 molecules and the structural basis of antigenic cross-reactivity. J. Allergy Clin. Immunol. 1993;92:707–716. doi: 10.1016/0091-6749(93)90014-7. [DOI] [PubMed] [Google Scholar]
- Hunt JH. Nourishment and the evolution of the social Vespidae. In: Ross KG, Matthews RW, editors. The social biology of wasps. Ithaca, NY: Cornell University Press; 1991. pp. 426–450. [Google Scholar]
- Ibrahim HR, Matsuzaki T, Aoki T. Genetic evidence that antibacterial activity of lysozyme is independent of its catalytic function. FEBS Lett. 2001;507:27–32. doi: 10.1016/s0014-5793(01)02872-1. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jang IH, Nam HJ, Lee WJ. CLIP-domain serine proteases in Drosophila innate immunity. Bmb Reports. 2008;41:102–107. doi: 10.5483/bmbrep.2008.41.2.102. [DOI] [PubMed] [Google Scholar]
- Jensen DV, Jones JC. The development of the salivary glands in Anopheles albimanus Wiedeman (Diptera, Culicidae) Ann. Ent. Soc. America. 1957;50:464–469. [Google Scholar]
- Justice RW, Dimitratos S, Walter MF, Woods DF, Biessmann H. Sexual dimorphic expression of putative antennal carrier protein genes in the malaria vector Anopheles gambiae. Insect Mol. Biol. 2003;12:581–594. doi: 10.1046/j.1365-2583.2003.00443.x. [DOI] [PubMed] [Google Scholar]
- Kanost MR. Serine proteinase inhibitors in arthropod immunity. Dev. Comp. Immunol. 1999;23:291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- Kolho KL, Saarinen R, Paju A, Stenman J, Stenman UH, Pitkaranta A. New insights into juvenile parotitis. Acta Paediatr. 2005;94:1566–1570. doi: 10.1080/08035250505100399. [DOI] [PubMed] [Google Scholar]
- Korayem AM, Fabbri M, Takahashi K, Scherfer C, Lindgren M, Schmidt O, Ueda R, Dushay MS, Theopold U. A Drosophila salivary gland mucin is also expressed in immune tissues: evidence for a function in coagulation and the entrapment of bacteria. Insect Biochem. Mol. Biol. 2004;34:1297–1304. doi: 10.1016/j.ibmb.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lancaster GI, Febbraio MA. Mechanisms of stress-induced cellular Hsp72 release: implications for exercise-induced increases in extracellular Hsp72. Exerc. Immunol. Rev. 2005;11:46–52. [PubMed] [Google Scholar]
- Lanfrancotti A, Lombardo F, Santolamazza F, Veneri M, Castrignano T, Coluzzi M, Arca B. Novel cDNAs encoding salivary proteins from the malaria vector Anopheles gambiae. FEBS Lett. 2002;517:567–571. doi: 10.1016/s0014-5793(02)02578-4. [DOI] [PubMed] [Google Scholar]
- Langner KFA, Darpel KE, Denison E, Drolet BS, Leibold W, Mellor PS, Mertens PPC, Nimtz M, Greiser-Wilke I. Collection and analysis of salivary proteins from the biting midge Culicoides nubeculosus (Diptera : Ceratopogonidae) J. Med. Entomol. 2007;44:238–248. doi: 10.1603/0022-2585(2007)44[238:caaosp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Calvo E, Marinotti O, James AA, Paskewitz SM. Characterization of the c-type lysozyme gene family in Anopheles gambiae. Gene. 2005;360:131–139. doi: 10.1016/j.gene.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Li B, Paskewitz SM. A role for lysozyme in melanization of Sephadex beads in Anopheles gambiae. J. Insect Physiol. 2006;52:936–942. doi: 10.1016/j.jinsphys.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Ligtenberg AJM, Veerman ECI, Nieuw Amerongen AV, Mollenhauer J. Salivary agglutinin/gilycoprotein-340/DMBT1: a single molecule with variable composition and with different functions in infection, inflammation and cancer. Biol. Chem. 2007;388:1275–1289. doi: 10.1515/BC.2007.158. [DOI] [PubMed] [Google Scholar]
- Liu F, Cui LW, Cox-Foster D, Felton GW. Characterization of a salivary lysozyme in larval Helicoverpa zea. Journal of Chemical Ecology. 2004;30:2439–2457. doi: 10.1007/s10886-004-7944-0. [DOI] [PubMed] [Google Scholar]
- Loker ES, Adema CM, Zhang SM, Kepler TB. Invertebrate immune systems - not homogeneous, not simple, not well understood. Immunol. Rev. 2004;198:10–24. doi: 10.1111/j.0105-2896.2004.0117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo F, Di Cristina M, Spanos L, Louis C, Coluzzi M, Arca B. Promoter sequences of the putative Anopheles gambiae apyrase confer salivary gland expression in Drosophila melanogaster. J. Biol. Chem. 2000;275:23861–23868. doi: 10.1074/jbc.M909547199. [DOI] [PubMed] [Google Scholar]
- Marinotti O, Calvo E, Nguyen QK, Dissanayake S, Ribeiro JMC, James AA. Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol. Biol. 2006;15:1–12. doi: 10.1111/j.1365-2583.2006.00610.x. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Andersen JF, Francischetti IM, Valenzuela JG, Schwan TG, Pham VM, Garfield MK, Hammer CH, Ribeiro JMC. Comparative sialomics between hard and soft ticks: Implications for the evolution of blood-feeding behavior. Insect Mol. Biol. 2008;38:42–58. doi: 10.1016/j.ibmb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinotti O, Nguyen QK, Calvo E, James AA, Ribeiro JMC. Microarray analysis of genes showing variable expression following a blood meal in Anopheles gambiae. Insect Mol. Biol. 2005;14:365–373. doi: 10.1111/j.1365-2583.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- McGettigan J, McLennan RKJ, Broderick KE, Kean L, Allan AK, Cabrero P, Regulski MR, Pollock VP, Gould GW, Davies SA, Dow JAT. Insect renal tubules constitute a cell-autonomous immune system that protects the organism against bacterial infection. Insect Biochem. Mol. Biol. 2005;35:741–754. doi: 10.1016/j.ibmb.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Megraw T, Kaufman TC, Kovalick GE. Sequence and expression of Drosophila Antigen 5-related 2, a new member of the CAP gene family. Gene. 1998;222:297–304. doi: 10.1016/s0378-1119(98)00489-2. [DOI] [PubMed] [Google Scholar]
- Moal VLL, Servin AL. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin. Microbiol. Rev. 2006;19:315–337. doi: 10.1128/CMR.19.2.315-337.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montfort WR, Weichsel A, Andersen JF. Nitrophorins and related antihemostatic lipocalins from Rhodnius prolixus and other blood-sucking arthropods. Biochim. Biophys. Acta. 2000;1482:110–118. doi: 10.1016/s0167-4838(00)00165-5. [DOI] [PubMed] [Google Scholar]
- Muta T, Hashimoto R, Miyata T, Nishimura H, Toh Y, Iwanaga S. Poclotting enzyme from horseshoe-crab hemocytes - cDNA cloning, disulfide locations, and subcellular-localization. J. Biol. Chem. 1990;265:22426–22433. [PubMed] [Google Scholar]
- Neira Oviedo MN, VanEkeris L, Corena-Mcleod MDP, Linser PJ. A microarray-based analysis of transcriptional compartmentalization in the alimentary canal of Anopheles gambiae (Diptera : Culicidae) larvae. Insect Mol. Biol. 2008;17:61–72. doi: 10.1111/j.1365-2583.2008.00779.x. [DOI] [PubMed] [Google Scholar]
- Owhashi M, Harada M, Suguri S, Omae H, Ishii A. Identification of an eosinophil chemotactic factor from anopheline mosquitoes as a chitinase family protein. Parasitol. Res. 2008;102:357–363. doi: 10.1007/s00436-007-0769-3. [DOI] [PubMed] [Google Scholar]
- Paskewitz SM, Andreev O, Shi L. Gene silencing of serine proteases affects melanization of Sephadex beads in Anopheles gambiae. Insect Biochem. Mol. Biol. 2006;36:701–711. doi: 10.1016/j.ibmb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC. Role of saliva in blood-feeding by arthropods. Annu. Rev. Entomol. 1987;32:463–478. doi: 10.1146/annurev.en.32.010187.002335. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Arca B, Lombardo F, Calvo E, Phan VM, Chandra PK, Wikel K. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC, Topalis P, Louis C. AnoXcel: an Anopheles gambiae protein database. Insect Mol. Biol. 2004;13:449–457. doi: 10.1111/j.0962-1075.2004.00503.x. [DOI] [PubMed] [Google Scholar]
- Santos A, Ribeiro JMC, Lehane MJ, Gontijo NF, Veloso AB, Santanna MRV, Araujo RN, Grisard EC, Pereira MH. The sialotranscriptome of the blood-sucking bug Triatoma brasiliensis (Hemiptera, Triatominae) Insect Biochem. Mol. Biol. 2007;37:702–712. doi: 10.1016/j.ibmb.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh D, Horii A, Ochiai M, Ashida M. Prophenoloxidase-activating enzyme of the silkworm, Bombyx mori - Purification, characterization, and cDNA cloning. J. Biol. Chem. 1999;274:7441–7453. doi: 10.1074/jbc.274.11.7441. [DOI] [PubMed] [Google Scholar]
- Shi L, Paskewitz SM. Identification and molecular characterization of two immune-responsive chitinase-like proteins from Anopheles gambiae. Insect Mol. Biol. 2004;13:387–398. doi: 10.1111/j.0962-1075.2004.00496.x. [DOI] [PubMed] [Google Scholar]
- Smith KE, VanEkeris LA, Okech BA, Harvey WR, Linser PJ. Larval anopheline mosquito recta exhibit a dramatic change in localization patterns of ion transport proteins in response to shifting salinity: a comparison between anopheline and culicine larvae. J. Exp. Biol. 2008;211:3067–3076. doi: 10.1242/jeb.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KR, James AA. A factor Xa-directed anticoagulant from the salivary glands of the yellow fever mosquito Aedes aegypti. Exp. Parasitol. 1995;81:321–331. doi: 10.1006/expr.1995.1123. [DOI] [PubMed] [Google Scholar]
- Strode C, Steen K, Ortelli F, Ranson H. Differential expression of the detoxification genes in the different life stages of the malaria vector Anopheles gambiae. Insect Mol. Biol. 2006;15:523–530. doi: 10.1111/j.1365-2583.2006.00667.x. [DOI] [PubMed] [Google Scholar]
- Stubbs MT, Morenweiser R, Sturzebecher J, Bauer M, Bode W, Huber R, Piechottka GP, Matschiner G, Sommerhoff CP, Fritz H, Auerswald EA. The three-dimensional structure of recombinant leech-derived tryptase inhibitor in complex with trypsin - Implications for the structure of human mast cell tryptase and its inhibition. J. Biol. Chem. 1997;272:19931–19937. doi: 10.1074/jbc.272.32.19931. [DOI] [PubMed] [Google Scholar]
- Szyperski T, Fernandez C, Mumenthaler C, Wuthrich K. Structure comparison of human glioma pathogenesis-related protein GliPR and the plant pathogenesis-related protein P14a indicates a functional link between the human immune system and a plant defense system. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2262–2266. doi: 10.1073/pnas.95.5.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa B, Kramer SD. The biochemistry of drug metabolism -An introduction -Part 2. Redox reactions and their enzymes. Chemistry & Biodiversity. 2007;4:257–405. doi: 10.1002/cbdv.200790032. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W - Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turillazzi S, Perito B, Pazzagli L, Pantera B, Gorfer S, Tancredi M. Antibacterial activity of larval saliva of the European paper wasp Polistes dominulus (Hymenoptera, Vespidae) Insectes Sociaux. 2004;51:339–341. [Google Scholar]
- Valenzuela JG, Charlab R, Gonzalez EC, de Miranda-Santos IKF, Marinotti O, Francischetti IMB, Ribeiro JMC. The D7 family of salivary proteins in blood sucking Diptera. Insect Mol. Biol. 2002;11:149–155. doi: 10.1046/j.1365-2583.2002.00319.x. [DOI] [PubMed] [Google Scholar]
- Van Den Abbeele, J., Calion G, Dierick JF, Moens L, De Ridder K, Coosemans M. The Glossina morsitans tsetse fly saliva: General characteristics and identification of novel salivary proteins. Insect Biochem. Mol. Biol. 2007;37:1075–1085. doi: 10.1016/j.ibmb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Van Steijn GJ, Amerongen AVN, Veerman ECI, Kasanmoentalib S, Overdijk B. Chitinase in whole and glandular human salivas and in whole saliva of patients with periodontal inflammation. Eur. J. Oral Sci. 1999;107:328–337. doi: 10.1046/j.0909-8836.1999.eos107503.x. [DOI] [PubMed] [Google Scholar]
- Verma PS, Balyan BS. Studies on the hydrogen-ion concentration and digestive enzymes in the mature larva of Platyedra gossypiella Saund. (Lepidoptera: Gelechidae) Indian J. Ent. 1972;34:136–141. [Google Scholar]
- Verma PS, Singh B, Agarwal VK. Physiology of digestion in the mature larva of Sylepta derogate Fabr. Indian J. Ent. 1977;39:132–138. [Google Scholar]
- Wang X, Rocheleau TA, Fuchs JF, Hillyer JF, Chen CC, Christensen BM. A novel lectin with a fibrinogen-like domain and its potential involvement in the innate immune response of Armigeres subalbatus against bacteria. Insect Mol. Biol. 2004;13:273–282. doi: 10.1111/j.0962-1075.2004.00484.x. [DOI] [PubMed] [Google Scholar]
- Wang XG, Zhao Q, Christensen BM. Identification and characterization of the fibrinogen-like domain of fibrinogen-related proteins in the mosquito, Anopheles gambiae, and the fruitfly, Drosophila melanogaster, genomes. BMC Genomics. 2005;6:114. doi: 10.1186/1471-2164-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr E, Aguilar R, Dong YM, Mahairaki V, Dimopoulos G. Spatial and sex-specific dissection of the Anopheles gambiae midgut transcriptome. BMC Genomics. 2007;8:37. doi: 10.1186/1471-2164-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei GX, Bobek LA. Human salivary mucin MUC7 12-mer-L and 12-mer-D peptides: Antifungal activity in saliva, enhancement of activity with protease inhibitor cocktail or EDTA, and cytotoxicity to human cells. Antimicrob. Agents Chemother. 2005;49:2336–2342. doi: 10.1128/AAC.49.6.2336-2342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YP, Retnakaran A, Krell PJ, Arif BM, Primavera M, Feng QL. Temporal, spatial and induced expression of chitinase in the spruce budworm, Choristoneura fumiferana. J. Insect Physiol. 2003;49:241–247. doi: 10.1016/s0022-1910(02)00271-8. [DOI] [PubMed] [Google Scholar]
- Zhou J, Liao M, Ueda M, Gong H, Xuan X, Fujisaki K. Sequence characterization and expression patterns of two defensin-like antimicrobial peptides from the tick Haemaphysalis longicornis. Peptides. 2007;28:1304–1310. doi: 10.1016/j.peptides.2007.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scatter and volcano plots of the transcriptomic comparison between larval salivary glands and whole larvae in An. gambiae.
Heat map showing enrichment of selected probes across all major compartments of the digestive tract.
qRT-PCR evaluation of immune-related transcript enrichment in salivary glands.
Comparison of the amino-acid sequences of putative salivary and putative immune chitinases of An. gambiae.
Sequence of PCR and qRT-PCR primers used in the study.
Probes present and significantly enriched in the larval salivary glands, and their associated functional annotation.
ENSEMBL identifiers associated with proteins detected by 2D gel electrophoresis / MS.