Abstract
The precise spatio-temporal dynamics of protein activity are often critical in determining cell behaviour, yet for most proteins they remain poorly understood; it remains difficult to manipulate protein activity at precise times and places within living cells. Protein activity has been controlled by light, through protein derivatization with photocleavable moieties1 or using photoreactive small molecule ligands2. However, this requires use of toxic UV wavelengths, activation is irreversible, and/or cell loading is accomplished via disruption of the cell membrane (i.e. through microinjection). We have developed a new approach to produce genetically-encoded photo-activatable derivatives of Rac1, a key GTPase regulating actin cytoskeletal dynamics3,4. Rac1 mutants were fused to the photoreactive LOV (light oxygen voltage) domain from phototropin5,6, sterically blocking Rac1 interactions until irradiation unwound a helix linking LOV to Rac1. Photoactivatable Rac1 (PA-Rac1) could be reversibly and repeatedly activated using 458 or 473 nm light to generate precisely localized cell protrusions and ruffling. Localized Rac activation or inactivation was sufficient to produce cell motility and control the direction of cell movement. Myosin was involved in Rac control of directionality but not in Rac-induced protrusion, while PAK was required for Rac-induced protrusion. PA-Rac1 was used to elucidate Rac regulation of RhoA in cell motility. Rac and Rho coordinate cytoskeletal behaviours with seconds and submicron precision7,8. Their mutual regulation remains controversial9, with data indicating that Rac inhibits and/or activates Rho10,11. Rac was shown to inhibit RhoA in living cells, with inhibition modulated at protrusions and ruffles. A PA-Rac crystal structure and modelling revealed LOV-Rac interactions that will facilitate extension of this photoactivation approach to other proteins.
Recent NMR studies by Harper et al. revealed the mechanism of a protein light switch in Avena sativa Phototropin16,12: a flavin-binding LOV2 domain interacts with a C-terminal helical extension (Jα) in the dark. Photon absorption leads to formation of a covalent bond between Cys450 and the flavin chromophore, causing conformational changes that result in dissociation and unwinding of the Jα helix. We fused the complete LOV2-Jα sequence (404–547) to the N-terminus of a constitutively active Rac1, anticipating that the LOV domain in its closed conformation would block the binding of effectors to Rac1, and that light-induced unwinding of the Jα helix would release steric inhibition, leading to Rac1 activation (Fig. 1a). Sampling of different junctional sequences in pull down assays revealed that connecting Leu546 of LOV2-Jα to Ile4 of Rac1 led to substantial reduction in Rac1 binding to its effector PAK (Fig. 1b and Supplementary Fig. S1a). To ensure that the photoactivatable Rac1 would induce no dominant negative effects and that its activity would not be subject to upstream regulation, mutations were introduced to abolish GTP hydrolysis and diminish interactions with nucleotide exchange factors, guanine nucleotide dissociation inhibitors (Q61L) and GTPase activating proteins (E91H and N92H) (Supplementary: Fig. S2 and “Characterization of Rac1 constructs”). This resulted in the photoactivatable analogue of Rac1 (PA-Rac1) used in the following studies. Pull down assays showed that PA-Rac1 has greatly reduced affinity for its effector protein PAK in the dark, as does a PA-Rac1 construct containing a light-insensitive LOV2 mutation (C450A)13. Effector binding was restored in a PA-Rac1 construct containing a LOV2 mutant (I539E)14 that mimics the unfolded ‘lit state’ (Fig. 1b and Supplementary Fig. S1b). Isothermal titration experiments indicated that the dark and lit state mutants of PA-Rac1 differed 10-fold in effector binding (200 nM versus 2 µM) (Supplementary Fig. S3 and Table S1), with lit state effector affinity similar to that of native Rac15.
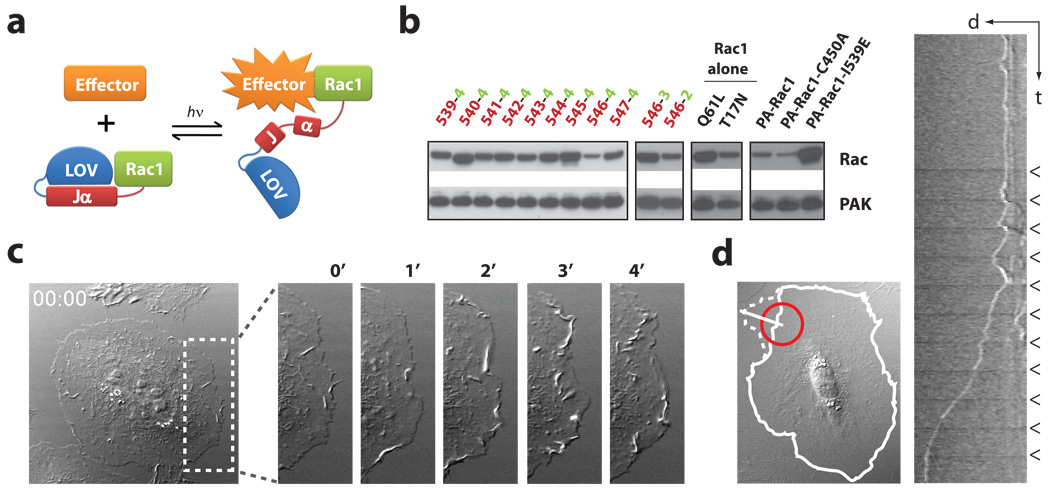
Figure 1. Engineering and in vivo characterization of a photoactivatable Rac1 (PA-Rac1).
a, Cartoon representation of PA-Rac1 design. b, Pulldown of PA-Rac1 constructs with PAK in the dark. Truncations of LOV and Rac at their linkage point were tested: 539–547, in red = terminal amino acid of Jα; 2–4, in green = first residue of Rac1. 546−4 showed the strongest inhibition; PA-Rac1 = 546−4, Q61L/E91H/N92H; -C450A, light-insensitive mutant; -I539E, lit state mutant. Pulldown by constitutively active (Q61L) and dominant negative (T17N) mutants are included for comparison with PA-Rac1. c, Whole cell irradiation of a HeLa cell expressing PA-Rac1. (minutes after irradiation, DIC, short axis of box = 20 µm). d, Spatial control of Rac1 activity. A 20-µm circle (red) was irradiated every 60 seconds in serum-starved MEF cells. Solid line = cell border at time 0, dotted line = 10 minutes after initial light pulse. Little movement of the cell border was detected, except adjacent to the point of irradiation. The kymograph (taken using white line, 20 µm), shows the initial formation of ruffles after each pulse, followed by protrusion (arrowheads = irradiation pulses).
Activation of PA-Rac1 was examined in HeLa cells expressing a YFP fusion of PA-Rac1 to gauge expression level. The cells remained quiescent when illuminated with wavelengths longer than flavin absorbance (515, 568 or 633 nm, data not shown), but within seconds after switching to 458 nm, lamellipodial protrusions and membrane ruffles appeared around the cell edges (Fig. 1c and Supplementary Movie S1). To show that this effect was due to PA-Rac1, kymograms were used to quantify maximum protrusion length; Irradiation of PA-Rac1 elicited protrusions that were four times as long as those seen in cells expressing either LOV domain alone or the light-insensitive PA-Rac1-C450A mutant (Supplementary Fig. S4). An important advantage of PA-Rac1 is its ability to precisely control the subcellular location of Rac activation. We first examined this in mouse embryo fibroblasts (MEF) stably expressing PA-Rac1, and cultured without serum to minimize cell activity prior to irradiation. Irradiation of 20 µm spots at the cell edge generated large protrusions clearly localized next to the point of irradiation (Fig. 1d and Supplementary Movie S2). Repeated irradiation led first to ruffles and then to protrusion. YFP-actin, YFP-PAK, and YFP-Arp3 revealed actin polymerization at the edge of the Rac-induced protrusions with associated translocation of downstream effectors, and induction of localized PAK phosphorylation was shown by immunostaining (Supplementary Fig S5,6 and Movies S3,4). Movement of a laser spot to different positions led to cessation of ruffling or protrusion at the initial irradiation position and new activities appearing where the laser spot was brought to rest (HeLa cells, Supplementary Movie S5), demonstrating reversible activation. In MEF cells, more prone to movement than HeLa, complex shape changes were produced by ‘painting’ the cell with the laser spot (Supplementary Movie S6). The area of protrusions in MEF cells was dependent on light dosage, indicating the valuable ability to control the level of Rac1 activation (Supplementary Fig. S7). PA-Rac1 diffusion was analyzed using FRAP (fluorescence recovery after photobleaching) and using PA-Rac1 tagged with photoactivatable GFP16 (Supplementary Fig. S8 and Movies S7,8), indicating that PA-Rac1 diffuses more slowly than cytosolic proteins, likely because it is membrane bound (10 µm spot, FRAP D=0.55 µm2/s or t1/2= 12.1s; PA-GFP t1/2=14.6 s). The half life of dark recovery for PA-Rac1 was determined to be 43s at room temperature. Simulation using this value indicated that, for two adjacent 10-µm spots, the unirradiated spot will achieve at most 7.5% the activation of the irradiated region (Supplementary Fig. S8c). Together these studies validate PA-Rac1 as a robust, genetically encoded and reversible caged protein effective in living cells.
We used PA-Rac1 to ask whether localized Rac activation is sufficient to specify cell polarity. In MEF cells, activating Rac1 at one spot near the cell edge not only generated protrusion locally, but also produced retraction on the opposite side of the cell (Fig. 2a and Supplementary Movie S9). To test whether this cross-cell coordination was due to a gradient of Rac1 activity, we fused the LOV domain to a dominant negative mutant of Rac1 using the same linkage as in PA-Rac1. Irradiation of this PA-Rac1-T17N led to nearby retraction rather than protrusion, and now generated protrusion in other areas of the cell (Fig 2a and Supplemental Movies S10,11). The ability of Rac1 alone to control polarized movement was confirmed by repeated irradiation at the cell edge, which could be used to produce prolonged cell movement by generating consistent coordinated extension and retraction (MEF cells: Fig. 2b and Supplemental Movie S12; HEK293 cells: Supplemental Movie S13). In contrast to MEF and HEK293 cells, HeLa cells showed localized protrusion but could not be induced to retract or move simply by activating Rac (Supplemental Movie S3), indicating that Rac-induced motility is subject to modulation by other pathways.
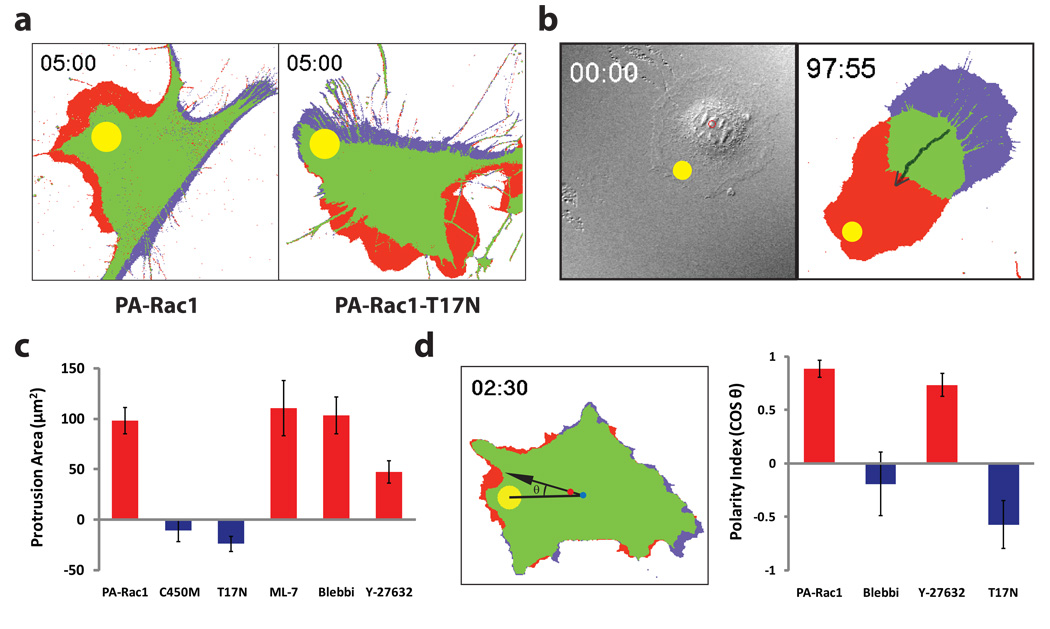
Figure 2. Localized activation or inactivation of PA-Rac1 induces myosin-dependent migration.
a, Protrusion/retraction map after a single pulse of activating illumination. MEFs expressing PA-Rac1 (left) generated protrusions at the site of irradiation (red) and retraction at the opposite side of the cell (blue) (in all 50 cells studied). Irradiation of the dominant negativeT17N mutant of PA-Rac1 (right) produced retraction near the point of irradiation, with protrusion in area(s) other than the site of irradiation (in all 25 cells studied). b, Repeated activation of PA-Rac1 at the cell edge induces directional migration. (MEF, 2-minute intervals, avg. 0.8 microns/pulse, n = 6). c, Localized activation of PA-Rac1 in the presence of ML-7 (MLCK inhibitor, 1 µM), Blebbistatin (Myosin II ATPase inhibitor, 1 µM), or Y-27632 (ROCK inhibitor, 10 µM). Protrusions analyzed as in panel a. d, Effect of myosin or ROCK inhibition on the ability of Rac1 to specify the direction of movement. The cosine of the angle between two lines (from the irradiation spot to the cell centroid at time 0, from the centroid at time 0 to the centroid at the end of the experiment) indicated how much the cell deviates from the direction specified by local irradiation. (panels c and d, n > 25; means +/− 95% confidence intervals; throughout figure 3 irradiation at 458 nm, Ø = 10 µm).
PA-Rac1 enabled control of Rac1 activity without the prior cellular compensation seen with other techniques, i.e. mutation or altered expression. Using this advantage, we examined the role of myosin, a key mediator of actin-based contractility, in Rac-induced motility. Global inhibition of myosin activity using the myosin ATPase inhibitor blebbistatin or the myosin light chain kinase inhibitor ML-7 strongly affected Rac’s ability to specify the direction of cell movement, but minimally affected Rac-induced protrusion (Fig. 2c,d). Myosin may mediate Rac’s control of directionality through induction of tail retraction17, contraction of the cell cortex to direct protrusive force18, or coupling of actin to adhesions differently at the front and rear19. In contrast, inhibition of PAK was found to strongly affect Rac-induced protrusion (Supplementary Fig. S9). Inhibition of the Rho-activated kinase ROCK using Y27632 suggested a role for ROCK in Rac-induced protrusion, but these results must be interpreted with caution due to known off-target effects20.
Where and how Rac regulates Rho in vivo remains largely unknown; this was examined by using PA-Rac1 together with a RhoA biosensor8. Localized activation of Rac1 led to immediate inhibition of RhoA, and this inhibition spread outward from the irradiated spot (Fig. 3a and Supplementary Movie S14). This was not simply an artifact of biosensor photobleaching, as irradiating the photo-inactive C450M mutant (Fig. 2c) of PA-Rac1 led to localized biosensor photobleaching and recovery, but no prolonged local inhibition or wave of inhibition (Fig. 3a). There were striking differences between constitutive MEF protrusions and protrusions induced by pulsed PA-Rac1 irradiation. In contrast to constitutive protrusions, RhoA activity was greatly reduced in protrusions induced by PA-Rac (Fig. 3b). Inhibition of RhoA appears to be compartmentalized or controlled kinetically when Rac is activated in the context of normal motility, as both active Rac and active Rho are seen at the leading edge7,8,21. PA-Rac activation led to large ruffles moving from the site of irradiation rearwards towards the nucleus (Supplementary Movie S15), suggesting that Rac regulates rearwards membrane flow. In control experiments, irradiation of cells expressing the photo-inactive C450M mutant did not produce polarized ruffling or show reduced RhoA activity (data not shown).
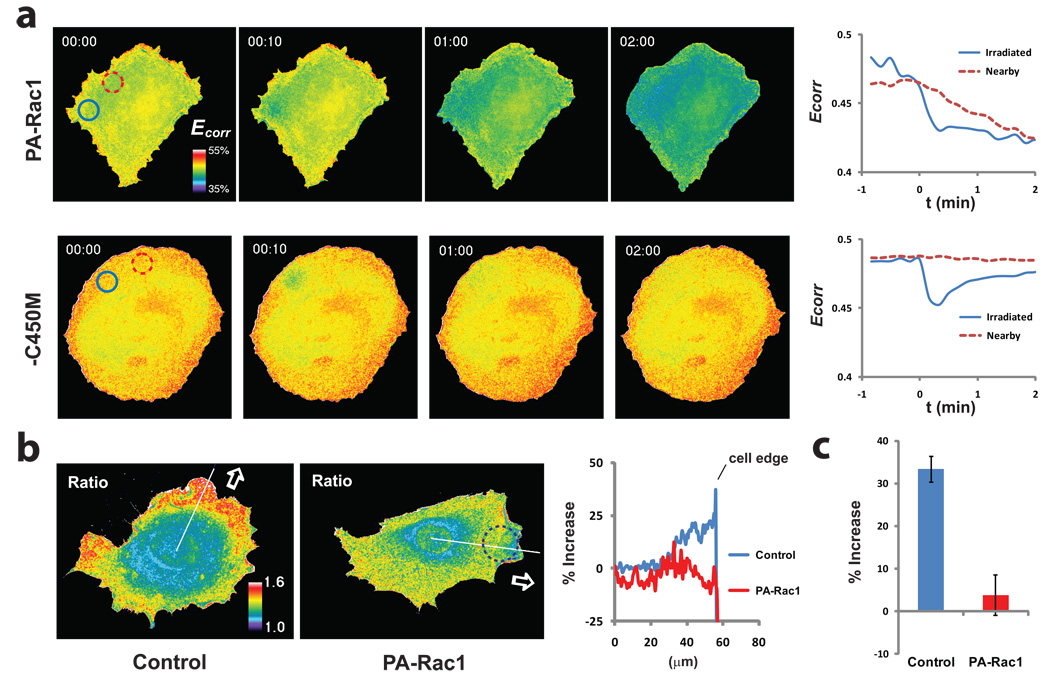
Figure 3. Inhibition of RhoA by PA-Rac1.
a, HeLa cells expressing RhoA biosensor and either PA-Rac1 or its C450M photo-inactive mutant, illuminated in a 10-µm circle with a single pulse of 473 nm light. Changes in the FRET efficiency (Ecorr) of the RhoA biosensor, indicative of RhoA activation, are shown in pseudocolor and as plots of average FRET efficiency within the irradiated circle (blue) and a nearby circle (red). In the PA-Rac1 cells, the irradiated spot showed bleaching of the biosensor followed by a relatively constant level of reduced RhoA activity. The nearby spot showed no bleaching, but a gradual decrease in RhoA activity reaching the low level achieved in the irradiated spot (n = 3 cells). In the control cells (C450M), the biosensor returned to near initial activation readouts after bleaching, and no change was seen in the nearby spot (n =3 cells). b, RhoA activation in constitutive pseudopods8,21 versus pseudopods induced by PA-Rac1 (473 nm, 20-µm circle shown, Supplemental Movie S15). The bar graph (c) shows the percent increase in biosensor FRET/CFP ratio in the region 1 micron from the cell edge versus the mean of the flat region at the left of the line scan. (means +/− 95% confidence intervals, 18 lines from 6 cells per bar).
To understand the structural basis of the PA-Rac1 switch for future application to other proteins, we performed Rosetta structure prediction simulations22 on several LOV2-Rac1 constructs, and determined high-resolution crystal structures of photo-active and inactive PA-Rac1 in the dark state. The crystal structures confirmed that the LOV domain occludes effector binding in the dark state (Fig. 4a and Supplementary Table S2). LOV-Jα adopted a closed conformation that superimposes with the recently published structure of isolated LOV-Jα23. In the conformational ensemble predicted by simulations of the dark state, the effector binding site of Rac was sterically blocked by the LOV domain in a majority of the low energy models (Supplementary Tables S3–5 and Figs. S10–13). Consistent with pull down assays (Fig. 1b and Supplementary Fig. S1a), adding or removing even one residue from the connection between LOV and Rac resulted in conformational ensembles with exposed effector binding sites. In the dark state, Rac was seen to form an extensive interface with the LOV domain (Fig. 4b), occluding Rac binding interactions. Given the substantial structural similarity between Rac1 and Cdc42, we hypothesized that the LOV domain could also be used to cage Cdc42. However, the linkage used for PA-Rac1 failed to reduce Cdc42 binding to PAK (Fig. 4c and Supplementary Fig. S1d). Using the PA-Rac1 crystal structure as a template, a model was built of the Cdc42-LOV dark state. At the interface between Rac and LOV a hydrophobic cluster is formed between residues Phe37 and Trp56 from Rac and Leu422, Pro423, Ile428, Tyr508 and Leu546 from LOV. Consistent with this being a weak, non-evolved interaction, most of the hydrogen bonding potential at the Rac-LOV interface is satisfied by buried and partially buried water molecules instead of inter-domain hydrogen bonds (Fig 4b). This interface model was used to identify a mutation to Cdc42, F56W at the Rac-LOV interface, that was predicted to stabilize the dark state. Pull down assays showed that this mutation substantially improves dark state inhibition of PAK binding, and produces differential affinity for Cdc42 effector in the dark versus the lit state (Fig. 4d). In living cells, irradiation of the mutated PA-Cdc42 led to production of filopodia and in some cases protrusions and/or ruffles, consistent with Cdc42 induction of filopodia and activation of Rac24 (Supplementary Fig. S14 and Supplementary Movie S16). These results argue that PA-Rac1 can serve as a blueprint for engineering other caged GTPases.
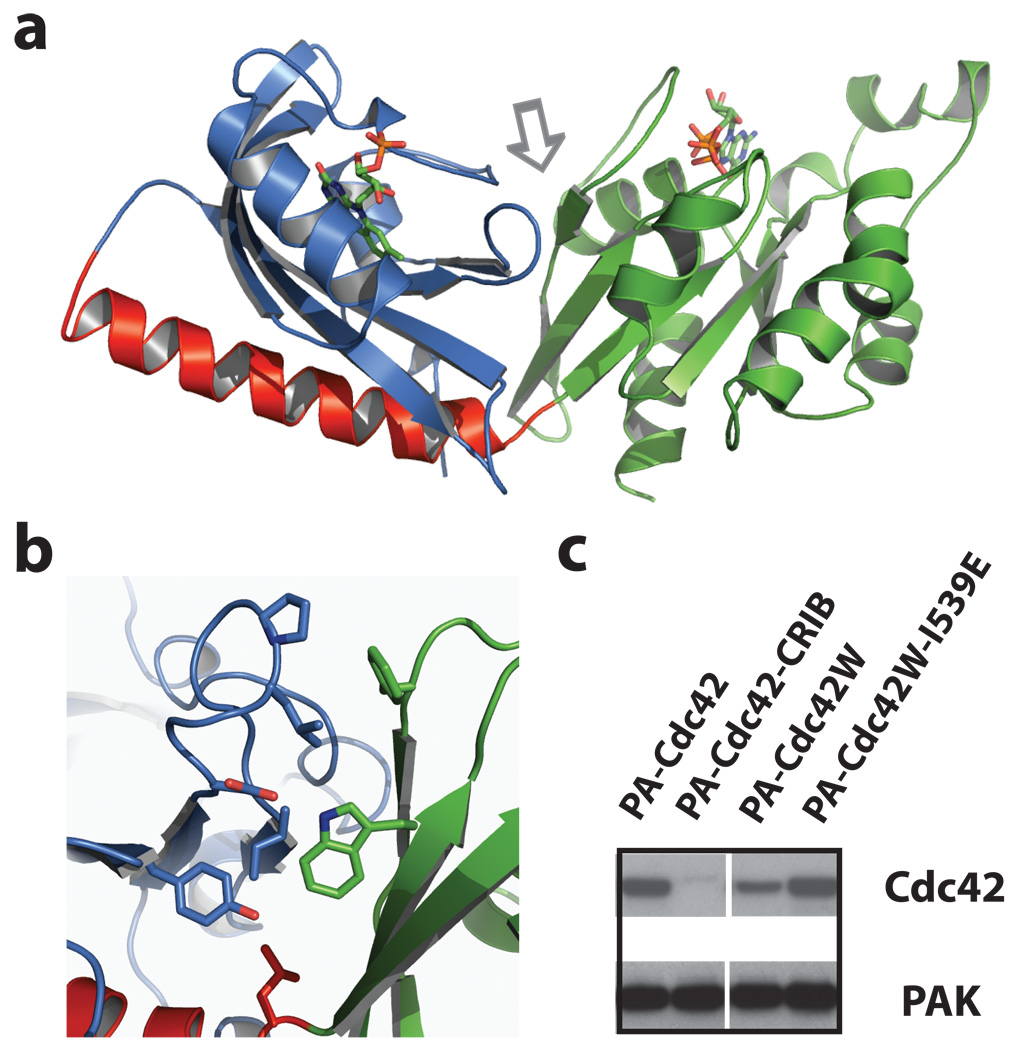
Figure 4. Crystallization and structural modelling of PA-Rac1.
a, Dark state crystal structure of PA-Rac1. Blue = LOV domain, red = Jα helix, and green = Rac1. b, Interacting residues at the LOV-Rac interface (arrow in panel a), including Trp56. c, Mutating Cdc42 to include the Trp involved in stabilizing the LOV2-Rac1 interaction substantially improved LOV inhibition of Cdc42. Lane 1, PA-Cdc42; linking LOV to Cdc42 using the same truncations that produced good inhibition for Rac does not inhibit Cdc42-PAK binding. Lane 2, PA-Cdc42-CRIB; covalently linking the CRIB domain of PAK to PA-Cdc42 blocks PAK binding. Lane 3, PA-Cdc42-F56W; introduction of the tryptophan substantially improves LOV inhibition of Cdc42 binding to PAK. Lane 4, lit state mutant of PA-Cdc42-F56W, showing that Cdc42 inhibition is sensitive to the lit/dark state of the LOV domain. Supplemental Movie S16 and Fig. S14 demonstrate the ability of PA-Cdc42-F56W to produce filopodia and protrusions in living cells.
In summary, we have engineered genetically-encoded photoactivatable Rac1 analogs that enable precise spatial and temporal control of Rac activity in live cells, with reversible activation at 458 or 473 nm. Localized Rac activation or deactivation was sufficient to generate polarized cell movement. Rac could be activated without cellular compensation, enabling us to probe the role of myosin and PAK in Rac-mediated motility. Spatially-regulated Rac inhibition of Rho was demonstrated in living cells. Structural studies indicate that a non-evolved interaction at the Rac-LOV interface can be engineered to cage other GTPases. This study and other recent work25–28 show that coupling genetically encoded light-modulated domains to other proteins provides a versatile new route to control protein activities in living cells.
METHODS SUMMARY
Imaging experiments were conducted on an Olympus FluoView 1000 laser scanning confocal microscope and an Olympus IX81-ZDC inverted microscope. Biosensor imaging was performed as previously described8,29. Simultaneous biosensor imaging and activation of PA-Rac was achieved using a MAG Biosystems FRAP-3D add-on (Photometrics) for galvanometer control of laser position. Detailed materials and methods are included in the supplementary information.
Supplementary Material
Acknowledgements
The authors are grateful for help and constructs from Winslow Briggs, Keith Moffat, Ashutosh Tripathy, Gary Bokoch and Ken Jacobson. Diffraction data were collected at the Swiss Light Source, beamline X10SA, Paul Scherrer Institute, Villigen, Switzerland. We thank the Dortmund-Heidelberg team for data collection, and Anuschka Pauluhn and Martin Fuchs for their support in setting up the beamline. This research was supported by the American Heart Association (YW) and the National Institutes of Health (KMH grants GM057464 and GM64346).
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
Author Information The structural coordinates of PA-Rac1 and its mutants have been submitted to the Protein Data Bank under accessions 2wkp (WT), 2wkq (C450A) and 2wkr (C450M).
References
- 1.Goeldner M, Givens R. Dynamic Studies in Biology: Phototriggers, Photoswitches and Caged Biomolecules. Wiley-VCH; 2005. [Google Scholar]
- 2.Fortin DL, et al. Photochemical control of endogenous ion channels and cellular excitability. Nat Methods. 2008;5:331–338. doi: 10.1038/nmeth.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Developmental Biology. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 5.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci U S A. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 7.Kraynov VS, et al. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 8.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 9.Burridge K, Wennerberg K. Rho and Rac Take Center Stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 10.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 11.Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol. 1999;147:1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao X, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-Jα photoswitch. Nature Chemical Biology. 2008;4:491. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 14.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jα helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 15.Thompson G, Owen D, Chalk PA, Lowe PN. Delineation of the Cdc42/Rac-binding domain of p21-activated kinase. Biochemistry. 1998;37:7885–7891. doi: 10.1021/bi980140+. [DOI] [PubMed] [Google Scholar]
- 16.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 17.Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. The Journal of Cell Biology. 2007;176:573. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu Rev Cell Dev Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- 19.Giannone G, et al. Lamellipodial Actin Mechanically Links Myosin Activity with Adhesion-Site Formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurokawa K, Matsuda M. Localized RhoA Activation as a Requirement for the Induction of Membrane Ruffling. Molecular Biology of the Cell. 2005;16:4294–4303. doi: 10.1091/mbc.E04-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 23.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 24.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, et al. Surface sites for engineering allosteric control in proteins. Science. 2008;322:438–442. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moglich A, Ayers RA, Moffat K. Design and Signaling Mechanism of Light-Regulated Histidine Kinases. J Mol Biol. 2008 doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proceedings of the National Academy of Sciences. 2008;105:10709. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung DW, Otomo C, Chory J, Rosen MK. Genetically encoded photoswitching of actin assembly through the Cdc42-WASP-Arp2/3 complex pathway. Proc Natl Acad Sci U S A. 2008;105:12797–12802. doi: 10.1073/pnas.0801232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hodgson L, Shen F, Hahn KM. Biosensors for characterizing the dynamics of Rho family GTPases in living cells. Current protocols in cell biology. doi: 10.1002/0471143030.cb1411s46. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.