Abstract
Hematopoietic stem cell (HSC) engraftment is a multistep process involving HSC homing to bone marrow, self-renewal, proliferation, and differentiation to mature blood cells. Here, we show that loss of p190-B RhoGTPase activating protein, a negative regulator of Rho GTPases, results in enhanced long-term engraftment during serial transplantation. This effect is associated with maintenance of functional HSC-enriched cells. Furthermore, loss of p190-B led to marked improvement of HSC in vivo repopulation capacity during ex vivo culture without altering proliferation and multilineage differentiation of HSC and progeny. Transcriptional analysis revealed that p190-B deficiency represses the up-regulation of p16Ink4a in HSCs in primary and secondary transplantation recipients, providing a possible mechanism of p190-B–mediated HSC functions. Our study defines p190-B as a critical transducer element of HSC self-renewal activity and long-term engraftment, thus suggesting that p190-B is a target for HSC-based therapies requiring maintenance of engraftment phenotype.
Introduction
Hematopoietic stem cells (HSCs) are defined by their unique ability to give rise to all mature immune and blood cell lineages while at the same time regenerating themselves in a process termed “self-renewal” to sustain hematopoiesis throughout life. In addition, HSCs traffic throughout the body and possess the ability to reconstitute all hematopoietic lineages on transplantation into lethally irradiated mice, characteristics that have been used in therapeutic stem cell transplantation.1–3 To maintain an adequate number of both mature blood cells and HSCs, the quality and quantity of HSC divisions must be tightly controlled.4,5 Several regulatory pathways that play a role in the maintenance of HSC functions have been identified; these include both cell-intrinsic and extrinsic factors. For example, signaling through integrins, stem cell factor, thrombopoietin, angiopoietin, and transforming growth factor-β regulate HSC properties.6–9 Cell-cycle regulators, p21Cip1, p16Ink4a, p18Ink4c, and p57Kip2, and proteins that control transcription, such as HoxB4, c-myc, FOXO, Zfx, Tel, Gfi-1, Pbx-1, or epigenetic factor, Bmi-1, and Ezh2,4,5,10–15 are essential for HSC functions. A fundamental issue in HSC biology is to understand how these programs are regulated and to exploit this knowledge for the development of HSC-based therapies for therapeutic purposes.
Members of the Rho GTPase family operate as molecular switches to effect signaling downstream of numerous receptors, including integrins, chemokines and cytokine receptors.16,17 Most canonical Rho GTPases cycle between an active guanosine triphosphate (GTP)–bound and an inactive guanosine diphosphate (GDP)–bound state. This GDP-GTP cycle is tightly regulated by 3 families of proteins. Guanine nucleotide exchange factors promote the exchange of GDP for GTP, whereas GTPase-activating proteins (GAPs) accelerate the rate of hydrolysis of GTP. In addition, guanine nucleotide dissociation inhibitors may interfere with GTP binding by preventing membrane localization of the protein. Of the 20 Rho GTPases currently known, the best studied Rho GTPases are Rho, Rac, and Cdc42, which are crucial regulators of cytoskeleton dynamics, cell migration, adhesion, and cell-cycle progression. As such, Rho GTPases regulate a broad variety of cellular processes in many mammalian cells, including in hematopoietic cells.16–23
Whereas the role of Rho GTPases in cell functions has begun to be understood, the role of GAPs and guanine nucleotide exchange factors in vivo has been understudied. Because more than 70 RhoGAPs have been identified in eukaryotes, the RhoGAPs outnumber the Rho GTPases that they regulate.24 Some GAPs show preferential tissue expression and appear to have tissue-specific functions. Moreover, each GAP can regulate a restricted number of Rho GTPase signaling pathways.25 Finally, the presence of several functional domains suggests that GAPs may mediate signaling pathways that are not limited to Rho GTPase activity.26 Therefore, defining the role of GAPs in vivo will probably help the identification of specific regulatory pathways that are critical for mammalian cell functions.
p190-B RhoGAP (hereafter p190-B) serves as negative regulator of Rho activity.27 Disruption of p190-B in gene-targeted mice has revealed defects in the central nervous system, thymus, and lung, which lead to perinatal lethality.28 Furthermore, p190-B has been implicated in regulating cell size during fetal development,28 adipogenesis-myogenesis cell fate determination,29 and mammary morphogenesis.30 Here, we examined the role of p190-B in HSC functions. Our study demonstrates that the loss of p190-B results in enhanced long-term HSC engraftment. This phenotype was correlated with maintenance of low expression of p16Ink4a, indicating a possible molecular mechanism by which p190-B may mediate these effects on HSC functions. Importantly, the absence of p190-B led to sustained HSC in vivo repopulation potential during ex vivo culture. Therefore, our study reveals p190-B as an important molecule limiting HSC self-renewal. In addition, loss of p190-B was associated with increased progenitor homing to the bone marrow (BM). p190-B might thus be a potential target to improve HSC-based therapies requiring maintenance of engraftment phenotype during in vitro manipulation or in the settings requiring engraftment of limited numbers of HSCs.
Methods
Mice
p190-B RhoGAP+/− mice (backcrossed into C57BL/6J N = 10) and B6.SJL-PtrcaPep3b/BoyJ (B6.BoyJ, CD45.1+) congenic mice were bred in house in a pathogen-free environment. Embryonic day 14.5 fetal livers (FLs) p190-B and wild type (WT) littermates were used for experiments. All studies were conducted with a protocol approved by the Animal Care Committee of Cincinnati Children's Hospital Medical Center.
Flow cytometric analysis and purification of HSC/P
Lineage staining of BM cells used a cocktail of biotinylated antimouse antibodies to anti-CD11b (M1/70), anti-B220 (RA3-6B2), anti-CD5 (53-7.3), anti-Gr-1 (RB6-8C5), anti-Ter119, and anti-CD8a (53-6.7). Lineage staining of FLs used a similar cocktail of biotinylated antibodies, except for CD11b. For detection and sorting, we used streptavidin-allophycocyanin (APC)–Cy7 or AlexaFluor 700. Directly conjugated antibodies were anti–Sca-1-phycoerythrin (PE)–Cy7 (clone D7; eBioscience), anti–c-kit-APC or APC-Cy7 (clone 2B8), anti–CD34-PE, anti–CD48-fluorescein isothiocyanate, anti–CD150-PE or APC (eBioscience), anti–CD45-APC. For congenic strain discrimination, anti–CD45.1-PE and anti–CD45.2-fluorescein isothiocyanate were used. Unless specified, all antibodies were from BD Biosciences PharMingen. Fluorescence-activated cell sorting (FACS) was performed on a FACSStarplus cell sorter, and analysis was performed with a Facscanto (BD Biosciences) or LSRII (BD Biosciences).
Cell-culture assays
Myeloid clonogenic progenitor assays, high-proliferation-potential (HPP) colony-forming cell, and cobblestone area-forming cell (CAFC) assays were performed as previously described.18,20,31
Competitive repopulation assays and serial transplantation
For competitive repopulation of total FL cells, 0.4 × 106 E14.5 FL cells from WT or p190-B−/− mice (CD45.2+) plus 1.6 × 106 CD45.1+ BM cells were injected into 8 to 10 lethally irradiated CD45.1+ congenic recipient mice per experiment. At 4 months after transplantation, 106 BM cells from primary recipients were harvested and used as donors for serial transplantation. For transplantation of lineage−Sca-1+c-Kit+ cells (LSK) FL cells, 1500 FL LSK (CD45.2+) cells plus 106 FL CD45.1+ cells were used. For transplantation of LSK cells after in vitro culture, 800 000 CD45.2+ cells plus 250 000 BM CD45.1+ cells were used. Reconstitution of CD45.2+ and lineage differentiation were monitored in peripheral blood (PB) and BM cells.
Ex vivo cell-cycle and -division analysis
For cell-cycle analysis, BM cells from serially transplanted animals were first stained with cell-surface markers, fixed and permeabilized, and then incubated with Hoechst 33342 (10 μg/mL, Invitrogen) and Pyronin Y (1 μg/mL; Sigma-Aldrich) for 20 minutes. For in vitro bromodeoxyuridine (BrdU) incorporation, we used the APC-BrdU Flow kit (BD Biosciences PharMingen) according to the manufacturer's instructions. BrdU (10 μM) was added to the culture medium for 30 minutes. The cells were then fixed and stained with anti-BrdU Ab, 7-amino-actinomycin D (7-AAD), and for LSK as described in “Flow cytometric analysis and purification of HSC/P.” For cell-division analysis, LSK FLs were stained with 0.5μM carboxyfluorescein succinimidyl ester (CFSE; Invitrogen) for 10 minutes at 37°C and then incubated with cytokines. Fluorescence intensity of CFSE was analyzed at 24, 36, and 48 hours of culture by FACS.
5-Fluorouracil
5-Fluorouracil (5-FU; 150 mg/kg) was administered by intraperitoneal injection. Differential counts and lineage reconstitution were analyzed in peripheral blood obtained by retro-orbital bleed at the time of injection and at weekly intervals for the next 5 weeks.
In vivo cell-cycle analysis
In vivo BrdU labeling was performed by injecting control animals and experimental animals 3 days after 5-FU treatment with 200 μL (1.8 mg BrdU/mL; Sigma-Aldrich) intraperitoneally and then giving drinking water containing 1 mg/mL BrdU and 5% glucose for 15 hours. BrdU staining was performed using BrdU-APC kit combining with adequate cell surface markers.
In vitro culture of HSCs
FL LSK cells (5 × 103 cells) were cultured in Stem-Span medium (Stem Cell Technology), containing 50 ng/mL stem cell factor, 50 ng/mL megakaryocyte growth and development factor (Amgen), 20 ng/mL interleukin-3 (IL-3), 50 ng/mL IL-11 (Genetic Institute Inc), and 50 ng/mL IL-6. Unless specified, all cytokines were from PeproTech. Cells were analyzed 6 to 7 days after culture and assessed for cell cycle, apoptosis, progenitor assay, and competitive repopulation assay.
Gene expression analysis
RNA was isolated using RNeasy Micro Kit (QIAGEN) from CD45.2+ LSK cells. First-strand complementary DNA synthesis was primed with random hexamers using the Sensiscript RT Kit (QIAGEN). Real-time quantitative polymerase chain reaction (PCR) was carried out in triplicate in an ABI Prism 7700 Sequence Detector using Taqman PCR Master Mix reagent (Applied Biosystems). Gene expressions are relative to β-actin. Primers and probes for p16Ink4a, p21Cip1, p27Kip1, and β-actin were from Applied Biosystems.
Cell homing and migration and adhesion assays
Statistical analysis
Data are expressed as the mean plus or minus SEM. Differences between groups were analyzed by an unpaired 2-sided t test. A probability value less than .05 was considered significant.
Results
Effect of p190-B deficiency on FL hematopoiesis
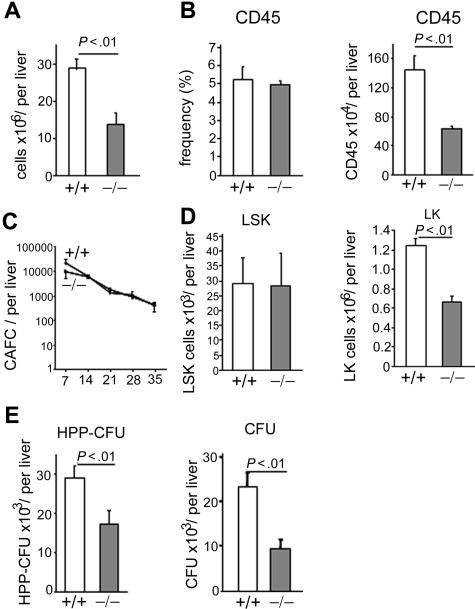
Because p190-B−/− embryos die before birth,28 FL tissues from day E14.5 WT and p190-B−/− embryos were used to investigate the effect of genetic deletion of p190-B on HSC/P functions (supplemental Figure 1A, available on the Blood website; see the Supplemental Materials link at the top of the online article). p190-B−/− FLs exhibited an increase in RhoA activity without altering the activity of Rac and Cdc42 (supplemental Figure 1B). p190-B−/− FLs exhibited a 2-fold decreased cellularity compared with WT controls (Figure 1A) because of an impairment of late erythropoiesis at this stage of development (K.S., S.E., H.X., J.S., T.K., D.A.W., and M.-D.F., manuscript in preparation). The total number of CD45+ cells per p190-B−/− FLs was also reduced (Figure 1B). However, the total number of primitive p190-B−/− HSC per liver, as assessed by day 35 CAFCs, was comparable with that of WT (Figure 1C). Immunophenotypic analysis of day 14.5 FLs indicated that the total number of HSC-enriched cells (LSK) in p190-B−/− FLs was similar to that of WT FLs. In contrast, the total number of p190-B−/− Lin−c-Kit+ (LK) cells was decreased (Figure 1D). The total number of HPP-colony-forming cells (a primitive progenitor with extensive proliferative capacity defined in vitro) and myeloid colony-forming unit (CFU) progenitors per liver were also decreased in p190-B−/− FL cells (Figure 1E). Of note, there was no difference in the relative distribution of multipotential, erythroid, and granulomonocytic colonies (not shown). Thus, p190-B deficiency is associated with a normal number of FL HSC-enriched population.
Figure 1.
Effect of p190-B deficiency on FL hematopoiesis. (A) FL cellularity of WT and p190-B−/− E14.5 embryos. Data shown are mean ± SD per liver (n = 24). (B) Frequency and total number of CD45+ cells per E14.5 FLs. Histograms show mean ± SD; n = 6. (C) Primitive progenitor cells evaluated using the CAFC assay. Total FL cells were cultured with FBMD-1 cells for 35 days. Frequency of CAFC for WT (+/+) and p190-B−/− (−/−) was determined on the day noted on the x-axis. Data represent total number of CAFC per liver, mean ± SEM (n = 3 independent experiments, n = 2 or 3 embryos per group in each experiment). (D) Total number of LSK and LK cells per liver, mean ± SD; n = 24. (E) Total number of HPP-CFU and CFU per liver (mean ± SD, n = 9-12, from 3 independent experiments).
p190-B deficiency enhances HSC long-term engraftment
To investigate the role of p190-B−/− in HSC functions, the capacity of p190-B−/− FLs to reconstitute hematopoiesis in lethally irradiated mice was analyzed in noncompetitive transplantation assay using lethally irradiated recipients. p190-B−/− FL cells were able to fully reconstitute irradiated mice, as assessed by PB chimerism (supplemental Figure 2A), lineage differentiation analysis in PB and in BM (supplemental Figure 2B-E), and by PB counts (supplemental Table 1). Of note, these mice also exhibited similar frequency of BM CFU (data not shown), indicating that the reduced number of myeloid progenitors seen in the FLs is not transplantable. Thus, this change could be developmentally regulated or the result of cell-extrinsic defects in the FL microenvironment.
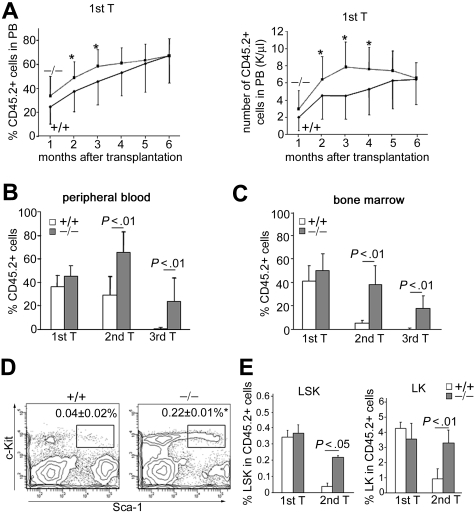
To further assess the function of p190-B−/− HSC/P, we performed 2 sets of competitive repopulation assay. In the first set, FL cells from each genotype were competed against congenic WT BM cells. In agreement with the finding that FL-derived HSCs possess extensive self-renewal capacity compared with BM-derived HSCs,32 the PB chimerism derived from transplanted WT FL cells increased over time and reached a level of approximately 60% 6 months after transplantation (Figure 2A). p190-B−/− FL cells reached this level of chimera earlier than WT controls (Figure 2A). Analysis of BM 4 months after transplantation indicated that BM chimerism in recipients of both genotypes were equivalent, including the LSK-defined primitive population. These data suggest modestly enhanced kinetics of engraftment of p190-B−/−-derived hematopoiesis compared with WT controls in irradiated recipients.
Figure 2.
Enhanced long-term engraftment capacity of p190-B−/− FL cells. (A) WT and p190-B−/− FL cells (0.4 × 106 CD45.2+ FL) plus 1.6 × 106 BM congenic competitor (CD45.1+) cells were injected into lethally irradiated recipient mice. The number of CD45.2+ versus CD45.1+ cells in the PB was determined between one and 6 months. Histograms show the percentage of CD45.2+ cells (left panel) and the absolute number of CD45.2+ cells (right panel) in the PB at the indicated time points after transplantation (mean ± SD; n = 24). *P < .05. (B-C) Serial transplantation of WT and p190-B−/− FL cells. Average percentage of CD45.2+ cells in PB (B) and BM (C) 4 months after transplantation of primary, secondary, and tertiary recipients. Data are mean ± SD; n = 6 to 10 (1 representative experiment from 3 independent experiments all with similar results). (D) Flow cytometric analysis dot plot of LSK frequency in CD45.2+ BM cells from secondary recipient. Data shown are the frequencies of LSK cells (mean ± SD; n = 6). *P < .01. (E) Frequency of LSK (left panel) and LK cells (right panel) in CD45.2+ BM cells of primary and secondary recipients 4 months after transplantation. Data are mean ± SD; n = 6 to 10 (1 representative experiment from 3 independent experiments all with similar results).
To investigate the long term self-renewal capacity of p190-B−/− HSC/P, serial transplantations of competitively transplanted animals were performed in which identical numbers of BM cells from recipient animals were used for serial transplantations at 4-month intervals. PB analysis indicated that donor-cell chimera was significantly higher in secondary and tertiary animals that received p190-B−/− cells compared with animals that received WT cells (Figure 2B). The level of contribution of p190-B−/− cells to the BM was maintained between the primary and secondary recipients and still easily detectable in tertiary recipients (Figure 2C). In contrast, the level of contribution of FL WT cells dramatically decreased with successive serial transplantation and was barely detectable in tertiary recipients. Further analysis revealed that secondary p190-B−/− recipients exhibited significantly higher BM LSK and LK frequencies in BM than recipients transplanted with WT FL cells (Figure 2D-E). p190-B−/− LSK and LK cells were still detectable in the BM of tertiary recipients, although at a lower frequency than in secondary recipients (data not shown). Because the BM cellularity was comparable between the genotypes, the total number of LSK and LK cells per femur was higher in p190-B−/−-transplanted secondary recipients than in WT control (data not shown), and remained in a range that was comparable with a nontransplanted control mouse. Importantly, p190-B−/− HSCs maintained multilineage differentiation of myeloid and lymphoid cells in secondary recipient mice equivalent to WT HSCs (supplemental Table 2). Persistent multilineage differentiation was observed in tertiary recipients transplanted with p190-B−/− cells, although the numbers of B220+ cells was decreased, a finding similar to that reported in aged mice.33 These data indicate maintenance of functional p190-B−/− HSCs in secondary recipients. Therefore, loss of p190-B confers enhanced serial transplantation capacity to HSCs, which results in the suppression of exhaustion of phenotypically defined HSC/P in secondary and tertiary recipients. Of note, the rapid exhaustion of WT FL HSCs during serial transplantation in competition with BM cells is consistent with a previous report showing that FL cells from C57Bl/6, which may contain an HSC subset with repopulation ability that exceeds that of BM HSCs in the initial transplantation,32,34 fail to expand in serial transplantation.34 As a result, the repopulation ability of FL HSCs in secondary recipients decreases 186-fold compared with 7.3-fold for BM HSCs.34 The fact that chimerism was higher in PB than in BM of WT secondary transplanted mice is probably the result of the presence of long-living lymphocytes in the PB, which constitute 60% of total white blood cells of mice.
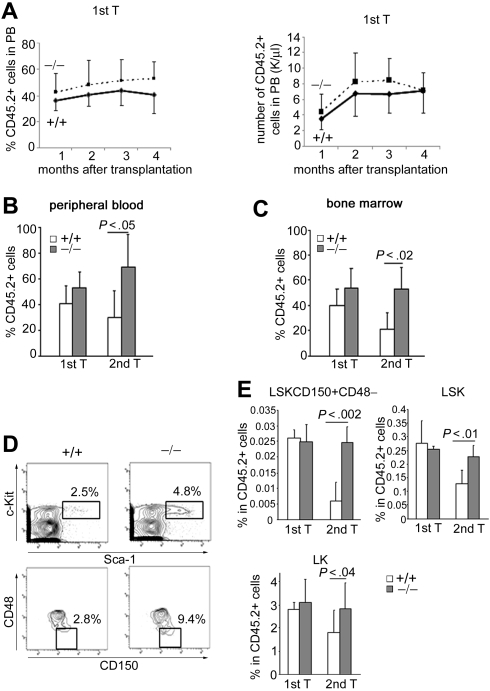
We noted that p190-B−/− FLs exhibited increased HSC-enriched cell frequency. To confirm that the increased engraftment seen in p190-B−/− FLs is the result of cell-intrinsic differences in the HSC population, we also performed serial transplantation assays using equivalent numbers of immunophenotypically defined LSK cells from E14.5 WT and p190-B−/− FL. In this set of experiments, we used congenic FL cells (CD45.1+) as competitors to avoid rapid HSC exhaustion. Consistent with the engraftment data with total FL cells, loss of p190-B conferred HSC engraftment advantage. Donor chimerism analyzed in the PB 4 months after transplantation was similar in primary recipients of WT and p190-B−/− LSK cells (Figure 3A). However, secondary recipients of p190-B−/− LSK cells exhibited significantly higher chimerism than WT, as analyzed in the PB (Figure 3B) and in the BM (Figure 3C), 4 to 6 months after transplantation. BM analysis revealed that secondary recipients of p190-B−/− LSK cells maintained a frequency of LSK cells and long-term HSCs (LT-HSCs) within this population (defined as LSKCD150+CD48− cells) to normal level, whereas these populations were significantly reduced in recipients of WT cells (Figure 3D-E). Finally, recipients transplanted with p190-B−/− cells exhibited normal lineage differentiation (not shown). Collectively, these findings indicate that p190-B deficiency preserves HSC “stemness” during serial transplantation and suggest increased self-renewal activity of p190-B−/− HSCs.
Figure 3.
Highly enriched p190-B−/− HSCs exhibit enhanced long-term engraftment. Competitive repopulation assay of purified LSK FL cells. LSK FL cells (1500) plus 1.0 × 106 FL competitor cells were injected into lethally irradiated recipients. (A) Percentage of CD45.2+ cells (left panel) and absolute number of CD45.2+ cells (right panel) in the PB at the indicated time points after transplantation (mean ± SD; n = 4). (B-C) Histograms show the percentage of CD45.2+ in the PB (B) and BM (C) 4 to 6 months after primary and secondary transplantation (mean ± SD; n = 6). (D) Flow cytometry analysis dot plot of LSKCD150+CD48− frequency in CD45.2+ BM cells from secondary recipient. (E) Frequency of LSKCD150+CD48−, LSK, and LK cells in CD45.2+ BM cells of primary and secondary recipients 4 to 6 months after transplantation. Data are mean ± SD; n = 6.
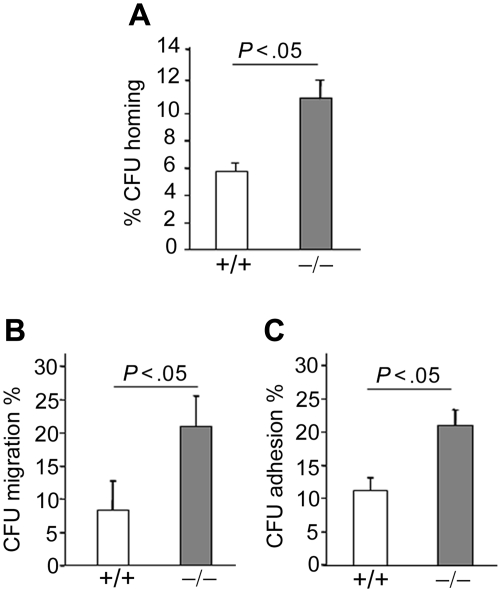
Homing to the BM is a critical first step in HSC and progenitor engraftment35 and is regulated in part by cytoskeleton, shape, and adhesion characteristics of the cell, all functions regulated by Rho GTPases. To assess CFU homing, we used BM cells of animals reconstituted with p190-B−/− and WT FL cells, which exhibited comparable frequency of LSK and LSKCD150+CD48− cells, respectively (supplemental Figure 2E). BM of irradiated recipient animals that received 5 × 106 BM cells of each genotype was harvested 16 hours after injection and assessed by CFU assay. p190-B−/− progenitors demonstrated an increase in CFU homing to BM compared with WT cells (Figure 4A). Hematopoietic progenitor cells (HPCs) home to the BM microenvironment by migrating across the endothelium in response to stromal cell-derived factor-1α (SDF-1α), a ligand for the chemokine receptor CXCR4,36 and depend on the function of α4β1 and α5β1 integrins.37,38 SDF-1α–induced migration was examined using transwell migration assays. p190-B−/− LSK cells exhibited significant increased SDF-1α–induced CFU migration compared with WT (Figure 4B). In addition, although the expression of both α4β1 and α5β1 integrins was equivalent between the genotypes (data not shown), p190-B−/− LSK cells showed increased CFU adhesion to recombinant fibronectin (FN CH-296, containing binding sites for both α4β1 and α5β1) compared with WT cells (Figure 4C). Thus, enhanced migration and adhesion may promote progenitor homing.
Figure 4.
Homing of p190-B−/− HPCs. (A) Homing of HPCs to BM. WT and p190-B−/− BM cells (5 × 106) were injected into lethally irradiated mice. Homing of HPCs, analyzed by CFU assay, was assessed 16 hours after transplantation. Data show the percentage of progenitors (CFU) in the BM divided by CFU activity of input cells (mean ± SEM; n = 3 independent experiments, n = 5 recipients/group). (B) Migration of HPCs. WT and p190-B−/− LSK FL cells were allowed to migrate toward SDF-1α. Data represent the percentage of migrated progenitors (CFU) divided by CFU activity of input LSK FL cells (mean ± SEM; n = 3 independent experiments). (C) Adhesion of HPCs to fibronectin fragment CH 296 (FN). WT and p190-B−/− LSK FL cells were allowed to adhere for 1 hour at 37°C to FN. Adherent cells were collected for CFU assays. Data represent the percentage of adherent progenitors (CFU) divided by CFU activity of input cells (mean ± SEM; n = 3 independent experiments).
p190-B deficiency does not grossly affect HSC survival or cell cycling
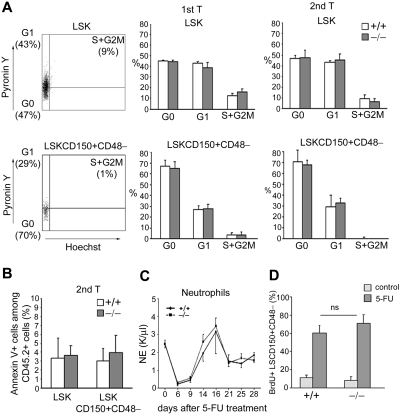
Efficient long-term hematopoietic reconstitution after transplantation is thought to depend on the self-renewal of a pool of quiescent HSCs.5,39–41 The frequency of HSC/P in cycle during serial transplantation was thus analyzed. Surprisingly, the frequencies of G0, G1, and S/G2M cells in donor (CD45.2+) LSK and LSKCD150+CD48− cells were comparable between WT and p190-B−/− HSC/P-transplanted primary and secondary recipients (Figure 5A), indicating no gross alteration of steady-state cell proliferation in the absence of p190-B. Annexin V staining of BM CD45.2+ LSK and LSKCD150+CD48− cells of secondary recipients did not reveal any differences in cell survival between p190-B−/− and WT (Figure 5B).
Figure 5.
p190-B−/− HSC/P exhibit normal cell-cycle rate and survival during serial transplantation. (A) Cell-cycle status of WT and p190-B−/− LSKCD150+CD48− cells. BM LSKCD150+CD48− cells of primary and secondary recipients were stained with Hoechst 33342 dye and Pyronin Y. Histograms represent the percentage of CD45.2+ LSKCD150+CD48− cells and LSK cells in G0, G1, and S/G2M phases of the cell cycle (mean ± SD; n = 5). (B) Apoptosis analysis of LSKCD150+CD48− cells. BM cells from secondary transplantation recipients were stained with annexin V and analyzed by flow cytometry. Data represent the percentage of CD45.2+ LSKCD150+CD48− and LSK annexin V+ cells (mean ± SD; n = 5). (C) PB recovery after a single dose of 5-FU. Kinetics of neutrophil PB counts are shown (mean ± SEM; n = 3 independent experiments, n = 5 mice per group and experiment). (D) The proliferation of LSCD150+CD48− cells was measured by in vivo BrdU incorporation over 15 hours. Histogram is percentage of LSCD150+CD48−, which incorporated BrdU (mean ± SD; n = 3).
To further investigate HSC proliferation under stress, the hematologic recovery of PB after treatment with 5-FU was analyzed, a treatment that depletes cycling cells. In this assay, the time to hematologic recovery after 5-FU depends on both the quantity and the preservation of a population of quiescent HSCs.42 To rule out a quantitative effect on the number of LSKs present in BM, we examined the hematologic response of animals reconstituted with FL cells from each genotype with a similar frequency of HSC/P (LSK) and LT-HSC (LSKCD150+CD48−) cells (supplemental Figure 2E). The recovery of neutrophils (Figure 5C) and platelets (not shown) after 5-FU treatment was comparable between WT and p190-B–deficient animals, which suggested equal HSC proliferation of WT and p190-B−/− cells on stress. We then analyzed LT-HSC cell-cycle entry by BrdU incorporation 3 days after 5-FU. In this experiment, we analyzed the LSCD150+CD48− population because c-Kit cell surface expression is down-regulated on functional HSCs in response to 5-FU.43 Consistent with the blood recovery, the frequency of cell-cycle entry of LSCD150+CD48− cells was similar between the genotypes (Figure 5D). Because HSC activity can be maintained or expended without changing the cycling rate of the cells,44,45 the maintenance of p190-B−/− HSC engraftment may be an indication of maintenance of HSC integrity as the cells divide to efficiently reconstitute the pool of HSC after engraftment. Overall, p190-B deficiency maintains HSC activity during serial transplantation, without grossly altering the measured cycling rate, survival, and differentiation potential of HSC/P.
p190-B−/− HSC/P maintain low expression of p16Ink4a during serial transplantation
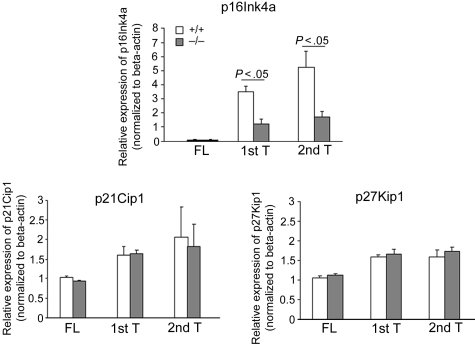
Previous studies have suggested that the exhaustion of HSCs during serial transplantation can depend, at least in part, on the up-regulation of the cyclin-dependent kinase inhibitor p16Ink4a.46 Interestingly, although p16Ink4a is a negative regulator of cell-cycle progression,47 repression of p16Ink4a expression in young HSCs has not been associated with increased cycling rate of HSCs.44,46,48 p16Ink4a mRNA was analyzed by quantitative real-time PCR in the donor-derived LSK population of serially transplanted animals. Analysis of a more HSC-enriched cell population was not possible because of the extremely small number of donor-derived cells. The expression of p16Ink4a was not detectable, in either WT or p190-B−/− FL LSK cells. Consistent with previously described data,46 expression of p16Ink4a increased in WT CD45.2+ BM LSK cells during serial transplantation. In contrast, CD45.2+ p190-B−/− BM LSK cells up-regulated p16Ink4a expression after transplantation but to a significantly lower level in both primary and secondary recipients compared with WT controls (Figure 6). In contrast but consistent with an unaffected stem and progenitor cell cycling, loss of p190-B had no impact on the expression of other cyclin-dependent kinase inhibitors, including p21Cip1 and p27Kip1 (Figure 6), which have been shown to play a role in HSC quiescence and HPC proliferation, respectively.39,49 These data indicate an inverse correlation between engraftment ability of WT and p190-B–deficient HSCs and the levels of p16Ink4a, suggesting a possible mechanistic relationship between p190-B and p16Ink4a in the preservation of stem cell functions that may be independent on the cell-cycle regulation.
Figure 6.
Maintenance of low expression of p16Ink4a in p190-B−/− HSC/P during serial transplantation. p16 Ink4a, p21Cip1, and p27Kip1 expression of FL LSK cells and donor-derived BM LSK cells during serial transplantation. FL LSK cells and CD45.2+ BM LSK cells from primary and secondary transplantation recipients 4 months after transplantation were purified by FACS sorting and used for RNA preparation. The relative mRNA expression level of each gene was examined by quantitative real-time PCR and normalized to β-actin expression. Experiments were performed in triplicate and in 2 independent transplants (mean ± SD of the replicate).
p190-B deficiency also enhances HSC/P reconstitution activity after in vitro culture
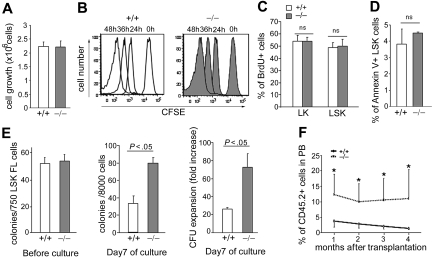
Therapeutic applications of HSCs require in vitro manipulation, and in the future ex vivo HSC expansion may be critical. We next investigated whether p190-B deficiency may preserve the repopulating capacity of HSC/P during ex vivo culture. WT and p190-B−/− LSK cells isolated from FLs were incubated with a cocktail of cytokines in serum-free medium for 7 days.50,51 The deficiency of p190-B was associated with no apparent growth advantage compared with WT cells (Figure 7A). Furthermore, cell division as assessed by CFSE labeling of WT and p190-B−/− LSK cells for the first 2 days of culture was comparable between each genotype (Figure 7B). As seen in primary cells during engraftment studies outlined in Figure 5, there was no detectable difference in cell-cycle progression and apoptosis of WT and p190-B−/− cells on culture, as assessed by BrdU incorporation, and annexin V and 7-AAD staining, respectively (Figure 7C-D). These data indicate that the loss of p190-B does not affect the growth, cell division, and survival of HSC/P. However, although freshly isolated LSK cells from WT and p190-B−/− mice exhibited comparable clonogenic capacity (Figure 7E) or repopulation ability (Figure 3), the frequency of CFU after 7 days in culture was 2-fold higher in p190-B−/− compared with WT cultures (Figure 7E). Competitive repopulation assays using cells cultured for 7 days ex vivo showed significantly higher repopulating activity measured after 4 months in mice that received p190-B−/− cells compared with WT cells (Figure 7F). The reconstitution data showed a 4.4-fold increase in the estimated frequency of repopulation units from p190-B−/− LSK culture compared with WT cells. These data are in agreement with an impact of p190-B on the maintenance of HSC functions without grossly altering cycling rate and survival of these cells and thus suggest potential enhanced HSC self-renewal divisions in the absence of p190-B. Therefore, p190-B signaling may be used in the future to develop new strategies of ex vivo HSC expansion.
Figure 7.
Ex vivo expansion of CFU and sustained repopulation activity by loss of p190-B. FL LSK cells were incubated in serum-free medium with stem cell factor, IL-3, IL-6, IL-11, and megakaryocyte growth and development factor for 7 days. (A) Total cell number at day 7 of culture enumerated by hemocytometer (mean ± SEM; n = 6 independent experiments). (B) Analysis of cell division of cultured FL LSK cells. FL LSK cells were labeled with fluorescent dye CFSE and fluorescence intensity of CFSE+ LSK cells was analyzed at 0, 24, 36, and 48 hours of culture. Data shown are the representative flow profiles of fluorescent intensity of CFSE (2 independent experiments). (C) Cell-cycle analysis of cultured FL LSK cells. After 6 days of culture, cells were labeled with BrdU in vitro for 30 minutes and stained for LSK. Data represent the percentage of BrdU+ cells in LK or LSK cell population (mean ± SD; n = 3; the difference is not statistically significant [ns]). (D) Apoptosis analysis of cultured FL LSK cells at day 6 of culture. The cells were stained with annexin V and 7-AAD. Histogram represents the percentage of annexin V+ cells in LSK cell population (mean ± SD; n = 3; the difference is not statistically significant [ns]). (E) CFU plating efficiency of FL LSK cells before and after in vitro culture. Data in 2 left panels represent the number of colonies/input cells on day 0 and day 7 during the culture. Data represent mean ± SEM (n = 3 independent experiments). Data on right panel represent CFU expansion of cultured FL LSK cells expressed as fold increase (mean ± SD; n = 3; 1 representative experiment from 3 independent experiments). (F) Competitive repopulation assay of cultured FL LSK cells. Cultured cells plus freshly isolated competitor BM cells were injected into lethally irradiated mice. Histogram represents the percentage of donor (CD45.2+) cells in the PB at the indicated time points after transplantation (mean ± SD; n = 5). *P < .05.
Discussion
Maintenance of the HSC pool after BM transplantation is essential to ensure long-term hematopoiesis. Here, we show that loss of p190-B protects a pool of functional HSCs from exhaustion during serial transplantation without altering proliferation and multilineage differentiation. Transcriptional analysis reveals a concomitant prevention of up-regulation of p16Ink4a expression after transplantation. Importantly, loss of p190-B leads to significant ex vivo expansion of primitive progenitors that sustain repopulation potential in vivo. Loss of p-190-B thus supports long-term hematopoiesis probably by favoring self-renewal divisions of HSCs. In addition, loss of p190-B is associated with increased homing that may contribute to enhanced engraftment. These findings define p190-B as a critical regulator of HSC/P functions probably via self-renewal activity while maintaining a balance between proliferation and differentiation and via homing.
Homing to the BM, which involves a series of chemotactic and adhesive events, is a critical first step in HSC and progenitor engraftment.35 Given the role of Rho GTPases and their regulators, including p190-B, in cytoskeletal reorganization,16,17 changes in CFU homing in the absence of p190-B were anticipated. Although our homing assay does not assess for HSC homing, enhanced homing of p190-B–deficient progenitors may also reflect HSC behavior. Because the hematopoietic reconstitution can depend on the initial dose of HSCs injected, this finding may contribute, to some extent, to the observed enhanced engraftment of p190-B–deficient cells. However, in our study, the engraftment of purified p190-B–deficient LSK cells in primary recipients was similar to WT LSK cells. In contrast, secondary recipients of p190-B–deficient cells showed dramatic enhanced engraftment compared with WT. Therefore, an additional mechanism other than homing may also contribute to the enhanced engraftment conferred by loss of p190-B.
To maintain long-term hematopoietic reconstitution after transplantation, the number of HSC cell divisions must be regulated followed by a return of a proportion of cells to quiescent HSCs that are capable of sustaining self-renewal capacity.4,5 Our data suggest that the absence of p190-B preserves HSC “stemness” both in vivo and in vitro while maintaining a balance between proliferation and multilineage differentiation. This effect may be mediated, at least in part, by controlling optimum levels of the cell-cycle regulator p16Ink4a (hereafter p16). Whereas p16 expression gradually increased in WT HSCs during serial transplantation, p190-B–deficient HSCs maintained lower levels of p16 after primary and secondary transplantations. The important role of p16 in HSC self-renewal has been recently emphasized.46,48,52,53 Notably, p16 specifically regulates stem cell fitness during serial transplantation, as increased p16 expression accompanying serial transplantation was associated with HSC exhaustion.46 In HSCs, p16 expression can be regulated by the p38 mitogen-activated protein kinase (MAPK) pathway. Elevated levels of phosphorylated p38MAPK are associated with increased p16 expression and HSC exhaustion during transplantation.46 Interestingly, the level of phosphorylated p38MAPK is substantially reduced in p190-B−/− embryonic tissues, including FL cells28 (and M.-D.F., unpublished data, May 2007). Although we were not able to analyze p38MAPK activity specifically in HSCs from serially transplanted animals, this pathway may establish a regulatory link between p190-B and p16 expression.
The mechanism whereby p190-B maintains HSC self-renewal remains to be defined. The fact that cell-cycle kinetics was not grossly affected in p190-B–deficient HSC/P was initially surprising given the importance of cell-cycle regulation in maintaining functional HSCs.5 Furthermore, Ink4 proteins are inhibitors of G1 phase progression.47 p16 can act as a repressor of HSC cell cycle, which has a negative impact on their engraftment potential.52 During aging, HSCs up-regulate their expression of p16 and exhibit reduced proliferation. This effect causes a decrease in engraftment potential as old p16-deficient HSC/P exhibit improved serial transplantation efficiency associated with increased HSC/P cell cycling, and also reduced apoptosis.52 However, the role of p16 in the functions of young HSCs during engraftment may be different. The increase in p16 expression observed during serial transplantation of young HSC/P does not lead to an accumulation of “senescent” HSCs (as seen during aging) but results in the exhaustion of quiescent HSCs.46 Furthermore, the loss of Bmi-1 expression, which derepresses p16 expression, leads to a marked depletion of HSCs without grossly altering cell-cycle kinetics of these cells.48,53 Finally, down-regulation of p16 via overexpression of Bmi-1 contributes to promoting HSC self-renewal but leaves their proliferation potential intact.44,48 Instead, ectopic Bmi-1 expression seems to modulate the propensity of HSCs to self-renew through symmetric division.44 Therefore, an interesting alternative would be an effect on preserving HSC function and/or integrity as the cells divide. Although other in vitro assays may directly assess self-renewal division, this hypothesis is supported by studies reported here showing that p190-B deficiency maintains in vivo repopulation activity during ex vivo culture without altering proliferation. It has been shown that HSC self-renewal divisions can be programmed during cell-cycle transit or during their dormancy stage, without altering cycling kinetics.45,44 Further investigations are necessary to establish the precise mechanism(s) of action of p190-B–mediated HSC functions.
It is worth noting that, whereas p190-B loss confers enhanced HSC and progenitor engraftment after transplantation, p190-B–deficient E14.5 FLs show impaired hematopoietic progenitor development. One way to explain this is that the fetal hematopoietic defect may be non–cell-autonomous. Hematopoiesis is maintained and regulated by specific microenvironment, including in the FL.54 A few transcription factors, such as Pitx2 and Xbp1, are known to control a normal liver hematopoiesis supportive microenvironment.55,56 For example, Pitx2-deficient embryos have hypoplastic livers with reduced numbers of hematopoietic cells. Pitx2-deficient FL cells are able to fully reconstitute hematopoiesis of irradiated hosts, but stromas established from Pitx2-deficient FLs do not have hematopoietic supportive capacity ex vivo.55 FL stromal cells probably express p190-B. The defect in p190-B–deficient hematopoietic progenitors in the FL may be secondary to impaired FL stromal cell functions. Alternatively, because hematopoiesis is developmentally regulated57 with fetal and adult hematopoietic progenitors exhibiting distinct proliferative capacity and lineage differentiation commitment,58 p190-B may control regulatory pathways that are specifically required for fetal but not adult hematopoietic progenitor development.
Our study significantly extends the functions previously attributed to p190-B. Initial studies have reported that homozygous deletion of p190-B results in central nervous system defects and perinatal lethality.28 p190-B regulates cell size and adipocyte/myocyte cell fate decision during development and cell growth in mammary gland.28–30 Our study did not reveal any defect in cell size (data not shown), growth, or cell differentiation associated with p190-B deficiency in hematopoietic cells, thus indicating cell context-dependency of p190-B functions. In most cases, p190-B functions have been attributed to activities of RhoA and the RhoA effector ROCK as p190-B deficiency is associated with high levels of RhoA activity28 and because treatment of p190-B–deficient cells with a ROCK inhibitor reverted some p190-B–mediated phenotype.28,29 However, whereas p190-B–deficient FL hematopoietic cells exhibit high RhoA activity,28 the contribution of RhoA and its effector ROCK in p190-B–mediated HSC self-renewal remains unclear. As shown in our previous studies, decreasing RhoA activity in hematopoietic cells, by the expression of dominant negative (DN) mutant RhoA, results in enhanced proliferation of myeloid progenitors and decreased HSC/P adhesion and SDF-1α–directed migration in vitro.19 This phenotype is associated with enhanced HSC engraftment, but the experimental approach suffers from a lack of specificity that is known when DN GTPases are expressed in mammalian cells. In the studies reported here, high RhoA activity generated through the deficiency of the regulatory protein p190-B is associated with enhanced HSC/P homing and migration, but no effect on proliferation. This phenotype is also associated with enhanced HSC engraftment. Although high or low RhoA activity each results in enhanced HSC engraftment, the mechanisms of these events appear quite distinct. Because of the existence of several GAPs inhibiting RhoA activity, p190-B probably regulates only some RhoA pathways.59 In addition, because of the presence of multiple protein motifs, p190-B may regulate some HSC functions that are not mediated by RhoA activity, as recently suggested in other cell types.26 In this regard, treatment of p190-B−/− LSK cells with a ROCK inhibitor does not appear to prevent the observed expansion ex vivo reported here (M.-D.F., unpublished data, March 2008). Conversely, DNRhoA may also alter the activity of others regulatory proteins in addition to RhoA that may not be affected in p190-B−/− HSCs. Thus, the contribution of RhoA and its effector ROCK in p190-B–mediated HSC self-renewal remains to be examined in more detail.
Several transcriptional regulators, epigenetic factors, and cell-cycle regulators have now been implicated in HSC self-renewal. Our study defines p190-B as an important transducer element limiting HSC self-renewal. Importantly, p190-B deficiency appears to markedly enhance HSC functions without grossly affecting their proliferative potential. Given that unregulated growth of immature cells may be deleterious in protocol of ex vivo HSC expansion and favor malignant development, our findings may be of particular significance for designing new strategies for the development of HSC-based therapeutic modalities.
Supplementary Material
Acknowledgments
The authors thank Dr Jose Cancelas (Division of Experimental Hematology and Cancer Biology, Cincinnati Children's Hospital Medical Center) for advice on flow cytometry and comments on the manuscript; Dr Heighton L. Grimes (Division of Immunobiology, Cincinnati Children's Hospital Medical Center) for reagents; Shelli Homan and Christina Sexton for animal husbandry; the Mouse Core at Cincinnati Children's Hospital Medical Center; Jeff Bailey and Victoria Summey for bone marrow transplantation; Dr Daniel Marmer and the Flow core at Cincinnati Children's Hospital Medical Center for assistance with cell sorting; and Amgen and Takara Bio for reagents.
The work was supported in part by a Board of Trustee Cincinnati Children's Hospital Medical Center Award (M.-D.F.), an American Heart Association Scientific and Development Grant Award (M.-D.F.), and the National Institutes of Health (DK62757) (D.A.W.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.X., a graduate student of D.A.W., designed and performed experiments, analyzed the data, and wrote draft of the manuscript; S.E. performed experiments and analyzed the data; H.G. performed experiments, analyzed the data, and edited the manuscript; K.S. and D.D. performed experiments; Y.Z. contributed to the design of experiments, analyzed the data, and edited the manuscript; J.S. contributed vital new reagents by providing the p190-B RhoGAP knockout mouse and edited the manuscript; E.F.S. contributed to the design of experiments, analyzed the data, and edited the manuscript; D.A.W. contributed to the supervision of the study and to the design of experiments, analyzed data, and edited the manuscript; and the experiments were performed in the laboratory of M.-D.F., who directed the overall research project, designed research and experiments, analyzed data, and wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of D.A.W. is Division of Hematology/Oncology, Children's Hospital Boston and Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA.
Correspondence: Marie-Dominique Filippi, Division of Experimental Hematology and Cancer Biology, S7.605, Cincinnati Children's Research Foundation, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: Marie-Dominique.Filippi@cchmc.org; or David A. Williams, Division of Hematology/Oncology, Children's Hospital Boston and Dana-Farber Cancer Institute, Harvard Medical School, 300 Longwood Ave, Karp 08125.3, Boston, MA 02115; e-mail: DAWilliams@childrens.harvard.edu.
References
- 1.Purton LE, Scadden DT. Limiting factors in murine hematopoietic stem cell assays. Cell Stem Cell. 2007;1(3):263–270. doi: 10.1016/j.stem.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Shizuru JA, Negrin RS, Weissman IL. Hematopoietic stem and progenitor cells: clinical and preclinical regeneration of the hematolymphoid system. Annu Rev Med. 2005;56:509–538. doi: 10.1146/annurev.med.54.101601.152334. [DOI] [PubMed] [Google Scholar]
- 3.Laird DJ, von Andrian UH, Wagers AJ. Stem cell trafficking in tissue development, growth, and disease. Cell. 2008;132(4):612–630. doi: 10.1016/j.cell.2008.01.041. [DOI] [PubMed] [Google Scholar]
- 4.Akala OO, Clarke MF. Hematopoietic stem cell self-renewal. Curr Opin Genet Dev. 2006;16(5):496–501. doi: 10.1016/j.gde.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9(2):115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 6.Harrison DE, Zsebo KM, Astle CM. Splenic primitive hematopoietic stem cell (PHSC) activity is enhanced by steel factor because of PHSC proliferation. Blood. 1994;83(11):3146–3151. [PubMed] [Google Scholar]
- 7.Kimura S, Roberts AW, Metcalf D, Alexander WS. Hematopoietic stem cell deficiencies in mice lacking c-Mpl, the receptor for thrombopoietin. Proc Natl Acad Sci U S A. 1998;95(3):1195–1200. doi: 10.1073/pnas.95.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsunaga T, Kato T, Miyazaki H, Ogawa M. Thrombopoietin promotes the survival of murine hematopoietic long-term reconstituting cells: comparison with the effects of FLT3/FLK-2 ligand and interleukin-6. Blood. 1998;92(2):452–461. [PubMed] [Google Scholar]
- 9.Arai F, Hirao A, Ohmura M, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118(2):149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Yamazaki S, Iwama A, Takayanagi S, et al. Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. EMBO J. 2006;25(15):3515–3523. doi: 10.1038/sj.emboj.7601236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blank U, Karlsson G, Karlsson S. Signaling pathways governing stem-cell fate. Blood. 2008;111(2):492–503. doi: 10.1182/blood-2007-07-075168. [DOI] [PubMed] [Google Scholar]
- 12.Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2(5):484–496. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galan-Caridad JM, Harel S, Arenzana TL, et al. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129(2):345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamminga LM, Bystrykh LV, de Boer A, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107(5):2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tothova Z, Kollipara R, Huntly BJ, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Hall A. G proteins and small GTPases: distant relatives keep in touch. Science. 1998;280:2074–2075. doi: 10.1126/science.280.5372.2074. [DOI] [PubMed] [Google Scholar]
- 17.Williams DA, Zheng Y, Cancelas JA. Rho GTPases and regulation of hematopoietic stem cell localization. Methods Enzymol. 2008;439:365–393. doi: 10.1016/S0076-6879(07)00427-2. [DOI] [PubMed] [Google Scholar]
- 18.Cancelas JA, Lee AW, Prabhakar R, Stringer KF, Zheng Y, Williams DA. Rac GTPases differentially integrate signals regulating hematopoietic stem cell localization. Nat Med. 2005;11(8):886–891. doi: 10.1038/nm1274. [DOI] [PubMed] [Google Scholar]
- 19.Ghiaur G, Lee A, Bailey J, Cancelas JA, Zheng Y, Williams DA. Inhibition of RhoA GTPase activity enhances hematopoietic stem and progenitor cell proliferation and engraftment. Blood. 2006;108(6):2087–2094. doi: 10.1182/blood-2006-02-001560. [DOI] [PubMed] [Google Scholar]
- 20.Gu Y, Filippi MD, Cancelas JA, et al. Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science. 2003;302(5644):445–449. doi: 10.1126/science.1088485. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Yang L, Filippi MD, Williams DA, Zheng Y. Genetic deletion of Cdc42GAP reveals a role of Cdc42 in erythropoiesis and hematopoietic stem/progenitor cell survival, adhesion, and engraftment. Blood. 2006;107(38):98–105. doi: 10.1182/blood-2005-05-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang FC, Atkinson SJ, Gu Y, et al. Rac and Cdc42 GTPases control hematopoietic stem cell shape, adhesion, migration, and mobilization. Proc Natl Acad Sci U S A. 2001;98(10):5614–5618. doi: 10.1073/pnas.101546898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L, Wang L, Geiger H, Cancelas JA, Mo J, Zheng Y. Rho GTPase Cdc42 coordinates hematopoietic stem cell quiescence and niche interaction in the bone marrow. Proc Natl Acad Sci U S A. 2007;104(12):5091–5096. doi: 10.1073/pnas.0610819104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99(2):67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 25.Hirose K, Kawashima T, Iwamoto I, Nosaka T, Kitamura T. MgcRacGAP is involved in cytokinesis through associating with mitotic spindle and midbody. J Biol Chem. 2001;276(8):5821–5828. doi: 10.1074/jbc.M007252200. [DOI] [PubMed] [Google Scholar]
- 26.Matheson SF, Hu KQ, Brouns MR, Sordella R, VanderHeide JD, Settleman J. Distinct but overlapping functions for the closely related p190 RhoGAPs in neural development. Dev Neurosci. 2006;28(6):538–550. doi: 10.1159/000095116. [DOI] [PubMed] [Google Scholar]
- 27.Burbelo PD, Miyamoto S, Utani A, et al. p190-B, a new member of the Rho GAP family, and Rho are induced to cluster after integrin cross-linking. J Biol Chem. 1995;270(52):30919–30926. doi: 10.1074/jbc.270.52.30919. [DOI] [PubMed] [Google Scholar]
- 28.Sordella R, Classon M, Hu KQ, et al. Modulation of CREB activity by the Rho GTPase regulates cell and organism size during mouse embryonic development. Dev Cell. 2002;2(2):553–565. doi: 10.1016/s1534-5807(02)00162-4. [DOI] [PubMed] [Google Scholar]
- 29.Sordella R, Jiang W, Chen GC, Curto M, Settleman J. Modulation of Rho GTPase signaling regulates a switch between adipogenesis and myogenesis. Cell. 2003;113(2):147–158. doi: 10.1016/s0092-8674(03)00271-x. [DOI] [PubMed] [Google Scholar]
- 30.Chakravarty G, Hadsell D, Buitrago W, Settleman J, Rosen JM. p190-B RhoGAP regulates mammary ductal morphogenesis. Mol Endocrinol. 2003;17(6):1054–1065. doi: 10.1210/me.2002-0428. [DOI] [PubMed] [Google Scholar]
- 31.Geiger H, True JM, Grimes B, Carroll EJ, Fleischman RA, Van Zant G. Analysis of the hematopoietic potential of muscle-derived cells in mice. Blood. 2002;100(2):721–723. doi: 10.1182/blood.v100.2.721. [DOI] [PubMed] [Google Scholar]
- 32.Rebel VI, Miller CL, Eaves CJ, Lansdorp PM. The repopulation potential of fetal liver hematopoietic stem cells in mice exceeds that of their liver adult bone marrow counterparts. Blood. 1996;87(8):3500–3507. [PubMed] [Google Scholar]
- 33.de Haan G, Nijhof W, Van Zant G. Mouse strain-dependent changes in frequency and proliferation of hematopoietic stem cells during aging: correlation between lifespan and cycling activity. Blood. 1997;89(5):1543–1550. [PubMed] [Google Scholar]
- 34.Yuan R, Astle CM, Chen J, Harrison DE. Genetic regulation of hematopoietic stem cell exhaustion during development and growth. Exp Hematol. 2005;33(2):243–250. doi: 10.1016/j.exphem.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 35.Lapidot T, Dar A, Kollet O. How do stem cells find their way home? Blood. 2005;106(6):1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- 36.Peled A, Grabovsky V, Habler L, et al. The chemokine SDF-1 stimulates integrin-mediated arrest of CD34(+) cells on vascular endothelium under shear flow. J Clin Invest. 1999;104(9):1199–1211. doi: 10.1172/JCI7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potocnik AJ, Brakebusch C, Fassler R. Fetal and adult hematopoietic stem cells require beta1 integrin function for colonizing fetal liver, spleen, and bone marrow. Immunity. 2000;12(6):653–663. doi: 10.1016/s1074-7613(00)80216-2. [DOI] [PubMed] [Google Scholar]
- 38.Williams DA, Rios M, Stephens C, Patel VP. Fibronectin and VLA-4 in haematopoietic stem cell-microenvironment interactions. Nature. 1991;352(6334):438–441. doi: 10.1038/352438a0. [DOI] [PubMed] [Google Scholar]
- 39.Cheng T, Rodrigues N, Shen H, et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287(5459):1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 40.Hock H, Hamblen MJ, Rooke HM, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431(7011):1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 41.Yilmaz OH, Valdez R, Theisen BK, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441(7092):475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 42.Yeager AM, Levin J, Levin FC. The effects of 5-fluorouracil on hematopoiesis: studies of murine megakaryocyte-CFC, granulocyte-macrophage-CFC, and peripheral blood cell levels. Exp Hematol. 1983;11(10):944–952. [PubMed] [Google Scholar]
- 43.Randall TD, Weissman IL. Phenotypic and functional changes induced at the clonal level in hematopoietic stem cells after 5-fluorouracil treatment. Blood. 1997;89(10):3596–3606. [PubMed] [Google Scholar]
- 44.Iwama A, Oguro H, Negishi M, et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21(6):843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Kent DG, Dykstra BJ, Cheyne J, Ma E, Eaves CJ. Steel factor coordinately regulates the molecular signature and biologic function of hematopoietic stem cells. Blood. 2008;112(3):560–567. doi: 10.1182/blood-2007-10-117820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito K, Hirao A, Arai F, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12(4):446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 47.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7(9):667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 48.Oguro H, Iwama A, Morita Y, Kamijo T, van Lohuizen M, Nakauchi H. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J Exp Med. 2006;203(10):2247–2253. doi: 10.1084/jem.20052477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng T, Rodrigues N, Dombkowski D, Stier S, Scadden DT. Stem cell repopulation efficiency but not pool size is governed by p27(kip1). Nat Med. 2000;6(11):1235–1240. doi: 10.1038/81335. [DOI] [PubMed] [Google Scholar]
- 50.Habibian HK, Peters SO, Hsieh CC, et al. The fluctuating phenotype of the lymphohematopoietic stem cell with cell cycle transit. J Exp Med. 1998;188(2):393–398. doi: 10.1084/jem.188.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nygren JM, Bryder D, Jacobsen SE. Prolonged cell cycle transit is a defining and developmentally conserved hemopoietic stem cell property. J Immunol. 2006;177(1):201–208. doi: 10.4049/jimmunol.177.1.201. [DOI] [PubMed] [Google Scholar]
- 52.Janzen V, Forkert R, Fleming HE, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443(7110):421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 53.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 54.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132(4):598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kieusseian A, Chagraoui J, Kerdudo C, et al. Expression of Pitx2 in stromal cells is required for normal hematopoiesis. Blood. 2006;107(2):492–500. doi: 10.1182/blood-2005-02-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reimold AM, Etkin A, Clauss I, et al. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14(2):152–157. [PMC free article] [PubMed] [Google Scholar]
- 57.Mikkola HK, Orkin SH. The journey of developing hematopoietic stem cells. Development. 2006;133(19):3733–3744. doi: 10.1242/dev.02568. [DOI] [PubMed] [Google Scholar]
- 58.Traver D, Miyamoto T, Christensen J, Iwasaki-Arai J, Akashi K, Weissman IL. Fetal liver myelopoiesis occurs through distinct, prospectively isolatable progenitor subsets. Blood. 2001;98(3):627–635. doi: 10.1182/blood.v98.3.627. [DOI] [PubMed] [Google Scholar]
- 59.Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13(1):13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.