Abstract
The tubby mouse is characterized by progressive retinal and cochlear degeneration and late-onset obesity. These phenotypes are caused by a loss-of-function mutation in the tub gene and are shared with several human syndromes, suggesting the importance of tubby protein in central nervous system (CNS) functioning. Although evidence suggests that tubby may act as a transcription factor mediating G-protein coupled receptor (GPCR) signaling, any downstream gene regulated by tubby has yet to be identified. To explore potential target genes of tubby with region-specific transcription patterns in the brain, we performed a microarray analysis using the cerebral cortex and hypothalamus of tubby mice. We also validated the changes of gene expression level observed with the microarray analysis using real-time RT-PCR. We found that expression of erythroid differentiation factor 1 (Erdr1) and caspase 1 (Casp1) increased, while p21-activated kinase 1 (Pak1) and cholecystokinin 2 receptor (Cck2r) expression decreased in the cerebral cortex of tubby mice. In the hypothalamic region, Casp 1 was up-regulated and µ-crystallin (CRYM) was down-regulated. Based on the reported functions of the differentially expressed genes, these individual or grouped genes may account for the phenotype of tubby mice. We discussed how altered expression of genes in tubby mice might be understood as the underlying mechanism behind tubby phenotypes.
Keywords: Tubby, Microarray, Gene expression, Cerebral cortex, Hypothalamus
INTRODUCTION
The tubby mouse, which arose from spontaneous mutation of the tub gene in the Jackson Laboratory, shows characteristic phenotypes including progressive retinal and cochlear degeneration, and late-onset obesity with insulin resistance due to a loss-of-function mutation in tub (Coleman and Eicher, 1990; Ohlemiller et al., 1995; Stubdal et al., 2000). Interestingly, the tubby mouse shares its combination of tubby phenotypes with several human syndromes such as Usher's syndrome (progressive neurosensory deficits) and Bardet Biedl syndrome (late-onset obesity and progressive retinal degeneration) (Katsanis et al., 2001; Kremer et al., 2006). Despite the importance of tubby protein, its functions and regulatory mechanisms are only just now beginning to be unraveled at the molecular level.
To date, several clues have suggested possible biochemical and physiological functions of tubby protein. Tubby is widely expressed in both the cochlea and retina as well as throughout the brain, including the hypothalamus, a brain region responsible for energy metabolism and possibly related to the obesity phenotype (Kleyn et al., 1996; Ikeda et al., 1999; Ikeda et al., 2002; Schwartz and Porte, 2005). At the cellular level, tubby is localized to the plasma membrane by the interaction of its carboxy-terminal domain with phosphorylated inositol lipids in the basal state (Santagata et al., 2001). Upon activation of the G-protein coupled receptor (GPCR) linked to the Gαq protein, phospholipase Cβ (PLCβ) is activated by Gαq, and then activated PLCβ hydrolyzes PIP2 into IP3, releasing tubby from the membrane and inducing its translocation into the nucleus (Santagata et al., 2001). In addition, the amino-terminal domain of tubby is able to activate transcription and the carboxy-terminus can bind to double-stranded DNA, suggesting that tubby might function as a transcription factor (Boggon et al., 1999). Nonetheless, no genes regulated by tubby have been identified so far.
In the present study, we examined gene expression patterns in the cerebral cortex and hypothalamus of tubby mice using microarray analysis to find genes that might be differentially expressed and related to tubby phenotypes. We employed real-time RT-PCR to validate candidate genes found in the microarray analysis. Ultimately, we identified multiple genes that were up- or down-regulated in tubby mice, reflecting their potential involvement in expressing tubby phenotypes.
METHODS
Cerebral cortex and hypothalamus collection in tubby mice
Two female and one male B6.Cg-Tubtub/J mice (the Jackson Laboratory) at an age of 32 weeks were used. Wild-type male mice from the colony of the same age were used as control mice. All animals were cared for according to NIH guidelines for the use of animals in research. Mice were sacrificed by CO2 overdose and whole brains were removed from the skull. The cerebral cortex and hypothalamus were isolated according to the protocol (Zapala et al., 2005). Briefly, each mouse brain was placed ventral side up. The hypothalamus was gently pinched out using curved forceps, leaving a small depression in the middle of the ventral side. To isolate the cerebral cortex, each mouse brain was placed dorsal side up and cut sagittally along the midline, leaving the cerebellum. The cerebral cortex was spread apart from the midbrain using two forceps. A portion of cerebral cortex was cut away with a scalpel blade. The isolated hypothalamus and cerebral cortex were immediately used for RNA preparation.
Microarray analysis
Total RNA was prepared from the cerebral cortex and hypothalamus of tubby and control mice using the RNeasy lipid tissue mini-kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. RNA samples were processed for hybridization to the microarray chip and data were acquired according to Illumina BeadStation 500X manuals. Briefly, biotinylated cRNAs were prepared using the Illumina Amplification Kit (Ambion, Austin, USA) and hybridized to the Sentrix Mouse Ref-8 v.2 Expression BeadChip (>24,000 probes) (Illumina, San Diego, CA, USA) for 16~18 h at 58℃, according to the manufacturer's instructions. Hybridization signals on the chip were analyzed by an Illumina bead array confocal scanner and Illumina Beadstudio v3.1.3 software (Gene Expression Module v3.3.8). The quality of hybridization and overall chip performance were monitored by visual inspection of both internal quality control checks and the raw scanned data. Raw data were transformed by logarithm and normalized by the Quantile Normalization method to remove systematic bias. Data were statistically analyzed by Avadis Prophetic version.3.3 (Strand Genomics, Bangalore, India) and presented as the mean of fold change. Genes with ≥2 or ≤-2 fold change in at least two independent comparisons among a total of three comparisons were identified. The fold change was calculated by dividing the gene expression level of the mutant by that of the wild-type. For the easy recognition of down-regulated genes in tubby mice, the negative reciprocals were listed when values were less than one (e.g., 0.5 was reported as -2). Among genes displaying sufficient fold changes, those relevant to tubby phenotypes were selected and annotated using gene symbols and definitions extracted from GenBank records.
Real-time quantitative reverse transcriptase-polymerase chain reaction (RT-PCR)
cDNA was synthesized by reverse transcription from 2 µg of extracted RNA to a final volume of 100 µl using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The amount of cDNA was measured by real-time RT-PCR, as described previously (Lee et al., 2007). Briefly, 20 µl PCR reactions were set up with 2 µl of synthesized cDNA and 10 µl of SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The final concentration of each primer (Table 1) was 250 nM. PCR was performed using an Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using the following cycle conditions: 10 min at 95℃, 40 cycles of 20 sec at 95℃ and 40 sec at 60℃, and 30 sec at 70℃. Triplicate real-time PCR reactions were executed for each sample and the obtained Ct values were averaged. According to the comparative Ct method, the expression levels of genes were presented as a fold change. Results were reported as mean±standard error of the mean (S.E.M). The values were normalized to β-actin level. Data were analyzed with Student's t-test, and significance was defined as p<0.05.
Table 1.
Sequences of primers used in real-time RT-PCR
RESULTS
Microarray analysis of gene expression in the cerebral cortex and hypothalamus of tubby mice
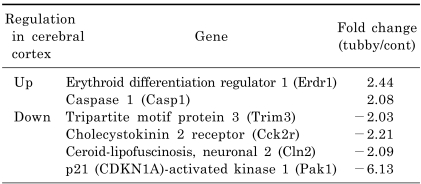
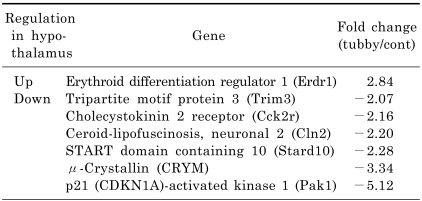
We dissected the cerebral cortex and hypothalamus from the brain of three age-matched pairs of tubby (two female and one male tubby mice) and control mice (all male control mice) at the age of 32 weeks. In order to compare the transcript profiling of these regions, total RNA was isolated and processed for microarray analysis. Processed cRNAs were hybridized to Illumina chips containing over 24,000 probes of mouse genes. Hybridization signals were analyzed and the changes in gene expression of each tubby mouse over its control mouse were presented as the mean of fold change. We screened over 18,000 genes and found 13 genes with ≥2-fold change in at least two independent experiments. Among them, we selected eight genes of interest that appeared to be related to tubby phenotypes. Five genes of interests, namely erythroid differentiation regulator 1 (Erdr1), tripartite motif protein 3 (Trim 3), cholecystokinin 2 receptor (Cck2r), ceroid-lipofuscinosis neuronal 2 (Cln2), and p21-activated kinase 1 (Pak1), were differentially expressed in both the cerebral cortex and hypothalamus of tubby mice (Table 2 and 3). In contrast, one gene, caspase 1 (Casp1), was up-regulated only in the cerebral cortex of tubby mice (Table 2) and two genes, TART domain containing 10 (Stard10) and µ-crystallin (CRYM), were down-regulated only in the hypothalamus (Table 3). Quantitatively, Erdr1 and Casp1 were up-regulated 2.44 and 2.08 fold, respectively, and Trim 3, Cck2r, Cln2, and Pak1 were down-regulated 2.03, 2.21, 2.09, and 6.13 fold, respectively, in the cerebral cortex of tubby mice compared to control mice (Table 2). In the hypothalamus of tubby mice, Erdr1 was up-regulated 2.84 fold and Trim3, Cck2r, Cln2, Stard10, CRYM, and Pak1 were down-regulated 2.07, 2.16, 2.20, 2.28, 3.34, and 5.12 fold, respectively, compared to control mice (Table 3).
Table 2.
A selection of genes up- or down-regulated in the cerebral cortex of tubby mice compared to control mice. Gene profiling was performed using microarray analysis. Expression level is presented as the mean of fold change (n=3). Negative fold change values indicate decreased gene expression in tubby mice
Table 3.
A selection of genes up- or down-regulated in the hypothalamus of tubby mice compared to control mice. Gene profiling was performed using microarray analysis. Expression level is presented as the mean of fold change (n=3). Negative fold change values indicate decreased gene expression in tubby mice
Validation of microarray data using real-time RT-PCR
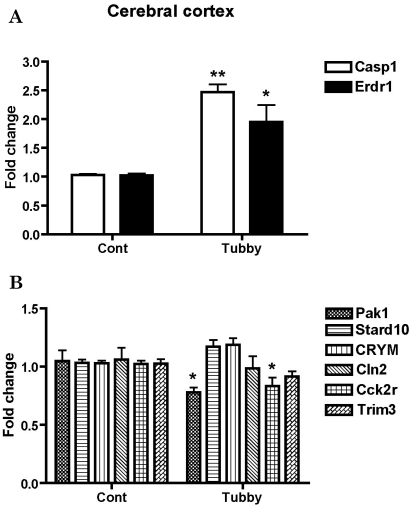
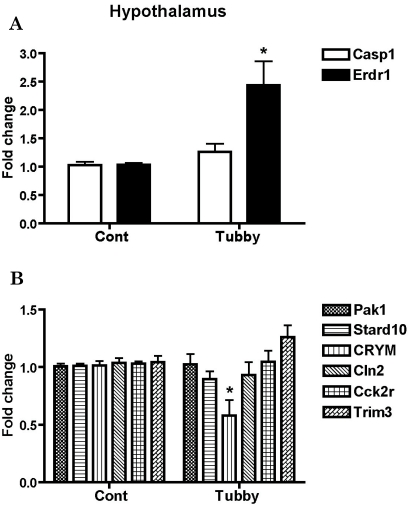
To validate the reliability of the results obtained from the microarray analysis, we performed real-time RT-PCR. cDNAs were synthesized from cerebral cortical and hypothalamic RNAs used in the microarray analysis. We designed PCR primers covering all candidate genes (Table 1) and performed real-time RT-PCR for all of the genes of interest using cDNAs from both the cerebral cortex and the hypothalamus of tubby mice to confirm region-specific expression patterns. The changes in mRNA levels were presented as a fold change. For the easy recognition of differentially regulated genes, genes were grouped into up-regulated (upper panel) and down-regulated (lower panel) genes primarily based on the findings in the cerebral cortex (Fig. 1 and 2). We confirmed that Casp1 and Erdr1 were up-regulated 2.47 and 1.95 fold, respectively, and Pak1 and Cck2r were down-regulated -1.28 and -1.20 fold, respectively, (28% and 17% lower than the control) in the cerebral cortex (Fig. 1). In addition, the expression of Erdr1 increased 2.43 fold, while CRYM expression decreased -1.72 fold (42% lower than the control) in the hypothalamus (Fig. 2). However, Stard10, CRYM, Cln2, and Trim3 did not show significant alteration of their RNA transcriptions in the cerebral cortex of tubby mice (Fig. 1). In the hypothalamus, expression levels of Casp1, Pak1, Stard10, Cln2, Cck2r, and Trim3 were not significantly changed (Fig. 2).
Fig. 1.
Validation of array-based gene expression profiles in the cerebral cortex of tubby mice using real-time RT-PCR. mRNA levels of genes were determined using real-time RT-PCR. Expression profiles of up-regulated (A) and down-regulated genes (B) are presented as fold changes compared to the cerebral cortex of control mice. Expression levels of β-actin were used to normalize the values. *p<0.05 and **p<0.01.
Fig. 2.
Validation of array-based gene expression profiles in the hypothalamus of tubby mice using real-time RT-PCR. mRNA levels of genes were determined using real-time RT-PCR. Hypothalamic Casp1 and Erdr1 expressions (A), the up-regulated genes in the cortex, and the expression profiles of other down-regulated genes in the hypothalamus (B) are represented as fold changes compared to the hypothalmus of control mice. Expression levels of β-actin were used to normalize the values. *p<0.05.
DISCUSSION
To our knowledge, this is the first attempt to screen genome-wide gene expression in tubby mice. Using real-time RT-PCR to validate genes selected in the microarray analysis, we found that Erdr1, Casp1, Pak1, Cck2r, and CRYM were differentially expressed in tubby mice. Although additional candidate genes were identified in the microarray analysis, real-time PCR showed that several genes, such as Trim3, Cln2, and Stard10, were false-positives. This discrepancy between microarray and real-time PCR data might be due to the low specificity of microarray analysis caused by less optimal bioinformatics and algorithms, indicating that real-time PCR remains the gold standard for gene expression measurement (Mackay et al., 2002; Wong and Medrano, 2005; Wang et al., 2006). In addition, technical limitations of microarrays can be caused by narrow dynamic range, fast signal saturations, and cross-hybridization, resulting in a certain level of fold change repression where fold changes of pair-wise tissues in microarrays are usually more depressed than those in real-time PCR (Wang et al., 2006). This limitation of microarrays could explain why we did not observe the reductions in pro-opiomelanocortin (POMC) and neuropeptide Y (NPY) expression in tubby mice previously identified by in situ hybridization (Guan et al., 1998). Although limitations clearly exist in microarray analysis, combining it with real-time PCR validation enabled us to overcome some of the shortcomings and more accurately measure changes in gene expression.
Erythroid differentiation regulator 1 (Erdr 1)
Among the up-regulated genes, strong increases in Erdr1 were confirmed by real-time RT-PCR (Fig. 1 and 2) in both the cerebral cortex and hypothalamus of tubby mice. Erdr1 is a secretory protein produced in many tissues, including the brain (Dormer et al., 2004b), that induces hemoglobin synthesis in erythroleukaemia cell lines and plays a role as a stress (e.g. heat, hydrogen peroxide, and trypsin etc)-related survival factor in haematopoietic progenitors (Dormer et al., 2004a; Dormer et al., 2004b). Erdr1 functions in other tissues remain unclear.
In tubby mice, vision and auditory deficits result from the progressive degeneration of sensory neurons in the retina and cochlea. The neuroprotective Wlds gene up-regulates the expression of an Erdr1-like gene in mice and human cells, indicating that Erdr1 might play a role in neuroprotection (Gillingwater et al., 2006). Further investigation is needed to examine how tubby might directly or indirectly regulate the increased expression of Erdr1 in response to neurodegenerating stress, although the response appears insufficient to protect the degeneration of certain types of neurons, particularly sensory neurons in the eye and ear, in tubby mice.
Caspase 1 (Casp1)
The expression of Casp1 was elevated only in the cerebral cortex of tubby mice. Casp1 is involved in processing pro-IL-1β, stimulating inflammatory processes (Martinon et al., 2002). It also mediates apoptosis of neuronal cells (Friedlander, 2000). In rats, Casp1 basal activity is higher in the hypothalamus than the frontal cortex, and administration of IL-1β-inducer 3,4-methylenedioxymethamphetamin (ecstasy) increases the activity of Casp1 in the frontal cortex, but not the hypothalamus (Orio et al., 2004; O'Shea et al., 2005), indicating that the difference between the cerebral cortex and hypothalamus might be a consequence of region-specific activation of different pathways leading to Casp1 expression. We also found that Casp1 was up-regulated only in the cerebral cortex of tubby mice, and not in the hypothalamus.
The inhibition of Casp1 slows the progression of various neurological disorders such as Huntington's disease and traumatic brain injury, and can also regulate the activity of caspase 3 (Casp3), another important mediator of apoptosis (Friedlander, 2000). The progressive death of photoreceptors in the retina and ensuing retinal degeneration is a typical phenotype of tubby mice (Ikeda et al., 1999; Stubdal et al., 2000). When Casp3 activity is inhibited, retinal degeneration in tubby mice is reduced (Bode and Wolfrum, 2003). In addition, light-induced retinal degeneration is mediated by a Casp1- and Casp3-dependent mechanism (Perche et al., 2007). Understanding how tubby regulates the expression of Casp1 could inform the exact mechanism of retinal degeneration in tubby mice.
p21-activated kinase 1 (Pak1)
Pak1, a serine/threonine protein kinase that mediates Rac1 and Cdc42 activity (Bokoch, 2003), has been proposed as an important regulator of neuronal polarity, cytoskeletal dynamics, and cell migration (Sells et al., 1997; Cau and Hall, 2005; Jacobs et al., 2007). Notably, Pak is required for regulating axon guidance and targeting of photoreceptors in Drosophila (Hing et al., 1999). In addition, Pak 1 has been implicated in the cell survival pathway through the direct phosphorylation of the death-promoting Bad protein, suggesting that Pak 1 could be neuroprotective by preventing the apoptosis of sensory neurons in control mice (Schurmann et al., 2000; Tang et al., 2000). Although the effect of tubby on axonal guidance or neuronal migration has not been studied so far, it would be intriguing to explore the role of tubby in regulating expression of Pak 1 and its possible involvement in the processes of neuronal migration or axonal outgrowth, as well as the degeneration of sensory neurons.
Cholecystokinin 2 receptor (Cck2r)
CCK is a peptide hormone in the gut and brain that plays various roles, including the regulation of feeding and digestion through CCK receptors subtype 1 (Cck1r) and 2 (Cck2r) (Crawley and Corwin, 1994). Cck1r is mostly localized to the gastrointestinal tract, while Cck2r is abundant in the brain (Hill et al., 1987a; Hill et al., 1987b). Obesity with hyperphagia and insulin resistance was recently reported in Cck2r deficient mice (Clerc et al., 2007). However, mechanisms of hyperphagia and obesity in Cck2r deficient mice are not fully elucidated. Therefore, we cannot exclude the possibility that Cck2r activity in brain regions other than hypothalamus, a traditional eating center, plays a regulatory role in hyperphagia and obesity of tubby mice, implying that the decrease of Cck2r expression in the cerebral cortex could be meaningful. For example, cholecystokinin induced satiety by interacting through Cck1r located in the hindbrain (Chandra and Liddle, 2007). On the other hand, the expression level of Cck2r is higher in the cerebral cortex than the hypothalamus (Gaudreau et al., 1983; Morency et al., 1994). Considering that the expression level of Cck2r mRNA was slightly decreased (about 17%) in the cerebral cortex of tubby mice (Fig. 1), it might be difficult to see the change in the expression level of Cck2r in the hypothalamus of tubby mice due to its low expression level and small number of samples in our experiment (Fig. 2). Tubby might have less effect on Cck2r expression than other candidate genes. However, the decreased expression of Cck2r in the cerebral cortex can be a clue to reveal an underlying mechanism of obesity in tubby mice. Further exploration of the exact mechanism by which tubby regulates Cck2r expression is needed.
µ-Crystallin (CRYM)
CRYM was down-regulated in the tubby mouse hypothalamus. CRYM is a nicotinamide adenine dinucleotide phosphate (NADPH)-dependent cytosolic 3,5,3'-triiodo-L-thyronine (T3) binding protein expressed in specific tissues including the brain, retina, and inner ear (Segovia et al., 1997; Abe et al., 2003; Suzuki et al., 2003). Although the exact functions and regulatory mechanisms of CRYM are not well understood, it has pivotal roles in organ development and cellular differentiation by affecting thyroid hormone concentration, which leads to the modification of thyroid hormone action (Suzuki et al., 2007). For example, families with hereditary nonsyndromic deafness carry a mutation of CRYM gene that prevents NADPH-dependent T3 binding to CRYM and causes severe deafness (Abe et al., 2003; Oshima et al., 2006). In addition, based on the critical role of thyroid hormone in retinal development (Roberts et al., 2006) and evidence that CRYM is associated with macular degeneration of the retina (Umeda et al., 2005), the observed down-regulation of CRYM expression in tubby mice suggests that CRYM is involved in the progression of retinal degeneration. Moreover, thyroid hormone is a positive regulator of tubby gene expression (Koritschoner et al., 2001). It could be that CRYM is down-regulated in the absence of tubby protein, leading to a reduction of thyroid hormone concentration in the cytoplasm. Accordingly, CRYM might be an important molecular link between thyroid hormone function and tubby-associated phenotypes, including hearing loss and visual dysfunction.
We used the cerebral cortex and hypothalamus of tubby mice for identifying target genes which can be regulated by tubby. Although we found several candidate genes in those brain tissues, some tubby phenotypes like progressive retinal degeneration and hearing loss are localized to the retina and cochlear tissue. Therefore, it will be interesting to further examine the local changes of candidate genes in the retina and cochlear tissues of tubby mice. Especially, Casp 1 and CRYM that seem to be related to the retinal and cochlear degeneration are worth examining their expression levels in the retinal and cochlear tissues of tubby mice.
In this study, we identified candidate target genes of tubby using microarray analysis and real-time RT-PCR. Based on their reported functions, the differentially expressed genes that we found are attractive target molecules that could be responsible for tubby mouse phenotypes such as obesity and retinal and cochlear degeneration. Combined with PCR validation, microarray analysis with transgenic mice represents a useful tool for identifying unknown downstream genes regulated by an interesting protein that induces special phenotypes. Our data provide deeper insight into the target genes of tubby protein and the pathogenesis of tubby phenotypes.
ACKNOWLEDGEMENTS
This work was supported by a Korea Research Foundation Grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF-2006-312-E00063) (to Y.S.A) and by the Korea Science and Engineering Foundation (KOSEF), funded by the Korean Government (MEST) (R11-2007-040-01006-0) (to C.H.K).
ABBREVIATIONS
- GPCR
G-protein coupled receptor
- Pak1
p21-activated kinase 1
- Erdr1
erythroid differentiation factor 1
- Casp1
caspase 1
- Cck2r
cholecystokinin 2 receptor
References
- 1.Abe S, Katagiri T, Saito-Hisaminato A, Usami S, Inoue Y, Tsunoda T, Nakamura Y. Identification of CRYM as a candidate responsible for nonsyndromic deafness, through cDNA microarray analysis of human cochlear and vestibular tissues. Am J Hum Genet. 2003;72:73–82. doi: 10.1086/345398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bode C, Wolfrum U. Caspase-3 inhibitor reduces apototic photoreceptor cell death during inherited retinal degeneration in tubby mice. Mol Vis. 2003;9:144–150. [PubMed] [Google Scholar]
- 3.Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 1999;286:2119–2125. doi: 10.1126/science.286.5447.2119. [DOI] [PubMed] [Google Scholar]
- 4.Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 5.Cau J, Hall A. Cdc42 controls the polarity of the actin and microtubule cytoskeletons through two distinct signal transduction pathways. J Cell Sci. 2005;118:2579–2587. doi: 10.1242/jcs.02385. [DOI] [PubMed] [Google Scholar]
- 6.Chandra R, Liddle RA. Cholecystokinin. Curr Opin Endocrinol Diabetes Obes. 2007;14:63–67. doi: 10.1097/MED.0b013e3280122850. [DOI] [PubMed] [Google Scholar]
- 7.Clerc P, Coll Constans MG, Lulka H, Broussaud S, Guigne C, Leung-Theung-Long S, Perrin C, Knauf C, Carpene C, Penicaud L, Seva C, Burcelin R, Valet P, Fourmy D, Dufresne M. Involvement of cholecystokinin 2 receptor in food intake regulation: hyperphagia and increased fat deposition in cholecystokinin 2 receptor-deficient mice. Endocrinology. 2007;148:1039–1049. doi: 10.1210/en.2006-1064. [DOI] [PubMed] [Google Scholar]
- 8.Coleman DL, Eicher EM. Fat (fat) and tubby (tub): two autosomal recessive mutations causing obesity syndromes in the mouse. J Hered. 1990;81:424–427. doi: 10.1093/oxfordjournals.jhered.a111019. [DOI] [PubMed] [Google Scholar]
- 9.Crawley JN, Corwin RL. Biological actions of cholecystokinin. Peptides. 1994;15:731–755. doi: 10.1016/0196-9781(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 10.Dormer P, Spitzer E, Moller W. EDR is a stress-related survival factor from stroma and other tissues acting on early haematopoietic progenitors (E-Mix) Cytokine. 2004a;27:47–57. doi: 10.1016/j.cyto.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Dormer P, Spitzer E, Frankenberger M, Kremmer E. Erythroid differentiation regulator (EDR), a novel, highly conserved factor I. Induction of haemoglobin synthesis in erythroleukaemic cells. Cytokine. 2004b;26:231–242. doi: 10.1016/j.cyto.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Friedlander R. Role of caspase 1 in neurologic disease. Arch Neurol. 2000;57:1273–1276. doi: 10.1001/archneur.57.9.1273. [DOI] [PubMed] [Google Scholar]
- 13.Gaudreau P, Quirion R, St-Pierre S, Pert CB. Characterization and visualization of cholecystokinin receptors in rat brain using [3H]pentagastrin. Peptides. 1983;4:755–762. doi: 10.1016/0196-9781(83)90032-3. [DOI] [PubMed] [Google Scholar]
- 14.Gillingwater TH, Wishart TM, Chen PE, Haley JE, Robertson K, MacDonald SH, Middleton S, Wawrowski K, Shipston MJ, Melmed S, Wyllie DJ, Skehel PA, Coleman MP, Ribchester RR. The neuroprotective WldS gene regulates expression of PTTG1 and erythroid differentiation regulator 1-like gene in mice and human cells. Hum Mol Genet. 2006;15:625–635. doi: 10.1093/hmg/ddi478. [DOI] [PubMed] [Google Scholar]
- 15.Guan XM, Yu H, Van der Ploeg LH. Evidence of altered hypothalamic pro-opiomelanocortin/neuropeptide Y mRNA expression in tubby mice. Brain Res Mol Brain Res. 1998;59:273–279. doi: 10.1016/s0169-328x(98)00150-8. [DOI] [PubMed] [Google Scholar]
- 16.Hill DR, Shaw TM, Woodruff GN. Species differences in the localization of 'peripheral' type cholecystokinin receptors in rodent brain. Neurosci Lett. 1987a;79:286–289. doi: 10.1016/0304-3940(87)90445-9. [DOI] [PubMed] [Google Scholar]
- 17.Hill DR, Campbell NJ, Shaw TM, Woodruff GN. Autoradiographic localization and biochemical characterization of peripheral type CCK receptors in rat CNS using highly selective nonpeptide CCK antagonists. J Neurosci. 1987b;7:2967–2976. doi: 10.1523/JNEUROSCI.07-09-02967.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hing H, Xiao J, Harden N, Lim L, Zipursky SL. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. 1999;97:853–863. doi: 10.1016/s0092-8674(00)80798-9. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda A, Naggert JK, Nishina PM. Genetic modification of retinal degeneration in tubby mice. Exp Eye Res. 2002;74:455–461. doi: 10.1006/exer.2001.1139. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda S, He W, Ikeda A, Naggert JK, North MA, Nishina PM. Cell-specific expression of tubby gene family members (tub, Tulp1,2, and 3) in the retina. Invest Ophthalmol Vis Sci. 1999;40:2706–2712. [PubMed] [Google Scholar]
- 21.Jacobs T, Causeret F, Nishimura YV, Terao M, Norman A, Hoshino M, Nikolic M. Localized activation of p21-activated kinase controls neuronal polarity and morphology. J Neurosci. 2007;27:8604–8615. doi: 10.1523/JNEUROSCI.0765-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katsanis N, Lupski JR, Beales PL. Exploring the molecular basis of Bardet-Biedl syndrome. Hum Mol Genet. 2001;10:2293–2299. doi: 10.1093/hmg/10.20.2293. [DOI] [PubMed] [Google Scholar]
- 23.Kleyn PW, Fan W, Kovats SG, Lee JJ, Pulido JC, Wu Y, Berkemeier LR, Misumi DJ, Holmgren L, Charlat O, Woolf EA, Tayber O, Brody T, Shu P, Hawkins F, Kennedy B, Baldini L, Ebeling C, Alperin GD, Deeds J, Lakey ND, Culpepper J, Chen H, Glücksmann-Kuis MA, Carlson GA, Duyk GM, Moore KJ. Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell. 1996;85:281–290. doi: 10.1016/s0092-8674(00)81104-6. [DOI] [PubMed] [Google Scholar]
- 24.Koritschoner N, Alvarez-Dolado M, Kurz S, Heikenwälder M, Hacker C, Vogel F, Muñoz A, Zenke M. Thyroid hormone regulates the obesity gene tub. EMBO Rep. 2001;2:499–504. doi: 10.1093/embo-reports/kve107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kremer H, van Wijk E, Marker T, Wolfrum U, Roepman R. Usher syndrome: molecular links of pathogenesis, proteins and pathways. Hum Mol Genet. 2006;15 Spec No 2:R262–R270. doi: 10.1093/hmg/ddl205. [DOI] [PubMed] [Google Scholar]
- 26.Lee JH, Lee J, Seo GH, Kim CH, Ahn YS. Heparin inhibits NF-kappaB activation and increases cell death in cerebral endothelial cells after oxygen-glucose deprivation. J Mol Neurosci. 2007;32:145–154. doi: 10.1007/s12031-007-0026-3. [DOI] [PubMed] [Google Scholar]
- 27.Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology. Nucleic Acids Res. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 29.Morency MA, Quirion R, Mishra RK. Distribution of cholecystokinin receptors in the bovine brain: a quantitative autoradiographic study. Neuroscience. 1994;62:307–316. doi: 10.1016/0306-4522(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 30.O'Shea E, Sanchez V, Orio L, Escobedo I, Green AR, Colado MI. 3,4-Methylenedioxymethamphetamine increases pro-interleukin-1beta production and caspase-1 protease activity in frontal cortex, but not in hypothalamus, of Dark Agouti rats: role of interleukin-1beta in neurotoxicity. Neuroscience. 2005;135:1095–1105. doi: 10.1016/j.neuroscience.2005.06.084. [DOI] [PubMed] [Google Scholar]
- 31.Ohlemiller KK, Hughes RM, Mosinger-Ogilvie J, Speck JD, Grosof DH, Silverman MS. Cochlear and retinal degeneration in the tubby mouse. Neuroreport. 1995;6:845–849. doi: 10.1097/00001756-199504190-00005. [DOI] [PubMed] [Google Scholar]
- 32.Orio L, O'Shea E, Sanchez V, Pradillo JM, Escobedo I, Camarero J, Moro MA, Green AR, Colado MI. 3,4-Methylenedioxymethamphetamine increases interleukin-1beta levels and activates microglia in rat brain: studies on the relationship with acute hyperthermia and 5-HT depletion. J Neurochem. 2004;89:1445–1453. doi: 10.1111/j.1471-4159.2004.02443.x. [DOI] [PubMed] [Google Scholar]
- 33.Oshima A, Suzuki S, Takumi Y, Hashizume K, Abe S, Usami S. CRYM mutations cause deafness through thyroid hormone binding properties in the fibrocytes of the cochlea. J Med Genet. 2006;43:e25. doi: 10.1136/jmg.2005.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perche O, Doly M, Ranchon-Cole I. Caspase-dependent apoptosis in light-induced retinal degeneration. Invest Ophthalmol Vis Sci. 2007;48:2753–2759. doi: 10.1167/iovs.06-1258. [DOI] [PubMed] [Google Scholar]
- 35.Roberts MR, Srinivas M, Forrest D, Morreale de Escobar G, Reh TA. Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc Natl Acad Sci U S A. 2006;103:6218–6223. doi: 10.1073/pnas.0509981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- 37.Schurmann A, Mooney AF, Sanders LC, Sells MA, Wang HG, Reed JC, Bokoch GM. p21-activated kinase 1 phosphorylates the death agonist bad and protects cells from apoptosis. Mol Cell Biol. 2000;20:453–461. doi: 10.1128/mcb.20.2.453-461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz MW, Porte D., Jr Diabetes, obesity, and the brain. Science. 2005;307:375–379. doi: 10.1126/science.1104344. [DOI] [PubMed] [Google Scholar]
- 39.Segovia L, Horwitz J, Gasser R, Wistow G. Two roles for mu-crystallin: a lens structural protein in diurnal marsupials and a possible enzyme in mammalian retinas. Mol Vis. 1997;3:9. [PubMed] [Google Scholar]
- 40.Sells MA, Knaus UG, Bagrodia S, Ambrose DM, Bokoch GM, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 41.Stubdal H, Lynch CA, Moriarty A, Fang Q, Chickering T, Deeds JD, Fairchild-Huntress V, Charlat O, Dunmore JH, Kleyn P, Huszar D, Kapeller R. Targeted deletion of the tub mouse obesity gene reveals that tubby is a loss-of-function mutation. Mol Cell Biol. 2000;20:878–882. doi: 10.1128/mcb.20.3.878-882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki S, Mori J, Hashizume K. Mu-crystallin, a NADPH-dependent T(3)-binding protein in cytosol. Trends Endocrinol Metab. 2007;18:286–289. doi: 10.1016/j.tem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki S, Mori J, Kobayashi M, Inagaki T, Inaba H, Komatsu A, Yamashita K, Takeda T, Miyamoto T, Ichikawa K, Hashizume K. Cell-specific expression of NADPH-dependent cytosolic 3,5,3'-triiodo-L-thyronine-binding protein (p38CTBP) Eur J Endocrinol. 2003;148:259–268. doi: 10.1530/eje.0.1480259. [DOI] [PubMed] [Google Scholar]
- 44.Tang Y, Zhou H, Chen A, Pittman RN, Field J. The Akt proto-oncogene links Ras to Pak and cell survival signals. J Biol Chem. 2000;275:9106–9109. doi: 10.1074/jbc.275.13.9106. [DOI] [PubMed] [Google Scholar]
- 45.Umeda S, Suzuki MT, Okamoto H, Ono F, Mizota A, Terao K, Yoshikawa Y, Tanaka Y, Iwata T. Molecular composition of drusen and possible involvement of anti-retinal autoimmunity in two different forms of macular degeneration in cynomolgus monkey (Macaca fascicularis) FASEB J. 2005;19:1683–1685. doi: 10.1096/fj.04-3525fje. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Barbacioru C, Hyland F, Xiao W, Hunkapiller KL, Blake J, Chan F, Gonzalez C, Zhang L, Samaha RR. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 2006;7:59. doi: 10.1186/1471-2164-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 48.Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, Tynan W, Broide RS, Helton R, Stoveken BS, Winrow C, Lockhart DJ, Reilly JF, Young WG, Bloom FE, Barlow C. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci U S A. 2005;102:10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]