Abstract
The mushroom Cordyceps militaris has been used for a long time in eastern Asia as a nutraceutical and in traditional Chinese medicine as a treatment for cancer patients. In the present study, a cytotoxic antifungal protease was purified from the dried fruiting bodies of C. militaris using anion-exchange chromatography on a DEAE-Sepharose column. Electrophoretic analyses indicated that this protein, designated C. militaris protein (CMP), has a molecular mass of 12 kDa and a pI of 5.1. The optimum conditions for protease activity were a temperature of 37℃ and pH of 7.0~9.0. The enzyme activity was specifically inhibited by the serine protease inhibitor phenylmethylsulfonyl fluoride. Amino acid composition of intact CMP and amino acid sequences of three major peptides from a tryptic digest of CMP were determined. CMP exerted strong antifungal effect against the growth of the fungus Fusarium oxysporum, and exhibited cytotoxicity against human breast and bladder cancer cells. These results indicate that C. militaris represents a source of a novel protein that might be applied in diverse biological and medicinal applications.
Keywords: Mushroom, Cordyceps militaris, Protease, Antifungal activity, Cytotoxicity
INTRODUCTION
Mushrooms contain numerous compounds that exhibit interesting biological activities, including antibacterial, antifungal, antiviral, antitumor, antiproliferative, immunomodulatory, lectin-like, protease, and nuclease activities (Ng 2004; Lindequist et al, 2005). The mushroom Cordyceps militaris is a species of entomogenous fungus that is parasitic on insects, attacking the underground pupae and larvae of lepidopteran insects. These fungi produce inhibitors of the phenoloxidase-activating systems of insects, and then they secrete various enzymes that facilitate the infection of parasites, which then proliferate within the insects (St Leger et al, 1987; Watanabe et al, 2006).
C. militaris has been used for a long time in eastern Asia as a nutraceutical and in traditional Chinese medicine as a treatment for cancer patients. The biological functions of its chemical constituents, including cordycepin (Cunningham et al, 1950), polysaccharide (Yu et al, 2004), and enzymes (Hattori et al, 2005; Kim et al, 2006) has been reported, and we recently purified a lectin from C. militaris that showed mitogenic activity against mouse splenocytes (Jung et al, 2007).
In the present study, we purified a novel protease from C. militaris, designated C. militaris protein (CMP), which exhibits antifungal activity and cytotoxicity against human cancer cells, and the molecular properties of CMP, including molecular mass, isoelectric point, number of subunits, amino acid composition, and partial amino acid sequence.
METHODS
Purification
Dried fruiting bodies (1 g) of C. militaris mushrooms were collected in Yangpyong, Korea, and homogenized and extracted overnight in 10 mM Tris-HCl (pH 8.0) at 4℃. The homogenate was dialyzed against the same buffer for 3 days, filtered using a 0.45 µm membrane filter, and subjected to centrifugation at 1,000 g for 30 min. The supernatant was then loaded onto an anion-exchange column (3×8 cm) of DEAE-Sepharose (Sigma-Aldrich, St Louis, MO, USA) that had been equilibrated in 20 mM Tris-HCl (pH 8.0), and the protein was eluted by stepwise increase of the NaCl concentration from 0 to 1.0 M in 10 mM Tris-HCl (pH 8.0). Eluted fractions (1 ml of each) were stored at 4℃. The protein concentration was determined by the method of Bradford (Bradford, 1976), using bovine serum albumin as the standard.
Electrophoretic analyses
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the method of Laemmli (Laemmli, 1970) using a 15% acrylamide slab gel in the presence and absence of 2-mercaptoethanol. The gel was stained with Coomassie Brilliant Blue R-250 or silver nitrate. Isoelectric focusing (IEF) was carried out on a gel using a 2% ampholyte IEF-ready gel (pH 3~10) according to the manufacturer's instructions (Bio-Rad, Hercules, CA, USA).
Gel-filtration chromatography
Gel-filtration chromatography was performed using a TSK-GEL G3000SW column (Tosoh, Tokyo, Japan) with a conventional high-performance liquid chromatography (HPLC) system (Shimadzu, Kyoto, Japan) equilibrated with 20 mM Tris-HCl (pH 8.0) containing 0.1 M NaCl at a flow rate of 0.5 ml/min, and monitored at 280 nm. Molecular mass markers (13.7~2,000 kDa) were used with gel-filtration calibration kit (Amersham Biosciences, Piscataway, NJ, USA).
Protease activity
Protease activity was measured using the EnzChek Protease Assay Kit (Invitrogen, Groningen, NL, USA) according to the manufacturer's instructions. This kit determines the rate of hydrolysis of casein labeled with pH-insensitive green-fluorescent BODIPY dye. CMP or bovine pancreatic trypsin (Sigma-Aldrich, St Louis, MO, USA) was incubated with 2 µM substrate at 37℃, and the appearance of hydrolyzed peptide labeled with fluorescent BODIPY dye was measured using a microplate reader (GENios-Pro, Tecan, Durham, NC, USA) with excitation at 492 nm and emission at 535 nm. Protease activity was expressed as the fluorescence change per minute, and the detection limit was defined as the amount of enzyme required to cause a 10% change in fluorescence compared to the control sample. The specific activity was expressed as the ratio of protease activity per protein (mg/ml). The kinetics parameters of the hydrolysis reaction were determined from Lineweaver-Burk plots.
Optimal pH and temperature and effect of inhibitors
CMP dissolved in neutral buffer was dialyzed first overnight against Milli-Q at 4℃, and then against buffers with pH values ranging from 4.0 to 11.0. The buffers contained 0.1 µM NaCl and 20 mM citric acid for pH 4.0, 5.0, and 6.0; 20 mM Tris-HCl for pH 7.0, 8.0, and 9.0; or 20 mM NaHCO3-NaOH for pH 10.0 and 11.0. The dialyzed CMP was then assayed for protease activity. In the neutral condition, protease activity was measured after incubating CMP for 1 h at 4, 25, 37, 45, 55, and 65℃. Phenylmethylsulfonyl fluoride (PMSF, in 1% dimethyl sulfoxide, DMSO), ethylenediaminetetraacetic acid (EDTA), and pepstatin A (in 1% DMSO) were used as protease inhibitors. Control samples contained either no protease inhibitors or 1% DMSO.
Amino acid composition
CMP (30 µg) was hydrolyzed in vacuo with 6 N HCl for 24 h at 110℃. For the analysis of cysteic acid and Trp, oxidized CMP (30 µg) treated with performic acid and hydrolyzed CMP (30 µg) with methanesulfonic acid were prepared, respectively. PITC-amino acids were analyzed by HPLC (Waters, Milford, MA, USA) with Nova-Pak C18 column (3.93×300 mm).
Amino acid sequence
CMP (20 µg) was applied to a 15% SDS-PAGE gel, electroblotted onto a PVDF membrane (Bio-Rad, Hercules, CA, USA), and analyzed using an Automatic Protein Sequencer (476A-01-120, Applied Biosystems, Foster city, CA, USA). In order to obtain the amino acid sequence, CMP was subjected to in-gel digestion with trypsin (Promega, Madison, WI, USA), and the digestion products were desalted and concentrated. MS/MS of peptides generated by the in-gel digestion was performed by nano-ESI on a mass spectrometer (Q-TOF2, Micromass, Milford, MA, USA). Product ions were analyzed using an orthogonal TOF analyzer fitted with a reflector, a microchannel plate detector, and a time-to-digital converter. The data were processed using a PC running Mass Lynx software on the Windows NT environment. Sequence homologues were searched for using BLAST (www.ncbi.nlm.nih.gov/BLAST.cgi).
Antifungal activity
Standardized fungal strains Fusarium oxysporum and Botrytis cinerea were purchased from the ATCC (American Type Culture Collection, Rockville, MD, USA). The antifungal activity was assessed as described previously (Moreno et al, 2003). Briefly, antifungal activity was measured under sterile conditions using Petri dishes containing 15 ml of potato dextrose agar (39 g/l). The mycelium of the fungi was inoculated in the middle of the plate. After 3~4 days, the discs were wetted with serial fivefold dilutions of CMP, after which the dishes were incubated at 37℃ for another 24~48 h.
Cytotoxicity against human cancer cells
The human cell lines MCF-7 (breast cancer), 5637 (bladder cancer), and A-549 (lung cancer) were purchased from the KCLB (Korean Cell Line Bank, Seoul, Korea). The cells were incubated at 6×104 cells/0.1 ml/well with 100 µl of serial threefold dilutions of CMP in 96-well culture plates (Costar, Cambridge, MA, USA) at 37℃ for 72 h in a humidified atmosphere of 5% CO2. The percentage viability of cancer cells was determined by performing a CCK assay (Dojindo, Kumamoto, Japan) according to a modified version of Mosmann's method (Mosmann, 1983) and calculated as described previously (Kim et al, 2004).
RESULTS
Purification and molecular characterization
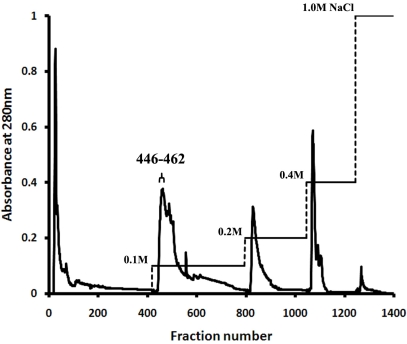
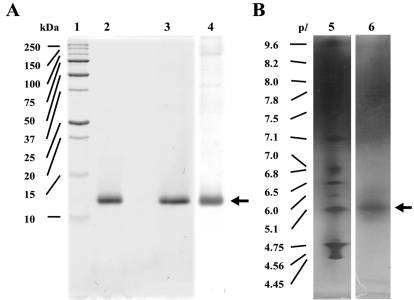
Crude protein extract from C. militaris was subjected directly to anion-exchange chromatography on a DEAE-Sepharose column. After washing the unbound fractions, stepwise elution with a buffer containing NaCl produced five major peaks (Fig. 1). SDS-PAGE analysis on a 15% gel revealed that fractions 446~462 eluted at 0.1 M NaCl contained CMP. These fractions showed a single band corresponding to a molecular mass of 12 kDa under both non-reducing (lane 2, 4) and reducing conditions (lane 3), regardless of whether the gel was stained with Coomassie Brilliant Blue R-250 (lane 2, 3) or silver nitrate (lane 4) (Fig. 2A). In IEF gel electrophoresis, the protein was focused into a single band with an estimated pI of 5.1 (lane 6) (Fig. 2B). The gel-filtration chromatography results indicate that CMP elutes as a single peak with a molecular mass of 25,006.7 Da for the native molecule, as calibrated with molecular mass markers from 13.7 to 2000 kDa (data not shown). The procedure resulted in a 17.4-fold purification of the protein; the yield was 3.4%, and the specific activity of the CMP was 652.0 (Table 1).
Fig. 1.
Elution profile of C. militaris protein extract applied to DEAE-Sepharose anion-exchange column chromatography, showing absorbance at 280 nm and NaCl concentration gradient.
Fig. 2.
(A) SDS-PAGE and (B) IEF gel of purified CMP. Lane 1, standard protein markers; lane 2, CMP without 2-mercaptoethanol; lane 3, CMP with 2-mercaptoethanol; lane 4, CMP stained with silver nitrate; lane 5, standard pI markers; lane 6, CMP.
Table 1.
Summary of CMP purification from C. militaris
aProtease activity is expressed as the fluorescence change per minute. bSpecific activity is expressed as the ratio of protease activity per protein (mg/ml).
Protease activity
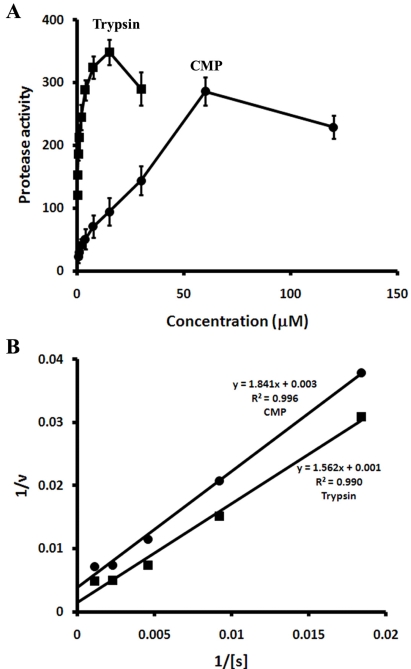
In the protease activity assay used in this study, the activity was proportional to the amount of hydrolyzed peptide resulting from hydrolysis of a casein substrate labeled with a pH-insensitive green-fluorescent BODIPY dye. The level of protease activity, quantified as the fluorescence change per minute, was proportional to the concentration of CMP, similar to the results obtained with trypsin (Fig. 3A). The detection limits of the protease assay were 0.5 and 0.1 µM for CMP and trypsin, respectively. The kinetics properties of CMP and trypsin were investigated by varying the concentration of casein from 1.7 to 870 µM. Substrate saturation curves for casein indicated that both CMP and trypsin followed simple Michaelis-Menten kinetics (Fig. 3B inset). The kinetics parameters of CMP and trypsin for casein calculated from the Lineweaver-Burk plots (Fig. 3B) were Km = 0.61 mM and Vmax/Km = 0.54, and Km = 1.56 mM and Vmax/Km = 0.64, respectively.
Fig. 3.
(A) Comparison of the protease activities of CMP (●) and trypsin (■) using casein as a substrate. Protease activity is expressed as the fluorescence change per minute. (B) Lineweaver-Burk plots of casein hydrolysis by CMP (●) and trypsin (■).
Optimal pH and temperature and effect of inhibitors on the protease activity
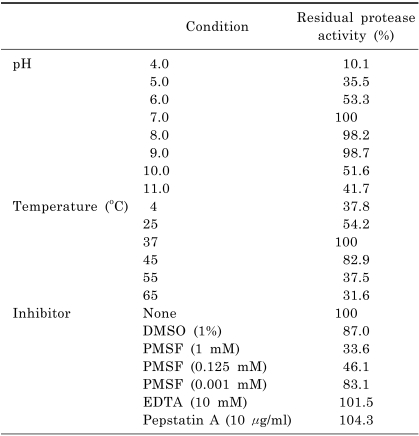
To determine the optimal reaction conditions for CMP, its protease activity was investigated over the temperature range of 4~65℃ and the pH range of 4.0~11.0 (Table 2), which revealed that the optimal conditions were a temperature of 37℃ and a pH of 7.0~9.0. The activity was reduced by nearly 50% at 25℃ and at pH values of 6.0, 10.0, and 11.0, and declined rapidly at temperatures below 4℃ or above 55℃, and for pH 4.0, and 5.0.
Table 2.
Effects of pH, temperature, and inhibitors on the protease activity of CMP
PMSF, which is a serine protease inhibitor, was the only effective inhibitor of protease activity. PMSF at 1 mM reduced CMP activity to 33.6% of the control, and the inhibition was proportional to the concentration of PMSF (Table 2). In contrast, neither EDTA nor pepstatin A affected the activity of CMP, even when they were used at the recommended concentrations of 10 mM and 10 µg/ml, respectively.
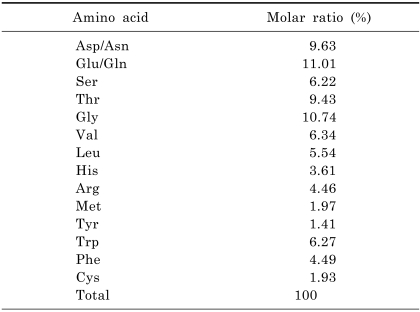
Amino acid composition
The amino acid composition of CMP showed relatively high contents of Glu/Gln (11.01%), Gly (10.74%), Thr (9.43%), Asp/Asn (9.63%), Val (6.34%), and Trp (6.27%), and lower contents of Met (1.97%), Cys (1.93%), and Tyr (1.41%) (Table 3).
Table 3.
Amino acid composition of CMP
Amino acid sequence
Since the N-terminal amino acid was blocked, the peptide sequences of trypsin-treated CMP could be analyzed. Peptides from a tryptic digest were analyzed by ESI-MS/MS, which revealed three major peaks at m/z 958.0, 910.5, and 641.9 (data not shown), identified as YQXXVTFXDF, VSXXGDSGVGGN, and NAFNDYTFK, respectively, where X indicates that the residue is either Ile or Leu (these are indistinguishable because they have the same molecular mass).
Antifungal activity
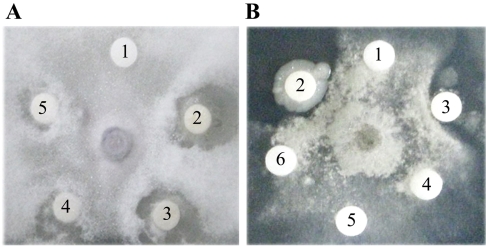
At 3~4 days after inoculation with fungi, the discs in the Petri dish plate were filled with various concentrations of CMP and further incubated at 37℃ for 24~48 h. Figure 4 shows that CMP exerted dose-dependent antifungal effects against F. oxysporum (Fig. 4A) that were stronger than those of negative controls containing buffer only. The plates exhibited clear zones of inhibition around the disc at a minimum concentration of 1.6 µM. However, the antifungal activity of CMP against B. cinerea (Fig. 4B) was relatively weak, with crescents appearing in the plate only once when the CMP concentration reached 40 µM.
Fig. 4.
Antifungal activity of CMP against the growth of (A) F. oxysporum and (B) B. cinerea. Discs contained either buffer alone (10 mM Tris-HCl with 0.1 M NaCl, pH 7.0) or following concentrations of CMP in this buffer: disc 1, 0 µM; disc 2, 200 µM; disc 3, 40 µM; disc 4, 8 µM; disc 5, 1.6 µM; and disc 6, 0.32 µM.
Cytotoxicity against human cancer cells
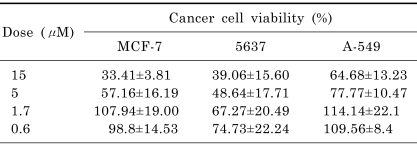
The cytotoxicities of CMP against MCF-7, 5637, and A-549 cells were investigated using the CCK assay (Table 4). CMP at 15 µM strongly inhibited the viability of MCF-7 cells (reduced to 33.41±3.81%, mean±SEM) with an IC50 of 9.3 µM and the viability of 5637 cells (reduced to 39.06±15.60%) with an IC50 of 8.1 µM, but was less effective against A-549 cells (reduced to 64.68±13.23%).
Table 4.
Cancer cell viability after treatment with CMP
Data are expressed as mean±SEM (n=3).
DISCUSSION
In the present study, we purified a protein, CMP, from the C. militaris mushroom and characterized its molecular properties. The yield of CMP was 3 mg from of the crude protein extract of 1 g of the dried fruiting bodies of C. militaris. CMP appeared as a single band on SDS-PAGE stained with silver nitrate and on the IEF gel, confirming that the purification procedure was able to isolate CMP from all other proteins. It is noteworthy that a single step of DEAE-Sepharose column chromatography was sufficient to purify CMP to homogeneity. The results of electrophoretic analyses and gel-filtration chromatography indicate that CMP is a 12 kDa protein that exists as a dimer whose subunits interact through noncovalent bonds.
The protease activity of CMP was proportional to its concentration, similar to the results obtained with trypsin, and the detection limit of the protease assay was fivefold higher for CMP than for trypsin. However, the Lineweaver-Burk plot indicated that the Km value and Vmax/Km ratio of CMP (0.61 mM and 0.54, respectively) for casein was slightly lower than those of trypsin (1.56 mM and 0.64, respectively).
In general, the molecular mass, optimum pH, and optimum temperature of mushroom proteases differ substantially from those of proteases of other fungal species (Ng, 2004). We found that the optimal conditions for CMP were a temperature of 37℃ and pH of 7.0~9.0. The test with inhibitors indicated that CMP possesses trypsin-like serine protease activity. Several mushroom proteases have been purified previously, and nearly all of them are metalloendoproteinases, aspartic endopeptidases, or prolyl endopeptidases (Ng, 2004). In contrast, there are only a few reports on serine proteases similar to CMP, based on the inhibition tests described here (Ng, 2004; Hattori et al, 2005).
Based on the first sequence of CMP (YQXXVTFXDF), it appears that CMP is a novel protease with no homology to other known proteins. Interestingly, database searches using BLAST indicated that the sequences of the other two peptides of CMP (VSXXGDSGVGGN and NAFNDYTFK) showed more than 70% homology to Rab family GTPase from Entamoeba histolytica (residues 6~17; VSLIGDSGVGKT) and exodeoxyribonuclease V alpha chain from Haemophilus influenzae (residues 273~281; NAFNDYTRY).
A 52-kDa fibrinolytic enzyme (Kim et al, 2006) and a 23-kDa protease (Hattori et al, 2005) were recently identified in C. militaris culture media. The molecular masses, purification methods, and amino acid sequences of these proteins differ substantially from those of CMP. In addition, however, it is not known whether those proteins have any activities in addition to acting as proteases. Our preliminary data indicate that this mushroom contains at least three more proteases that have yet to be studied (data not shown).
The CMP purified in the present study was found to inhibit the growth of F. oxysporum in a dose-dependent manner. The minimum concentration required for other proteins, which were obtained from other mushrooms, to exhibit antifungal activity have been reported to the range from 10 to 100 µM (Nagai & Ng, 2003; Nagai et al, 2005), which indicates that CMP (with a minimum required concentration of 1.6 µM) exerts strong effects against F. oxysporum. Moreover, CMP also exhibits cytotoxicity toward human breast and bladder cancer cells, although the underlying mechanisms are unclear. These results indicate that C. militaris represents a source of a novel protein that might be useful in diverse biological and medicinal applications.
There have been reports on the antifungal activity of a protease obtained from bacteria (Yen et al, 2006) and of protease inhibitors obtained from plants (Joshi et al, 1998; Vernekar et al, 1999), and on a cytotoxic antifungal peptide obtained from invertebrates (Li et al, 1995) or synthetically (Hong et al, 1999). However, to our knowledge, the present CMP is the first reported protein with cytotoxic antifungal protease activity obtained from a mushroom. Typical reported activities of other antifungal proteins, such as glucanase, chitinase, and chitin-binding activities, were not detected for CMP (data not shown).
In conclusion, CMP is a 12 kDa protein that exists as a dimer whose subunits interact through noncovalent bonds. Even though the partial amino acid sequence and secondary structure of CMP are quite different from those of trypsin, CMP appears to be a trypsin-like serine protease. However, unlike trypsin, CMP has multiple functions not only as a protease but also in exerting antifungal effect and cytotoxicity against human cancer cells. Our results suggest that CMP is necessary for this entomogenous fungus to inhibit foreign fungal or cellular infections and to hydrolyze cuticle components including mainly proteins, and ultimately survive within the insect. These functions of CMP also suggest that the mushroom C. militaris could be used as a nutraceutical for cancer patients, and also as a fungicide or for meat tenderizer because of its protease activity, as has been suggested previously (Ng, 2004).
ACKNOWLEDGEMENT
This work was supported by the Chung-Ang University Research Scholarship Grants in 2007.
ABBREVIATIONS
- CMP
Cordyceps militaris protein
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- HPLC
high-performance liquid chromatography
- PMSF
phenylmethylsulfonyl fluoride
- DMSO
dimethyl sulfoxide
- EDTA
ethylenediaminetetraacetic acid
References
- 1.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham KG, Manson W, Spring FS, Hutchinson SA. Cordycepin, a metabolic product isolated from cultures of Cordyceps militaris (Linn.) Link. Nature. 1950;166:949. doi: 10.1038/166949a0. [DOI] [PubMed] [Google Scholar]
- 3.Hattori M, Isomura S, Yokoyama E, Ujita M, Hara A. Extracellular trypsin-like proteases produced by Cordyceps militaris. J Biosci Bioeng. 2005;100:631–636. doi: 10.1263/jbb.100.631. [DOI] [PubMed] [Google Scholar]
- 4.Hong SY, Oh JE, Lee KH. In vitro antifungal activity and cytotoxicity of a novel membrane-active peptide. Antimicrob Agents Chemother. 1999;43:1704–1707. doi: 10.1128/aac.43.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joshi BN, Sainani MN, Bastawade KB, Gupta VS, Ranjekar PK. Cysteine protease inhibitor from pearl millet: a new class of antifungal protein. Biochem Biophys Res Commun. 1998;246:382–387. doi: 10.1006/bbrc.1998.8625. [DOI] [PubMed] [Google Scholar]
- 6.Jung EC, Kim KD, Bae CH, Kim JC, Kim DK, Kim HH. A mushroom lectin from ascomycete Cordyceps militaris. Biochim Biophys Acta. 2007;1770:833–838. doi: 10.1016/j.bbagen.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Kim BS, Oh KT, Cho DH, Kim YJ, Koo WM, Kong KH, Kim HH. A sialic acid-binding lectin from the legume Maackia fauriei: comparison with lectins from M. amurensis. Plant Science. 2004;167:1315–1321. [Google Scholar]
- 8.Kim JS, Sapkota K, Park SE, Choi BS, Kim S, Nguyen TH, Kim CS, Choi HS, Kim MK, Chun HS, Park Y, Kim SJ. A fibrinolytic enzyme from the medicinal mushroom Cordyceps militaris. J Microbiol. 2006;44:622–631. [PubMed] [Google Scholar]
- 9.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Matsunaga S, Fusetani N. Halicylindramides A-C, antifungal and cytotoxic depsipeptides from the marine sponge Halichondria cylindrata. J Med Chem. 1995;38:338–343. doi: 10.1021/jm00002a015. [DOI] [PubMed] [Google Scholar]
- 11.Lindequist U, Niedermeyer TH, Julich WD. The pharmacological potential of mushrooms. Evid Based Complement Alternat Med. 2005;2:285–299. doi: 10.1093/ecam/neh107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreno AB, del Pozo AM, Borja M, Segundo BS. Activity of the antifungal protein from Aspergillus giganteus against Botrytis cinerea. Phytopathology. 2003;93:1344–1353. doi: 10.1094/PHYTO.2003.93.11.1344. [DOI] [PubMed] [Google Scholar]
- 13.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 14.Nagai PHK, Ng TB. Lentin, a novel and potent antifungal protein from shitake mushroom with inhibitory effects on activity of human immunodeficiency virus-1 reverse transcriptase and proliferation of leukemia cells. Life Sciences. 2003;73:3363–3374. doi: 10.1016/j.lfs.2003.06.023. [DOI] [PubMed] [Google Scholar]
- 15.Nagai PHK, Zhao Z, Ng TB. Agrocybin, an antifungal peptide from the edible mushroom Agrocybe cylindracea. Peptides. 2005;26:191–196. doi: 10.1016/j.peptides.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Ng TB. Peptides and proteins from fungi. Peptides. 2004;25:1055–1073. doi: 10.1016/j.peptides.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 17.St Leger RJ, Charnley AK, Cooper RM. Characterization of cuticle-degrading proteases produced by the entomopathogen Metarhizium anisopliae. Arch Biochem Biophys. 1987;253:221–232. doi: 10.1016/0003-9861(87)90655-2. [DOI] [PubMed] [Google Scholar]
- 18.Vernekar JV, Ghatge MS, Deshpande VV. Alkaline protease inhibitor: a novel class of antifungal proteins against phytopathogenic fungi. Biochem Biophys Res Commun. 1999;262:702–727. doi: 10.1006/bbrc.1999.1269. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe N, Hattori M, Yokoyama E, Isomura S, Ujita M, Hara A. Entomogenous fungi that produce 2,6-pyridine dicarboxylic acid (dipicolinic acid) Biosci Bioeng. 2006;102:365–368. doi: 10.1263/jbb.102.365. [DOI] [PubMed] [Google Scholar]
- 20.Yen YH, Li PL, Wang CL, Wang SL. An antifungal protease produced by Pseudomonas aeruginosa M-1001 with shrimp and crab shell powder as a carbon source. Enzyme Microb Technol. 2006;39:311–317. [Google Scholar]
- 21.Yu R, Song L, Zhao Y, Bin W, Wang L, Zhang H, Wu Y, Ye W, Yao X. Isolation and biological properties of polysaccharide CPS-1 from cultured Cordyceps militaris. Fitoterapia. 2004;75:465–472. doi: 10.1016/j.fitote.2004.04.003. [DOI] [PubMed] [Google Scholar]