Abstract
Background
Allergic asthma results from inappropriate Th2-mediated inflammation. Both IL-4 and IL-13 contribute to asthma pathogenesis, but IL-4 predominantly drives Th2 induction, while IL-13 is necessary and sufficient for allergen-induced AHR and goblet cell hyperplasia. Although these 2 cytokines share signaling components, the molecular mechanisms by which they mediate different phases of the allergic asthma response remain elusive.
Objective
We sought to clarify the role(s) of IL-4 and IL-13 in asthma pathogenesis.
Methods
We employed DNA Affymetrix microarrays to profile pulmonary gene expression in BALB/c mice inoculated intratracheally with ragweed pollen (RWP), house dust mite (HDM), IL-4, IL-13, or both cytokines. IL-13 dependence was confirmed by comparing pulmonary gene expression in HDM-inoculated WT and IL-13KO mice.
Results
A signature gene expression profile consisting of 23 genes was commonly induced by inoculation with HDM, RWP, or IL-4 plus IL-13. Although rIL-4 and rIL-13 treatment induced an overlapping set of genes, IL-4 uniquely induced 21 genes, half of which were IFN response genes and half were genes important in immunoregulation. IL-13 uniquely induced 8 genes, most of which encode proteins produced by epithelial cells.
Conclusions
IL-4 and IL-13 together account for most allergen-induced pulmonary genes. Selective IL-4 induction of IFN-γ-response genes and other genes that may negatively regulate allergic inflammation may partially explain the greater importance of IL-13 in the effector phase of allergic airway disease.
Clinical Implication
The identification of genes selectively induced by individual cytokines, especially IL-13, may provide novel therapeutic targets for the treatment of asthma.
Keywords: Th2 cytokines, microarrays, allergic asthma, mouse
Introduction
The dramatic increase in asthma incidence lends urgency to the quest for new therapeutic targets1. Although asthma etiology is multi-factorial, it is thought to arise largely from maladaptive inflammatory responses in genetically susceptible individuals to common aeroallergens. More specifically, allergic asthma is mediated by Th2-polarized CD4+ T lymphocyte secretion of IL-4, IL-5 and IL-13, which stimulate airway hyperresponsiveness (AHR), pulmonary eosinophilia, elevated serum IgE, sub-epithelial fibrosis and goblet cell metaplasia2, 3. However, the precise molecular mechanisms by which Th2 cytokines mediate allergic responses are still poorly understood.
Although numerous studies support a role for IL-4 in the initiation of the immune responses that lead to asthma4, 5, IL-4 is not required for AHR or goblet cell metaplasia6-8. However, components of the IL-4 receptor signaling cascade that are also activated by IL-13 (IL-4Rα, STAT6 and IL-13Rα1) are essential for both disease development and maintenance9, 10,11 and the importance of IL-13 in the effector phase of pulmonary allergy has been demonstrated in several ways. Specific blockade of IL-13 in allergen-challenged mice reverses AHR and mucus production12,13. Acute IL-13 administration and transgenic pulmonary IL-13 overexpression stimulate many features of the allergic phenotype12, 13, 14. Allergen-immunized IL-13 deficient mice fail to develop AHR and goblet cell metaplasia and adoptive transfer of antigen-specific Th2 cells generated from IL-13 deficient mice fails to elicit AHR in recipient mice, despite considerable production of IL-4 and IL-5 and significant airway inflammation15. Thus, collectively, the current literature suggests that while IL-4 is essential for the initial development and expansion of Th2 responses, IL-13 is essential for the effector phase of the response. The present study seeks to clarify why IL-13 contributes uniquely to the effector phase of airway allergy even though IL-4 and IL-13 both signal by binding to the type 2 IL-4R complex composed of the IL-4Rα and IL-13Rα1 chains. To this end, we conducted a comprehensive gene profiling experiment to: 1) define the gene expression patterns associated with allergen challenge in the mouse lung; and 2) to define the overlapping or unique pathways regulated by IL-4 and IL-13. Our studies demonstrate that IL-4 and IL-13 together induce most pulmonary genes that are activated by inhaled allergens and show that most genes activated by one of these cytokines are also activated by the other. They also identify, however, sets of genes that are uniquely activated by IL-4 or IL-13 and provide a possible basis for the dominance of IL-13 in the effector phase of airway allergy by suggesting that some genes that are uniquely activated by IL-4 may inhibit allergic airway inflammation.
Methods
Animals
Four week old female BALB/c mice were purchased from Jackson Laboratories (Bar Harbor, ME). All mice were housed under laminar flow hoods in an environmentally controlled specific pathogen-free animal facility. The studies reported conformed to the principles for laboratory animal research, as outlined by the Animal Care and Use Committee of Cincinnati Children's Hospital Medical Center.
Allergen and Cytokine Treatment Protocols
Mice were sensitized by an intraperitoneal injection of 150 μg of endotoxin free-RWP protein or HDM (Greer Laboratories Lenoir, NC) plus alum or PBS on days 0 and 3. On days 10 and 17, mice were anesthetized with ketamine and xylazine (45 and 8 mg/kg body weight, respectively) and challenged intratracheally with 40 μl of PBS (control) or PBS that contained 200 μg of either RWP or HDM. Lungs were harvested at 72 h (RWP, HDM) after the last allergen challenge. The timing and doses of rIL-4 and rIL-13 administration were those that induced allergic phenotypic changes similar to those observed with allergen challenges in vivo 16. Thus, mice were inoculated daily by intratracheal challenge with either PBS for 10 d, IL-4 (2 μg) for 10 d, IL-4 for 10 d with IL-13 during the last 3 d, or PBS for 7 d, followed by IL-13 for 3 days, as previously described16. Lungs from cytokine-treated mice were harvested at 72 h.
Microarray Assays
RNA was isolated from whole lungs of mice and hybridized to Affymetrix U74v2 GeneChips as previously described16,17; see on-line repository for details.
Quantitative real-time RT-PCR
IL-13 specific genes were validated in a separate set of mice by RT-PCR as previously described18, see on-line repository for details.
Results
Comparison of Allergen and Th2 Cytokine-Induced Gene Expression Patterns
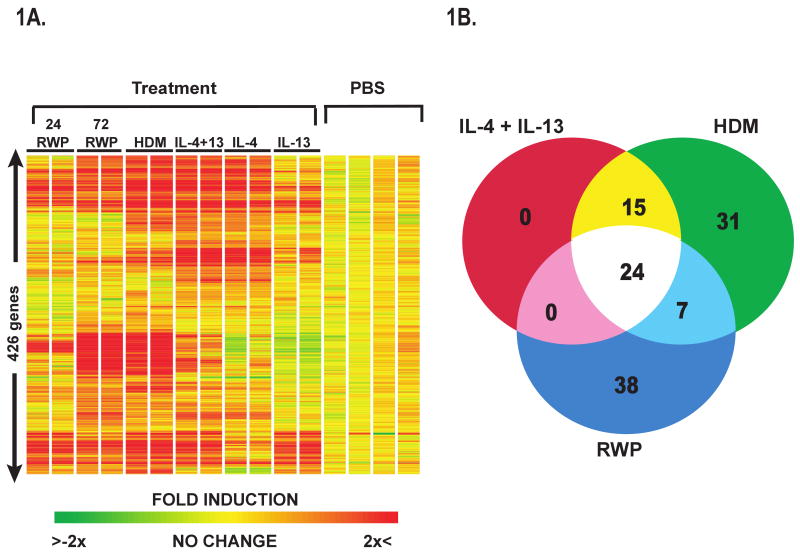
Initial experiments compared gene expression in whole lungs isolated from mice challenged with either PBS, RWP, or HDM, or with rIL-4, rIL-13 or rIL-4+rIL-13. Of the approximate 19,207 unique genes and 7,600 expressed sequence tags (EST's) represented by the 45,000 probes on the array set, expression of 1813 gene transcripts were found to be significantly different in the lungs of allergen or cytokine-treated lungs as compared to their corresponding control groups. Hierarchical cluster representation of a subset of these genes (426 genes) revealed both similarities and differences in gene expression patterns between the allergen-sensitized and - challenged and cytokine-treated mice (Figure 1A). Venn analysis was performed on a set of 115 unique genes that represented the compilation of differentially expressed genes (≥ 2-fold change) from each of these 3 treatments (Figure 1B). Zbtb16 was the only gene downregulated by a factor >2 in the allergen or cytokine treated groups. Comparison of the two 72 h allergen-treated groups with the rIL-4 +rIL-13-treated group revealed significant overlap between the three treatment groups with 24 genes being induced >2 fold by each treatment (Table I). Not surprisingly, 24 out of the 39 genes induced >2 fold by the combined cytokine treatment (Table I) were induced by both allergens. These genes could be grouped into a few broad classifications including: epithelial cell products (Itlnb, Muc5ac, Retnlb), ion channels (Clca3), chemokines (Ccl8, Ccl9, Ccl11), complement components (C1qa, C1qg), inflammatory mediators (Chi3l3, Chia, Ear2, Itgax, Scin), proteases or protease inhibitors (MMP12, Serpina3g), immunoglobulin receptors (FcR11b), and arginine metabolism (Arg 1, Gatm). The remaining 15 genes that were significantly induced by combined rIL-4/rIL-13 treatment and by HDM, but not RWP included: Slc5a1, Adam8, Ccr5, cathepsin Z, CD83, F10 (coagulation factor X), cholesterol 25-hydroxylase, interferon regulatory factor 4, insulin-like growth factor 1, insulin-like growth factor binding protein 3, latent transforming growth factor beta binding protein 4, tissue factor 2, arachidonate 15-lipoxygenase, paroxonase 1, and activating transcription factor 3. There were seven genes induced by both allergens that were not induced by rIL-4 + rIL-13 (Table IE, in the online respository). These included Ig μ, γ and ε chains, S100 calcium binding protein A8, and interleukin-1 receptor-like 1. It is tempting to speculate that these genes are important in early immunoglobulin production and in the case of IL-1RL1, may promote initiation of immune responses via acting as a cofactor for DC or T cell activation. Taken together these results suggest that combined IL-4 and IL-13 treatment recapitulates many of the molecular changes occurring following allergen challenge, confirming the pivotal role of these cytokines in disease pathogenesis.
Figure 1. Gene Expression Changes in the Lungs of Allergen- and Cytokine-Treated Mice.
A. Hierarchical clustering of gene expression data from HDM or RWP allergen exposed, rIL-4 and rIL-13-treated, and PBS- exposed Balb/c wild-type mice revealed that 426 genes were significantly different (p<0.05) between treatment groups and PBS controls. Data shown are two independent samples per treatment B. Venn analysis of gene expression changes showed that 23 genes were shared following all 3 exposures.
Table I. Shared Gene Expression Patterns Between Allergens (HDM and RWP) and Combined IL-4 and IL-13 Treatment.
| Symbol | Description | RWP | HDM | IL-4+ IL13 |
|---|---|---|---|---|
| Secreted Epithelial Products | ||||
| Itlnb | intelectin b | 3.7 | 5.7 | 3.5 |
| Muc5ac | mucin 5, subtypes A and C, tracheobronchial/gastric | 2.1 | 3.0 | 2.4 |
| Retnlb | resistin like beta | 2.5 | 6.3 | 2.8 |
| Ion Channels/ Transporters | ||||
| Clca3 | chloride channel calcium activated 3 | 41.3 | 26.9 | 29.0 |
| Chemokines | ||||
| Ccl8 | chemokine (C-C motif) ligand 8 | 12.8 | 23.4 | 9.9 |
| Ccl9 | chemokine (C-C motif) ligand 9 | 2.7 | 5.7 | 5.0 |
| Ccl11 | small chemokine (C-C motif) ligand 11 | 3.6 | 8.4 | 4.1 |
| Classical Complement Pathway | ||||
| C1qa | complement component 1, q subcomponent, alpha polypeptide | 2.7 | 3.5 | 2.6 |
| C1qg | complement component 1, q subcomponent, gamma polypeptide | 2.3 | 2.9 | 2.5 |
| Inflammation | ||||
| Chi3l3 | chitinase 3-like 3 | 9.9 | 10.7 | 5.2 |
| Chia | chitinase, acidic | 8.2 | 9.9 | 8.2 |
| Ear2 | Eosinophil-associated, ribonuclease A family, member 2 | 2.0 | 2.3 | 2.5 |
| Itgax | integrin alpha X | 2.2 | 2.5 | 3.1 |
| Scin | scinderin | 2.1 | 3.1 | 2.1 |
| Protease Pathways | ||||
| Mmp12 | matrix metalloproteinase 12 | 7.2 | 16.6 | 22.1 |
| Serpina3g | serine (or cysteine) proteinase inhibitor, clade A, member 3G | 3.3 | 6.3 | 5.2 |
| Arginine Metabolism | ||||
| Arg1 | arginase 1, liver | 3.2 | 9.0 | 12.6 |
| Gatm | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 2.1 | 4.2 | 3.7 |
| Receptors | ||||
| Fcgbp | RIKEN cDNA A430096B05 gene | 4.8 | 5.8 | 2.7 |
| Fcgr2b | Fc receptor, IgG, low affinity IIb | 2.4 | 5.4 | 3.5 |
| Pigr | polymeric immunoglobulin receptor | 3.3 | 4.3 | 2.8 |
| Others | ||||
| Fbp1 | fructose bisphosphatase 1 | 2.1 | 2.7 | 6.2 |
| Zbtb16 | Zinc finger and BTB domain containing 16 | -4.4 | -2.2 | -2.1 |
24 genes induced in the lung by all 3 exposures, as determined via Venn Analysis (Fig1B). Values represent the mean (N=2 independent samples) fold-changes between treatment and corresponding PBS-controls. Gene expression differences between treatment and corresponding control groups for each of the 3 treatments were determined via Student's t- test with p≤0.05. The overlapping gene lists were further filtered by fold induction of ≥2.HDM-house dust mite, RWP, ragweed pollen protein.
Differences in Gene Expression Patterns Between Ragweed and House Dust Mite Exposure
To determine the universality of the gene expression patterns induced in the lung by different allergens, we compared RWP-induced gene expression patterns with those induced by HDM, 72 h after allergen challenge. We have previously shown that both allergen exposure regimes induced AHR, goblet cell metaplasia, IgE production, and eosinophilic inflammation19. As expected, RWP and HDM both induced numerous gene expression changes in the lung. RWP induced differential expression of 538 genes (Student's t-test, p ≤0.05), of which 68 were ≥ 2-fold different (Table EII, in the online respository) from PBS control. HDM allergen exposure induced changes in expression of 356 genes (Student's t-test, p ≤0.05), of which 76 were ≥ 2-fold different (Table EIII, in the online respository), from PBS. From these independent sets of differentially expressed genes, we identified 76 genes whose expression by Venn analysis is >2 fold different between HDM and RWP (Table EIV, in the online respository, Figure 1B). In general, RWP is a less potent stimulus than HDM 72 h post-inoculation.
IL-4-Dependent Gene Expression Changes
Although IL-4 is thought to be essential in the initiation of Th2-mediated immune responses, several lines of evidence suggest that IL-4 can induce, but is not essential in the development of airway hyperresponsiveness or mucus cell metaplasia 6-8, while IL-13 has been shown to be sufficient and essential for allergen induction of these disease features. To determine the individual role(s), of IL-4 and IL-13 to the allergic phenotype, we sought to determine the potential contribution of each cytokine to the allergen-induced gene changes. To this end, we examined the gene expression patterns in lungs from mice exposed to either rIL-4 or rIL-13. Of the 1813 most variable genes comprising the main dataset, 455 genes were induced by rIL-4 (Student's t-test, p ≤0.05). Of these, 41 genes were expressed ≥2-fold more than the PBS controls. 21 of these 41 genes were also induced by IL-13 (Table II). The remaining 20 genes were determined to be uniquely IL-4-dependent (Table III). These included genes for chemokines, signaling mediators, immune response and apoptosis regulation (Table III). Most interestingly, the majority of the IL-4-specific genes are known to be IFN-γ-inducible, and most likely represent a footprint of IFN-γ, a cytokine induced by IL-4, but not IL-13 20 that suppresses AHR and goblet cell hyperplasia. The IL-4-stimulated IFN-γ inducible genes included: immunity-related GTPase family member M, interferon inducible GTPase, interferon gamma induced GTPase, and interferon activated gene 202B (Table III). Although we did not observe significant elevations in IFN-γ by microarray analyses, we observed significant elevations in the levels of IFN-γ mRNA in the lungs of IL-4-treated (10.5 fold, P<0.001), but not IL-13-treated (1.3 fold) mice, when compared to those of PBS-treated mice when assessed by RT-PCR.
Table II. IL-4-Induced Gene Expression Patterns Shared by IL-13 in the Mouse Lung.
| Symbol | Description | IL-4+13 | IL-4 | IL-13 |
|---|---|---|---|---|
| Epithelial Products | ||||
| Arg1 | arginase 1, liver | 12.6 | 8.6 | 4.4 |
| Clca3 | chloride channel calcium activated 3 | 29.0 | 24.7 | 25.7 |
| Chemokines | ||||
| Ccl7 | chemokine (C-C motif) ligand 7 | 3.2 | 2.0 | 2.5 |
| Ccl8 | chemokine (C-C motif) ligand 8 | 9.9 | 8.7 | 2.9 |
| Ccl9 | chemokine (C-C motif) ligand 9 | 5.0 | 4.3 | 2.2 |
| Signaling | ||||
| Gbp4 | Guanylate nucleotide binding protein 4 | 2.5 | 2.4 | 2.1 |
| Tgtp | T-cell specific GTPase | 3.2 | 3.4 | 2.7 |
| Inflammation | ||||
| Chi3l3 | chitinase 3-like 3 | 5.2 | 5.2 | 10.2 |
| Chia | chitinase, acidic | 8.2 | 4.2 | 4.5 |
| Ear1 | eosinophil-associated, ribonuclease A family, member 1 | 2.3 | 2.1 | 2.9 |
| Ear2 | Eosinophil-associated, ribonuclease A family, member 2 | 2.5 | 2.5 | 2.9 |
| Host Immune Reponse | ||||
| Ifi16 | Interferon, gamma-inducible protein 16 | 2.9 | 2.4 | 2.3 |
| Ifi47 | interferon gamma inducible protein 47 | 2.4 | 2.4 | 2.3 |
| Protease Pathways | ||||
| Mmp12 | matrix metalloproteinase 12 | 22.1 | 21.6 | 4.1 |
| Serpina3g | serine (or cysteine) proteinase inhibitor, clade A, member 3G | 5.2 | 4.2 | 3.1 |
| Adam8 | a disintegrin and metalloprotease domain 8 | 2.5 | 2.3 | 2.2 |
| Matrix Homeostasis | ||||
| Ctsk | cathepsin K | 3.0 | 2.9 | 2.3 |
| Immunoglobulins | ||||
| Fcgbp | Fc fragment of IgG binding protein | 2.7 | 2.0 | 2.5 |
| Lilrb4 | leukocyte immunoglobulin-like receptor, subfamily B, member 4 | 3.1 | 2.6 | 2.3 |
Values represent mean fold-changes (N=2 samples/treatment) from corresponding PBS-treated controls. Gene expression differences were determined via Student's t- test with p<0.05, and further filtered by a fold induction ≥2.
Table III. Unique IL-4 and IL-13-Dependent Gene Expression Patterns in the Mouse Lung.
| Symbol | Description | IL-4+13 | IL-4 | IL-13 |
|---|---|---|---|---|
| Unique IL-4-inducible genes | ||||
| Chemokines | ||||
| Ccl17 | chemokine (C-C motif) ligand 17 | 4.3 | 5.8 | 1.4 |
| Ccr5 | chemokine (C-C motif) receptor 5 | 5.4 | 4.0 | 1.1 |
| Host Immune Reponses | ||||
| Atf3 | activating transcription factor 3 | 3.0 | 2.5 | 1.4 |
| Cd83 | CD83 antigen | 2.2 | 2.1 | 1.2 |
| Fcgr2b | Fc receptor, IgG, low affinity IIb | 3.5 | 2.9 | 1.4 |
| Igtp | interferon gamma induced GTPase | 2.5 | 2.9 | 1.1 |
| Iigp1 | interferon inducible GTPase 1 | 3.4 | 3.6 | 1.2 |
| Iigp1 | interferon inducible GTPase 1 | 2.4 | 2.5 | 1.1 |
| Lilrb4 | leukocyte immunoglobulin-like receptor, subfamily B, member 4 | 2.7 | 2.4 | 1.4 |
| Arginine Metabolism | ||||
| Gatm | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 3.7 | 2.7 | 1.2 |
| Inflammation | ||||
| Itgax | integrin alpha X | 3.1 | 2.9 | 1.4 |
| Vnn1 | vanin 1 | 1.8 | 2.3 | 1.1 |
| Stress response | ||||
| Hspa1a | heat shock protein 1A | 1.2 | 2.4 | 1.5 |
| Hspa1b | heat shock protein 1B | 1.3 | 2.9 | 1.5 |
| Complement Pathway | ||||
| C1qa | complement component 1, q subcomponent, alpha polypeptide | 2.6 | 2.4 | -1.1 |
| C1qg | complement component 1, q subcomponent, gamma polypeptide | 2.5 | 2.3 | 1.0 |
| Cfp | complement factor properdin | 2.0 | 2.0 | 1.0 |
| Lipid Metabolites/ Mediators | ||||
| Ch25h | cholesterol 25-hydroxylase | 2.9 | 2.0 | 1.3 |
| Coagulation | ||||
| F10 | coagulation factor X | 2.3 | 2.2 | 1.3 |
| Others | ||||
| Fbp1 | fructose bisphosphatase 1 | 6.2 | 6.3 | 1.5 |
| Unique IL-13-inducible genes | ||||
| Ion Transporters | ||||
| Slc5a1 | solute carrier family 5 (sodium/glucose cotransporter), member 1 | 2.6 | 1.3 | 3.0 |
| Other Epithelial Products | ||||
| Agr2 | anterior gradient 2 (Xenopus laevis) | 3.2 | 1.6 | 3.9 |
| Itln2 | intelectin 2 | 3.5 | 1.5 | 2.5 |
| Retnlb | resistin like beta | 2.8 | 1.4 | 2.3 |
| Sprr2a | small proline-rich protein 2A | 3.4 | 0.9 | 4.0 |
| Lysine Metabolism | ||||
| Aass | aminoadipate-semialdehyde synthase | 1.7 | 1.0 | 2.1 |
| Chemokines | ||||
| Ccl11 | small chemokine (C-C motif) ligand 11 | 4.1 | 1.0 | 2.9 |
| Actin Binding | ||||
| Scin | Scinderin | 2.2 | 1.4 | 2.0 |
Values represent mean fold-changes (N=2 samples/treatment) from corresponding PBS-treated controls. Gene expression differences were determined via Student's t- test with p<0.05, and further filtered by a fold induction ≥2.
IL-13 Dependent Gene Expression Changes
Of the 1813 most variable genes comprising the main dataset, 231 genes were differentially expressed in response to IL-13, as compared to PBS (Student's t-test, p ≤0.05). Of these, 19 genes were ≥ 2-fold different from PBS and 8 were induced at least 2-fold more by IL-13 than IL-4 (Table III). Of these 8, the majority were epithelial cell products including: the eosinophil specific chemokine, Ccl11; Itln2, a newly described lectin; Retnlb, a goblet cell specific gene; Sprr2a, an epithelial cell gene associated with squamous cell changes; the ion transporter Slc5a1; Aass, a gene involved in arginine metabolism; Scin, a molecule known to bind actin; and Agr2, an estrogen receptor-responsive gene.
Confirmation of IL-13-Dependent Gene Expression in IL-13 Deficient Mice
To verify the IL-13 dependence of the gene expression patterns determined to be selective for IL-13, we compared gene expression in the lungs of individual WT and IL-13KO mice treated with PBS or HDM (3 mice/group). Gene expression analyses identified 372 gene transcripts that were differentially expressed between HDM treated wild-type mice and IL-13 deficient mice (Student's t-test, p ≤0.05), of which 68 were ≥ 2-fold different (Table IV). This comparison identified many of the same, predominantly epithelial cell products and ion transporters that were preferentially induced by IL-13 in WT mice, including: Ccl11, Sprr2a, Retnlb, Itln2, Agr2, Slc5a1, Scin, Aass. Interestingly, several genes were found to be increased in the IL-13 deficient mice including: IL1rl1, Edem1, Rrm2, CD209e, Ccl12, and Mgmg (macrophage galactose N-acetylgalactosamine specific lectin-1). Whether these changes are due to compensatory effects of IL-13 gene deletion or to active repression of these genes by IL-13 in vivo is currently unknown.
Table IV. Gene Expression Patterns in Lungs of IL-13 Deficient Mice.
| Symbol | Description | KO HDM | WT HDM |
|---|---|---|---|
| Secreted Epithelial Products | |||
| Agr2 | anterior gradient 2 (Xenopus laevis) | 1.4 | 4.5 |
| Itlna | intelectin a | 1.3 | 8.6 |
| Retnlb | resistin like beta | 1.1 | 5.8 |
| Scgb3a2 | secretoglobin, family 3A, member 2 | -1.5 | -3.9 |
| Ion Channels /Transporters | |||
| Atp1a3 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | 1.4 | 2.7 |
| Clca3 | chloride channel calcium activated 3 | 1.2 | 4.7 |
| Fxyd4 | FXYD domain-containing ion transport regulator 4 | 1.4 | 3.7 |
| Kcnj15 | potassium inwardly-rectifying channel, subfamily J, member 15 | -1.1 | 2.3 |
| Slc5a1 | solute carrier family 5 (sodium/glucose cotransporter), member 1 | 1.2 | 2.6 |
| Chemokines | |||
| Cxcl1 | chemokine (C-X-C motif) ligand 1 | 1.2 | 2.3 |
| Ccl11 | small chemokine (C-C motif) ligand 11 | 1.1 | 12.7 |
| Host Immune Response | |||
| Csf2ra | colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) | 1.4 | 2.3 |
| Fpr-rs2 | formyl peptide receptor, related sequence 2 | 1.2 | -2.4 |
| Klf4 | Kruppel-like factor 4 (gut) | -1.5 | -2.1 |
| Ltb | lymphotoxin B | 1.3 | 2.6 |
| Pigr | polymeric immunoglobulin receptor | 1.2 | 7.6 |
| Scin | scinderin | 1.3 | 3.4 |
| Tnfrsf9 | tumor necrosis factor receptor superfamily, member 9 | 1.2 | 2.1 |
| Tnfaip8 | tumor necrosis factor, alpha-induced protein 8 | 1.0 | 2.2 |
| Inflammation | |||
| Chi3l1 | chitinase 3-like 1 | 1.1 | 2.1 |
| Ear2 | eosinophil-associated, ribonuclease A family, member 2 | 1.5 | 2.5 |
| Ear3 | eosinophil-associated, ribonuclease A family, member 3 | 1.3 | 2.4 |
| Fcer2a | Fc receptor, IgE, low affinity II, alpha polypeptide | 1.2 | 2.6 |
| Il33 | interleukin 33 | 1.3 | 2.9 |
| Olr1 | oxidized low density lipoprotein (lectin-like) receptor 1 | 1.2 | 2.5 |
| Matrix Homeostasis | |||
| Col6a2 | procollagen, type VI, alpha 2 | 1.0 | 2.1 |
| Tnc | tenascin C | 1.4 | 2.5 |
| Lysine Metabolism | |||
| Aass | aminoadipate-semialdehyde synthase | 1.1 | 2.5 |
| Signaling | |||
| Adra2a | adrenergic receptor, alpha 2a | 1.3 | 3.2 |
| Gpr35 | G protein-coupled receptor 35 | 1.2 | 2.1 |
| Gla | galactosidase, alpha | 1.5 | 2.5 |
| Gclc | glutamate-cysteine ligase, catalytic subunit | 1.2 | 2.5 |
| Guca2a | guanylate cyclase activator 2a (guanylin) | 1.1 | 2.1 |
| Hrb | HIV-1 Rev binding protein | -1.5 | -2.6 |
| Mod1 | malic enzyme, supernatant | -1.1 | 2.2 |
| Prkcb1 | protein kinase C, beta 1 | 1.4 | 2.0 |
| Ppp1r9a | protein phosphatase 1, regulatory (inhibitor) subunit 9A | -1.4 | -2.0 |
| Rgs4 | regulator of G-protein signaling 4 | 1.3 | 2.1 |
| Rbbp4 | retinoblastoma binding protein 4 | -1.1 | 2.0 |
| Rbp4 | retinol binding protein 4, plasma | -1.5 | 3.2 |
| LOC6404 41 | similar to thrombospondin 1 | 1.3 | 2.4 |
| Thbs1 | thrombospondin 1 | 1.0 | 2.5 |
| Tmepai | transmembrane, prostate androgen induced RNA | 1.2 | 2.0 |
| Regulation of Gene Expression | |||
| Atf3 | activating transcription factor 3 | 1.3 | 2.2 |
| Eif3m | eukaryotic translation initiation factor 3, subunit M | 1.0 | -2.0 |
| Ubtf | upstream binding transcription factor, RNA polymerase I | 1.3 | 6.5 |
| Protease Pathways | |||
| Capn9 | calpain 9 (nCL-4) | 1.1 | 2.7 |
| Corin | corin | 1.1 | 2.4 |
| Cell Growth | |||
| Arrdc3 | arrestin domain containing 3 | -1.5 | -2.1 |
| BC004044 | cDNA sequence BC004044 | 1.3 | 2.4 |
| Egln3 | EGL nine homolog 3 (C. elegans) | 1.2 | 2.1 |
| Efnb2 | ephrin B2 | -1.4 | -2.2 |
| Fgfbp1 | fibroblast growth factor binding protein 1 | -1.5 | -2.2 |
| Gadd45g | growth arrest and DNA-damage-inducible 45 gamma | 1.2 | 2.3 |
| Tfpi2 | tissue factor pathway inhibitor 2 | 1.2 | 2.6 |
| Electron Transport | |||
| EG668771 | 3-phosphoglycerate dehydrogenase | 1.1 | 2.6 |
| Ckmt1 | creatine kinase, mitochondrial 1, ubiquitous | 1.0 | 2.3 |
| Fmo3 | flavin containing monooxygenase 3 | -1.3 | -2.3 |
| Hba-a1 | hemoglobin alpha, adult chain 1 | 1.3 | 3.9 |
| Tdo2 | tryptophan 2,3-dioxygenase | 1.2 | 4.0 |
| Other | |||
| Bex2 | brain expressed X-linked 2 | 0.7 | -2.2 |
| Cttnbp2nl | CTTNBP2 N-terminal like | 0.7 | -2.1 |
| 1190002H 23Rik | RIKEN cDNA 1190002H23 gene | 0.7 | -2.5 |
| 4833422F 24Rik | RIKEN cDNA 4833422F24 gene | 1.3 | 2.5 |
| 3200002M 19Rik | RIKEN cDNA 3200002M19 gene | 1.0 | 2.2 |
Values represent mean fold-changes (N=3 independent samples per condition) over corresponding PBS-treated controls. Gene expression differences were determined via Student's t- test with p<0.05, and further filtered by a fold induction ≥2.
Verification of Gene Expression Changes in Whole Lung by Quantitative RT-PCR
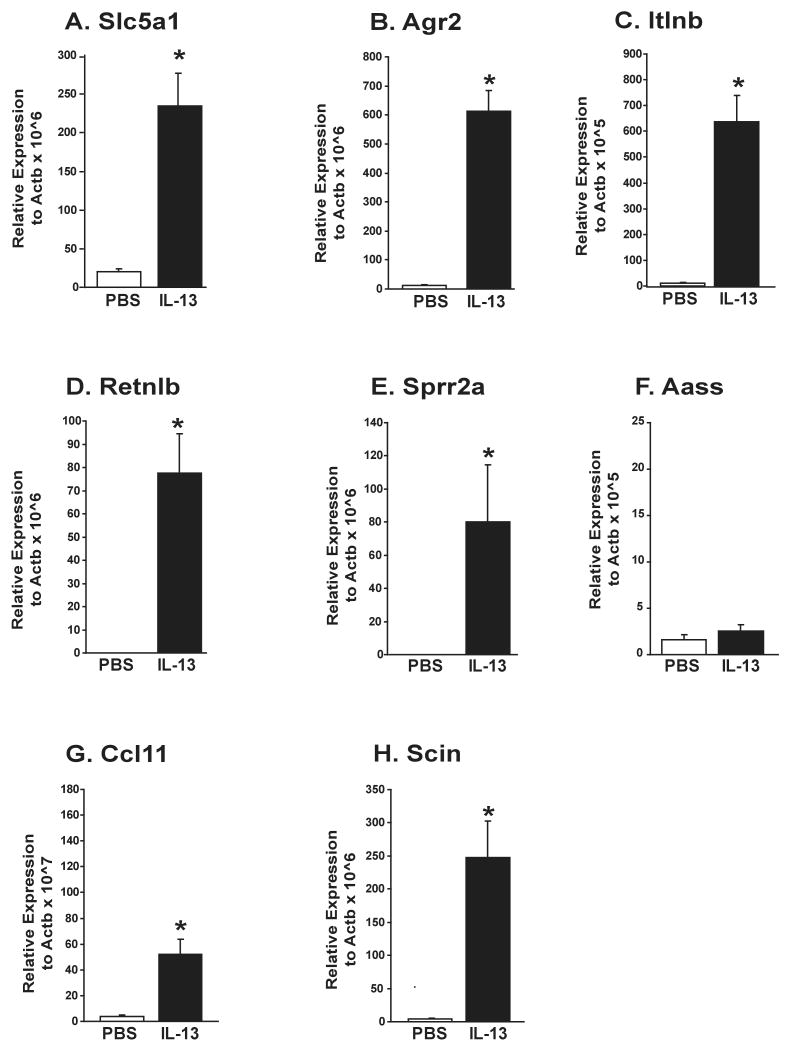
IL-13-induced pulmonary gene expression changes detected by the Affymetrix GeneChips were verified using quantitative real-time PCR for mice treated with IL-13 or PBS. As shown in Figure 2, all of the gene expression patterns identified to be IL-13 dependent via microarray analyses were reproduced with quantitative real-time PCR, with the exception of one (Aass), whose expression was determined to be unchanged with IL-13. The expression of Slc5a1, Agr2, Itlnb, Retnlb, Sprr2a, Ccl11, and Scin were all upregulated in response to IL-13 cytokine treatment. The reproducibility of gene expression results confirm that our global gene profiling approach accurately reflects the complex pattern of genes expressed in the lung following allergen exposure or cytokine treatment.
Figure 2. Verification of IL-4 and IL-13-Dependent Gene Expression Changes by Quantitative Real-Time PCR.
Gene expression changes identified in microarray analyses were verified in the whole lungs of wild-type Balb/c mice treated with A) IL-13 or B) IL-4. * denotes genes significantly different (p<0.05) from controls.
Discussion
The present study employed Affymetrix microarray technology to comprehensively profile gene expression in the allergic lung to gain insight into the molecular mechanisms underlying the pathogenesis of asthma and to identify novel targets for therapeutic development. Our specific objectives were to define the patterns of expression associated with allergen challenge in the mouse lung and determine the relative contribution of IL-4 and IL-13 to this pattern. To this end, we identified an “asthma signature” gene expression profile consisting of 23 genes that was induced in the mouse lung by exposure to two common real world allergens (HDM, RWP) as well as the combination of rIL-4 and rIL-13. This signature profile included genes encoding chemokines, components of the complement activation pathway, arginine metabolism, immunoglobulins, epithelial specific gene products, and proteases. These genes included both a group of asthma-related genes that have been previously described in the literature, including those specific for arginase21-23, members of the chitinase family of enzymes22,25, intelectin26, mucins27, and gob-5/ Clca328, as well as a group of novel candidate genes (Agr2, scin) and were similar to genes previously found to be induced in lungs by inoculation with other allergens, including ovalbumin17 and aspergillus29. Although HDM and RWP induced pulmonary gene expression patterns that were predominantly similar, HDM appears to be more potent than RWP, with >2-fold greater induction of several genes. Other differences in gene expression between RWP and HDM at 72 hr represented differences in the kinetics of the response to each allergen as most of these genes not induced by RWP at 72 hrs were significantly induced at 24 hrs by RWP (Supp). Finally, although the combined cytokine treatment faithfully recapitulated most of the allergen-induced gene expression pattern, some genes induced by both allergens were not induced by the combined IL-4 and IL-13 cytokine treatments. These genes appear to be important in the initiation of immune responses as they encode genes important in innate immune responses (Il1rl1, S100a8), and immunoglobulin synthesis. Taken together these findings support previous reports that both IL-4 and IL-13 are essential contributors to allergen-induced asthma.
One of the most important findings of the current study is the identification of unique IL-4 and IL-13 induced gene expression profiles. Not surprising, when gene expression patterns induced by rIL-4 and rIL-13 treatment of mice were compared, we found that for the most part, the patterns induced by each cytokine were overlapping (Table II). This observation is consistent with their use of a shared signaling receptor composed of IL-13Rα1 and IL-4Rα chains. However, we also found that both IL-4 and IL-13 induced unique, non-overlapping gene expression patterns (Table III). Specifically, intratracheal inoculation of mice with rIL-4 induced a unique set of genes not induced by rIL-13, that is largely comprised of IFN-γ inducible genes (Ifi202b, Ifi204, Ifit3) of currently unknown function, as well as genes important in immunoregulation (C1qa, granzyme A, Stat1). These IL-4 specific gene expression changes likely reflect the ability of IL-4, but not IL-13, to signal through the type 1 IL-4 receptor, which is composed of the γc and IL-4Rα chains. These results suggest that through IL-4's unique binding to the type 1 IL-4R, it may be able to regulate the level of the pro-allergic signal provided through the type 2 IL-4R by activating a counterregulatory immune response. In support of this hypothesis, we found that rIL-4, but not rIL-13, induced significant elevations in lung IFN-γ gene expression. Moreover, we have previously reported that IL-4 is able to inhibit the induction of several IL-13-induced genes, through a process involving γc16. These observations provide important insight into the differences observed in the contribution of these two molecules to the allergic diathesis and may also provide a plausible explanation for their apparent duplication during evolution.
Identification of IL-13 selective genes was based upon both IL-13 cytokine treatment and allergen challenge of IL-13-deficient mice. Based on the criteria that a gene was significantly induced by rIL-13, not by rIL-4 and that it was not significantly induced in the lungs of allergen-challenged IL-13 deficient mice, we identified 8 genes we refer to as IL-13-selective genes. This gene set contained mainly products of the epithelium (Ccl11, Sprr2a, Retnlb, Itln2, Agr2, Slc5a1, Scin, Aass). The fact that the IL-13-selective genes likely represent an epithelial specific gene program is consistent with the previous demonstration by Kuperman and colleagues30 that reconstitution of STAT6 in the airway epithelium of STAT6 deficient mice is sufficient to mediate IL-13 induced AHR and mucus cell changes. Induction of these genes by IL-13 may set into motion a series of changes that either directly alter epithelial function and/or indirectly regulate other pulmonary cell types by the release of mediators from epithelium. Of the IL-13 selective genes, Ccl11 (eotaxin), the eosinophil specific chemokine, has been shown to play an important role in allergic asthma31. Likewise upregulation of Sprr2a (small proline-rich protein) expression has been previously reported in the context of allergic inflammation32, and while the relevance of this gene in asthma pathogenesis is currently unknown, the metaplastic changes that occur in the epithelium as part of the airway remodeling process may explain the upregulation of these Sprr genes33. In addition, Sprr proteins have been reported to migrate to the nucleus and influence gene expression and cellular differentiation34. Both of these effects may influence airway responsiveness, and the latter effect has the potential to influence goblet cell differentiation and mucus production. Retnlb (is a goblet cell specific protein that is induced in the intestinal mucosa upon bacterial infection35 and in the lung by both allergens and IL-1336. Agr2 is a sex hormone-responsive gene known to be overexpressed in various cancers, including that of the lung37. Its relevance to the allergic response is not currently known. Itln2 (intelectin-2) is a Ca+2-dependent secreted lectin with affinity for galactofuranosyl moieties found in bacterial cell wall preparations, suggesting a role in recognition of bacterial pathogens and innate immunity38. Its expression has been previously reported in the allergic lung26, as well as the goblet and Paneth cells of the jejunum after parasite infection39. Interestingly, murine strains (C57BL/10) that demonstrate a profound inability to expel the intestinal parasite, Trichuris muris, lack this gene, while it is present and upregulated during parasite infection in mice which readily clear the parasite (Balb/c)40. Further studies are required to determine its contribution to asthma pathogenesis. Slc5a1 is a sodium/ glucose transporter and its altered expression could contribute to dysregulated airway epithelial polarity, ultimately contributing to mucus production and/ or enhanced susceptibility to inappropriate host immune responses41. Scinderin or adseverin is a pH and Ca2+ regulated member of the gelsolin super family of actin filament severing proteins that function to specifically cleave actin filaments to permit vesicles to dock during regulated secretion42. Its relevance to allergic airway disease is unknown, but synthetic peptides corresponding to its actin binding domains inhibit mucin secretion43. The Aass gene transcribes a protein that catalyzes the first two steps in the lysine degradation pathway44. While it is not clear what role this gene plays in allergic inflammation, it may play a role though its regulation of the arginine pathway that has been previously implicated in asthma21.
Although the ability of IL-4 to induce a unique profile of genes is easily explained by it's signaling through the type 1 IL-4R, the mechanisms through which IL-13 may induce a unique set of genes are less clear. Four mechanisms may contribute. First, higher affinity of IL-13 than IL-4 for the type 2 IL-4R may allow IL-13 to induce signaling pathways that are not activated by IL-448. Secondly, IL-4 signaling through the type 1 IL-4R may activate signaling pathways or stimulate production of cytokines, such as IFN-γ and IL-10, that inhibit some of the effects of signaling through the type 2 IL-4R. In this regard, at least some of the differences in IL-4 -vs. IL-13-induced gene expression are no longer found when these cytokines are administered to γc-deficient mice, which lack the type 1 IL-4R16. Thirdly, IL-13, but not IL-4 may signal in some circumstances through cell membrane IL-13Rα245 and soluble complexes of IL-13 with the soluble form of IL-13Rα2 may also have proinflammatory effects46. Fourthly, more IL-13 than IL-4 is produced in the lungs in response to allergen adminstration11. A predominantly quantitative explanation for a difference in IL-4 vs. IL-13 effects cannot account for the differences we see in cytokine-induced gene expression in mice stimulated with similar quantities of these cytokines, but probably explains, to some extent, their different contributions to allergen-induced AHR and goblet cell hyperplasia, inasmuch as IL-4 induces both phenomena in the absence of IL-1347.
In summary, our results identify a group of asthma signature expression changes in the allergic lung following exposure to relevant human aeroallergens and suggest unique roles for IL-4 and IL-13 in asthma pathogenesis. Specifically, we provide evidence that IL-4, in addition to its proallergic effects, may limit allergic responses by stimulating a counterregulatory pathway through activation of the type 1 IL-4 receptor, which is not stimulated by IL-13. This novel observation, taken together with evidence of greater potency and expression of IL-13 than IL-4 in signaling through the type 2 IL-4R and for possible signaling by IL-13 through an IL-4R-independent IL-13R may explain the greater importance of IL-13 than IL-4 during the effector phase of asthma. This information may be particularly relevant given the development of therapeutics for asthma that selectively target IL-13 or simultaneously target IL-4 and IL-13.
Supplementary Material
Acknowledgments
Sources of Funding: PO1 HL076383 to MWK and FDF; HL67736-08 (MWK), AI052099 (FDF); the CONICIT and Universidad Centroccidental Lisandro Alvarado (Venezuela) to JS.
Abbreviations used
- IL
interleukin
- AHR
airway hyperresponsiveness
- HDM
house dust mite
- RWP
ragweed pollen protein
- RT-PCR
real time polymerase chain reaction
- IgE
immunoglobulin E
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akinbami L. The state of childhood asthma, United States, 1980-2005. Adv Data. 2006:1–24. [PubMed] [Google Scholar]
- 2.Wills-Karp M. Immunologic basis of antigen-induced airway hyperresponsiveness. Annu Rev Immunol. 1999;17:255–81. doi: 10.1146/annurev.immunol.17.1.255. [DOI] [PubMed] [Google Scholar]
- 3.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Immunol Rev. 2004;202:175–90. doi: 10.1111/j.0105-2896.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 4.Coyle AJ, Le Gros G, Bertrand C, Tsuyuki S, Heusser CH, Kopf M, et al. Interleukin-4 is required for the induction of lung Th2 mucosal immunity. Am J Respir Cell Mol Biol. 1995;13:54–9. doi: 10.1165/ajrcmb.13.1.7598937. [DOI] [PubMed] [Google Scholar]
- 5.Rankin JA, Picarella DE, Geba GP, Temann UA, Prasad B, DiCosmo B, et al. Phenotypic and physiologic characterization of transgenic mice expressing interleukin 4 in the lung: lymphocytic and eosinophilic inflammation without airway hyperreactivity. Proc Natl Acad Sci U S A. 1996;93:7821–5. doi: 10.1073/pnas.93.15.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogan SP, Matthaei KI, Young JM, Koskinen A, Young IG, Foster PS. A novel T cell-regulated mechanism modulating allergen-induced airways hyperreactivity in BALB/c mice independently of IL-4 and IL-5. J Immunol. 1998;161:1501–9. [PubMed] [Google Scholar]
- 7.Cohn L, Tepper JS, Bottomly K. IL-4-independent induction of airway hyperresponsiveness by Th2, but not Th1, cells. J Immunol. 1998;161:3813–6. [PubMed] [Google Scholar]
- 8.Tanaka H, Nagai H, Maeda Y. Effect of anti-IL-4 and anti-IL-5 antibodies on allergic airway hyperresponsiveness in mice. Life Sci. 1998;62:PL169–74. doi: 10.1016/s0024-3205(98)00047-2. [DOI] [PubMed] [Google Scholar]
- 9.Gavett SH, O'Hearn DJ, Karp CL, Patel EA, Schofield BH, Finkelman FD, et al. Interleukin-4 receptor blockade prevents airway responses induced by antigen challenge in mice. Am J Physiol. 1997;272:L253–61. doi: 10.1152/ajplung.1997.272.2.L253. [DOI] [PubMed] [Google Scholar]
- 10.Kuperman D, Schofield B, Wills-Karp M, Grusby MJ. Signal transducer and activator of transcription factor 6 (Stat6)-deficient mice are protected from antigen-induced airway hyperresponsiveness and mucus production. J Exp Med. 1998;187:939–48. doi: 10.1084/jem.187.6.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munitz A, Brandt EB, Mingler M, Finkelman FD, Rothenberg ME. Distinct roles for IL-13 and IL-4 via IL-13 receptor a1 and the type II IL-4 receptor in asthma pathogenesis. Proc Natl Acad Sci USA. 2008;105:7240–5. doi: 10.1073/pnas.0802465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, et al. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258–61. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 13.Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–3. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, Zhang Y, Elias JA. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–88. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter DM, McIntire JJ, Berry G, McKenzie AN, Donaldson DD, DeKruyff RH, et al. Critical role for IL-13 in the development of allergen-induced airway hyperreactivity. J Immunol. 2001;167:4668–75. doi: 10.4049/jimmunol.167.8.4668. [DOI] [PubMed] [Google Scholar]
- 16.Finkelman FD, Yang M, Perkins C, Schleifer K, Sproles A, Santeliz J, et al. Suppressive effect of IL-4 on IL-13-induced genes in mouse lung. J Immunol. 2005;174:4630–8. doi: 10.4049/jimmunol.174.8.4630. [DOI] [PubMed] [Google Scholar]
- 17.Follettie MT, Ellis DK, Donaldson DD, Hill AA, Diesl V, DeClercq C, Sypek JP, Dorner AJ, Wills-Karp M. Gene expression analysis in a murine model of allergic asthma reveals overlapping disease and therapy dependent pathways in the lung. Pharmacogenomics J. 2006;6:141–52. doi: 10.1038/sj.tpj.6500357. [DOI] [PubMed] [Google Scholar]
- 18.Lewkowich IP, Herman NS, Schleifer KW, Dance MP, Chen BL, Dienger KM, Sproles AA, Shah JS, Kohl J, Belkaid Y, Wills-Karp M. CD4+CD25+ T cells protect against experimentally induced asthma and alter pulmonary dendritic cell phenotype and function. J Exp Med. 2005;202:1549–61. doi: 10.1084/jem.20051506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santeliz JV, Van Nest G, Traquina P, Larsen E, Wills-Karp M. Amb a 1-linked CpG oligodeoxynucleotides reverse established airway hyperresponsiveness in a murine model of asthma. J Allergy Clin Immunol. 2002;109:455–62. doi: 10.1067/mai.2002.122156. [DOI] [PubMed] [Google Scholar]
- 20.Morris SC, Orekhova T, Meadows MJ, Ruwe SM, Yang J, Finkelman FD. IL-4 induces in vivo production of IFN-γ by NK and NKT cells. J Immunol. 2006;176:5299–305. doi: 10.4049/jimmunol.176.9.5299. [DOI] [PubMed] [Google Scholar]
- 21.Zimmerman N, King NE, Laporte J, Yang M, Mishra A, Pope SM, Muntel EE, Witte DP, Pegg AA, Foster PS, Hamid Q, Rothenberg ME. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest. 2003;111:1863–74. doi: 10.1172/JCI17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang M, Rangasamy D, Matthaei KI, Frew AJ, Zimmmermann N, Mahalingam S, et al. Inhibition of arginase I activity by RNA interference attenuates IL-13-induced airways hyperresponsiveness. J Immunol. 2006;177:5595–603. doi: 10.4049/jimmunol.177.8.5595. [DOI] [PubMed] [Google Scholar]
- 23.Morris CR, Poljakovic M, Lavrisha L, Machado L, Kuypers FA, Morris SM., Jr Decreased arginine bioavailability and increased serum arginase activity in asthma. Am J Respir Crit Care Med. 2004;170:148–53. doi: 10.1164/rccm.200309-1304OC. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, et al. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–82. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 25.Webb DC, McKenzie AN, Foster PS. Expression of the Ym2 lectin-binding protein is dependent on interleukin (IL)-4 and IL-13 signal transduction: identification of a novel allergy-associated protein. J Biol Chem. 2001;276:41969–76. doi: 10.1074/jbc.M106223200. [DOI] [PubMed] [Google Scholar]
- 26.Kuperman DA, Lewis CC, Woodruff PG, Rodriguez MW, Yang YH, Dolganov GM, et al. Dissecting asthma using focused transgenic modeling and functional genomics. J Allergy Clin Immunol. 2005;116:305–11. doi: 10.1016/j.jaci.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Martin LD, Minnicozzi M, Greenfeder S, Fine J, Pettersen CA, Chorley B, Adler KB. Enhanced expression of mucin genes in a guinea pig model of allergic asthma. Am J Respir Cell Mol Biol. 2001;25:644–51. doi: 10.1165/ajrcmb.25.5.4485. [DOI] [PubMed] [Google Scholar]
- 28.Nakanishi A, Morita S, Iwashita H, Sagiya Y, Ashida Y, Shirafuji H, Fujisawa Y, Nishimura O, Fujino M. Role of gob-5 in mucus overproduction and airway hyperresponsiveness in asthma. Proc Natl Acad Sci USA. 2001;98:5175–80. doi: 10.1073/pnas.081510898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmermann N, Mishra A, King NE, Fulkerson PC, Doepker MP, Nikolaidis NM, Kindinger LE, Moulton EA, Aronow BJ, Rothenberg ME. Transcript signatures in experimental asthma: identification of STAT6-dependent and -independent pathways. J Immunol. 2004;172:1815–24. doi: 10.4049/jimmunol.172.3.1815. [DOI] [PubMed] [Google Scholar]
- 30.Kuperman DA, Huang X, Koth LL, Chang GH, Dolganov GM, Zhu Z, et al. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat Med. 2002;8:885–9. doi: 10.1038/nm734. [DOI] [PubMed] [Google Scholar]
- 31.Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–74. doi: 10.1146/annurev.immunol.24.021605.090720. [DOI] [PubMed] [Google Scholar]
- 32.Zimmermann N, Doepker MP, Witte DP, Stringer KF, Fulkerson PC, Pope SM, et al. Expression and regulation of small proline-rich protein 2 in allergic inflammation. Am J Respir Cell Mol Biol. 2005;32:428–35. doi: 10.1165/rcmb.2004-0269OC. [DOI] [PubMed] [Google Scholar]
- 33.Tesfaigzi J, Th'ng J, Hotchkiss JA, Harkema JR, Wright PS. A small proline-rich protein, SPRR1, is upregulated early during tobacco smoke-induced squamous metaplasia in rat nasal epithelia. Am J Respir Cell Mol Biol. 1996;14:478–86. doi: 10.1165/ajrcmb.14.5.8624253. [DOI] [PubMed] [Google Scholar]
- 34.Tesfaigzi J, Carlson DM. Expression, regulation, and function of the SPR family of proteins: a review. Cell Biochem Biophys. 1999;30:243. doi: 10.1007/BF02738069. [DOI] [PubMed] [Google Scholar]
- 35.Artis D, Wang ML, Keilbaugh SA, He W, Brenes M, Swain GP, Knight PA, Donaldson DD, Lazar MA, Miller HR, Schad GA, Scott P, Wu GD. RELMbeta/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc Natl Acad Sci USA. 2004;101:13596–600. doi: 10.1073/pnas.0404034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishra A, Wang M, Schlotman J, Nikolaidis NM, DeBrosse CW, Karow ML, Rothenberg ME. Resistin-like molecule-beta is an allergen-induced cytokine with inflammatory and remodeling activity in the murine lung. Am J Physiol Lung Cell Mol Physiol. 2007;292:L305–13. doi: 10.1152/ajplung.00147.2007. [DOI] [PubMed] [Google Scholar]
- 37.Fritzsche FR, Dahl E, Dankof A, Burkhardt M, Pahl S, Peterson I, Dietel M, Kristiansen G. Expression of AGR2 in non small cell lung cancer. Histol Histopathol. 2007;22:703–8. doi: 10.14670/HH-22.703. [DOI] [PubMed] [Google Scholar]
- 38.Tsuji S, Uehori J, Matsumoto M, Suzuki Y, Matsuhisa A, Toyoshima K, et al. Human intelectin is a novel soluble lectin that recognizes galactofuranose in carbohydrate chains of bacterial cell wall. J Biol Chem. 2001;276:23456–63. doi: 10.1074/jbc.M103162200. [DOI] [PubMed] [Google Scholar]
- 39.Pemberton AD, Knight PA, Wright SH, Miller HR. Proteomic analysis of mouse jejunal epithelium and its response to infection with the intestinal nematode, Trichinella spiralis. Proteomics. 2004;4:1101–8. doi: 10.1002/pmic.200300658. [DOI] [PubMed] [Google Scholar]
- 40.Pemberton AD, Knight PA, Gamble J, Colledge WH, Lee JK, Pierce M, et al. Innate BALB/c enteric epithelial responses to Trichinella spiralis: inducible expression of a novel goblet cell lectin, intelectin-2, and its natural deletion in C57BL/10 mice. J Immunol. 2004;173:1894–901. doi: 10.4049/jimmunol.173.3.1894. [DOI] [PubMed] [Google Scholar]
- 41.Wright EM, Turk E. The sodium/glucose cotransport family SLC5. Pflugers Arch. 2004;447:510–8. doi: 10.1007/s00424-003-1063-6. [DOI] [PubMed] [Google Scholar]
- 42.Ehre C, Rossi AH, Abdullah LH, DePestel K, Hill S, Olsen JC, Davis CW. Barrier role of actin filaments in regulated mucin secretion from airway goblet cells. Am J Physiol Cell Physiol. 2005;288:C46–56. doi: 10.1152/ajpcell.00397.2004. [DOI] [PubMed] [Google Scholar]
- 43.Praphanphoj V, Sacksteder KA, Gould SJ, Thomas GH, Geraghty MT. Identification of the alpha-aminoadipic semialdehyde dehydrogenase-phosphopantetheferase gene, the human ortholog of the yeast LYS5 gene. Mol Genet Metab. 2001;72:336–42. doi: 10.1006/mgme.2000.3138. [DOI] [PubMed] [Google Scholar]
- 44.LaPorte SL, Juo ZS, Vaclaikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–72. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13Ra2 receptor is involved in induction of TGF-B1 production and fibrosis. Nat Med. 2006;12:99–106. doi: 10.1038/nm1332. [DOI] [PubMed] [Google Scholar]
- 46.Khodoun M, Lewis C, Yang J, Orekhova T, Potter C, Wynn T, Mentink-Kane M, Khurana Hershey GK, Wills-Karp M, Finkelman FD. Differences in expression, affinity, and function of soluble (s)IL-4Ra and sIL-13Ra2 suggest opposity effects on allergic responses. J Immunol. 2007;179:6429–38. doi: 10.4049/jimmunol.179.10.6429. [DOI] [PubMed] [Google Scholar]
- 47.Perkins C, Wills-Karp M, Finkelman FD. IL-4 indues IL-13-independent allergic airway inflammation. J Allergy Clin Immunol. 2006;118:410–9. doi: 10.1016/j.jaci.2006.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.