Abstract
We aimed to examine the physical interaction between CtBPs and KLF4 and the potential importance of this interaction. Co-immunoprecipitation indicated that CtBP1 indeed interacted with KLF4. This was supported by the co-localization of both KLF4 and CtBP1 in the promoter regions of KLF4 downstream target genes. In addition, overexpression of CtBP1 significantly decreased KLF4-mediated transcriptional activation in both an artificial (pGL5) and genuine (IAP and Keratin-4) reporter system. Mutations in the potential CtBP binding motif in KLF4 were accompanied by loss of the inhibitory effect of CtBP1 in the reporter assay and of the physical interaction with CtBP1. Overall, our results suggest that CtBPs attenuate KLF4-mediated transcriptional activation through the physical interaction with KLF4.
Keywords: CtBPs, KLF4, transcription, co-immunoprecipitation, chromatin immunoprecipitation, transient co-transfection
1. Introduction
Regulation of gene expression is a fundamental cellular process. Critical to the control of gene expression are transcription factors that bind to specific DNA sequences and subsequently modulate gene transcription. A family of the C2H2-zinc finger proteins exhibits homology to the Drosophila melanogaster segmentation gene product, Krüppel 3, hence the name Krüppel-like factors (KLFs). KLFs play important roles in many fundamental biologic processes, including development, proliferation, differentiation and apoptosis (1). One member of this family, Krüppel-like factor 4 (KLF4), also known as gut-enriched Krüppel-like factor (GKLF), was found to be highly expressed in epithelial cells of many tissues, including small intestine and skin (2). In these tissues, by directly binding to CACCC motif in the promoter region of its target genes, KLF4 transcriptionally regulates its target genes resulting in inhibition of proliferation and promotion of terminal differentiation (3, 4).
C-terminal binding proteins (CtBPs) family proteins are modulators of several essential cellular processes (5). Vertebrate genomes code for two related proteins, CtBP1 and CtBP2. CtBP family proteins are highly conserved. The founding member of this family, CtBP1 was discovered as a cellular protein that interacted with the C-terminus of adenovirus E1A proteins (6). CtBPs function as transcriptional co-repressors. A large number of sequence-specific DNA-binding transcriptional repressors of Drosophila and mammalian species mediate their activity in a CtBP-dependent manner (7). At present, more than thirty different vertebrate transcriptional regulators have been reported to modulate their activity through recruitment of CtBPs.
The various DNA-binding repressors recruit CtBP through a conserved CtBP-binding motif, a Pro-X-Asp-Leu-Ser (PXDLS) CtBP interaction domain (Fig. 1A) originally identified in adenovirus E1A protein (8). A PYDLA motif was also found in the N-terminal region of human KLF4 protein (aa53-57, Fig. 1A). In the present study, we aimed to examine the physical interaction between CtBPs and KLF4 and the potential importance of this interaction. Our findings suggest that CtBPs attenuate KLF4-mediated transcriptional activation, and this attenuation is most likely fulfilled through directly interacting with KLF4 via PYDLA motif.
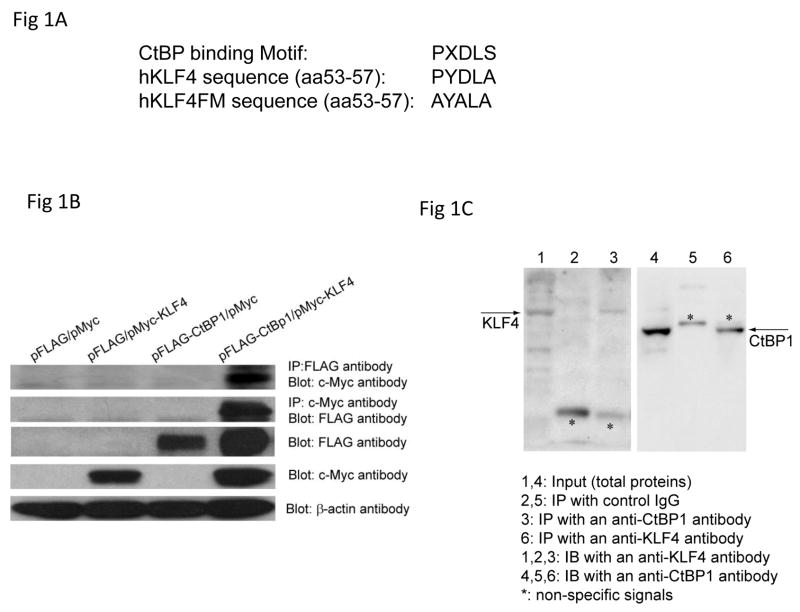
Figure 1.
CtBP1 associated with KLF4. A: The sequences of CtBP consensus binding motif, potential hKLF4 CtBP binding motif and the mutant hKLF4 binding motif. B. Co-IP and Western Blot analysis to examine the physical interaction between pMyc-KLF4 and pFLAG-CtBP1. Transient transfections using AGS cells, Co-IP, and Western blotting were performed as described in Materials and Methods. C. Endogenous Co-IP and Western Blot analysis. Protein extracts from AGS cells and anti-KLF4 and anti-CtBP1 antibodies were used for the assay.
2. Materials and Methods
2.1 Cell culture
AGS cells and KLF4 Mouse embryo fibroblasts (MEFs) (KLF4+/−, KLF4 −/−) cells or CtBP MEFs (CtBP+/−, CtBP−/−) cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) similarly as described (9). Human colon cancer HCT116 cells were grown in complete McCoy’s medium.
2.2 Plasmids
Plasmids used for artificial promoter reporter assay were as follows: pGL5-Luc reporter plasmid, pM vector, pM/KLF4, pFLAG vector, pFLAG-CtBP1, pFLAG-CtBP2 (from Dr. G. Chinnadurai), and pM/KLF4FM, a mutated version of KLF4 that harbors point mutations in potential CtBP binding motif (Fig. 1A), IAP and KRT luciferase reporter constructs (from Drs C. Liu and A. Rustgi respectively), pcDNA 3/Myc-His B vector (pMyc), pMyc-KLF4, and pMyc-KLF4FM. All the cloned plasmids have been sequence confirmed.
2.3. Transient transfection and co-immunoprecipitation assays
Plasmids were transfected into indicated cells using Lipofectamine 2000 (Invitrogen). Proteins were extracted, followed by Western blotting analysis using mouse anti-FLAG tag (SIGMA), rabbit anti-Myc tag (ABR), rabbit anti-human β-actin (SIGMA) antibodies. Endogenous Co-IP experiments were similarly performed using an anti-KLF4 antibody (Santa Cruz) and an anti-CtBP1 antibody (Santa Cruz) together with IgG controls.
2.4. Chromatin immunoprecipitation (ChIP) assays
CHIP assays using AGS or KLF4 MEFs cells were performed as previously described (9). Extracted DNA samples from transfected AGS cells were used to amplify promoter fragments of KLF4 target genes. The promoter fragments of target human genes were amplified and the primer pairs were as follows: CCND1 (Upper: 5’-TCTACACCCCCAACAAAACCAA-3’; Lower: 5’-ACTCTTCGGGCTGCCTTCCTAC-3’), p21 (Upper: 5’-GACCGGCTGGCCTGCTGGAACT-3’; Lower: 5’-GCACGCTTGGCTCGGCTCGGCTCTGG-3’), IAP (Upper: 5’-GGG CCC ATG GAA AAC AGA CTC A-3’; Lower: 5’-AGACGCGTTGCCACTCCTTCAT-3’), Laminin α 1 (Upper: 5’-GGCAAACAAAGTCGGGAACAAG-3’; Lower: 5’-TAGGAGGTGGGC AGAGAAGGTG-3’), HDC (Upper: 5’-GAACTGAGGGCTCTTTTACG-3’; Lower: 5’-CAGTGTGGGCCCTTTATTTA-3’ ) and Keratin-4 (Upper: 5′-GATCGCCACCTACCGCAAACTG-3′; Lower: 5’-GAGCCGGAGCCAAAGCCACTAC-3’). Extracted DNA samples from transfected KLF4 MEFs cells were used to amplify promoter fragments of KLF4 targets mouse genes. The primer pairs used were as follows: CCND1 (Upper: 5’-CCCCCAGCGAGGAGGAA-3’; Lower: 5’-AGGCGGACCCATTGCTTAGA-3’), IAP (Upper: 5’-CACCCAACCTGCAGGAGACAT A -3′; Lower: 5’-GCTAGGGAACCAAGGCACCAG-3′), p21 (Upper: 5’-CTG CCT CCC GAG TGC TGT G -3′; Lower: 5’-GGG GCC CCG ATG GTA CCG -3’), HDC (Upper: 5’-TGGCAATTCTTCCCCCTTACG -3′; Lower: 5’-GCTCCTGCCCTGGCTTCTCTAT-3’) and Keratin-4 (Upper: 5′ CTTGGCCTGGGTAGCGGTTTTT 3′; Lower: 5’ TGATGGTGGCGGAAGAGGTGAT3’). PCR products were analyzed on 2.0% agarose gels containing ethidium bromide (0.25μg/ml).
2.5. Transient cotransfection and dual luciferase assays
HCT116 cells were used for transient co-transfection and dual luciferase assay using a previous published protocol (9).
2.6. Total RNA isolation and quantitative Real-Time PCR
CtBP MEFs cells were grown to 80% confluence in 6-well plates and then the culture medium was replaced by DMEM without FBS for another 1h, 2h, 4h and 8h, respectively. RNA samples were then prepared, followed by the complimentary DNA (cDNA) synthesis, and quantitative real-time PCR (qRT-PCR) analysis using the following primer pairs of mouse origin: KLF4-5RT (5′ CAAGTCCCGCCGCTCCATTACCAA 3′) and KLF4-3RT (5′ CCACAGCCGTCCCAGTCACAGTGG 3′) for KLF4 gene, IAP-5RT (5′-CGCCTATCTCTGTGGGGTCAAG -3′) and IAP-3RT (5′-TAGGTGCCGGCTGGAGAGG -3′) for the IAP gene, Keratin-4-RT5 (5′ CTTGGCCTGGGTAGCGGTTTTT 3′) and Keratin-4-RT3 (5′ TGATGGTGGCGGAAGAGGTGAT 3′) for Keratin-4 gene, CCND1-RT5 (5′ AGCCCCAACAACTTCCTCTCCT 3′) and CCND1-RT3 (5′ TTCCCCCTCCTCCTCAGTGG 3′) for CCND1 gene, and GAPDH-5RT (5′ GACATCAA GAAGGTGGTGAAGC 3′) and GAPDH-3RT (5′ GTC CACCACCCTGTTGCTGTAG 3′) for GAPDH gene. Transcript abundances were first normalized to the levels of GAPDH RNA, and then to their own levels at the 0 time point in the presence of CtBP.
3. Results and Discussion
3.1 KLF4 interacts with CtBP1
Two Krüppel like family members, BKLF/KLF3 and KBKF3/KLF8 have been reported to interact with CtBP through a PVDLT motif, which is highly similar to CtBP consensus binding motif PXDLS (10, 11). Sequence analysis revealed a potential CtBP binding motif at the N-terminal of KLF4 (PYDLA, Fig 1A). To test the putative KLF4 and CtBP1 interaction, Co-immunoprecipitation was performed. AGS cells, human gastric cancer cells where KLF4 is expressed, were cotransfected with pMyc-KLF4 and pFLAG-CtBP1 expression vectors. Cell lysates from transfected cells were then immunoprecipitated with an anti-Myc antibody and an anti-FLAG antibody followed by immunoblotting with an anti-Flag antibody and an anti-Myc antibody respectively. Immunoprecipation of pMyc-KLF4 was accompanied by detection of pFLAG-CtBP1, and vice versa (Fig 1B). Consistently, endogenous Co-IP also showed the physical association between KLF4 and CtBP1 (Fig 1C). Taken together, these results indicated that CtBP1 interacts with KLF4.
3.2 KLF4 and CtBP1 co-localized in promoter regions of some KLF4 downstream target genes
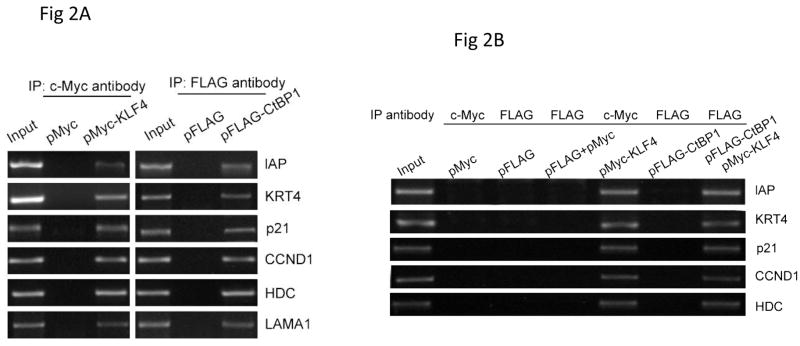
CtBPs have been shown to act as transcriptional corepressor of many transcription factors that are involved in important signaling transduction pathways, including transforming growth factor beta (TGF-beta) signaling (12, 13) and Notch signaling (14). If CtBP1 indeed interacts with KLF4, it is expected that both proteins colocalize on the promoter regions of KLF4 downstream target genes. To test this possibility, chromatin immunoprecipitation assays were performed using a colon cancer cell line (HCT 116 cells). These cells were transfected with pMyc-KLF4 or pFlag-CtBP1 together with vector controls, followed by ChIP assays using an anti-Myc or an anti-Flag antibody. As shown in Fig. 2A, DNA fragments of promoter regions of some KLF4 human target genes, including Intestinal alkaline phosphatase (IAP) (15), Keratin-4 (KRT4) (16), p21 (17), Cyclin D1 (CCND1) (18), Histidine decarboxylase (HDC) (19), and Laminin α1 (LAMA1) (20), were amplified from both Myc IPs using pMyc-KLF4-transfected elutes and Flag IPs using pFLAG-CtBP1-transfected elutes. Under the same condition, no bands were seen from control vectors-transfected elutes, indicating that KLF4 and CtBP1 both are localized on the promoter regions of these KLF4 target genes. To further examine the binding of CtBP1 with the promoters are dependent on KLF4, KLF4 knockout MEFs were used for CHIP assays. As shown in Fig. 2B, whereas DNA fragments of promoter regions of some KLF4 mouse target genes, including IAP, KRT4, p21, CCND1, and HDC, were amplified after KLF4 transfection. These fragments were barely seen from Flag IPs using pFLAG-CtBP1-transfected elutes without KLF4 transfection. However, after pMyc-KLF4 and pFLAG-CtBP1 cotransfection, these bands are robustly amplified, indicating that CtBP1 binding with these promoters are KLF4-dependent, which further supports the interaction between CtBP1 and KLF4.
Figure 2.
CtBP1 co-localized with KLF4 on the promoters of KLF4 downstream target genes. A. AGS cells were transfected by pMyc-KLF4, pFLAG-CtBP1, and the respective vectors. CHIP assays were performed to examine the occupancy of KLF4 and CtBP1 on the promoters of KLF4 downstream target genes, including IAP, KRT4, p21, CCND1, HDC and LAMA1. (B). pMyc-KLF4, pFLAG-CtBP1 KLF4, and the respective vectors with different combinations were transfected into KLF4 null MEFs for CHIP assays. Occupancy of CtBP1 on the promoters of IAP, KRT4, p21, CCND1, and HDC were examined to test the dependence of KLF4.
3.3. CtBPs attenuated KLF4-mediated transcriptional activation
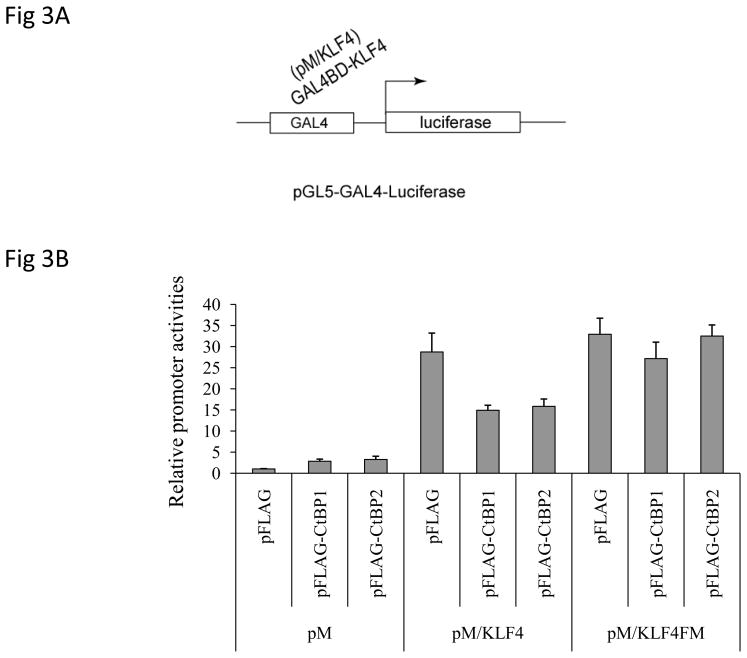
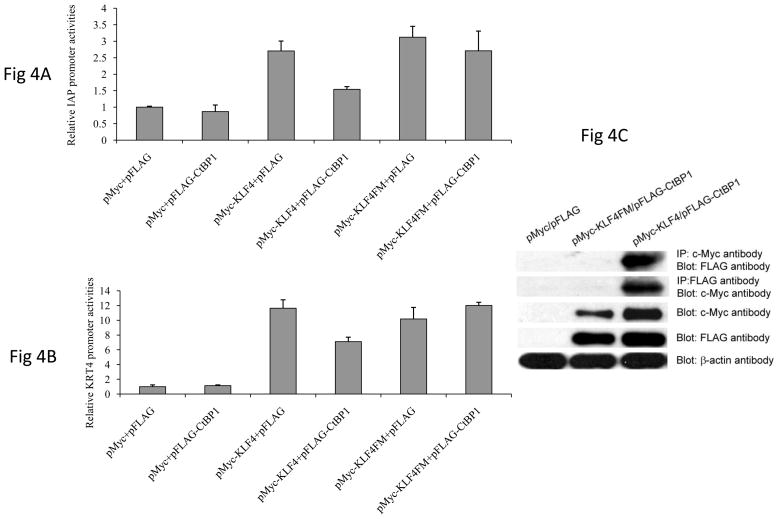
To analyze the role of CtBP1 in KLF4-mediated gene transcription regulation, we first used an artificial reporter system, where KLF4 was recruited to the promoter region to activate transcription (Fig 3A) (9), to perform a reporter assay. Since CtBP1 and CtBP2 have overlapping functions (21), we used both CtBP1 and CtBP2 expression constructs. As seen in Fig 3B, while CtBP1 and CtBP2 slightly activated the promoter activity, they significantly compromised KLF4-mediated transcriptional activation (about 50% decrease). Interestingly, when we introduced mutations in CtBP binding motif within KLF4, it did not significantly influence KLF4-induced transcriptional activation. However, either CtBP1 or CtBP2 attenuated KLF4-mediated activity, suggesting that CtBP binding motif in KLF4 is critical for CtBPs to regulate the activity of KLF4. Similarly, when the reporter constructs of two KLF4 target genes, including IAP and KRT4, were used in the same assay, CtBP1 significantly compromised KLF4-mediated reporter activation. In addition, when the same mutant KLF4, which harbors mutations in CtBP binding motif, was used, CtBP1 no longer inhibited KLF4-mediated reporter activation (Fig 4A and 4B). All these data suggest that CtBP inhibited KLF4-mediated transcriptional activation through CtBP binding motif. In order to examine if the mutant KLF4 used in the reporter assay lost its ability to interact with CtBP1, co-immunoprecipitation assay was performed. As shown in Fig 4C, while wild type pMyc-KLF4 was co-immunoprecipitated with pFLAG-CtBP1, the mutant KLF4 was not able to do so, indicating that mutations in CtBP binding motif in KLF4 abolished the physical interaction between KLF4 and CtBP1.
Figure 3.
CtBPs attenuated KLF4-mediated transcriptional activation. (A): A diagram of an artificial pGL5-Luciferase reporter system was shown, where KLF4 is recruited to the proximal promoter region to activate transcription. (B): Overexpression of CtBP1 and CtBP2 significantly reduced KLF4-mediated transcriptional activation. Transient co-transfections with pGL5 reporter construct and pM vector, pM/KLF4, pM/KLF4 and pFLAG-CtBP1 and pFLAG-CtBP2 with different combination were performed in HCT116 cells, followed by dual luciferase assay as described in Materials and Methods. Means ± SD for 3 independent experiments were shown.
Figure 4.
CtBPs attenuated KLF4-mediated IAP and KRT4 reporter activation. Similar to Fig 3B except IAP promoter reporter (A) or KRT4 reporter (B) construct was used instead of pGL5 reporter. C. Mutations in CtBP binding motif in KLF4 abolished interaction between KLF4 and CtBP1. Wild type (pMyc-KLF4) and mutant KLF4 (pMyc-KLF4M) that harbors mutations in CtBP binding motif were cotransfected with pFLAG-CtBP1 and co-immunoprecipatation assays were performed to examine the interaction between KLF4 and CtBP1 as described in Figure 1B.
3.4. Expression of KLF4 downstream target genes is dependent on the status of CtBPs
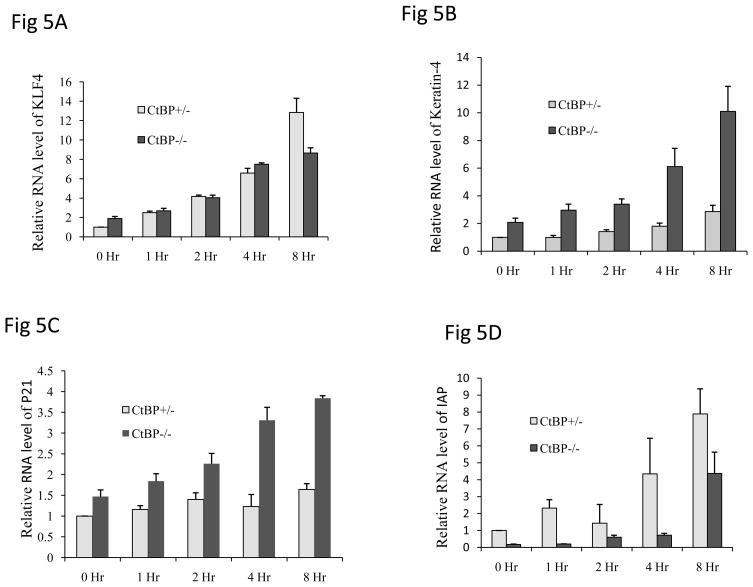
Overexpression of CtBP1 or CtBP2 or in combination of both in vitro resulted in no significant changes of KLF4 expression (data not shown). We then serum starved the cells, which is an efficient way to induce KLF4 expression. In order to further examine the importance of physical interaction between KLF4 and CtBPs, we measured the expression of KLF4 downstream target genes by quantitative RT-PCR (qRT-PCR) after KLF4 upregulation and in the presence and absence of CtBP. CtBP+/− MEFs and CtBP−/− (CtBP1-CtBP2 -) MEFs were serum-starved for 1h, 2h, 4h and 8h. The RNA samples from these serum-starved and normally cultured cells were used in qRT-PCR to study the changes in RNA levels of KLF4 and some of its target genes. KLF4 RNA levels of both CtBP−/− and CtBP+/− MEFs were steadily increased when cells were serum-starved from 1h toward 8h, with the highest RNA level at 8h time point (4.59 fold and 12.83 fold increase respectively when compare to that of 0h time point, Fig. 5A). Consistent with the upregulation of KLF4 upon serum starvation, KRT4 mRNA levels were similarly increased from 1hr to 8hr serum starvation. However, while the basal levels of KLF4 and KRT4 are very similar when cells are not serum starved in both CtBP−/− and CtBP+/− cells, with serum starvation, the fold increase of KRT4 in CtBP−/− MEFs are much higher than CtBP+/− MEFs (Fig 5B), that is significantly different from KLF4 increase upon serum starvation. It appears that fold increase are very similar in both CtBP−/− and CtBP+/− MEFs (Fig 5A). Although there might be other mechanisms for this difference, based on our studies we would argue that CtBPs downregulate KLF4-mediated transcriptional activation. In the absence of CtBP, this inhibitory effort is relieved, resulting in the upregulation of the downstream target genes such as KRT4. A similar observation was also seen for p21, another KLF4 target gene (Fig 5C). It should be noted that not all KLF4 downstream target gene behave the same as KRT4 and p21, such as IAP and CCND1 (Fig 5D and data not shown), which suggests the complexity of transcriptional regulation mediated by KLF4. For example, KLF3, another known CtBP binder (22), is activated by KLF4 (23) which also binds CtBP from our current study. It is likely that the CtBP-dependent effects may be mediated through KLF3 rather than KLF4. On the other hand, we can not exclude the possibility that KLF4 mediates the repression. Furthermore, KLF4 downstream target genes are not necessarily solely, or exclusively KLF4 targets. KLF3, which can be activated by KLF4, also represses KLF8, and KLF8 is also involved in the regulation of KLF4, so the situation is complex, and it may be difficult to separate the effects (24, 25). The existence of such cross-regulation and potential redundancy between KLF family members makes it difficult to precisely explain how KLF4 represses gene expression through CtBPs.
Figure 5.
Expression of KLF4 downstream target genes is dependent on the status of CtBPs. CtBP+/- and CtBP−/− MEFs were cultured in the presence and absence of serum for 1 to 8 hrs. Total RNA samples were prepared at each time point followed by quantitative RT-PCR analysis to examine the expression levels of KLF4 (A), KRT4 (B), p21 (C), and IAP (D). Transcript abundances were normalized to levels of GAPDH RNA and were expressed as relative values. Means ± SD for 3 independent experiments were shown.
In summary, our current study provides evidence of directly interaction between KLF4 and CtBPs. It appears that this interaction is critical for CtBPs to attenuate KLF4-mediated transcriptional activation, since mutations within the CtBP binding motif in KLF4 not only abrogated the physical interaction between KLF4 and CtBP1, but also abolished the inhibitory effort of CtBPs. At the present time, the physiological importance of the interaction remains unknown. However, given the importance of the downstream target of KLF4, including p21 and CCND1, in the cell cycle control and tumor progression (26), it is likely that CtBP-mediated attenuation of KLF4 activity plays an critical role in these processes. Recently, KLF4 has been shown to have important functions in stem cell biology (27, 28). On the other hand, CtBP partially mediated the effect of TCF3 in maintaining pluripotency and self-renewal of mouse embryonic stem cells (29). It will be very interesting to examine if the regulation of KLF4 by CtBPs plays any roles in stem cell self-renewal or differentiation.
Acknowledgments
This work was supported by an NIH/NIDDK grant to WA (1K01DK069489). pFLAG-CtBP1 and pFLAG-CtBP2 constructs were provided by Dr. G Chinnadurai at Saint Louis University. KLF4 null MEFs (mouse embryonic fibroblasts) were provided by Dr. Vincent Yang at Emory University. CtBP MEFs were provided by Dr. Jeffery D. Hildebrand at University of Pittsburgh. IAP reporter construct was provided by Dr. Chunming Liu at the University of Texas Medical Branch. Keratin 4 (KRT4) reporter construct was provided by Dr. Anil Rustgi at University of Pennsylvania.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Structured summary:
MINT-7261981, MINT-7261995:
KLF4 (uniprotkb:O43474) physically interacts (MI:0915) with CTBP1 (uniprotkb:Q13363) by anti tag coimmunoprecipitation (MI:0007)
MINT-7262008, MINT-7262023:
CTBP1 (uniprotkb:Q13363) physically interacts (MI:0915) with KLF4 (uniprotkb:O43474) by anti bait coimmunoprecipitation (MI:0006)
References
- 1.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 2.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaubert J, Cheng J, Segre JA. Ectopic expression of kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 2003;130:2767–2777. doi: 10.1242/dev.00477. [DOI] [PubMed] [Google Scholar]
- 5.Chinnadurai G. The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res. 2009;69:731–734. doi: 10.1158/0008-5472.CAN-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 8.Schaeper U, Boyd JM, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc Natl Acad Sci U S A. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ai W, Zheng H, Yang X, Liu Y, Wang TC. Tip60 functions as a potential corepressor of KLF4 in regulation of HDC promoter activity. Nucleic Acids Res. 2007;35:6137–6149. doi: 10.1093/nar/gkm656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Vliet J, Turner J, Crossley M. Human Kruppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic Acids Res. 2000;28:1955–1962. doi: 10.1093/nar/28.9.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner J, Crossley M. Basic Kruppel-like factor functions within a network of interacting haematopoietic transcription factors. Int J Biochem Cell Biol. 1999;31:1169–1174. doi: 10.1016/s1357-2725(99)00067-9. [DOI] [PubMed] [Google Scholar]
- 12.Izutsu K, Kurokawa M, Imai Y, Maki K, Mitani K, Hirai H. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor beta signaling. Blood. 2001;97:2815–2822. doi: 10.1182/blood.v97.9.2815. [DOI] [PubMed] [Google Scholar]
- 13.Lin X, Liang YY, Sun B, Liang M, Shi Y, Brunicardi FC, Feng XH. Smad6 recruits transcription corepressor CtBP to repress bone morphogenetic protein-induced transcription. Mol Cell Biol. 2003;23:9081–9093. doi: 10.1128/MCB.23.24.9081-9093.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knochel W, Borggrefe T. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinnebusch BF, Siddique A, Henderson JW, Malo MS, Zhang W, Athaide CP, Abedrapo MA, Chen X, Yang VW, Hodin RA. Enterocyte differentiation marker intestinal alkaline phosphatase is a target gene of the gut-enriched Kruppel-like factor. Am J Physiol Gastrointest Liver Physiol. 2004;286:G23–30. doi: 10.1152/ajpgi.00203.2003. [DOI] [PubMed] [Google Scholar]
- 16.Okano J, Opitz OG, Nakagawa H, Jenkins TD, Friedman SL, Rustgi AK. The Kruppel-like transcriptional factors Zf9 and GKLF coactivate the human keratin 4 promoter and physically interact. FEBS Lett. 2000;473:95–100. doi: 10.1016/s0014-5793(00)01468-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, Biggs JR, Kraft AS, Yang VW. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shie JL, Chen ZY, Fu M, Pestell RG, Tseng CC. Gut-enriched Kruppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 2000;28:2969–2976. doi: 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ai W, Liu Y, Langlois M, Wang TC. Kruppel-like factor 4 (KLF4) represses histidine decarboxylase gene expression through an upstream Sp1 site and downstream gastrin responsive elements. J Biol Chem. 2004;279:8684–8693. doi: 10.1074/jbc.M308278200. [DOI] [PubMed] [Google Scholar]
- 20.Piccinni SA, Bolcato-Bellemin AL, Klein A, Yang VW, Kedinger M, Simon-Assmann P, Lefebvre O. Kruppel-like factors regulate the Lama1 gene encoding the laminin alpha1 chain. J Biol Chem. 2004;279:9103–9114. doi: 10.1074/jbc.M305804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;22:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel S, Xi ZF, Seo EY, McGaughey D, Segre JA. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc Natl Acad Sci U S A. 2006;103:18668–18673. doi: 10.1073/pnas.0608658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei H, Wang X, Gan B, Urvalek AM, Melkoumian ZK, Guan JL, Zhao J. Sumoylation delimits KLF8 transcriptional activity associated with the cell cycle regulation. J Biol Chem. 2006;281:16664–16671. doi: 10.1074/jbc.M513135200. [DOI] [PubMed] [Google Scholar]
- 25.Eaton SA, Funnell AP, Sue N, Nicholas H, Pearson RC, Crossley M. A network of Kruppel-like Factors (Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J Biol Chem. 2008;283:26937–26947. doi: 10.1074/jbc.M804831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murakami Y, Sekiya T. Accumulation of genetic alterations and their significance in each primary human cancer and cell line. Mutat Res. 1998;400:421–437. doi: 10.1016/s0027-5107(98)00031-1. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, McClintick J, Zhong L, Edenberg HJ, Yoder MC, Chan RJ. Murine embryonic stem cell differentiation is promoted by SOCS-3 and inhibited by the zinc finger transcription factor Klf4. Blood. 2005;105:635–637. doi: 10.1182/blood-2004-07-2681. [DOI] [PubMed] [Google Scholar]
- 28.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 29.Tam WL, Lim CY, Han J, Zhang J, Ang YS, Ng HH, Yang H, Lim B. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26:2019–2031. doi: 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]