Abstract
β-2-microglobulin (β2m) deposits as amyloid fibrils in the musculoskeletal system of patients undergoing long-term dialysis treatment as a result of kidney failure. Previous work has shown that Cu(II) binding causes β2m to organize into native-like dimers and tetramers that precede amyloid formation. Cu(II) is then released from higher order oligomers before mature Cu(II)-free amyloid fibrils are formed. While some of the Cu(II)-induced structural changes that enable β2m self assembly are starting to be revealed, the details of how the Cu(II) binding site evolves from the monomer to the dimers and tetramers are not known. Here, we report results from three mass spectrometry (MS) based methods that provide insight into the changing Cu-β2m interactions. We find that monomeric β2m binds Cu(II) via the N-terminal amine, the amide of Gln2, His31, and Asp59. In the dimer and tetramer, Asp59 is no longer bound to Cu(II), but the other residues still comprise a well-defined albeit weaker binding site that is better able to release Cu(II). Consistent with this is the observation that a fraction of the tetrameric species no longer binds Cu(II) at this weakened binding site, which agrees with a previous report that suggested the tetramer as the first Cu(II)-free oligomer. Our results also provide some insight into structural changes caused by Cu(II) binding that facilitate oligomer formation. Specifically, binding by Asp59 in the monomer requires significant movement of this residue, and we propose that this repositioning is important for establishing a pair of dimer-stabilizing salt bridges between this residue and Lys19. We also find evidence that Cu(II) binding in the N-terminal region of the monomer repels Arg3, which likely allows this residue to form a pair of dimer-stabilizing salt bridges with Glu16. Overall, our measurements suggest that the previously proposed conformational switch caused by Cu(II) binding includes not only a cis-trans isomerization at Pro32 but also the repositioning of residues that are critical for the formation of new electrostatic interactions.
β-2-microglobulin (β2m) is a 12 kDa subunit of the class I major histocompatibility complex and is a structural unit essential for the cell-surface expression of this complex. During normal turnover, β2m is released into serum and is eventually catabolized by the kidney. In patients undergoing hemodialysis as a result of kidney failure, β2m concentrations become elevated in the serum, and after as little as 18 months, β2m amyloids begin to form in the joints of these patients [1]. The cause of β2m amyloid formation in vivo is not precisely known, but several means of causing fibril formation in vitro have been identified. β2m amyloid fibrils can be generated under acidic conditions (pH < 3.6) (2), by removing the first six N-terminal amino acids (3), by mixing the protein with collagen at pH = 6.4 (4), by sonicating the protein in the presence of sodium dodecyl sulfate at pH = 7.0 (5), and by incubating the protein at physiological conditions in the presence of stoichiometric amounts of Cu(II) (6,7).
We have become interested in this latter means of initiating β2m fibril formation for a number of reasons. Several proteins are known to form amyloid fibrils in the presence of divalent metals, especially Cu(II), so Cu(II)-protein interactions seem to represent one of the general motifs for stimulating amyloid formation [8–12]. For β2m, the case has been made [6,13] that Cu(II) could be the initiating factor in vivo because of the elevated Cu concentrations that hemodialysis patients experience and the recent observation that Cu plays a catalytic role in β2m fibril formation [14]. A catalytic role for Cu is suggested by recent studies that found Cu binding to be essential for initiating oligomer formation under near-physiological conditions (i.e. pH 7.4, 37 °C, 150 mM ionic strength) in vitro but not essential for maintaining the stability of tetramers or hexamers and completely absent in the eventual amyloid fibrils [14,15]. While an in vivo role for Cu(II) in β2m amyloid formation is not confirmed, the metal does represent a discrete way to trigger amyloid formation so that the intermediates that precede the fibrils can be more easily studied. The added appeal of studying Cu as a trigger is that the structural changes brought about by Cu binding are subtle and the pre-fibril oligomers have native-like structures [16]. Intriguingly, very similar divalent metals, such as Ni(II), do not cause β2m to self assemble [16], indicating that the mode of Cu binding is unique enough to cause just the right structural changes to facilitate oligomerization and fibril formation.
Given the importance of β2m fibril formation and the unique ways in which Cu catalyzes this reaction, the structural details of how Cu binds the monomer and causes the necessary structural changes for oligomerization are important to determine. Several studies have provided partial insight into Cu-β2m interactions, and there seems to be some agreement that His31 is an important binding site in the monomer [7,17,18], although the N-terminus [17], His51 [18], and His13 [18] have also been suggested as binding sites. Much of the binding information for the wild-type protein has come from NMR studies [7,18]. So, unfortunately the data are somewhat ambiguous because of the non-specific paramagnetic broadening that can occur during NMR analyses of Cu complexes [19]. A recent crystal structure of the hexamer of the H13F mutant of β2m, however, provides atomic level information that offers some clues about the possible Cu binding site in the monomer [20]. In this mutant hexamer, Cu is bound to His31, the N-terminus, and backbone carbonyls near the N-terminus. While this crystal structure provides great atomic-level information about the possible structural changes associated with or caused by Cu binding, this structure does not directly answer questions about the Cu binding site in the monomer for at least two reasons. First, this structure is not of the wild-type protein, and mutation of a possible metal binding site (His13) might cause the binding site of this hexameric mutant to differ from the wild-type monomer. Second, two separate studies have shown that wild-type oligomers tetrameric and/or hexameric in size lose Cu before progressing to form amyloids [14,15]. Given these reasons, more direct evidence about Cu binding in the monomer is needed, and such information should provide additional insight into the potential structural changes caused by Cu binding.
In this manuscript, we apply three mass spectrometry (MS) based methods to identify the specific Cu binding sites in the wild-type monomer and oligomers of β2m. These methods have certain advantages over NMR for determining the β2m-Cu binding sites. First, unlike NMR, these MS based methods are not subject to paramagnetic effects that can complicate data interpretation. Second, the measurements can be done under amyloid forming conditions but at very low protein concentrations where oligomerization does not yet occur, thereby ensuring information about the monomeric form of the protein. Third, the specificity of MS allows us to obtain information about the Cu binding site in the β2m oligomers under conditions in which a mixture of oligomers is present. The first MS method is one developed in our group that uses metal catalyzed oxidation (MCO) reactions to site-specifically oxidize the amino acids bound to redox-active metals and tandem MS (MS/MS) to identify these modified residues [21–25]. We have shown that the MCO/MS method can readily identify the Cu(II) binding residues in peptides and proteins [21–23,25], and we have even provided a partial measure of the binding site in β2m [17]. The current results employ an improved MCO method [22] that has allowed for a more detailed characterization of the monomeric binding site. The second MS based method uses ‘detuned’ MCO reaction conditions to oxidize additional residues that are near Cu. We have recently shown that when the MCO reaction conditions are appropriately varied less specific oxidation can be achieved so that amino acids within about 10 Å of the Cu center can also be oxidized [26]. The third MS method is a covalent labeling method that relies on diethylpyrocarbonate (DEPC) and a succinimide derivative to modify residues with nucleophilic groups (e.g. His, Lys). As with the MCO/MS approach, MS/MS is used to identify the modified residues. These covalent labeling reactions can be used to identify Cu(II) binding sites because reactivity with these reagents is significantly decreased when residues are bound to Cu [25,27,28].
Upon applying these three MS based methods, we find some agreement with previously suggested Cu binding sites, but we also gain new insight into the residues bound to and near Cu that likely play an important role in mediating the structural changes that allow oligomer formation. We also find that the Cu binding site changes, as the new oligomers are formed, in a way that is consistent with the metal being released in the higher order oligomers. Overall, this work highlights not only some of the specific structural changes caused by Cu binding to β2m but also identifies general ways that divalent metal binding could perturb protein structure to facilitate protein aggregation.
Materials and Methods
Materials
Human β-2-microglobulin (β2m) was obtained from Fitzgerald Industries International, Inc. (Concord, MA). Diethylpyrocarbonate (DEPC), sulfo-N-hydroxysuccinimide acetate (NHSA), imidazole, dithiothreitol (DTT), copper(II) sulfate (CuSO4), 3-morpholinopropanesulfonic acid (MOPS), potassium acetate, sodium persulfate, and sodium ascorbate were obtained from Sigma-Aldrich (St. Louis, MO). Tris(hydroxymethyl)-aminomethane (Tris) was purchased from EM Science (Gladstone, NJ). Ammonium acetate, methanol, acetonitrile, and acetic acid were obtained from Fisher Scientific (Fair Lawn, NJ). Urea was purchased from Mallinckrodt Chemicals (Phillipsburg, NJ). Trypsin was from Promega (Madison, WI), and chymotrypsin was purchased from Roche Diagnostics (Indianapolis, IN). Centricon molecular weight cutoff (MWCO) filters were obtained from Millipore (Burlington, MA). Deionized water was prepared from a Millipore (Burlington, MA) Simplicity 185 water purification system.
Formation of β2m Oligomers and Fibrils
We and others have shown that discrete oligomers precede β2m amyloid fibrils when monomeric β2m is incubated under near-physiological conditions in the presence of Cu(II) [14–16]. To obtain these pre-fibril oligomers, a sample solution containing 100 µM β2m, 200 µM copper(II) sulfate, 150 mM potassium acetate, 500 mM urea and 25 mM MOPS (pH 7.4) was incubated at 37 °C. Control experiments without Cu(II) were also performed in which a solution containing 100 µM β2m, 10 mM ethylenediamine tetraacetic acid (EDTA), 150 mM potassium acetate, 500 mM urea, and 25 mM MOPS (pH 7.4) were also incubated at 37 °C. EDTA was added to prevent any association between trace amounts of Cu(II) and β2m. Under these control conditions, no oligomers or fibrils are formed.
Metal-catalyzed Oxidation (MCO) Reactions
The normal MCO reactions of monomeric β2m were performed in the presence of 2.5 μM β2m, 2.5 μM CuSO4, 1.0 mM sodium ascorbate, 0.1 mM sodium persulfate, 25 mM MOPS, 150 mM potassium acetate, and 500 mM urea at pH 7.4. The reactions were initiated by the addition of sodium persulfate and were stopped after 30 min by the addition of 1% (by volume) of glacial acetic acid. The samples were then immediately desalted using a 10,000 MWCO filter and reconstituted with a 25 mM solution Tris-HCl (pH =7) in preparation for proteolytic digestion. Samples that contained β2m oligomers were subjected to the MCO reactions at different time points after initiating the amyloid fibril formation reaction as described above. MCO reagents were added to aliquots taken from the fibril-forming reaction solution to achieve final concentrations of 1.0 mM for sodium ascorbate and 0.1 mM for sodium persulfate. The reactions were stopped after 30 min by the addition of 1% (by volume) of glacial acetic acid. The samples were then prepared for proteolytic digestion as before. The detuned MCO reactions were performed for both the monomeric and oligomeric forms of β2m under the conditions described above except that 100 µM ascorbate and 0.1, 0.2, 0.3, 0.4, or 0.5 mM persulfate were added.
Covalent Modification Reactions
The covalent modification reactions with DEPC or NHSA were performed for 1 min at 37 °C with 100 µM β2m, 200 µM CuSO4, 25 mM MOPS, 150 mM potassium acetate, and 500 mM urea at pH 7.4 and were initiated by adding various molar excesses of each reagent. Different concentrations (0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.8, 1.0, and 1.5 mM) of each reagent were added to generate second order reaction plots so that reaction rate constants for each modified residue could be determined and the protein structure integrity could be ensured [29]. The total reaction volume was 50 µL. CuSO4 was the last reagent added before DEPC or NHSA, and the protein and Cu(II) were allowed to equilibrate for 2 minutes prior to addition of the covalent labeling reagent. The covalent modification reactions with DEPC or NHSA were quenched after 1 min by adding 10 mM imidazole or 10 mM Tris, respectively. The covalently modified samples were then desalted using a 10,000 MWCO filter and reconstituted with deionized water in preparation for proteolytic digestion.
Proteolytic Digestion
Before the addition of trypsin and chymotrypsin, 80 µL solutions containing 1 µg/µL of the unmodified or modified protein in 25 mM Tris-HCl (pH = 7) and 1 mM CaCl2 were incubated with 10 µL of acetonitrile at 40 °C for 45 minutes. Also, to reduce the disulfide bond in β2m, the proteins were reacted with 10 mM DTT at 40 °C for 45 minutes prior to addition of the acetonitrile. Trypsin and chymotrypsin (both 0.5 µg/µL) were then added to yield final enzyme:substrate ratios of 1:20. The protein samples were then digested at 37 °C for 16 hours. The enzymes were inactivated by adding 2 µL of acetic acid, and the samples were immediately frozen at −10 °C and analyzed within 24 hours.
Liquid Chromatography and Mass Spectrometry
MS was used to (i) monitor β2m modifications and identify the amino acids that were modified during the MCO and covalent labeling reactions and (ii) monitor the formation of β2m oligomers after initiating the fibril-forming reaction. To monitor β2m modifications and identify the modified amino acids in β2m, a Bruker Esquire-LC (Billerica, MA) quadrupole ion trap mass spectrometer, which is equipped with an electrospray ionization (ESI) source, was used. Typically, the electrospray needle voltage was kept at 3–3.5 kV, and the capillary temperature was set to 300 °C. The voltages for the transfer optics between the electrospray source and the ion trap were optimized for maximum signal, with typical skimmer 1 and capillary offset values of 30–40 V and 50–60 V, respectively. For direct injection experiments similar source conditions were used, but sample was delivered at 1 µL/min using a syringe pump. Tandem mass spectra were acquired using isolation widths of 1.0 Da and excitation voltages between 0.6 and 1.0 V. Peptide sequences were determined from the tandem MS (MS/MS) data via de novo sequencing or with the help of BioTools™ (Bruker Daltonics, Billerica, MA).
For samples in which β2m was proteolytically digested before mass spectral analysis, high performance liquid chromatography (HPLC) was used to separate the peptide fragments for on-line analysis by MS and MS/MS. For the HPLC experiments, an HP1100 (Agilent, Wilmington, DE) HPLC system with a C18 column (15 cm ×2.1 mm, 5 µm particle size, Supelco, St. Louis, MO) was used. Tryptic fragments were eluted using a linear gradient of methanol containing 0.1% acetic acid that increased from 5% to 90% methanol over 30 min at a flow rate of 0.250 mL/min. The LC effluent was split at a ratio of 1:4 with the smaller fraction of the split flow being fed into the mass spectrometer.
To monitor the oligomers formed during the amyloid reaction, a JMS-700 MStation (JEOL, Tokyo, Japan) double-focusing mass spectrometer equipped with a standard ESI source was used as described previously [14]. The following source conditions were used: desolvating plate temperature, 100 °C; orifice temperature, 140 °C; orifice potential, 0 V; ring lens potential, 100 V. Just prior the ESI-MS analysis, the incubated β2m sample was desalted using a 5 ml HiTrap desalting column from Amersham Biosciences (Piscataway, NJ) that had an exclusion limit of 5,000 Da. The detailed procedure was described previously [14], but two things are important to note here. First, the desalting procedure is very fast and only 2 min elapsed between taking an aliquot from the incubated β2m sample and the acquisition of the ESI mass spectrum. Second, not only did the desalting column remove excess salt that would compromise subsequent ESI-MS analyses, but it also reduced the total concentration of the protein to about one tenth of its original concentration, which is important for minimizing non-specific complex formation during the ESI process.
Thioflavin T Fluorescence
Fluorescence experiments to monitor the rate of β2m dimer formation were performed using a QuantaMaster 4 SE spectrofluorometer (Photon Technology International, Lawrenceville, NJ). A solution containing 100 µM β2m, 80 µM thioflavin T (ThT), 500 mM Urea, 25 mM MOPS and 150 mM potassium acetate was initially equilibrated at 37 °C, and after the addition of 200 µM CuSO4, the fluorescent enhancement of ThT was measured. Measurements were taken at excitation and emission wavelengths of 437 and 483 nm, respectively.
Results
Copper binding to β2m monomer – MCO/MS
Under solution conditions that lead to β2m amyloid formation, identifying the Cu(II) binding site in the monomer is challenging because the protein readily oligomerizes within 30 min of adding Cu(II) (Figure 1), and our MCO/MS approach typically requires a 15 to 30 min reaction. We speculated that lower concentrations of β2m might slow initial dimer formation so that we could apply the MCO approach to probe the Cu-monomer binding site. Even though it was previously shown that the rate of β2m oligomer formation was independent of protein concentration down to 25 µM [16], we found by MS that no statistically significant dimer is formed in the first 30 min at protein concentrations below 5 µM (Figure 1, inset).
Figure 1.
ESI mass spectrum of a desalted β2m sample 30 minutes after beginning the amyloid forming reaction with a protein concentration of 100 µM. The spectrum shows that a small percentage of dimer has already formed. (Inset) Electrospray ionization mass spectrum of a desalted β2m sample 30 minutes after beginning the amyloid-forming reaction with a protein concentration of 5 µM. This spectrum shows that no statistically significant dimer formation at the lower concentration.
Given the low propensity for dimer formation at concentrations below 5 µM, we decided to perform the MCO/MS measurements at Cu(II) and β2m concentrations of 2.5 µM. Based on a previously reported Cu(II)-β2m Kd value of ~ 2.5 µM [6], at these concentrations approximately one-half of the protein should bind Cu(II) at equilibrium. After performing the 30-min MCO reactions, we observe products corresponding to both oxygen additions and protein cleavages (Figure 2) that are absent during control experiments in which the protein is reacted with the MCO reagents in the absence of Cu(II). The addition of up to three oxygen atoms is reproducibly observed, whereas cleavages corresponding to losses of 14, 30, 87, 97, and 113 are consistently observed. Because the peak corresponding to the third oxygen addition is similar in yield to the single oxygen addition in the control experiment (Figure 2, black trace), we conclude that only a total of two oxygen atoms are incorporated into the protein as a result of the MCO process.
Figure 2.
ESI mass spectrum around the +11 charge state of β2m before (black) and after (red) the MCO reaction. Legend: + 1 O = the addition of 16 Da; +2 O = the addition of 32 Da; A = loss of 14 Da; B = loss of 30 Da; C = loss of 87 Da; D = loss of 97 Da; E = loss of 113 Da. Note that before the MCO reaction the protein is already oxidized about 30 – 35% at Met99.
Identifying the specific amino acids that are modified by the MCO reactions, and thus gaining insight into the Cu(II)-monomer binding site, cannot be done directly from the mass spectrum in Figure 2. Instead, proteolytic digestion and LC-MS/MS analyses are required. After comparison of the proteolytic fragments from the modified protein with the fragments from the unmodified protein, 10 new peptide fragments are observed (Table 1). The addition of 16 Da, which is observed in five of the peptides in Table 1, is the most common modification that has been observed in previous MCO/MS studies [21–25]. A mass increase of 16 Da usually corresponds to hydroxylation of an amino acid side chain, except in the case of His residues where the formation 2-oxo-histidine results in carbonyl formation on the His side chain [30,31]. While these fragments indicate regions of the protein that are bound to Cu(II), MS/MS is necessary to pinpoint the modified amino acids. An example of how MS/MS is used to pinpoint the oxidation site is shown in the tandem mass spectrum of [Val27-Lys41+16+3H]3+ (Figure 3). A series of unoxidized y ions from y2 to y10, oxidized y ions from y11 to y14, unoxidized b ions from b2 to b4, and oxidized b ions from b5 to b13 indicate unambiguously that His31 is oxidized during the MCO reactions. MS/MS also identifies oxidized His31 in [Ser20-Lys41+16+3H] 3+, oxidized Ile1 in [Ile1-Arg3+16+H]+, and oxidized Trp60 in [Val49-Tyr63+16+2H]3+ and [Asp59-Tyr63+16+H]+.
Table 1.
New peptide fragments observed after proteolytic digestion of the MCO-modified monomer of β2m.
| Fragment | Observed m/z (charge state) |
|---|---|
| Ile1-Arg3 − 1 | 415.3 (+1) |
| Ile1-Arg3+16 | 432.3 (+1) |
| Gln2-Arg3 | 303.2 (+1) |
| Gln2-Arg3 − 1 | 302.2 (+1) |
| Val27-Lys41 + 16 | 836.4 (+2), 558.3 (+3) |
| Ser20-Lys41 + 16 | 1257.1 (+2), 838.4 (+3) |
| Val49-Tyr63 + 16 | 621.6 (+3) |
| Val49-Tyr63 − 30 | 606.3 (+3) |
| Asp59-Tyr63 + 16 | 733.3 (+1) |
| Asp59-Tyr63 − 30 | 687.3 (+1) |
Figure 3.
Tandem mass spectrum of [Val27-Lys41+16+3H]3+, showing oxidation occurs at His31. The peaks labeled with asterisks correspond to oxidized product ions.
Peptide fragments that show mass losses are also evident from the LC-MS/MS analyses. MS/MS of [Ile1-Arg3 - 1+H]+ indicates that the modification corresponding to a mass decrease of 1 Da occurs at Ile1. This modification most likely corresponds to an oxidative deamination reaction (Scheme 1) that has been observed for reactions of hydroxyl radicals with amino acids and peptides [32] and has also been reported for MCO reactions of proteins that bind Ni at the N-terminal amine [33]. Ions from two peptide fragments, [Val49-Tyr63 - 30+2H]3+ and [Asp59-Tyr63 - 30+H]+, show a loss of 30 Da, which is consistent with an oxidative decarboxylation (Scheme 2) reaction occurring at an acidic side chain. Such reactions have been observed for other peptides during Cu(II)-initiated MCO reactions [34,35] and from acidic peptides reacting with hydroxyl radicals [36]. The tandem mass spectrum of [Val49-Tyr63 - 30+2H]3+ was not conclusive enough to confirm the site of the 30 Da loss, but the 30 Da loss from [Asp59-Tyr63 - 30+H]+ comes from the Asp59 side chain. The failure to measure a 30 Da loss from Ile46-Lys58 or Val49-Lys58 rules out Glu50 or Asp53 as the possible site of modification in [Val49-Tyr63 - 30+2H]3+, making Asp59 the likely modification site in this peptide. Of importance is the fact that of the 14 total measured peptides that contain acidic residues, only the two peptides that contain Asp59 show any evidence of an oxidative decarboxylation reaction. Two other peptides that arise from MCO-induced cleavages, which are absent in the control experiment, are [Gln2-Arg3+H]+ and [Gln2-Arg3 - 1+H]+.The [Gln2-Arg3+H]+ peptide is consistent with the 113 loss from the whole protein and indicates a MCO-induced cleavage of the Ile1-Gln2 peptide bond, which is well established for peptides and proteins reacting with hydroxyl radicals [32]. The loss of 1 Da from the Gln2-Arg3 fragment may arise from a reaction similar to that seen in Scheme 1 but where the initial H abstraction occurs at the α carbon of Gln2.
Scheme 1.
Mechanism for the oxidative deamination of the N-terminus that occurs during the MCO reaction.
Scheme 2.
Mechanism for the oxidative decarboxylation of Asp59 that occurs during the MCO reaction.
No peptide fragments corresponding to losses of 14, 87, or 97 Da could be found, but there are good reasons for the absence of two of these. About 30–35% of the protein is found to be non-specifically oxidized at Met99 in the control experiment. If the loss of 14 Da from the whole protein arises from proteins that are oxidized at Met99 and undergo an oxidative decarboxylation reaction at Asp59, then one would not expect to see a peptide fragment missing 14 Da since no measured peptide fragments contain both Asp59 and Met99. The loss of 97 Da can be explained in a similar way. Oxidation at Met99 and cleavage of the peptide bond between Ile1 and Gln2 would give rise to a net mass change of 97 Da, but no peptide fragment can contain both the N-terminus and Met99. We were unable to find a peptide fragment corresponding to a loss of 87 Da, but we speculate that this mass loss could arise from cleavage of the Ile1 side chain.
When the above MCO/MS data is considered in its entirety, the N-terminus, the amide bond between Ile1 and Gln2, His31, Asp59, and Trp60 are all modified, suggesting that these residues are near or part of the Cu(II) binding site.
Copper binding to β2m monomer – Covalent Labeling
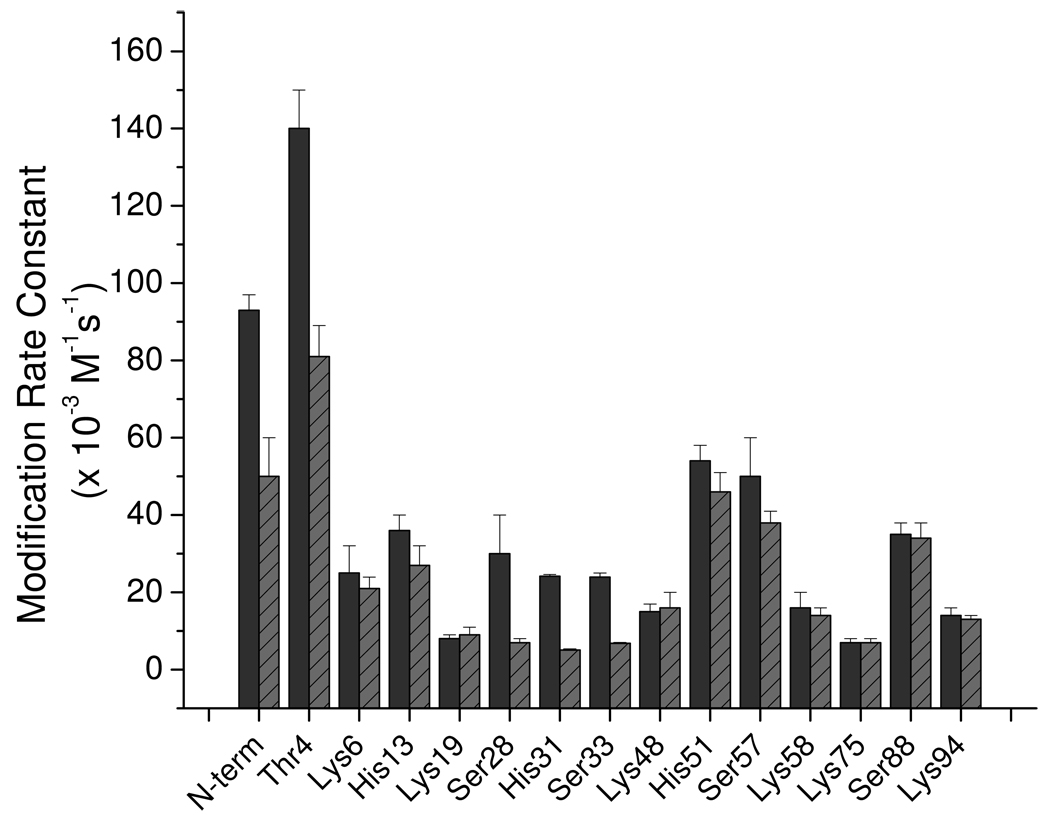
As a complement to the MCO reactions, we also monitored the reactivity of Cu-bound monomeric β2m with two covalent labeling reagents – diethylpyrocarbonate (DEPC) and sulfo-N-hydroxysuccinimide acetate (NHSA). DEPC can react with solvent exposed His, Tyr, Thr, and Ser residues [37], but this reaction is significantly slowed when these residues, especially His, are bound to Cu [25,27,28]. Likewise, NHSA can react with the N-terminus and Lys residues when these sites are solvent exposed [37], but its reactivity is reduced when these residues are protected, for example, by Cu binding. Figure 4 displays the measured 2nd order reaction rate constants for the reactive residues in the presence and absence of Cu(II). The calculated solvent accessible surface areas for all of these residues are listed in Table S1 in the Supporting Information. Of the residues that react with DEPC, Thr4, Ser28, His31, and Ser33 show the greatest drops in reactivity when Cu(II) is present. These residues show an average drop in reactivity of about 70%; no other DEPC-modifiable residue changes in reactivity more than 25%. The significant drop in reactivity of Thr4, Ser28, His31, and Ser33 can be rationalized by the nearby presence of Cu. Importantly, His13 and His51 do not show any significant changes in reactivity. Of the residues that react with NHSA, only the N-terminus is found to decrease in reactivity when Cu(II) is present, and this is probably due to the binding of Cu to the nearby N-terminus. Taken together, the DEPC and NHSA covalent labeling data are consistent with the MCO/MS data, indicating that at least the N-terminal amine and His31 are bound to Cu(II).
Figure 4.
Second order reaction rate constants for residues modified by DEPC and NHSA in the absence (solid lines) and presence (striped lines) of Cu(II). The reaction rate constants for the N-terminus and the lysine residues correspond to the reactions of these residues with NHSA. The rate constants for His, Thr, and Ser residues correspond to the reactions of these residues with DEPC.
Copper binding to β2m monomer – Detuned MCO/MS
Another MCO-related approach was used to examine the residues in the vicinity of the Cu binding site. We recently demonstrated that a so-called ‘detuned’ MCO/MS method could oxidize amino acid residues within about 10 Å of Cu(II) [26], thereby providing additional structural information about the residues around the metal. In this method, the protein is controllably oxidized to a greater extent by increasing the added oxidant and decreasing the amount of ascorbate that acts as a radical scavenger as well as a reducing agent. As described previously [26], the proper controls, such as dose-response plots (see Figure S1 in Supporting Information), are needed to ensure the integrity of the Cu binding site during the detuned reactions. After performing the detuned MCO reactions, the protein is digested and analyzed by LC-MS/MS in a manner identical to that described above for the tuned MCO reactions. Upon performing the detuned MCO reactions and performing the LC-MS/MS analyses, the same 10 modified peptides shown in Table 1 are observed, but oxidation at an additional four amino acids is also measured. The newly oxidized amino acids are Phe30, Pro32, Phe62, and His84.
Copper binding to β2m oligomers – MCO/MS
As stated in the introduction, previous results indicate that Cu(II) is necessary to initiate the β2m amyloid formation reaction, but eventually some Cu-free oligomers and fibrils are formed. To understand the evolution of the Cu(II) binding site, we used the MCO/MS method to identify the residues bound to Cu in the oligomers. Complete isolation of the dimer and tetramer was not possible without some dissociation back to the monomer or a lower-order oligomer, so the MCO reactions had to be performed at different times after initiating the amyloid-forming reaction. In previous work, we showed via dynamic light scattering, size-exclusion chromatography, and MS that β2m oligomers are formed sequentially with the dimer first appearing between 30 minutes and 1 hour and the initial form of the tetramer reproducibly appearing after approximately 24 hours [14]. Given this known temporal progression of the oligomers, we performed the MCO reactions at several time points after initiating the amyloid-forming reaction and used LC-MS/MS to identify the resulting modification sites. Table 2 shows the percent modification observed for Ile1, His31, Asp59, Trp60, His51, and His84 for amyloid formation reaction times ranging from 0 min to 4 days. Table 3 shows the percentages of the oligomers present at the same reaction times as measured by native MS.
Table 2.
The percentage of modified residues observed in the normal MCO reactions of the fibril-forming β2m samples as a function of time after the addition of Cu.
| Incubation Time | N-terminusa | Ile1-Gln2b | His31c | Asp59d | Trp60e | His84f | His51g |
|---|---|---|---|---|---|---|---|
| 0 min | 96 ± 2 | 49 ± 4 | 25 ± 6 | 72 ± 2 | 12 ± 4 | 0 | 0 |
| 30 min | 96 ± 1 | 59 ± 3 | 26 ± 4 | 67 ± 2 | 7 ± 3 | 16 ± 4 | 0 |
| 1 hr | 96 ± 2 | 69 ± 4 | 22 ± 3 | 59 ± 4 | 8 ± 2 | 23 ± 4 | 0 |
| 24 hr | 96 ± 2 | 72 ± 3 | 23 ± 3 | 45 ± 2 | 8 ± 2 | 23 ± 4 | 15 ± 2 |
| 48 hr | 94 ± 1 | 73 ± 12 | 26 ± 5 | 39 ± 1 | 8 ± 3 | 21 ± 5 | 24 ± 2 |
| 72 hr | 93 ± 2 | 64 ± 3 | 22 ± 5 | 37 ± 2 | 5 ± 3 | 21 ± 6 | 24 ± 2 |
| 96 hr | 94 ± 2 | 62 ± 5 | 24 ± 5 | 36 ± 4 | 5 ± 2 | 21 ± 4 | 24 ± 3 |
The percent modification for this residue was obtained by dividing the ion peak area of [Ile-Arg3-1]+ by the sum of the areas of this peak and the peak corresponding to its unmodified form.
The percent modification for this residue was obtained by dividing the ion peak area of [Gln2-Arg3-1]+ by the sum of the areas of this peak and the peak of [Ile1-Arg3]+.
The percent modification for this residue was obtained by dividing the ion peak area of [Val27-Lys41+16]3+ by the sum of the areas of this peak and the peak corresponding to its unmodified form.
The percent modification for this residue was obtained by dividing the ion peak area of [Val49-Tyr63-30]3+ by the sum of the areas of this peak and the peak corresponding to its unmodified form.
The percent modification for this residue was obtained by dividing the ion peak area of [Asp59-Tyr63+16]+ by the sum of the areas of this peak and the peak corresponding to its unmodified form.
The percent modification for this residue was obtained by dividing the ion peak area of [Val82-Lys91+16]2+ by the sum of the areas of this peak and the peak corresponding to its unmodified form.
The percent modification for this residue was obtained by dividing the ion peak area of [Val49-Lys58+16]2+ by the sum of the areas of this peak and the peak corresponding to its unmodified form.
Table 3.
The percentage of each oligomer measured by MS under fibril-forming conditions as a function of time.
| Incubation Time | Monomer | Dimer | Tetramer | Hexamer |
|---|---|---|---|---|
| 0 min | 100 | 0 | 0.0 | 0 |
| 30 min | 94.8 | 5.2 | 0.0 | 0.0 |
| 1 hr | 89.6 | 10.4 | 0.0 | 0.0 |
| 24 hr | 76.8 | 16.9 | 6.3 | 0.0 |
| 48 hr | 72.8 | 18.4 | 8.8 | 0.0 |
| 72 hr | 60.0 | 22.1 | 11.7 | 6.2 |
| 96 hr | 54.6 | 20.6 | 13.2 | 11.6 |
Examination of the data in table 2 indicates that the sites and extents of oxidation that result from the MCO reaction change during the course of the amyloid-forming reaction, suggesting that the residues bound to Cu(II) are changing as different oligomers are formed. Three sets of observations are noteworthy. First, the extent of oxidation to Ile1, Gln2, and His31 remain approximately constant or slightly decrease throughout the course of the amyloid-forming reaction, suggesting that these residues are bound to Cu(II) in all the oligomers. Second, the modification extents for Asp59 and Trp60 decrease as soon as the dimer is formed and continue to decrease as the tetramer and hexamer are formed. Because the percent modification decrease of these residues tracks with the percent decrease of the monomer, it seems reasonable that these residues are only bound to (or near) Cu in the monomer. Third, His84 and His51 become oxidized only after the dimer and tetramer, respectively, are present. These observations suggest that they bind Cu(II) in these oligomers but not in the monomer. The oxidation extent for these two residues, however, does not continue to increase. In both cases, the percent oxidation levels off or, in the case of His84, slightly decreases despite the continual increase in the percentage of tetramer and hexamer.
Copper binding to β2m oligomers – Detuned MCO/MS
Using DEPC and NHSA to monitor the Cu(II)-oligomer binding sites was not performed because the oligomer protein-protein interactions would also affect the labeling pattern, thereby making it difficult to determine the specific effect of Cu binding on reactivity. The detuned MCO/MS method, however, was employed. As described above, when only the monomer is present, Phe30, Pro32, Phe62, and His84 are modified in addition to the putative Cu binding sites at Ile1, Gln2, His31, Asp59, and Trp60. Unfortunately, no new sites of oxidation are observed in the oligomers, and most of the residues oxidized in the monomer remain oxidized in the oligomers. The trends in the modification extents for each of the binding residues are also about the same as in the tuned MCO experiments (Supporting Information, Table S2). The only new piece of information obtained from these experiments is that oxidation at Phe62 disappears as soon as the dimer is formed.
Discussion
Cu(II) binding to β2m causes the protein to initially form oligomers (i.e. dimers, tetramers, and hexamers) and to eventually form amyloid fibrils [14–16]. It has been shown that Cu binding to His31 and the N-terminal region of the protein likely causes a cis-trans isomerization at Pro32 and other compensating structural changes that make oligomerization possible [20,38]. The structural changes that are caused by Cu binding, however, have been mostly inferred from crystal structures of oligomeric forms of β2m mutants. Because these oligomers are formed from mutant sequences and are stable enough to crystallize, they may not be perfect representations of the transient oligomers that precede β2m amyloid fibril formation. Consequently, we set out to obtain additional insight into the Cu(II) binding sites of the monomeric and oligomeric forms of the wild-type protein. Using three complementary covalent labeling approaches with MS detection, we have been able to characterize how the Cu(II) binding site evolves from the monomer to the dimer and tetramer. Three general observations improve our understanding of Cu(II) binding and the associated structural changes that enable oligomer formation: (1) Four residues bind Cu(II) in the monomer; (2) Cu(II) binding to the monomer causes nearby structural changes that facilitate dimer formation; and (3) the Cu(II) binding site changes as oligomers are formed.
Four residues bind Cu(II) in the monomer
The MCO reactions of the Cu(II)-bound monomer result in seven identifiable modifications that collectively give a clear and reasonable picture of the Cu(II) binding site in monomeric β2m. The observed modifications are: (i) Ile1 oxidation, (ii) Ile1 deamination, (iii) cleavage of the peptide bond between Ile1 and Gln2, (iv) Gln2 deamination, (v) His31 oxidation, (vi) Asp59 decarboxylation, and (vii) Trp60 oxidation. Our previous work has shown that under the appropriate MCO reaction conditions only residues that are part of a cooperative Cu(II) binding site are oxidized; transient interactions do not lead to identifiable modifications [17,21–26]. Thus, we conclude from these data and the considerations below that Cu(II) binds simultaneously to the N-terminal amine of Ile1, the amide nitrogen of Gln2, His31, and Asp59. Each of these functional groups is known to bind Cu(II) in other proteins, and these functional groups are in close proximity to one another in the metal-free form of β2m. Thus, no major protein structural reorganization is necessary to accommodate this binding mode as has been observed by CD and NMR [6,16]. Interestingly, the identified residues comprise a site that is very similar to the well-known Cu binding motif, XXH, [39] in which Cu binds to a terminal amine, backbone amides, and the imidazole of a His residue. Possible similarities to the binding mode of the prion protein have also been suggested [40]. The detuned MCO/MS experiments are consistent with a Cu(II) binding site that is located near the N-terminal region of the protein as all the additional residues oxidized in the monomer during the detuned experiments are clustered around the N-terminus (Figure S2, Supporting Information). Furthermore, the covalent labeling experiments with DEPC and NHSA are very consistent with the identified Cu(II) binding site. The only residues that show a significant drop in reactivity are the N-terminus, Thr4, Ser28, His31, and Ser33, all of which are part or near the identified binding residues. A previous study has suggested that His13 and His51 bind Cu(II) [18], but we find no evidence from the MCO method, the detuned MCO method, or the covalent labeling experiments to indicate that these residues bind Cu in the monomer.
Trp60 is also oxidized to a small extent (~12%) in the monomer, but this residue is unlikely to be bound to Cu because tryptophan residues very infrequently bind transition metal ions. Moreover, of all amino acid residues, Trp has the second highest reaction rate constant with hydroxyl radicals [41], suggesting that if it was bound to Cu its extent of oxidation would be much higher. Indeed, Trp residues react with hydroxyl radicals two times faster than His residues and almost 200 times faster than Asp residues [41]. So, we conclude that Trp60’s proximity to Asp59 and ease with which it can be oxidized probably explains its relatively low level of oxidation. We observed something similar in our recent MCO studies of the Cu-bound form of the prion protein [25] in which proximate Trp residues were oxidized to a small degree even though these residues are not bound to Cu in this protein. Readily measurable modification of Asp59 argues strongly for this residue to be part of the Cu binding site because even under stronger oxidizing conditions (e.g. detuned MCO conditions) Asp residues are resistant to modification unless very close to the metal [26].
Our MCO/MS measurements for the monomer are consistent with previous studies of Cu(II) binding to β2m but do indicate the involvement of a previously uncharacterized residue, namely Asp59. Recently, Miranker and co-workers crystallized a hexamer of the H13F mutant of β2m (PDB ID: 3CIQ) that has a Cu bound to each protein subunit [20]. The Cu binding site is identical in each subunit and consists of the backbone amide of Met0, the backbone amide and carbonyl from Ile1, and His31. Our measurements indicate a slightly different Cu(II) binding site in the wild-type monomer. These slightly conflicting results are easily rationalized by considering the fact that the Cu(II) binding site probably changes to release Cu(II) as higher-order β2m oligomers are formed.
Cu(II) binding to Asp59 is somewhat surprising, but we speculate that its presence in the binding site has important implications for the eventual release of Cu from the β2m oligomers. Aspartate is the third most common amino acid found in transition metal binding sites, but a recent survey of the Protein Data Bank found that Asp (and Glu) residues are >10 times less likely to bind to Cu(II) than other first-row transition metal ions [42]. When carboxylates do bind Cu(II) in proteins, the average Cu(II)-ligand bond length is almost 0.3 Å greater than for Cu(II)-nitrogen bonds, which indicates the relatively weak interactions between Cu(II) and this functional group. Cu binding to Asp59 might then cause the Cu-β2m interactions to be strong enough to induce structural changes that lead to oligomer formation but weak enough that the resulting oligomers can eventually release Cu(II) and progress to form Cu(II)-free amyloid fibrils.
Cu(II) binding to the monomer causes nearby structural changes that facilitate dimer formation
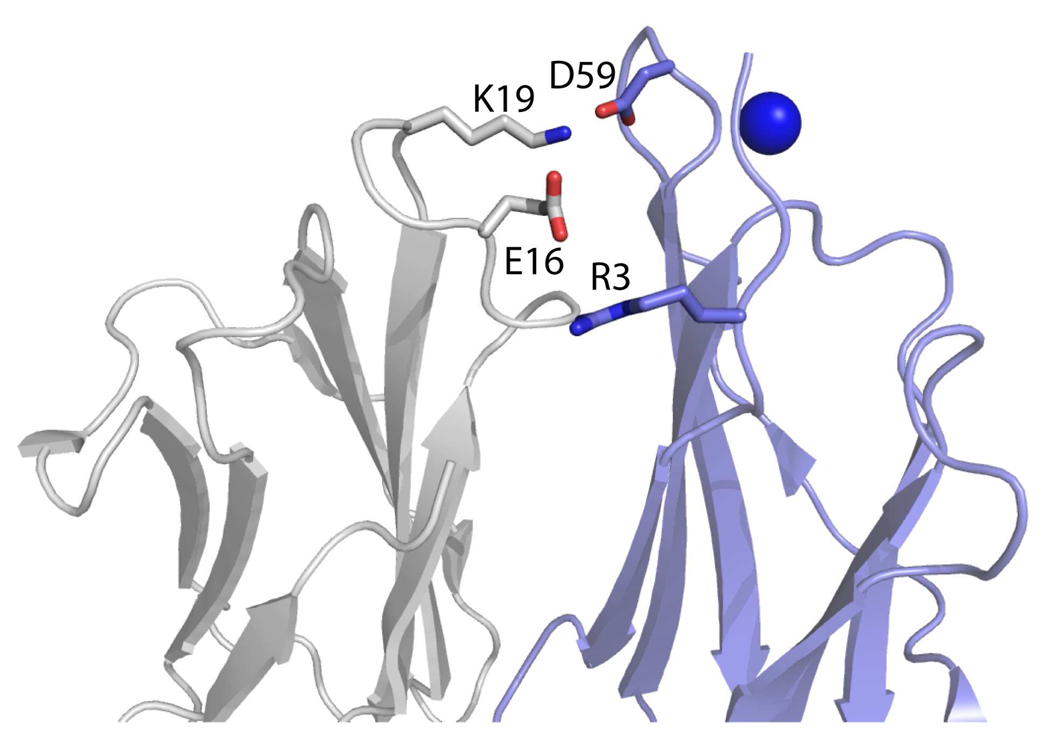
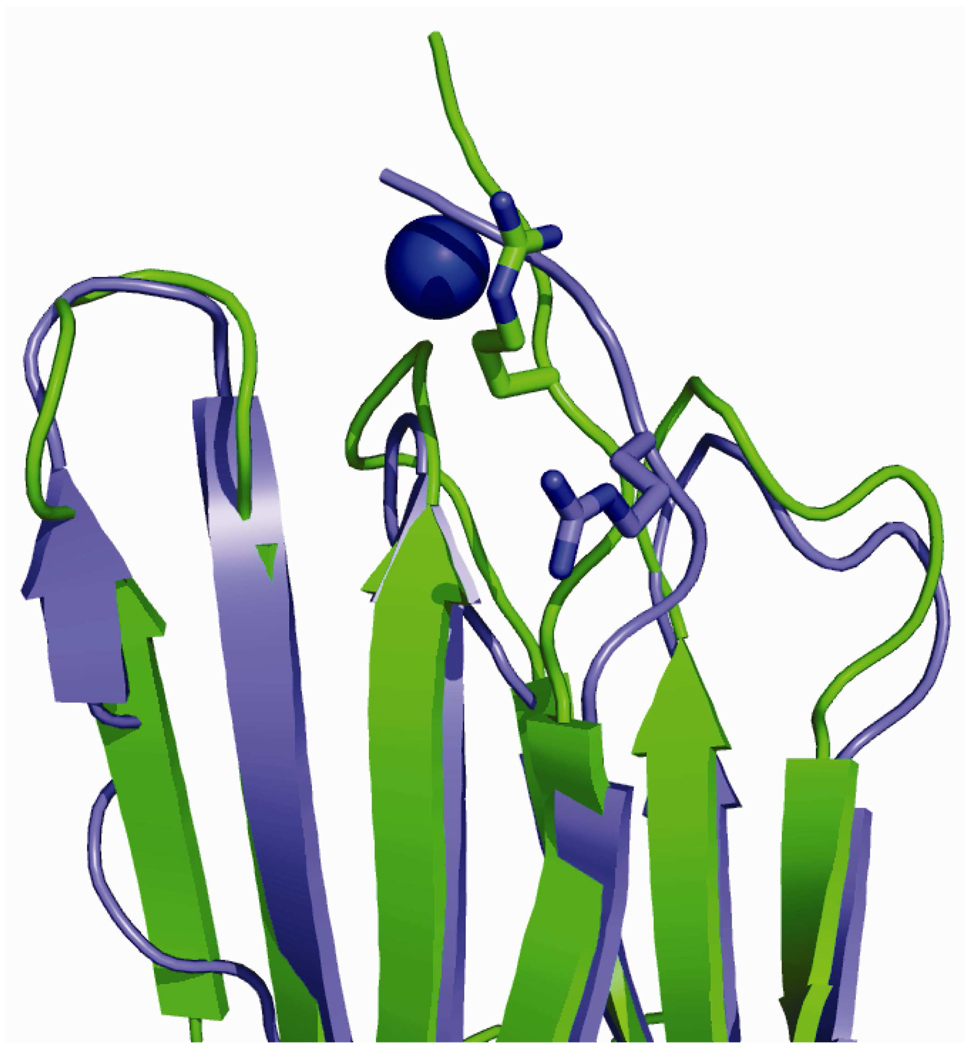
Cu(II) binding to Asp59 requires a substantial repositioning of this residue’s side chain that we propose is important for dimer formation. In both NMR and crystal structures of monomeric β2m [43,44], the side chain of Asp59 points away from the N-terminal strand (Figure 5); however, upon binding Cu(II) this residue must be repositioned to point toward the N-terminal amine and the amide between Ile1 and Gln2 to effectively bind Cu. Interestingly, a close examination of the crystal structure of the H13F hexamer of β2m [20] reveals that the side chain of Asp59 has rotated greater than 100° and is located closer to the N-terminus than in the monomer (Figure 5). Furthermore, inspection of the subunit contacts between chains B and C in the crystal structure of the H13F hexamer of β2m reveals that Asp59 of one protein subunit is about 2.8 Å from Lys19 of another protein subunit (Figure 6). This interaction is symmetric, giving rise to a pair of Asp59/Lys19 residues that likely form salt bridges. We predict that the interface between chains B and C in the hexamer may represent the interactions present in the dimer. If so, then the salt bridges formed between the two Asp59/Lys19 pairs are probably important for stabilizing the dimer. Because formation of these salt bridges requires Asp59 to be significantly repositioned (Figure 5), we speculate that Cu(II) binding to Asp59 then acts to relay this residue from its position in the monomer to a new position that allows it to more readily form a dimer-stabilizing intermolecular salt bridge with Lys19.
Figure 5.
A comparison of the crystal structure of monomeric β2m (green, PDB ID: 1LDS) and one of the subunits from the H13F hexamer (blue, PDB ID: 3CIQ) that illustrates the significant repositioning of Asp59.
Figure 6.
Interface between chains B and C in the H13F hexamer (PDB ID: 3CIQ), showing the intersubunit salt bridges involving Asp59/Lys19 and Arg3/Glu16.
Our data also suggests that Cu(II) binding to monomeric β2m repositions Arg3 to facilitate dimer formation. In previous work, we demonstrated that our detuned MCO/MS approach can oxidize most residues that are within approximately 10 Å of Cu. If Cu is hypothetically placed at the midpoint between the Ile1-Gln2 peptide bond and His31 in the crystal structure of β2m (PDB ID: 1LDS), then the side chains of the following 11 residues are found to be within about 10 Å of Cu: Ile1, Gln2, Arg3, Phe30, His31, Pro32, Trp60, Phe62, His84, Val85, and Thr86. The detuned MCO/MS data indicate that all of these residues except Arg3, Val85, and Thr86 are modified. Failure to find evidence for Val85 and Thr86 modification is not that surprising because these residues are about 10 Å from the chosen Cu binding site and are only mildly reactive with radicals [41]. Failure to modify Arg3, though, is surprising. The side chain of this residue is 5 to 8 Å from the chosen Cu site, and Arg reacts readily with radicals [41]. Indeed, in previous work, we found that Arg residues are easily deguanidated during the detuned MCO reactions [26]. A reasonable explanation for the failure to modify Arg3 is that Cu(II) binding repels Arg3 so that it is repositioned greater than 10 Å from the metal. Examination of the subunit contacts between chains B and C in the crystal structure of the H13F hexamer is consistent with this expectation and shows that the guanidinium group of Arg3 is approximately 11 Å from Cu. Moreover, Arg3 is now located about 3.8 Å from Glu16 of another protein subunit (Figure 6), suggesting that these residues may form an intermolecular salt bridge. Just as with the Asp59/Lys19 salt bridge, this interaction is symmetric, thereby giving rise to a pair of salt bridges that again are probably important for the stability of the dimer. Thus, we hypothesize that electrostatic repulsion between Cu(II) and Arg3 likely forces Arg3 to adopt a new position that allows it to readily form a dimer-stabilizing intermolecular salt bridge with Glu16 (Figure 7).
Figure 7.
A comparison of the crystal structure of monomeric β2m (green, PDB ID: 1LDS) and one of the subunits from the H13F hexamer (blue, PDB ID: 3CIQ) that illustrates the repositioning of Arg3.
To test the importance of these putative salt bridges on dimer formation, we monitored the rate of dimer formation as a function of ionic strength. Previously, it was indicated that the dimer exhibits amyloid characteristics and changes the fluorescence response of the dye thioflavin T [14,16]. Thus, we used this dye to monitor the rate of dimer formation and found that the rate (Figure S3 in Supporting Information) increases as the solution ionic strength decreases, which is consistent with the importance of salt bridges in the formation of the dimer. If, as we predict, the interface of chains B and C of the H13F hexamer represents the dimer interface, this trend supports the idea that Cu binding causes Asp59 and Arg3 to be repositioned to facilitate these salt bridge interactions.
Our MCO/MS measurements reveal two possible rearrangements that are caused by Cu(II) binding and facilitate dimer formation, but our measurements are also consistent with other postulated rearrangements that are important for oligomer formation. Evidence has been provided that Cu binding facilitates the cis-trans isomerization of Pro32 and causes Phe30 to rotate from the hydrophobic core to the solvent accessible surface of the protein [20]. If Cu is again placed at the midpoint between the Ile1-Gln2 peptide bond and His31 in the monomeric structure, Pro32 is about 5 Å from the metal. The cis-trans isomerization, however, would place the side chain of Pro32 further from Cu. Even so, Pro32 can still be oxidized to a small extent (~ 1.5 – 2 %). Phe30 is significantly further (~10 Å) from the hypothetical Cu site in the monomeric structure, which would suggest that it would not be oxidized very extensively. This residue, however, is oxidized quite readily in the detuned experiments (~ 15%). Even if one considers the fact that Phe residues are very reactive with radicals [41], this significant degree of oxidation suggests that this residue is closer than 10 Å to the Cu binding site. Indeed, if the H13F hexamer structure is used as an indicator of the proximity of Phe30 to Cu, then the rotation of Phe30 caused by Cu binding positions this residue about 5 Å from the metal, which is more consistent with its level of oxidation during the detuned MCO/MS experiments.
The Cu(II) binding site changes as oligomers are formed
The pattern of oxidation that is observed as different oligomers become populated in solution (Table 2) suggests that the initial Cu binding mode that is identified in the monomer changes as dimers and tetramers are formed. The oligomers cannot be fractionated and interrogated separately by the MCO/MS method, so to investigate the Cu(II) binding sites in the dimer and tetramer we must correlate the oxidation changes for individual residues at different time points with separate experiments that monitor the temporal progression of the oligomers in solution. When we do this, we find that the modification levels for Ile1, Gln2, and His31 remain approximately constant or slightly decrease as dimers and tetramers form in solution. We conclude from these data that these residues remain Cu binding sites in the oligomers. In contrast, the percent modifications for Asp59 and Trp60 decrease as soon as dimers are formed. Moreover, the modification extents for these residues decrease at about the same rate that the monomer concentration decreases in solution, suggesting that Asp59 is no longer bound to Cu(II) in the dimer or tetramer. These observations are consistent with Asp59 forming dimer-stabilizing salt bridges with Lys19. Coinciding with the initial drop in reactivity of Asp59 is the oxidation of His84; however, the oxidation level of this residue does not continually increase as more tetramer is formed. This result along with the previous observation that Cu(II) is eventually released from β2m oligomers causes us to speculate that His84 is a binding site only in the dimer and is released upon tetramer formation. Perhaps Cu(II) binding to His84 pays some of the enthalpic cost associated with the release of Asp59, but the additional intermolecular interactions that arise as the tetramer is formed compensate for the loss of His84 as a Cu(II) ligand in this oligomer. In addition, the conformational changes that lead to tetramer formation may weaken the Cu(II)-His84 as a prelude to the eventual release of the metal.
The appearance of His51 as a Cu(II) binding site coincides with the formation of the tetramer, but it is not reasonable to conclude that His51 is repositioned to be part of the Cu(II) binding site that includes Ile1, Gln2, and His31. In both the wild-type monomer and the H13F mutant crystal structures, His51 is greater than 20 Å from the Cu(II) binding site. Thus, a major structural reorganization would be necessary to move this residue in proximity to Ile1, Gln2, and His31, but β2m oligomers are found to be native-like by CD and NMR measurements [16], ruling out such a major structural change. A more likely explanation is that Cu(II) has been released from its initial binding site in some fraction of the oligomers, and instead of being completely free in solution it binds weakly to His51. Two previous experiments indicate that Cu-free oligomers are formed prior to β2m amyloid formation [14,15]. In our previous studies [14], we identified a tetramer as the first Cu(II)-free oligomer to be formed but found evidence for two types of tetramers – one that required Cu(II) for stability and one that did not. One way to rationalize the His51 binding site is to consider that Cu release is part of a stepwise process in which the first tetramer is converted to the second Cu(II)-free tetramer. The second tetramer forms enough new protein-protein interactions to pay the enthalpic cost of Cu(II) release, or the second tetramer undergoes conformational changes that weaken the Cu(II) binding site. Either way, Cu(II) release becomes possible, and some of the released Cu(II) is able to bind His51 and facilitate oxidation of this residue during the MCO reactions.
Summary and Conclusions
The results described here give us additional insight into how Cu(II) facilitates the β2m structural changes that lead to oligomer formation. In the monomer, Cu(II) binds to the N-terminus, a backbone amide between Ile1 and Gln2, His31, and Asp59. Binding to these residues causes several structural changes. We propose that movement of Asp59 and Arg3 allows two pairs of dimer-stabilizing salt bridges to be formed that involve Asp59/Lys19 and Arg3/Glu16. In effect, Cu(II) may act to relay Asp59 so that it can undergo a significant reorientation from its position in the Cu-free monomer to its position in the dimer. In contrast, Arg3 is repelled by Cu(II) binding, which enables it to form a salt bridge with Glu16. Movement of these two residues, along with the cis-trans isomerization of Pro32 and exposure of Phe30 to solvent, may therefore be part of the switch postulated by Miranker and co-workers that enables Cu(II) to initiate dimer formation [20], which then begins the pathway toward amyloid fibril formation. The formation of these salt bridges is highly probable because electrostatic interactions are usually important contributors to the specificity and stability of protein-protein complexes [45–47].
The evolution of the Cu(II) binding site also provides some molecular insight into how Cu(II) is eventually released from the oligomers and thus absent in the mature fibrils. Because Cu(II) may relay Asp59 so that it can form a salt bridge, this residue is not part of the binding site in the dimer. Instead, it appears that His84 replaces Asp59 as one of the Cu(II) binding residues in the dimer. His84, however, does not remain a binding site in the tetramer, suggesting that further conformational changes that are needed to form the tetramer weaken the Cu(II) binding site. Additional conformational changes evidently weaken the Cu(II) binding site further and provide enough favorable oligomer interactions that the metal is no longer needed to stabilize a second form of the tetramer, as was observed previously [14]. Some free Cu(II) is released into solution where His51, which could be solvent exposed, can act as a weak binding site for Cu(II). This binding site, however, is not essential for tetramer stability as the addition of EDTA can remove this Cu without dissociating the tetramer [14,15].
In a broader context, our results provide some additional details about the molecular role that Cu(II) can play in amyloid-forming systems. Divalent transition metals, especially Cu(II), appear to be one of the general ways to cause protein amyloid formation. Prior work shows that proteins such as α-synuclein, immunoglobulin light chain, Aβ, and even possibly the prion protein form amyloid aggregates in the presence of metal ions. In the present work we find that metal binding to certain residues and repulsion of other residues can perturb protein structure in a way that enables aggregation. Further studies of Cu-β2m interactions and the resulting aggregation may provide even more insight into the possible roles that divalent metals play in protein aggregation.
Supplementary Material
Abbreviations
- β2m
β-2-microglobulin
- NMR
nuclear magnetic resonance
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- MCO
metal catalyzed oxidation
- DEPC
diethylpyrocarbonate
- NHSA
sulfo-N-hydroxysuccinimide acetate
- DTT
dithiothreitol
- MOPS
3-morpholinopropanesulfonic acid
- Tris
Tris(hydroxymethyl)-aminomethane
- MWCO
molecular weight cutoff
- EDTA
ethylenediamine tetraacetic acid
- ESI
electrospray ionization
- HPLC
high performance liquid chromatography
- ThT
thioflavin T
- CD
circular dichroism
Footnotes
This material is based upon work supported by the National Institutes of Health Grant RO1 GM 075092
Supporting Information Available A table containing the calculated solvent accessible surface areas for all the modified amino acids; a table showing the oxidation extent of amino acids during the detuned MCO reaction; a figure showing the appropriate persulfate concentrations for the detuned MCO reaction; a figure illustrating the β2m residues oxidized during the detuned MCO reaction; and a plot showing the rate of β2m dimer formation as a function of ionic strength are found in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Ayers DC, Athanasou NA, Woods CG, Duthie RB. Dialysis arthropathy of the hip. Clin. Orthop. Relat. Res. 1993;290:216–224. [PubMed] [Google Scholar]
- 2.McParland VJ, Kad NM, Kalverda AP, Brown A, Kirwin-Jones P, Hunter MG, Sunde M, Radford SE. Partially unfolded states of β2-microglobulin and amyloid formation in vitro. Biochemistry. 2000;39:8735–8746. doi: 10.1021/bi000276j. [DOI] [PubMed] [Google Scholar]
- 3.Esposito G, Michelutti R, Verdone G, Viglino P, Hernandez H, Robinson CV, Amoresano A, Dal Piaz F, Monti M, Pucci P, Mangione P, Stoppini M, Merlini G, Ferri G, Bellotti V. Removal of the N-terminal hexapeptide from human β2- microglobulin facilitates protein aggregation and fibril formation. Prot. Sci. 2000;9:831–845. doi: 10.1110/ps.9.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Relini A, Canale C, De Stefano S, Rolandi R, Giorgetti S, Stoppini M, Rossi A, Fogolari F, Corazza A, Esposito G, Gliozzi A, Bellotti V. Collagen plays an active role in the aggregation of β2-microglobulin under physiopathological conditions of dialysis-related amyloidosis. J. Biol. Chem. 2006;281:16521–16529. doi: 10.1074/jbc.M513827200. [DOI] [PubMed] [Google Scholar]
- 5.Ohhashi Y, Kihara M, Naiki H, Goto Y. Ultrasonication-induced amyloid fibril formation of β2-microglobulin. J. Biol. Chem. 2005;280:32843–32848. doi: 10.1074/jbc.M506501200. [DOI] [PubMed] [Google Scholar]
- 6.Morgan CJ, Gelfand M, Atreya C, Miranker AD. Kidney dialysis-associated amyloidosis: a molecular role for Cu(II) in fiber formation. J. Mol. Biol. 2001;309:339–345. doi: 10.1006/jmbi.2001.4661. [DOI] [PubMed] [Google Scholar]
- 7.Eakin CM, Knight JD, Morgan CJ, Gelfand MA, Miranker AD. Formation of a copper specific binding site in non-native states of β-2-microglobulin. Biochemistry. 2002;41:10646–10656. doi: 10.1021/bi025944a. [DOI] [PubMed] [Google Scholar]
- 8.Bush AI, Tanzi RE. The galvanization of beta-amyloid in Alzheimer’s disease. Proc. Natl. Acad. Sci USA. 2002;99:7317–7319. doi: 10.1073/pnas.122249699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uversky VN, Li J, Fink AL. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. a possible molecular link between Parkinson's disease and heavy metal exposure. J. Biol. Chem. 2001;276:44284–44296. doi: 10.1074/jbc.M105343200. [DOI] [PubMed] [Google Scholar]
- 10.Wadsworth JD, Hill AF, Joiner S, Jackson GS, Clarke AR, Collinge J. Strain-specific prion-protein conformation determined by metal ions. Nature Cell Biol. 1999;1:55–59. doi: 10.1038/9030. [DOI] [PubMed] [Google Scholar]
- 11.Jobling MF, Huang X, Stewart LR, Barnham KJ, Curtain C, Volitakis I, Perugini M, White AR, Cherny RA, Masters CL, Barrow CJ, Collins SJ, Bush AI, Cappai R. Copper and zinc binding modulates the aggregation and neurotoxic properties of the prion peptide PrP106-126. Biochemistry. 2001;40:8073–8084. doi: 10.1021/bi0029088. [DOI] [PubMed] [Google Scholar]
- 12.Davis DP, Gallo G, Vogen SM, Dul JL, Sciarretta KL, Kumar A, Raffen R, Stevens FJ, Argon Y. Both the environment and somatic mutations govern the aggregation pathway of pathogenic immunoglobulin light chain. J. Mol. Biol. 2001;313:1021–1034. doi: 10.1006/jmbi.2001.5092. [DOI] [PubMed] [Google Scholar]
- 13.Eakin CM, Miranker AD. From chance to frequent encounters: origins of β2-microglobulin fibrillogenesis. Biochim. Biophys. Acta. 2005;1753:92–99. doi: 10.1016/j.bbapap.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Antwi K, Mahar M, Srikanth R, Olbris MR, Tyson JF, Vachet RW. Cu(II) organizes β-2-microglobulin oligomers but is released upon amyloid formation. Prot. Sci. 2008;17:748–759. doi: 10.1110/ps.073249008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calabrese MF, Miranker AD. Formation of a stable oligomer of β-2 microglobulin requires only transient encounter with Cu(II) J. Mol. Biol. 2007;367:1–7. doi: 10.1016/j.jmb.2006.12.034. [DOI] [PubMed] [Google Scholar]
- 16.Eakin CM, Attenello FJ, Morgan CJ, Miranker AD. Oligomeric assembly of native-like precursors precedes amyloid formation by β-2 microglobulin. Biochemistry. 2004;43:7808–7815. doi: 10.1021/bi049792q. [DOI] [PubMed] [Google Scholar]
- 17.Lim J, Vachet RW. Using mass spectrometry to study copper-protein binding under native and non-native conditions: β-2-microglobulin. Anal. Chem. 2004;76:3498–3504. doi: 10.1021/ac049716t. [DOI] [PubMed] [Google Scholar]
- 18.Villanueva J, Hoshino M, Katou H, Kardos J, Hasegawa K, Naiki H, Goto Y. Increase in the conformational flexibility of β2-microglobulin upon copper binding: a possible role for copper in dialysis-related amyloidosis. Prot. Sci. 2004;13:797–809. doi: 10.1110/ps.03445704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ubbink M, Worrall JAR, Canters GW, Groenen EJJ, Huber M. Paramagnetic Resonance of Biological Metal Centers. Annu. Rev. Biophys. Biomol. Struct. 2002;31:393–422. doi: 10.1146/annurev.biophys.31.091701.171000. [DOI] [PubMed] [Google Scholar]
- 20.Calabrese MF, Eakin CM, Wang JM, Miranker AD. A regulatable switch mediates self-association in an immunoglobulin fold. Nature Struct. Mol. Biol. 2008;15:965–971. doi: 10.1038/nsmb.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim J, Vachet RW. Development of a methodology based on metal-catalyzed oxidation reactions and mass spectrometry to determine the metal binding sites in copper metalloproteins. Anal. Chem. 2003;75:1164–1172. doi: 10.1021/ac026206v. [DOI] [PubMed] [Google Scholar]
- 22.Bridgewater JD, Vachet RW. Metal-catalyzed oxidation reactions and mass spectrometry: The role of ascorbate and different oxidizing agents in determining Cu-protein binding sites. Anal. Biochem., 2005. 2005;341:122–130. doi: 10.1016/j.ab.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 23.Bridgewater JD, Vachet RW. Using microwave-assisted metal-catalyzed oxidation reactions and mass spectrometry to increase the rate at which the copper-binding sites of a protein are determined. Anal. Chem. 2005;77:4649–4653. doi: 10.1021/ac0502551. [DOI] [PubMed] [Google Scholar]
- 24.Bridgewater JD, Lim J, Vachet RW. Transition metal-peptide binding studied by metal-catalyzed oxidation reactions and mass spectrometry. Anal. Chem. 2006;78:2432–2438. doi: 10.1021/ac051983r. [DOI] [PubMed] [Google Scholar]
- 25.Srikanth R, Wilson J, Burns CS, Vachet RW. Identification of Cu(II) coordinating residues in the prion protein by metal-catalyzed oxidation mass spectrometry: Evidence for multiple isomers at low Cu(II) loadings. Biochemistry. 2008;47:9258–9268. doi: 10.1021/bi800970m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bridgewater JD, Lim J, Vachet RW. Using metal-catalyzed oxidation reactions and mass spectrometry to identify amino acid residues within 10 Ǻ of the metal in Cu-binding proteins. J. Am. Soc. Mass Spectrom. 2006;17:1552–1559. doi: 10.1016/j.jasms.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Qin KF, Yang Y, Mastrangelo P, Westaway D. Mapping Cu(II) binding sites in prion proteins by diethyl pyrocarbonate modification and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometric footprinting. J. Biol. Chem. 2002;277:1981–1990. doi: 10.1074/jbc.M108744200. [DOI] [PubMed] [Google Scholar]
- 28.Narindrasorasak S, Kulkarni P, Deschamps P, She YM, Sarkar B. Characterization and copper binding properties of human COMMD1 (MURR1) Biochemistry. 2007;46:3116–3128. doi: 10.1021/bi0620656. [DOI] [PubMed] [Google Scholar]
- 29.Mendoza VL, Vachet RW. Protein surface mapping using diethylpyrocarbonate with mass spectrometric detection. Anal. Chem. 2008;80:2895–2904. doi: 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uchida K, Kawakishi S. Ascorbate-mediated specific oxidation of the imidazole ring in a histidine derivative. Bioorg. Chem. 1989;17:330–343. [Google Scholar]
- 31.Schöneich C. Mechanisms of metal-catalyzed oxidation of histidine to 2-oxo-histidine in peptides and proteins. J. Pharm. Biomed. Analysis. 21:1093–1097. doi: 10.1016/s0731-7085(99)00182-x. [DOI] [PubMed] [Google Scholar]
- 32.Garrison WM. Reaction mechanisms in the radiolysis of peptides, polypeptides, and proteins. Chem. Rev. 1987;87:381–398. [Google Scholar]
- 33.Levine J, Etter J, Apostol I. Nickel-catalyzed N-terminal oxidative deamination in peptides containing histidine at position 2 coupled with sulfite oxidation. J. Biol. Chem. 1999;274:4848–4857. doi: 10.1074/jbc.274.8.4848. [DOI] [PubMed] [Google Scholar]
- 34.Kowalik-Jankowska T, Ruta M, Wisniewska K, Lankiewicz L, Dyba M. Products of Cu(II)-catalyzed oxidation in the presence of hydrogen peroxide of the 1–10, 1–16 fragments of human and mouse β-amyloid peptide. J. Inorg. Biochem. 2004;98:940–950. doi: 10.1016/j.jinorgbio.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Inoue K, Nakagawa A, Hino T, Oka H. Screening assay for metal-catalyzed oxidation inhibitors using liquid chromatography-mass spectrometry with an N-terminal β-amyloid peptide. Anal. Chem. 2009;81:1819–1825. doi: 10.1021/ac802162n. [DOI] [PubMed] [Google Scholar]
- 36.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 37.Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev. 2009 doi: 10.1002/mas.20203. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eakin CM, Berman AJ, Miranker AD. A native to amyloidogenic transition regulated by a backbone trigger. Nature Struct. Mol. Biol. 2006;13:202–208. doi: 10.1038/nsmb1068. [DOI] [PubMed] [Google Scholar]
- 39.Harford C, Sarkar B. Amino terminal Cu(II)- and Ni(II)-binding (ATCUN) motif of proteins and peptides: Metal binding, DNA cleavage, and other properties. Acc. Chem. Res. 1997;30:123–130. [Google Scholar]
- 40.Calabrese MF, Miranker AD. Metal binding sheds light on mechanisms of amyloid assembly. Prion. 2009;3:1–4. doi: 10.4161/pri.3.1.8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buxton GV, Greenstock CL, Helman WP, Ross AB. Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms, and hydroxyl radicals in aqueous solution. J. Phys. Chem. Ref. Data. 1988;17:513–886. [Google Scholar]
- 42.Zheng H, Chruszcz M, Lasota P, Lebioda L, Minor W. Data mining of metal ion environments present in protein structures. J. Inorg. Biochem. 2008;102:1765–1776. doi: 10.1016/j.jinorgbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trinh CH, Smith DP, Kalverda AP, Phillips SEV, Radford SE. Crystal structure of monomeric human β-2-microglobulin reveals clues to its amyloidogenic properties. Proc. Natl. Acad. Sci. USA. 2002;99:9771–9776. doi: 10.1073/pnas.152337399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verdone G, Corazza A, Viglino P, Pettirossi F, Giorgetti S, Mangione P, Andreola A, Stoppini M, Bellotti V, Esposito G. The solution structure of human β2- microglobulin reveals the prodromes of its amyloid transition. Prot. Sci. 2002;11:487–499. doi: 10.1110/ps.29002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu D, Lin SL, Nussinov R. Protein binding versus protein folding: The role of hydrophilic bridges in protein associations. J. Mol. Biol. 1997;265:68–84. doi: 10.1006/jmbi.1996.0712. [DOI] [PubMed] [Google Scholar]
- 46.Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein-protein interactions. Curr. Opin. Struct. Biol. 2000;10:153–159. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 47.Keskin O, Gursoy A, Ma B, Nussinov R. Principles of protein-protein interactions: What are the preferred ways for proteins to interact? Chem. Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.