Abstract
There is emerging recognition of a novel fuel and redox sensing regulatory program that controls cellular adaptation via non-histone protein lysine-residue acetyl post-translation modifications. This program functions in tissues with high energy demand and oxidative capacity and is highly enriched in the heart. Deacetylation is regulated by NAD+-dependent activation of the sirtuin family of proteins while acetyltransferase modifications are controlled by less clearly delineated acetyltransferases. Subcellular localization specific protein targets of lysine-acetyl modification have been identified in the nucleus, cytoplasm and mitochondria. Despite distinct subcellular localizations, these modifications appear, in large part, to modify mitochondrial properties including respiration, energy production, apoptosis and anti-oxidant defenses. These mitochondrial regulatory programs are important in cardiovascular biology, although how protein acetyl modifications effects cardiovascular pathophysiology has not been extensively explored. This review will introduce the role of non-histone protein lysine-residue acetyl modifications, discuss their regulation and biochemistry and present the direct and indirect data implicating their involvement in the heart and vasculature.
Keywords: Sirtuins, Mitochondrial Metabolism, Apoptosis, Redox Stress, Heart, Vascular Biology
In the year of the bicentennial celebration of Darwin's birth, the pursuit of regulatory programs that underpin ‘adaptations to confer survival’ remains an important area of scientific investigation. Adaptation to acute stressors, as opposed to evolutionary ‘pressures’, would require rapidly inducible ‘stress-sensing systems’ that could initiate biological modifications, enabling survival advantage. As mitochondria are central to important cellular functions, including essential pathways for energy production, reactive oxygen species (ROS) signaling, calcium homeostasis and apoptosis, it would not be surprising if mitochondrial adaptations are instrumental in this ameliorative reprogramming. The heart has a high density of mitochondria with robust energetic demands and the concept of mitochondrial adaptation in the cardiovascular system to resist biomechanical stressors is well recognized.1-4 To date, the most extensively explored sensing program delineated in the heart is controlled by AMP protein kinase and this signaling network is responsive to energy depletion and rising AMP levels.5 A more recently identified nutrient and redox sensing regulatory program, exemplified by protein lysine-residue acetyl modification, is beginning to be investigated as an additional homeostatic control program.
Lysine-residue acetylation is a reversible post-translational modification (PTM) orchestrated by a diverse family of structurally unrelated enzymes collectively known as acetyltransferases. This modification usually involves the covalent transfer of an acetyl group from acetyl-coenzyme A to a ε-amino group on lysine. The reverse reaction is driven by deacetylase enzymes. These lysine-modifications have been most extensively explored in the regulation of histones to fine-tune gene transcription, and these regulatory programs are dynamically controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs). The role of acetylation/deacetylation in the modification of histones has recently been well described6 and is not the focus of this review. Initially identified in 1997, the first non-histone protein to exhibit acetyl-lysine modification was p53.7 A steady increase in the number of non-histone lysine acetylation modifications are being discovered and may play a diverse role in diffuse biological processes.8 Coupled to this, the enzymes controlling these PTMs are not confined to HATs and HDACs but include an expanding array of lysine acetyltransferases and deacetylases.
The modulation of mitochondrial biology and function via lysine acetylation involves proteins residing in the nucleus (mitochondrial regulatory proteins), cytoplasm (chaperones) and within mitochondria themselves. Together, these PTMs of target proteins contribute towards alterations in numerous mitochondrial functions including the regulation of mitochondrial biogenesis, apoptosis, thermogenesis, metabolism and possibly in the contribution of mitochondrial function to longevity.9 Although this field was initially identified and characterized in non-vertebrate eukaryotes, we focus on mammalian biology. In this review we explore: (1) the enzymes that orchestrate these modifications; (2) identify the substrates of these enzymes; (3) investigate the regulation of lysine acetylation; (4) describe the cardiovascular functional consequences of these regulatory events and (5) highlight potential areas of importance that need to be investigated to expand our understanding of the role of non-histone lysine acetylation and deacetylation on mitochondrial biology and cardiovascular homeostasis.
Non-Histone Lysine Acetyltransferases
Acetylation, as a process to regulate the availability of DNA for transcription, has been described for over forty years.10 HATs remove the positive charge of lysines in the histone tail, altering its interaction with DNA and allowing access to other DNA-associated proteins.11, 12 Since the discovery of non-histone lysine-acetylation,7 the number of candidate proteins documented has grown steadily, with a recent proteomics study identifying 195 new acetylated proteins in mammalian tissue.13 Strikingly, in this study, 133 proteins with acetylated lysine residues were concentrated in mitochondria.13 These data suggest that ~20% of the mitochondrial proteome may be targeted for acetyl modification. This concept is further supported by the finding that protein acetylation is similarly abundant in prokaryotes.14 The functional characterization of the role of lysine-residue acetylation in the vast majority of these proteins has yet to be determined.
The non-histone lysine acetyltransferases can be loosely grouped into three main families, though there are several other identified acetyltransferases that fall outside these defined categories.15, 16 These include the Gcn5-related N-acetyltransferase (GNAT), the MYST and the p300/CBP families.
The Gcn5-related N-acetyltransferase (GNAT) family contains HAT1, the first identified histone acetyltransferase,17 along with Gcn5/PCAF and multiple N-acetyltransferase (NAT) proteins. The GNAT family share 3-4 motifs involved in acetyl-CoA binding and catalysis, with nuclear-localized proteins and a bromodomain to facilitate DNA binding.18, 19 A role of Gcn5 relating to mitochondrial function is inferred by the acetylation and inactivation of peroxisome proliferator gamma coactivator 1 (PGC-1) family members, which are known to be master regulators in mitochondrial biogenesis and mediators of mitochondrial metabolism.20 Gcn5, activated by the steroid receptor coactivator SRC-3, acetylates the PGC1α and β, which inhibits their regulatory control of mitochondrial content and metabolic functioning.21-23
The MYST family, named after the original members MOZ, YBF2, SAS2 and TIP60, share both a ~240 amino acid core acetyl-CoA-binding domain and a C2HC zinc-finger domain.24 MYST proteins are found throughout the Eukaryota and are predominantly involved in histone acetylation, although some members are also involved in the regulation of transcription factors such as p53.25 Tat-interactive protein 60 (TIP60) is transiently expressed in heart tissues during early embryonic development, suggesting that acetylation is involved in regulating cardiac myocyte differentiation.26
Finally, there is the p300/CBP (CREB binding protein) family, which are large nuclear proteins that function as transcriptional co-regulators which have intrinsic histone acetyltransferase activity.27, 28 It has subsequently been shown that these large multi-domain proteins additionally have non-histone acetyltransferase activity and are capable of PGC-1α acetylation.29 The presence of p300 is required for the correct formation of several mouse tissues, with a single mutant allele being sufficient to produce major defects in heart structure and coronary vascularization.30 p300 also acetylates the early embryonic transcription factor GATA-4 which functions in differentiating embryonic stem cells into cardiac myocytes and in the development of cardiac hypertophy.31, 32 Collectively, it appears that the lysine-acetyltransferases are crucial for several steps in cardiac development, and may play a role in mitochondrial biology via this regulation of PGC-1. Interestingly, despite the prevalence of acetylated lysine residues on numerous mitochondrial proteins, to date mitochondrial enriched lysine-acetyltransferases have not been identified and are therefore not discussed further in this review.
Lysine Deacetylase Enzymes
The mammalian deacetylases are grouped according to phylogenetic analysis and sequence homology. The mammalian class I and II enzymes are nuclear and cytosolic-nuclear localized enzymes respectively that predominantly function as HDACs. To date, one HDAC has sequence similarity to both class I and II enzymes and has been designated as a class IV enzyme. Class I, II and IV enzymes have zinc-dependent deacetylase activity.33 The sirtuins are designated as class III deacetylases and are NAD+-dependent enzymes. The founding member of these enzymes is yeast Sir2, which silences chromatin via deacetylation of histones.34 Sir2 enzymes have been shown to mediate lifespan extension in yeast, worms and flies and are postulated to function, in part, via the modulation of mitochondrial function.9 Mammals have 7 sirtuin enzymes designated as SIRT1 through SIRT7. These have distinct tissue distributions and subcellular localizations which together probably contribute to their distinct biological functions 35. The mammalian sirtuins are further phylogenetically divided into five subclasses based on the homology of their 250 amino acid core domain.36 The mitochondrial enriched SIRT3 clusters with SIRT1 and SIRT2 in subclass I. These three enzymes show closest homology to yeast Sir2 and exhibit the most robust deacetylase activity. The additional mitochondrial enriched sirtuins SIRT4 and SIRT5 are assigned to subclasses II and III, and exhibit predominant ADP-ribosyltransferase and deacetylase activity respectively.37, 38
NAD+ Biochemistry
As sirtuin activity is dependent on NAD+, it has now been established that sirtuin activation is directly linked to the energetic and redox status of the cell as measured by the ratio of NAD+:NADH, by the absolute levels of NAD+, NADH, and by the NAD+ catabolite nicotinamide.39-41 Interestingly, nicotinamide itself inhibits sirtuin activity and nicotinamide-depletion during NAD biosynthesis inversely activates sirtuins.42
The NAD biosynthetic pathways are distinct in prokaryotes and invertebrates compared to vertebrates (reviewed43). We only briefly review vertebrate biochemistry here. De novo biosynthesis using tryptophan and nicotinic acid as precursors is the minor pathway for NAD generation. However, this pathway is induced by exercise and following the administration of peroxisome proliferator activated receptor alpha (PPARα) agonists.44, 45 The predominant pathway to generate NAD involves the salvage of NAD using nicotinamide as the precursor. In mammals there are two intermediary steps in NAD generation, initiated by the conversion of nicotinamide to nicotinamide mononucleotide (NMN) via the nicotinamide phosphoribosyltransferase (NAMPT) enzyme. Nicotinamide/nicotinic acid mononucleotide adenylyltransferase (NMNAT) then converts NMN to NAD. These biochemical pathways are most well characterized in the nucleus, and are pivotal for the activity of SIRT1.46 Moreover, NAMPT has been identified as the rate-controlling step in NAD biosynthesis in that overexpression of Nampt but not Nmnat increased cellular NAD levels.46
The investigation into the biology of NAD in the mitochondria has begun to be explored, and the identification of a mitochondrial-enriched NMNAT isoform supports the concept of subcellular compartment specific functioning of NAD biosynthesis.47 Moreover, mitochondrial NAD+ levels can now be measured by mass spectroscopy.48 Using this novel technique, the metabolic stress of fasting has been shown to increase mitochondrial NAMPT and to concomitantly induce mitochondrial NAD+ levels.48
Sirtuin Biochemistry
Sirtuins are known to deacetylate lysine residues on histone and non-histone proteins.49, 50 This deacetylation of target residues, coupled to the cleavage of NAD+ results in the generation of nicotinamide and the metabolite O-acetyl-ADP-ribose (OAADPr). The deacetylation of lysine residues on non-histone proteins modulates their cognate target protein function, as evidenced by the activation of Peroxisome proliferator activator receptor Gamma Coactivator - 1 alpha (PGC-1α) following its deacetylation by SIRT1.29, 51 Two potential biological consequences of lysine deacetylation include the unmasking of lysine to facilitate other PTMs, and a putative biological role following the generation of OAADPr.
In terms of PTMs, the lysine residue is highly promiscuous, with the potential to undergo, for example, acetylation, methylation, ubiquitination or SUMOylation on their ε-amino group (reviewed16). This multi-potential capacity for individual residue modification, the potential competition between these distinct PTMs at the same site and the emerging data to support cross-talk between multiple protein residue modifications highlights the emerging complexity of intramolecular signaling that may well govern biological processes.16 The role of lysine acetylation and/or deacetylation in this complex interplay is a challenging concept to be explored.52
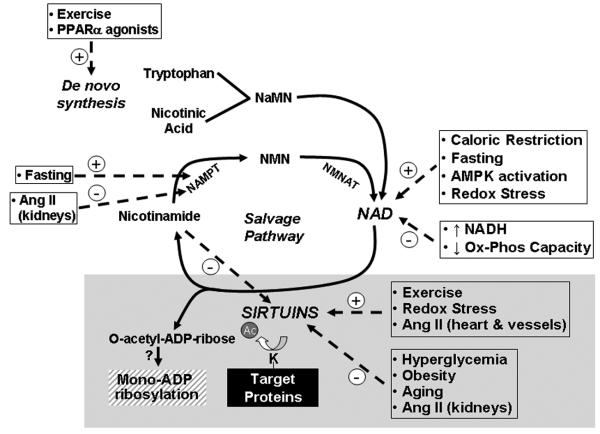
Interestingly the deacetylation metabolite OAADPr itself may directly facilitate post-translational modulatory effects following the enzymatic transfer of ADP-ribosyl groups to proteins.53 Although mono-ADP-ribosylation enzymes have been characterized in prokaryotes, and in extracellular compartments in eukaryotes, identification of intracellular enzymes in eukaryotes has been elusive.54 Sirtuins may possess the dual functionality, as both enzymatic deacetylases, and as mono-ADP ribosyltransferases using OAADPr as substrate for this PTM. To date, the mitochondrial- and nuclear- enriched SIRT4 and SIRT6 proteins respectively, exhibit ADP-ribosyltransferase activities.37, 55 However, the rate-constant of sirtuin ADP-ribosyltransferase activity is 5000 fold lower than classical bacterial enzymatic rates, and 500 times weaker than their deacetylase activities.56 These data question the physiological role of sirtuins in mono-ADP-ribosylation, however, the development of tools to investigate this biochemistry and to identify the target proteome57 should facilitate major advances in our understanding of this component of NAD and sirtuin biology in the near future. A diagram depicting NAD biosynthesis pathways are shown as the top half of figure 1.
Figure 1.
Biochemistry of NAD biosynthesis and sirtuin activity. The de novo pathway is the minor pathway and the salvage pathway the major pathway. The solid lines depict the biochemical pathways. The white boxes with their hatched lines represent biochemical and physiologic mediators of the NAD biosynthesis and sirtuin activation pathways. The black box shows the consequences of sirtuin mediated deacetylation. The hatched box represents a hypothetical modification that has not been robustly characterized. Abbreviations: NaMN – nicotinic acid mononucleotide; NMN – nicotinamide mononucleotide; NAMPT – nicotinamide phosphoribosyltransferase; NMNAT – nicotinamide/nicotinic acid mononucleotide adenyltransferase.
Sirtuin Subcellular Localization
The subcellular localization of the sirtuins is probably a pivotal feature in dictating their biological targets. Of the sirtuins known to modulate mitochondrial biology, SIRT1 has been established to predominantly reside and function in the nucleus, SIRT2, in the cytoplasm, SIRT4 in the mitochondria matrix and SIRT5 in the inner mitochondrial membrane58 or matrix.38 These locations are not exclusive and may be dynamic under specific conditions. For example, SIRT1 is exclusively nuclear during cardiac embryogenesis and then displays both nuclear and cytoplasm postnatal localization.59 Similarly, the subcellular localization of SIRT3 is predominantly in the mitochondrial matrix,60-62 although some studies suggest that SIRT3 is exclusively mitochondrial 63 while others show nuclear and cytosolic locations in whole tissue preparations64 and following overexpression studies.65-67 Whether changes in the subcellular localization of SIRT3 are associated with biological stressors, are tissue specific62, 64 or result from the genetic manipulation studies is not completely resolved. Nevertheless, the capacity of SIRT3 to alter its subcellular localization with compartment distinct functions is an intriguing concept that warrants further investigation.
Biological Triggers Orchestrating Sirtuin Activity
The biochemical pathways operational in sirtuin regulation suggest that metabolic cues are integral to their activity. Moreover, as the ratio of NAD:NADH is an important regulator of the cellular redox state, oxidative stress signaling may similarly be implicated in the regulation of sirtuin activity. More recently, the AMP-activated protein kinase (AMPK), the prototypic fuel sensing signaling kinase has also been found to modulate sirtuin activity.
Caloric restriction, which promotes cell survival and longevity functions, in part, through increasing NAD+ levels. As such, the modulation of sirtuin levels and activity has been investigated in response to this nutrient deficit. SIRT1, SIRT2 and SIRT3 levels and activity are induced in multiple organs in response to caloric restriction.67-69 However, the uniformity of this response has been questioned with the demonstration that both the ratio of NAD:NADH and SIRT1 levels are decreased in the liver in response to caloric restriction.70 Similarly, at least in pancreatic β-cells, SIRT4 activity is diminished in response to caloric restriction.37
Other modifications of nutrient exposure are also implicated in the regulation of sirtuins. In contrast to chronic caloric restriction, fasting acutely increases NAD:NADH ratio in the liver, activating SIRT1.71 A consequence of fasting induced SIRT1 induction is the deacetylation and activation of PGC-1α and PGC-1β leading to the activation of mitochondrial metabolism.22 Conversely, elevated glucose has been shown to downregulate SIRT1 in skeletal muscle and in hepatocytes, 71 while insulin, with or without elevated glucose, similarly downregulates SIRT1.68, 72 Although nutrient mediated modulation of the mitochondrial enriched sirtuins has not been extensively studied, SIRT3 is downregulated in skeletal muscle in mice with streptozotocin induced severe hyperglycemia73 and in brown adipose tissue in various murine genetic models of obesity.67 In human subjects SIRT3 levels have been shown to be diminished in skeletal muscle of sedentary older individuals, while endurance training ameliorates this effect.74
By virtue of the fact that sirtuins are activated by changes in the cellular redox state as reflected by their induction with higher levels of oxidized NAD+ or a change in the ratio of NAD+ to its reduced NADH form, imply these enzymes as redox sensitive. To date, all three subclass I sirtuins i.e. SIRT1, SIRT2 and SIRT3 have been shown to be induced by oxidative stressors.64, 69, 75, 76 Interestingly, with respect to cardiac biomechanical stressors, pressure overload and angiotensin II have been shown to increase SIRT1 and SIRT3 levels in the heart and cardiomyocytes respectively.64, 75
A more global role of AMPK in modulating the sirtuins has not been established. However, AMPK activates SIRT1 by increasing intracellular NAD+ levels77 and conversely SIRT1 deacetylates and activates the AMPK kinase LKB1.78 These observations suggest integrated biological effects of these two major nutrient and redox stress sensors in the cell. How these collectively modulate mitochondrial deacetylases and organelle homeostasis, however, has not been well characterized. The biochemistry of Sirtuin activation and the biological triggers modulating this program are schematized in the lower half of figure 1.
Modulation of mitochondrial function via nuclear protein lysine-acetylation
The control of mitochondrial function is not restricted to regulatory events within the mitochondria itself as exemplified by inter-genomic regulation between the nuclear and mitochondrial genomes and due to nuclear regulation of the intrinsic mitochondrial apoptotic program. Thus, it stands to reason that acetyl modification of non-histone lysine residues of nuclear proteins may be operational in the control of mitochondrial integrity and function.
The nuclear enriched SIRT1 has been shown to deacetylate numerous transcription factors and transcriptional co-activators that are known to control mitochondrial function. This includes the deacetylation and activation of PGC-1α. Activation of this transcriptional co-activator is know to upregulate mitochondrial biogenesis and increase mitochondrial metabolism with tissue specific preference.20, 79, 80 Furthermore, SIRT1-mediated deacetylation of FoxO proteins has been linked to the preferential activation of stress resistant targets81, 82 including the induction of mitochondrial antioxidant defenses. The deacetylation of p53 inactivates this transcription factor, thereby attenuating its pro-apoptotic action. Finally, the nuclear encoded mitochondrial transcription factor A (TFAM) has been shown to undergo acetyl PTM, however, the regulatory enzymes controlling this reaction and its functional significance, remains to be shown.83 In adipose tissue SIRT3 overexpression has also been shown to upregulate PGC-1α and promote mitochondrial uncoupling.67 However, the veracity of this data has been questioned due to the lack of alteration in thermogenesis in SIRT3 knockout mice exposed to hypothermic stress.62
Known Mitochondrial Targets of Lysine Acetylation/Deacetylation
The suggestion that mitochondria contain acetyltransferase and deacetylase enzymes was recognized in 1962, with the demonstration that isolated mitochondria could reversibly acetylate carnitine.84 The biology and multiple enzymes involved in carnitine ester formation has now been firmly established.85 However, the biochemistry and functional role of mitochondrial protein lysine acetylation is less well characterized and has become a recent focus of study in the context of the identification of mitochondrial enriched lysine-deacetylases.
Whether the mitochondrial enriched sirtuins (SIRT3, 4 and 5) function as deacetylases has recently been explored, using genetic deletion of each of these three genes to investigate global hepatic mitochondrial protein lysine acetylation.62 While the SIRT4 and SIRT5 knockouts showed little change relative to the control mice, the SIRT3 knockout show enhanced mitochondrial protein acetylation, suggesting that SIRT3 is a major mitochondrial deacetylase.62 Despite these post-translational changes, no obvious basal phenotype is evident in the SIRT3−/− mice. However, using affinity purification and mass spectroscopic analysis multiple SIRT3 interaction proteins have begun to be identified and a majority of these putative interacting proteins reside and function in the mitochondria.86
The functional characterization of individual target proteins is now being actively investigated. The mitochondrial enzyme acetyl-CoA synthetase 2 (AceCS2) was the first target of SIRT3 to be identified and partially characterized.87, 88 ACS2 is inactivated following acetylation and rapidly reactivated by SIRT3-mediated deacetylation.87, 88 Interestingly, AceCS2 is abundant in the murine heart and skeletal muscle and has been shown to be induced during ketosis.89 This implies that AceCS2 is involved in acetate conversion for energy production under ketogenic conditions. The functional characterization of the modulation of AceCS2 by SIRT3 in the heart has yet to be explored. Two additional mitochondrial matrix proteins have been identified as substrates of SIRT3 and in both instances lysine deacetylation results in increased enzyme activity. These include glutamate dehydrogenase (GDH) which facilitates the oxidative deamination of glutamate to alpha-ketoglutarate, and the citric acid cycle enzyme isocitrate dehydrogenase 2 (ICDH2).58, 62 Interestingly GDH is also a substrate for SIRT4 and this interaction results in the ADP-ribosylation and inactivation of GDH.37 The functional significance of the opposing effect of SIRT3 and SIRT4, on GDH activity, requires further characterization. The most recent functional characterization of a mitochondrial matrix metabolic target of the sirtuins is the interaction with, deacetylation of, and activation of the urea cycle enzyme carbamoyl phosphate synthetase 1 (CPS1) by SIRT5.38 This enzymes functions to detoxify and dispose of ammonia and in the SIRT5 knockout mouse, fasting results in elevated serum ammonia levels.38
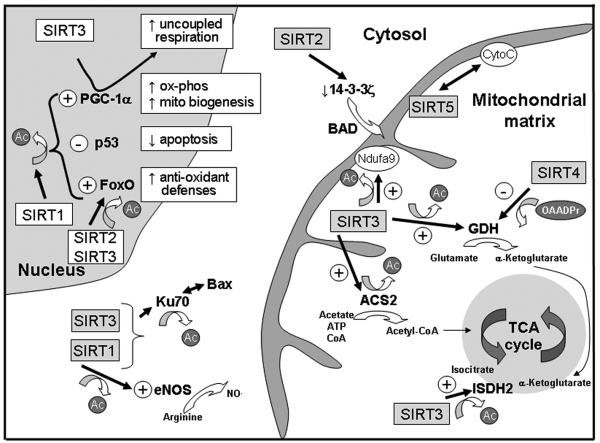
SIRT3 has also been shown to physically interact with NDUF9A, a subunit of complex I of the electron transfer chain.90 The interaction of SIRT3 with this inner mitochondrial membrane protein results in its deacetylation and activation and the genetic depletion of SIRT3 accordingly compromises complex I activity and ultimately mitochondrial oxygen consumption and ATP production.90 SIRT5 has also been shown to associate with an electron transfer chain protein, i.e. cytochrome c, although the functional consequences of this interaction has not to date been established.58 Figure 2 schematizes the currently identified nuclear, cytosolic and mitochondrial proteins that are targets of sirtuin deacetylation and PTM.
Figure 2.
Schematic of sirtuin targets that are proposed to be operational in the modulation of mitochondrial function in the cardiovascular system. Note the SIRT5 target carbamoyl phosphate synthetase 1 is not shown in this schematic as the detoxification of ammonia is not thought to be operational in the cardiovascular system. A single sided arrow indicates a functional interaction. A double sided arrows represents a protein:protein interaction.
Although a role of sirtuins in modulating outer mitochondrial membrane proteins has not been functionally characterized, it has recently been shown that long chain acyl CoA synthetase isolated from the outer membrane, also undergoes lysine-residue acetylation.91
Roles of Mitochondrial Protein Lysine Acetylation in the Heart
To date, the interrogation of the function of sirtuins in the heart has been limited, especially with respect to mitochondrial enriched sirtuins. However, prior studies linking sirtuins to metabolism, apoptosis, autophagy and aging suggest that altering sirtuin activity may modulate cardiovascular function and the responses to pathophysiologic stressors. This inference is further supported by the induction of NAMPT in rat cardiomyocytes in response to both hypoxia and serum deprivation.48 In that study, NAMPT mediated protection against cell death via mitochondrial NAD+ salvage was dependent on the presence of SIRT3 and SIRT4.48
Cardiac SIRT1 levels are induced by pressure overload, in response to the systemic administration of the oxidative stressor paraquat, with heart failure and during aging.75, 92 Together, these changes suggest that SIRT1 is modulated in response to cardiac biomechanical stressors. The first study to functionally characterize sirtuin function in the heart employed SIRT1 transgenic mice.75 Low to moderate-overexpression attenuates age-associated cardiac fibrosis, apoptosis, hypertrophy and cardiac dysfunction. Furthermore, modest SIRT1 induction protects against cardiac oxidative stress. The mitochondrial salutary effects associated with this SIRT1 induction include: increased FoxO dependent catalase expression; elevated cellular ATP levels and increased mitochondrial citrate synthase activity.75 A gene-dose effect of SIRT1 is evident in that transient overexpression of SIRT1 in cardiomyocytes prevents apoptosis via deacetylation and inactivation of p53,92 but robust SIRT1 induction in transgenic mice results in increased cardiac apoptosis, fibrosis and hypertrophy.75
Anoxia-reoxygenation stress in cardiac derived H9c2 cells results in the induction of the cytoplasm enriched sirtuin, SIRT2.76 Conversely, we have previously observed, in unpublished data, that cardiac SIRT2 gene expression is diminished by the cardioprotective program induced by delayed ischemic preconditioning.93 To characterize the stress-response to SIRT2 downregulation we used siRNA to genetically deplete SIRT2 in H9c2 myoblasts.76 SIRT2 depletion upregulates the cytosolic chaperone 14-3-3ζ, which in turn sequesters the pro-apoptotic mitochondrial protein BAD in the cytosol and augments tolerance against anoxia-reoxygenation induced cell death.76 Interestingly, the biology of SIRT2 in cellular homeostasis is not exclusively determined by levels of this deacetylase but also appears to have opposing functions under basal and stress conditions.69 These divergent phenotypes are illustrated by SIRT2-mediated induction of mitochondrial manganese superoxide dismutase via FOX3a deacetylation with the subsequent attenuation of reactive oxygen species levels at baseline. In contrast, under conditions of increased oxidative stress, SIRT2 promotes cell death in parallel with the induction of the pro-apoptotic protein Bim.69 Collectively these studies show that despite its extra-mitochondrial localization, SIRT2 modulates mitochondrial function via the modulation of mitochondrial pro-apoptotic proteins and via mitochondrial antioxidant enzyme regulation.
In rat neonatal cardiomyocytes SIRT3 levels are increased in response to H2O2, the alkylating agent MNNG, serum-starvation, and in response to phenylephrine and Angiotensin II. 64 This regulation shows similarity to SIRT1 and suggests stress-responsive functioning. In parallel, SIRT3 overexpression in cardiomyocytes enhanced resilience to genotoxic and oxidative stress. 64 An identified target for this ameliorative function is the deacetylation of Ku70. Ku70 is predominantly localized to the nucleus, although it is evident as a smaller cytoplasmic pool.94 The cytoplasmic pool is proposed to sequester the pro-apoptotic Bax protein and prevent its translocation to the mitochondria.94 During stress Ku70 is acetylated, which facilitates the release of Bax to promote mitochondrial-mediated apoptosis.95 Conversely, SIRT3 and SIRT1 both deacetylate Ku70, sequestering Bax in the cytosol and reducing genotoxic cell death.64 A recent study shows that SIRT3 knockout mice, display increased cardiac hypertrophy and interstitial fibrosis with an increased susceptibility to the development of angiotensin II-induced hypertrophy.96 In parallel, the cardiac restricted transgene overexpression an SIRT3 isoform ameliorates this phenotype. The mechanism of SIRT3 mediated protection here is shown to be via Foxo3a mediated induction of anti-oxidant defense enzymes suggesting, in part, a nuclear regulatory role of SIRT3 in cardiac stress-mediated adaptation.96
For completeness sake, we note that a cardiac phenotype is also evident in SIRT7 knockout mouse although whether this is associated with modulation in mitochondrial biology or function has not been investigated.97 SIRT7 resides in the nucleoli and following its genetic depletion mice develop and die from cardiac hypertrophy and an inflammatory cardiomyopathy.97 The absence of SIRT7 in primary murine cardiomyocytes enhances p53 acetylation, additionally leading to increased apoptosis and increased susceptibility to oxidative and genotoxic stressors.97
Potential Roles of Mitochondrial Protein Lysine Acetylation in Vascular Biology
Sirtuin function in the vasculature has not been directly explored; however, sirtuin biology is beginning to be investigated in biological programs that in part modulate vascular function, including nitric oxide biology, angiotensin signaling and hypoxia responsiveness.
Nitric oxide (NO) is a multifunction signaling molecule with diverse vascular functions which include the modulating of blood vessel tone, leukocyte adhesion, platelet activation and the development of atherosclerosis.98 Interestingly, NO also regulates mitochondrial biology through the modulation of the mitochondrial biogenesis regulatory program and via the regulation of electron transfer chain flux.99-101 The salutary effects of calorie restriction on mitochondrial biogenesis and metabolism has been shown to be dependent on NO, and this molecule is furthermore implicated in the transcriptional induction of SIRT1.102 SIRT1 itself deacetylates and activates endothelial NOS (eNOS), which via the generation of NO promotes endothelial dependent vasodilation.103 Together these regulatory events align with the known blood pressure lowering effect associated with caloric restriction,104 although whether mitochondrial biology per se and/or NO are important in these biological effects is yet to be determined.
Renin-angiotensin system (RAS) activation contributes to cardiovascular and renal disease resulting in significant morbidity and mortality. Deletion of the Ang-II type 1a receptor (AT1R) in mice extends their lifespan in association with blood pressure reduction and decreased cardiac hypertrophy and fibrosis.105, 106 Interestingly AT1R deletion leads to the upregulation of genes encoding for SIRT3 and NAMPT in the kidney.105 Consistent with this but in contrast to the heart, angiotensin II administration downregulates Sirt3 gene expression in cultured proximal tubule epithelial cells.105 As the kidney is central in angiotensin II induced hypertension,107 whether the modulation of SIRT3 and by inference mitochondrial biology plays a role in this pathophysiology is unknown. Interestingly and of relevance to the modulation of blood pressure, the overexpression of SIRT1 in vascular smooth muscle cells results in the downregulation of AT1R.108 In parallel, the SIRT1 activator resveratrol represses AT1R gene transcription and this pharmacologic compound blunts angiotensin II-induced hypertension in mice.108 Taken together, these data reveal a complex interaction between SIRT1 and SIRT3 and the RAS system, with apparent target organ specific effects. Although intriguing, the associations between sirtuins and hypertension-target organ biology are observational, and mechanistic studies need to be performed to understand the role of deacetylases and the mitochondria in this disease process.
Hypoxia is a potent trigger for the modulation of vascular tone and to promote angiogenesis. The latter program is orchestrated by the induction of hypoxia inducible factors (HIFs). HIFs are transcription factors that, under hypoxic conditions, regulate the shift in cell metabolism to glycolysis; augment cell survival through induction of anti-oxidant systems such as Heme-oxygenase-1 and superoxide dismutase (SOD); and initiate the angiogenesis regulatory program. This leads to improved mitochondrial function and cell survival.109 Hypoxia-induced redox stress stimulates SIRT1 activity, which in turn deacetylates and activates HIF-2α.41 In keeping with the role of HIF activation, this SIRT1 mediated deacetylation results in the upregulation of, for example, the mitochondrial anti-oxidant enzyme SOD2, and the angiogenesis regulatory factor VEGF.41 These data suggest that through its redox-sensing capacity, SIRT1 can modulate vascular biology via the activation of HIF-2α.
Taken together, these studies suggest that SIRT1 and SIRT3 have modulatory effects on numerous integrated biological programs governing vascular health and adaptation. The study into the mechanisms orchestrating these effects, and the determination of the physiologic role of the sirtuin program in vascular pathophysiology appears to be a promising area for future investigation. Table 1 summarizes all of the currently identified sirtuin protein targets and their function in the modulation of mitochondrial biology.
Table 1.
Summary of subcellular targets of sirtuin deacetylation or ribosylation* and proposed functional consequences
| Sirtuin | Target | Function |
|---|---|---|
|
Nuclear Targets |
||
| SIRT1 | PGC-1α | Promotes mitochondrial biogenesis |
| Foxo proteins | Upregulation of anti-oxidant defense programs | |
| eNOS | ? vasodilation and/or mitochondrial biogenesis effects | |
| p53 | Inactivated by deacetylation – diminishes apoptosis | |
| HIF-2α | Upregulates anti-oxidant enzymes and VEGF | |
| SIRT2 | Foxo3a | Activates anti-oxidant defense programs |
| Foxo3a | Activates Bim to promote apoptosis under redox stress | |
| 14-3-3 ζ | Downregulation - Bad translocation - mitochondrial apoptosis | |
| SIRT3 | Foxo3a | Upregulates anti-oxidant enzymes |
| PGC-1α | Upregulates genes encoding mitochondrial proteins | |
|
Cytosolic Targets |
||
| SIRT1 | Ku70 | Sequestered Bax in cytosol to inhibit mitochondrial apoptosis |
| LKB1 | Activates AMP kinase | |
| SIRT3 | Ku70 | Sequestered Bax in cytosol to inhibit mitochondrial apoptosis |
|
Mitochondrial Targets |
||
| SIRT3 | AceCS-2 | Acetate conversion to acetyl-CoA |
| GDH | Activation - deaminates glutamate to α-KG for TCA cycle | |
| NDUF9a | Activation of complex I of the ETC | |
| ISDH2 | Facilitates TCA cycling | |
| SIRT4* | GDH | Inactivation by ADP-ribosylation |
| SIRT5 | Cytochrome c | ? Promotes electron flux through ETC |
| CPS1 | Detoxification of ammonia via urea cycle |
Conclusions/Future Directions
In this review we discuss how the nutrient and redox sensing sirtuins modulate mitochondrial function via predominant nuclear (SIRT1), cytosolic (SIRT2) and mitochondrial (SIRT3) mediated effects. Their roles in cardiac biology have been limited to date, showing that: (1) the induction of SIRT1 mediated mitochondrial antioxidant and anti-apoptotic effects, (2) anti-apoptotic effects of SIRT2 deletion in cardiac derived cell lines and (3) SIRT3 mediated anti-apoptotic effects in response to oxidative and genotoxic stress in primary cardiomyocytes. A direct role of sirtuins in the vasculature has not been established but the augmentation of SIRT1 is associated with: (1) the downregulation of angiotensin receptor type 1, (2) lowering of blood pressure and (3) with the induction of HIF-2α mediated angiogenesis signaling and the upregulation of mitochondrial anti-oxidant enzymes. In addition, in the long-lived ATR1 knockout mice, which have low blood pressure, the genes encoding for SIRT3 and NAMPT are upregulated in the kidney. Collectively these data suggest a functional role of sirtuin biology in the modulation of cardiovascular pathophysiologic adaptations to stressors. Further and more direct in-vivo studies to investigate this biology are warranted.
The recognition that a large number of proteins that reside in the mitochondria undergo lysine-acetylation PTMs,13 and that their target proteins include numerous proteins controlling fatty acid oxidation (FAO), the tricarboxylic acid cycle (TCA) and the electron transfer chain (ETC) question whether these PTMs play an important role in cardiac substrate preference and utilization. Substrate preference and utilization and the efficient integration between metabolic pathways, for example coordination between the TCA cycle with the ETC,110 and the modulation of anaplerosis111 have begun to be recognized as important modulators of cardiac ischemia tolerance,112, 113 cardiac hypertrophy and the transition to heart failure.114, 115 Whether lysine-acetyl PTMs are important and operational in the modulation and integration of these metabolic fluxes are important questions that have not been addressed.
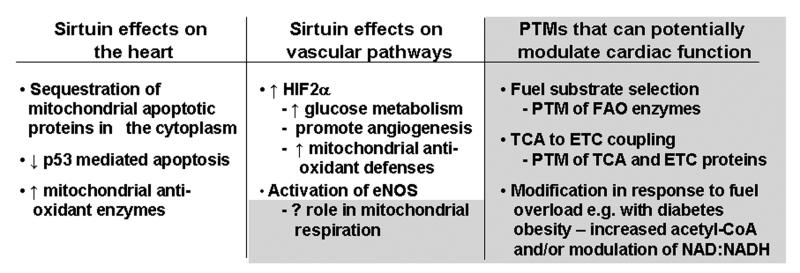
An additional area of research that has yet to be functionally characterized, but may be important, is the role of sirtuin biology in type 2 diabetes mellitus and obesity associated cardiovascular perturbations. Diabetes and obesity both associate with nutrient excess, mitochondrial dysfunction, and a predisposition to cardiovascular pathology.116, 117 Gene expression analysis shows that SIRT3 levels are diminished with obesity and hyperglycemia. Moreover, our understanding of how nutrient excess modulates biological function was recently enhanced with the recognition that elevated glucose could promote acetyl modification of proteins.118 Here, increasing concentrations of glucose promote histone acetylation through ATP citrate lyase mediated conversion of citrate to the acetylation precursor acetyl CoA.118 Whether excess nutrients modulates non-histone protein lysine acetylation via increasing acetyl CoA in distinct subcellular distributions remains to be determined119 and specifically to be evaluated in the cardiovascular system. Figure 3, schematizes both the identified pathways modified by sirtuin biology in the cardiovascular system and highlights areas of interest that have yet to be explored.
Figure 3.
Potential role of sirtuin mediated modulation of mitochondrial biology in the cardiovascular system. The direct and indirect effects of mitochondrial function that have been characterized are shown under the heart and vascular pathways columns. The potential effects of mitochondrial metabolic protein modifications are shown in the column to the right. How these may effect fuel substrate use and selection and the adaptations to direct cardiac stressors and to metabolic stresses on the heart and vasculature have yet to be ascertained. The speculative functions of acetyl-lysine PTM's is highlighted by their grey background.
The pharmacologic manipulation of the sirtuins as a potential therapeutic strategy is being investigated. The first compound explored was the plant-derived polyphenol resveratrol (3,5,4′-trihydroxystilbene) which has been shown to upregulate both SIRT1 and AMPK.120-122 and to induce mitochondrial biogenesis.122 Consistent with its known pleiotropic effects resveratrol administration confers protection against cardiac ischemia-reperfusion injury.120, 123 Recently SIRT1-specific small molecule activators have been identified and are shown to promote fatty acid oxidation and mitochondrial function.124 Whether these specific SIRT1 activators can directly ameliorate cardiac stress-tolerance and whether these effects include the modulation in mitochondrial function need to be investigated. No specific pharmacologic modulators of SIRT3 have been described, however numerous SIRT2 inhibitors have been identified.125-127 One of these inhibitors has been shown to prevent the development of Parkinson's-like disease in neuronal cells126 and it would be intriguing to investigate this compound in the context of cardiac redox stress.
Although, we have discussed the actions of sirtuins slanted towards the modulation of mitochondrial function, this family of proteins has a myriad of additional functions including for example roles in gene transcription,128 autophagy129 and the circadian rhythm.130 Hence, although our understanding of this important non-histone lysine-residue PTM should develop with ongoing investigations, we must be cognizant of the fact that the complexity and hierarchy of this biology is putatively more complex than the mitochondrial-centric focus elaborated on in this review.
In conclusion, fuel and redox sensing are important cellular commodities required to adapt to biomechanical and metabolic stressors. The PTM of non-histone lysine residues by acetylation/deacetylation appears to contribute to these sensing programs in the orchestration of important nuclear, cytosolic and mitochondrial responses to augment tolerance to injury. The investigation into this field is in its early stages and although many gaps exist in our knowledge, initial studies suggest that the modulation of this regulatory program may be important in controlling cardiovascular stress adaptation. Additionally, the emerging data supporting integration between sirtuin and AMPK adaptive programs may shed insight into the complexity of intracellular metabolic sensing and responses to nutrient and redox stress. Overall, these findings advance the observations of Charles Darwin from more than 150 years ago, which were initially directed at whole organism adaptations over the long-term, to the modern interrogation of subcellular adaptive reprogramming to acute-stressors. The recent identification and ongoing characterization of this acetylation-dependent stress-adaptive programming should further enlighten us to the myriad of innate programs exploited by nature to advance survival whether acutely and/or for the propagation and survival of species.
Acknowledgements
None
Source of Funding
This study was funded by the Divisions of Intramural Research of the NHBLI of the National Institutes of Health, Bethesda, MD, USA.
Non-Standard Abbreviations and Acronyms
- AceCS2
acetyl-coA synthetase 2
- ADP
adenosine diphosphate
- AMP
adenosine monophosphage
- AMPK
AMP-kinase
- CREB
cyclic AMP response element binding protein
- CPS1
carbamoyl phosphate synthetase 1
- eNOS
endothelial nitric oxide synthase
- ETC
electron transfer chain
- FAO
fatty acid oxidation
- Foxo
forkhead box O transcription factors
- GDH
glutamate dehydrogenase
- GNAT
Gcn5-related N-acetyltransferase
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HIF-2α
hypoxia inducible factor alpha
- ISDH2
isocitrate dehydrogenase 2
- MnSOD
manganese superoxide dismutase
- NAD
nicotinamide adenine dinucleotide
- NAMPT
nicotinamide phosphoribosyltransferase
- NMN
nicotinamide mononucleotide
- NMNAT
NMN adenylyltransferase
- LKB1
serine-threonine liver kinase B1
- OAADPr
O-acetyl-ADP-ribose
- PTM
post translational modification
- PPAR
peroxisome proliferator activated receptor
- PGC-1
PPAR gamma coactivator 1
- ROS
reactive oxygen species
- TCA
tricarboxylic acid
- TIP60
Tat-interactive protein 60.
Footnotes
Disclosures
None.
References
- 1.Murgia M, Giorgi C, Pinton P, Rizzuto R. Controlling metabolism and cell death: at the heart of mitochondrial calcium signalling. J Mol Cell Cardiol. 2009;46:781–8. doi: 10.1016/j.yjmcc.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy E, Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sack MN. Mitochondrial depolarization and the role of uncoupling proteins in ischemia tolerance. Cardiovasc Res. 2006;72:210–9. doi: 10.1016/j.cardiores.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Sack MN. Innate short-circuiting of mitochondrial metabolism in cardiac hypertrophy: identification of novel consequences of enhanced anaplerosis. Circ Res. 2009;104:717–9. doi: 10.1161/CIRCRESAHA.109.195495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young LH, Li J, Baron SJ, Russell RR. AMP-activated protein kinase: a key stress signaling pathway in the heart. Trends Cardiovasc Med. 2005;15:110–8. doi: 10.1016/j.tcm.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–87. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 7.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 8.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–98. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 9.Guarente L. Mitochondria--a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–6. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964;51:786–94. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol. 2008;18:682–9. doi: 10.1016/j.sbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–72. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 13.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–18. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics. 2009;8:215–25. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis. Curr Opin Genet Dev. 2001;11:155–61. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
- 16.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–61. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–7. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 18.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–51. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Fletcher CM, Zhou J, Allis CD, Wagner G. Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A. Nature. 1999;400:86–9. doi: 10.1038/21922. [DOI] [PubMed] [Google Scholar]
- 20.McLeod CJ, Pagel I, Sack MN. The mitochondrial biogenesis regulatory program in cardiac adaptation to ischemia--a putative target for therapeutic intervention. Trends Cardiovasc Med. 2005;15:118–23. doi: 10.1016/j.tcm.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, Schoonjans K, Puigserver P, O'Malley BW, Auwerx J. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1{alpha} Proc Natl Acad Sci U S A. 2008;105:17187–92. doi: 10.1073/pnas.0808207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly TJ, Lerin C, Haas W, Gygi SP, Puigserver P. GCN5-mediated transcriptional control of the metabolic coactivator PGC-1beta through lysine acetylation. J Biol Chem. 2009;284:19945–52. doi: 10.1074/jbc.M109.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–38. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Neal KC, Pannuti A, Smith ER, Lucchesi JC. A new human member of the MYST family of histone acetyl transferases with high sequence similarity to Drosophila MOF. Biochim Biophys Acta. 2000;1490:170–4. doi: 10.1016/s0167-4781(99)00211-0. [DOI] [PubMed] [Google Scholar]
- 25.Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 26.Kim MS, Merlo X, Wilson C, Lough J. Co-activation of atrial natriuretic factor promoter by Tip60 and serum response factor. J Biol Chem. 2006;281:15082–9. doi: 10.1074/jbc.M513593200. [DOI] [PubMed] [Google Scholar]
- 27.Eckner R. p300 and CBP as transcriptional regulators and targets of oncogenic events. Biol Chem. 1996;377:685–8. [PubMed] [Google Scholar]
- 28.McKenna NJ, Xu J, Nawaz Z, Tsai SY, Tsai MJ, O'Malley BW. Nuclear receptor coactivators: multiple enzymes, multiple complexes, multiple functions. J Steroid Biochem Mol Biol. 1999;69:3–12. doi: 10.1016/s0960-0760(98)00144-7. [DOI] [PubMed] [Google Scholar]
- 29.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 30.Shikama N, Lutz W, Kretzschmar R, Sauter N, Roth JF, Marino S, Wittwer J, Scheidweiler A, Eckner R. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 2003;22:5175–85. doi: 10.1093/emboj/cdg502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawamura T, Ono K, Morimoto T, Wada H, Hirai M, Hidaka K, Morisaki T, Heike T, Nakahata T, Kita T, Hasegawa K. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. J Biol Chem. 2005;280:19682–8. doi: 10.1074/jbc.M412428200. [DOI] [PubMed] [Google Scholar]
- 32.Takaya T, Kawamura T, Morimoto T, Ono K, Kita T, Shimatsu A, Hasegawa K. Identification of p300-targeted acetylated residues in GATA4 during hypertrophic responses in cardiac myocytes. J Biol Chem. 2008;283:9828–35. doi: 10.1074/jbc.M707391200. [DOI] [PubMed] [Google Scholar]
- 33.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–18. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–35. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 35.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–12. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 36.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–8. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 37.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–70. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for lifespan extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–8. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 40.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–90. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 41.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–93. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 42.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 43.Revollo JR, Grimm AA, Imai S. The regulation of nicotinamide adenine dinucleotide biosynthesis by Nampt/PBEF/visfatin in mammals. Curr Opin Gastroenterol. 2007;23:164–70. doi: 10.1097/MOG.0b013e32801b3c8f. [DOI] [PubMed] [Google Scholar]
- 44.Ito Y, Yonekura R, Maruta K, Koike T, Nakagami Y, Shibata K, Saito K, Nagamura Y. Tryptophan metabolism was accelerated by exercise in rat. Adv Exp Med Biol. 2003;527:531–5. doi: 10.1007/978-1-4615-0135-0_61. [DOI] [PubMed] [Google Scholar]
- 45.Shin M, Ohnishi M, Sano K, Umezawa C. NAD levels in the rat primary cultured hepatocytes affected by peroxisome-proliferators. Adv Exp Med Biol. 2003;527:653–8. doi: 10.1007/978-1-4615-0135-0_76. [DOI] [PubMed] [Google Scholar]
- 46.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–63. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 47.Berger F, Lau C, Dahlmann M, Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem. 2005;280:36334–41. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- 48.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de CR, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–11. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–82. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohas LM, St-Pierre J, Uldry M, Jager S, Handschin C, Spiegelman BM. A fundamental system of cellular energy homeostasis regulated by PGC-1alpha. Proc Natl Acad Sci U S A. 2007 May 8;104(19):7933–8. doi: 10.1073/pnas.0702683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–9. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di GM, Dani N, Stilla A, Corda D. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2005;272:4565–75. doi: 10.1111/j.1742-4658.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 54.Corda D, Di GM. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22:1953–8. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–20. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 56.Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogs and 32-P-NAD. Biochemistry. doi: 10.1021/bi802093g. 2009-epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 57.Dani N, Stilla A, Marchegiani A, Tamburro A, Till S, Ladurner AG, Corda D, Di GM. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc Natl Acad Sci U S A. 2009;106:4243–8. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 59.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–32. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 60.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–57. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99:13653–8. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr., Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–14. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cooper HM, Spelbrink JN. The human SIRT3 protein deacetylase is exclusively mitochondrial. Biochem J. 2008;411:279–85. doi: 10.1042/BJ20071624. [DOI] [PubMed] [Google Scholar]
- 64.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28:6384–401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res Commun. 2008;366:174–9. doi: 10.1016/j.bbrc.2007.11.122. [DOI] [PubMed] [Google Scholar]
- 66.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–8. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–7. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 68.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de CR, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–2. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 69.Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–14. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 70.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–7. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 72.Pagel-Langenickel I, Bao J, Joseph JJ, Schwartz DR, Mantell BS, Xu X, Raghavachari N, Sack MN. PGC-1alpha integrates insulin signaling, mitochondrial regulation, and bioenergetic function in skeletal muscle. J Biol Chem. 2008;283:22464–72. doi: 10.1074/jbc.M800842200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yechoor VK, Patti ME, Ueki K, Laustsen PG, Saccone R, Rauniyar R, Kahn CR. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc Natl Acad Sci U S A. 2004;101:16525–30. doi: 10.1073/pnas.0407574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lanza IR, Short DK, Short KR, Raghavakaimal S, Basu R, Joyner MJ, McConnell JP, Nair KS. Endurance exercise as a countermeasure for aging. Diabetes. 2008;57:2933–42. doi: 10.2337/db08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–21. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 76.Lynn EG, McLeod CJ, Gordon JP, Bao J, Sack MN. SIRT2 is a negative regulator of anoxia-reoxygenation tolerance via regulation of 14-3-3 zeta and BAD in H9c2 cells. FEBS Lett. 2008;582:2857–62. doi: 10.1016/j.febslet.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–60. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem. 2008;283:27628–35. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Finck BN, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) regulatory cascade in cardiac physiology and disease. Circulation. 2007;115:2540–8. doi: 10.1161/CIRCULATIONAHA.107.670588. [DOI] [PubMed] [Google Scholar]
- 80.Lin J, Tarr PT, Yang R, Rhee J, Puigserver P, Newgard CB, Spiegelman BM. PGC-1beta in the regulation of hepatic glucose and energy metabolism. J Biol Chem. 2003;278:30843–8. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 81.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 82.van der HA, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–9. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 83.Dinardo MM, Musicco C, Fracasso F, Milella F, Gadaleta MN, Gadaleta G, Cantatore P. Acetylation and level of mitochondrial transcription factor A in several organs of young and old rats. Biochem Biophys Res Commun. 2003;301:187–91. doi: 10.1016/s0006-291x(02)03008-5. [DOI] [PubMed] [Google Scholar]
- 84.Bremer J. Carnitine in intermediary metabolism. Reversible acetylation of carnitine by mitochondria. J Biol Chem. 1962;237:2228–31. [PubMed] [Google Scholar]
- 85.Zammit VA. Carnitine acyltransferases: functional significance of subcellular distribution and membrane topology. Prog Lipid Res. 1999;38:199–224. doi: 10.1016/s0163-7827(99)00002-8. [DOI] [PubMed] [Google Scholar]
- 86.Law IK, Liu L, Xu A, Lam KS, Vanhoutte PM, Che CM, Leung PT, Wang Y. Identification and characterization of proteins interacting with SIRT1 and SIRT3: implications in the anti-aging and metabolic effects of sirtuins. Proteomics. 2009;9:2444–56. doi: 10.1002/pmic.200800738. [DOI] [PubMed] [Google Scholar]
- 87.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–5. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–9. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fujino T, Kondo J, Ishikawa M, Morikawa K, Yamamoto TT. Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J Biol Chem. 2001;276:11420–6. doi: 10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]
- 90.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–52. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Distler AM, Kerner J, Lee K, Hoppel CL. Post-translational modifications of mitochondrial outer membrane proteins. Methods Enzymol. 2009;457:97–115. doi: 10.1016/S0076-6879(09)05006-X. [DOI] [PubMed] [Google Scholar]
- 92.Alcendor RR, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J. Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ Res. 2004;95:971–80. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- 93.McLeod CJ, Jeyabalan AP, Minners JO, Clevenger R, Hoyt RF, Jr., Sack MN. Delayed ischemic preconditioning activates nuclear-encoded electron-transfer-chain gene expression in parallel with enhanced postanoxic mitochondrial respiratory recovery. Circulation. 2004;110:534–9. doi: 10.1161/01.CIR.0000136997.53612.6C. [DOI] [PubMed] [Google Scholar]
- 94.Sawada M, Sun W, Hayes P, Leskov K, Boothman DA, Matsuyama S. Ku70 suppresses the apoptotic translocation of Bax to mitochondria. Nat Cell Biol. 2003;5:320–9. doi: 10.1038/ncb950. [DOI] [PubMed] [Google Scholar]
- 95.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–38. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 96.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. doi: 10.1172/JCI39162. 2009 epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102:703–10. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 98.Forstermann U, Munzel T. Endothelial nitric oxide synthase in vascular disease: from marvel to menace. Circulation. 2006;113:1708–14. doi: 10.1161/CIRCULATIONAHA.105.602532. [DOI] [PubMed] [Google Scholar]
- 99.Brown GC, Cooper CE. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome oxidase. FEBS Lett. 1994;356:295–8. doi: 10.1016/0014-5793(94)01290-3. [DOI] [PubMed] [Google Scholar]
- 100.Nisoli E, Falcone S, Tonello C, Cozzi V, Palomba L, Fiorani M, Pisconti A, Brunelli S, Cardile A, Francolini M, Cantoni O, Carruba MO, Moncada S, Clementi E. Mitochondrial biogenesis by NO yields functionally active mitochondria in mammals. Proc Natl Acad Sci U S A. 2004;101:16507–12. doi: 10.1073/pnas.0405432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shiva S, Sack MN, Greer JJ, Duranski M, Ringwood LA, Burwell L, Wang X, MacArthur PH, Shoja A, Raghavachari N, Calvert JW, Brookes PS, Lefer DJ, Gladwin MT. Nitrite augments tolerance to ischemia/reperfusion injury via the modulation of mitochondrial electron transfer. J Exp Med. 2007;204:2089–102. doi: 10.1084/jem.20070198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–9. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 103.Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, DeRicco J, Kasuno K, Irani K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 2007;104:14855–60. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sasaki S, Higashi Y, Nakagawa K, Kimura M, Noma K, Sasaki S, Hara K, Matsuura H, Goto C, Oshima T, Chayama K. A low-calorie diet improves endothelium-dependent vasodilation in obese patients with essential hypertension. Am J Hypertens. 2002;15:302–9. doi: 10.1016/s0895-7061(01)02322-6. [DOI] [PubMed] [Google Scholar]
- 105.Benigni A, Corna D, Zoja C, Sonzogni A, Latini R, Salio M, Conti S, Rottoli D, Longaretti L, Cassis P, Morigi M, Coffman TM, Remuzzi G. Disruption of the Ang II type 1 receptor promotes longevity in mice. J Clin Invest. 2009;119:524–30. doi: 10.1172/JCI36703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gembardt F, Heringer-Walther S, van Esch JH, Sterner-Kock A, van VR, Le TH, Garrelds IM, Coffman TM, Danser AH, Schultheiss HP, Walther T. Cardiovascular phenotype of mice lacking all three subtypes of angiotensin II receptors. FASEB J. 2008;22:3068–77. doi: 10.1096/fj.08-108316. [DOI] [PubMed] [Google Scholar]
- 107.Crowley SD, Gurley SB, Herrera MJ, Ruiz P, Griffiths R, Kumar AP, Kim HS, Smithies O, Le TH, Coffman TM. Angiotensin II causes hypertension and cardiac hypertrophy through its receptors in the kidney. Proc Natl Acad Sci U S A. 2006;103:17985–90. doi: 10.1073/pnas.0605545103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miyazaki R, Ichiki T, Hashimoto T, Inanaga K, Imayama I, Sadoshima J, Sunagawa K. SIRT1, a longevity gene, downregulates angiotensin II type 1 receptor expression in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2008;28:1263–9. doi: 10.1161/ATVBAHA.108.166991. [DOI] [PubMed] [Google Scholar]
- 109.Loor G, Schumacker PT. Role of hypoxia-inducible factor in cell survival during myocardial ischemia-reperfusion. Cell Death Differ. 2008;15:686–90. doi: 10.1038/cdd.2008.13. [DOI] [PubMed] [Google Scholar]
- 110.Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, Lopaschuk GD, Muoio DM. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 111.Pound KM, Sorokina N, Ballal K, Berkich DA, Fasano M, Lanoue KF, Taegtmeyer H, O'Donnell JM, Lewandowski ED. Substrate-enzyme competition attenuates upregulated anaplerotic flux through malic enzyme in hypertrophied rat heart and restores triacylglyceride content: attenuating upregulated anaplerosis in hypertrophy. Circ Res. 2009;104:805–12. doi: 10.1161/CIRCRESAHA.108.189951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Opie LH, Sack MN. Metabolic plasticity and the promotion of cardiac protection in ischemia and ischemic preconditioning. J Mol Cell Cardiol. 2002;34:1077–89. doi: 10.1006/jmcc.2002.2066. [DOI] [PubMed] [Google Scholar]
- 113.Ussher JR, Lopaschuk GD. The malonyl CoA axis as a potential target for treating ischaemic heart disease. Cardiovasc Res. 2008;79:259–68. doi: 10.1093/cvr/cvn130. [DOI] [PubMed] [Google Scholar]
- 114.Sack MN, Kelly DP. The energy substrate switch during development of heart failure: gene regulatory mechanisms. Int J Mol Med. 1998;1:17–24. doi: 10.3892/ijmm.1.1.17. [DOI] [PubMed] [Google Scholar]
- 115.Sharma N, Okere IC, Duda MK, Chess DJ, O'Shea KM, Stanley WC. Potential impact of carbohydrate and fat intake on pathological left ventricular hypertrophy. Cardiovasc Res. 2007;73:257–68. doi: 10.1016/j.cardiores.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Boudina S, Sena S, O'Neill BT, Tathireddy P, Young ME, Abel ED. Reduced mitochondrial oxidative capacity and increased mitochondrial uncoupling impair myocardial energetics in obesity. Circulation. 2005;112:2686–95. doi: 10.1161/CIRCULATIONAHA.105.554360. [DOI] [PubMed] [Google Scholar]
- 117.Sack MN. Type 2 diabetes, mitochondrial biology and the heart. J Mol Cell Cardiol. 2009;46:842–9. doi: 10.1016/j.yjmcc.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rathmell JC, Newgard CB. Biochemistry. A glucose-to-gene link. Science. 2009;324:1021–2. doi: 10.1126/science.1174665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le CD, Shaw RJ, Navas P, Puigserver P, Ingram DK, de CR, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci U S A. 2007;104:7217–22. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 123.Goh SS, Woodman OL, Pepe S, Cao AH, Qin C, Ritchie RH. The red wine antioxidant resveratrol prevents cardiomyocyte injury following ischemia-reperfusion via multiple sites and mechanisms. Antioxid Redox Signal. 2007;9:101–13. doi: 10.1089/ars.2007.9.101. [DOI] [PubMed] [Google Scholar]
- 124.Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–58. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 125.Gutierrez M, Andrianasolo EH, Shin WK, Goeger DE, Yokochi A, Schemies J, Jung M, France D, Cornell-Kennon S, Lee E, Gerwick WH. Structural and Synthetic Investigations of Tanikolide Dimer, a SIRT2 Selective Inhibitor, and Tanikolide seco-Acid from the Madagascar Marine Cyanobacterium Lyngbya majuscula. J Org Chem. 2009 doi: 10.1021/jo900578j. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, Kazantsev AG. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317:516–9. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 127.Zhang Y, Au Q, Zhang M, Barber JR, Ng SC, Zhang B. Identification of a small molecule SIRT2 inhibitor with selective tumor cytotoxicity. Biochem Biophys Res Commun. 2009;386:729–33. doi: 10.1016/j.bbrc.2009.06.113. [DOI] [PubMed] [Google Scholar]
- 128.Feige JN, Auwerx J. Transcriptional targets of sirtuins in the coordination of mammalian physiology. Curr Opin Cell Biol. 2008;20:303–9. doi: 10.1016/j.ceb.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–9. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]