Abstract
Development of effective topical microbicides for the prevention of HIV-1 sexual transmission represents a primary goal for the control of the AIDS pandemic. The viral co-receptor CCR5, used by the vast majority of primary HIV-1 isolates, is considered a primary target molecule. RANTES and its derivatives are the most suitable protein-based compounds to fight HIV-1 via CCR5 targeting. Yet, receptor activation should be avoided to prevent pro-inflammatory effects and possibly provide anti-inflammatory properties. C1C5 RANTES is a chemokine mutant that exhibits high anti-HIV-1 potency coupled with CCR5 antagonism. However, the need for the formation of an N-terminal intramolecular disulphide bridge between non-natural cysteine residues at position 1 and 5 represents a challenge for the correct folding of this protein in recombinant expression systems, a crucial step towards its development as a microbicide against HIV-1. We report here a rare case of superior folding in a prokaryote as compared to an eukaryotic expression system. Production of C1C5 RANTES was highly impaired in CHO cells, with a dramatic yield reduction compared to that of wild type RANTES and secretion of the molecule as disulphide-linked dimer. Conversely, a human vaginal isolate of Lactobacillus jensenii engineered to secrete C1C5 RANTES provided efficient delivery of the monomeric protein. This and other reports on successful secretion of complex proteins indicate that lactic acid bacteria are an excellent system for the expression of therapeutic proteins, which can be used as a platform for the engineering of conceptually novel RANTES mutants with potent anti-HIV-1 activity.
Keywords: lactobacilli, mammalian cells, protein folding, RANTES, anti-HIV-1 microbicide
Introduction
Lactobacilli are Gram-positive facultative anaerobic bacteria. Several strains of the Lactobacillus species (spp.)1 belong to the human commensal microbiota, which abundantly populate the gastrointestinal tract and the vagina. Together with other lactic acid bacteria (Lactococcus spp.), lactobacilli are often referred to as probiotics, since they play an important role in host physiology, immune response development, and defense against pathogenic bacteria, fungi, and viruses [1]. Due to their ability to colonize different mucosal compartments of the body and to express exogenous recombinant proteins, lactobacilli were recently used as delivery vehicles for mucosal vaccines or therapeutic molecules for allergic and gastrointestinal diseases. Preliminary results obtained in animal models indicate that lactobacilli delivery can be a realistic therapeutic option in humans [2].
Sexual transmission is the major route of infection for the human immunodeficiency virus type 1 (HIV-1) and the most difficult to control [3]. In women, infection occurs mainly in the cervico-vaginal mucosa layer where CD4+ T cells, Langherans cells and macrophages express the HIV-1 receptors CD4 and CCR5 [4]. At present, the best strategies to block HIV-1 mucosal transmission imply the development of vaccines and topical microbicides capable of preventing viral infection [5]. While an effective anti-HIV-1 vaccine is still being pursued, topical microbicides seem to be a more realistic option, although an efficacious microbicide is not available yet [6,7]. In order to assure the possibility to provide prevention globally, anti-HIV-1 microbicides need to be inexpensive to manufacture, stable, safe and effective at reducing the transmission of HIV-1 from an infected partner. Although a large number of promising protein-based HIV-1 inhibitors can potentially be developed, their production represents a major problem in terms of availability of efficient expression systems and manufacturing costs [3,8,9]. A potential solution to this issue resides in the development of ‘live microbicides’ based on genetically modified lactobacilli that produce and release potent protein-based HIV-1 inhibitors in vivo and in situ [10]. Of special interest is the possibility to enhance the function of vaginal microbiota by genetic engineering, in order to establish a protective low-cost shield against HIV-1. To date, several strains of lactobacilli have been engineered to produce different anti-HIV-1 proteins or peptides, including HIV-1 entry and fusion inhibitors [11–17]. Interestingly, regardless of the pathology to be tackled,lactic acid bacteria have proven to be very efficient in secreting rather complex protein moieties, such as multi-domain proteins and proteins containing intramolecular disulphide bonds [2]. Identical considerations can be made for the bacterial surface anchorage of recombinant proteins, a process that follows the same biosynthetic pathway of protein secretion.
RANTES is the most potent natural inhibitor of HIV-1 [18] and several full length and short peptide derivatives have been engineered that enhanced different aspects and properties of its anti-HIV-1 action [19]. Moreover, a chemically-modified RANTES variant provided the in vivo proof of principle for the use of RANTES derivatives as topical microbicides [20]. Therefore, RANTES represents the most important protein-based CCR5-targeting inhibitor of HIV-1 entry [19]. The classic source of recombinant RANTES is E. coli, where the protein is produced as inclusion bodies that, upon refolding procedures, provide the folded, active chemokine. Attempts have been made to improve this expression system to attain folded RANTES molecules, avoiding the refolding process [21,22]. However, given the increasing prominence RANTES is gaining in the field of HIV-1 prevention, the need for a simple and efficient expression system capable to provide a reliable platform for the screening of several derivatives becomes a key issue to the discovery and development of new potent HIV-1 blockers based on RANTES. In other words, a fundamental requirement for the ideal expression/screening platform would be the quick availability of the active protein, i.e., secretion in the culture medium. Aside of the HIV-1 field, a recombinant cell system producing RANTES derivatives would be important also for therapeutic aspects related to the chemokine involvement in several inflammatory conditions.
In this perspective and with the aim of further evaluating the protein folding properties of lactic acid bacteria, we report here a case of superior protein folding capacity by Lactobacillus jensenii (a human vaginal strain) as compared to the mammalian Chinese hamster ovary (CHO) cell line. C1C5 RANTES, a RANTES mutant in which serine residues in position 1 and 5 were substituted by cysteine, yielding a potent anti-HIV-1 CCR5 antagonist [23], is reported here as a probe to attest such folding efficiency. We found that CHO cells secreted C1C5 RANTES at very low levels and mostly as an intermolecular disulphide-linked dimer, while L. jensenii was able to efficiently secrete the protein in its monomeric form with the expected C1C5 intramolecular disulphide bond formed. These results indicate that lactobacilli represent an efficient and convenient system to screen new generation RANTES mutants in search of potential therapeutic leads.
Material and methods
Plasmid construction and cell culture
CHO-K1 cells (ATCC CCL-61) were stably transfected using the pcDNA3.1+ vector (Invitrogen). An EcoRI fragment encoding human RANTES full-length sequence (including the secretion leader sequence) was derived by RT-PCR amplification from total RNA of in vitro-activated primary human T cells [24] and cloned into pcDNA3.1+ to obtain pcDNA3-RANTES. C1C5 RANTES was obtained by the QuikChange site-directed mutagenesis kit (Stratagene) from pcDNA3-RANTES using specific primers (C1C5-forward 5’-CCTGCATCTGCCTGCCCATATTCCTGTGACACCACACCC-3’ and C1C5-reverse 5’-GGGTGTGGTGTCACAGGAATATGGGCAGGCAGATGCAGG-3’, bases in bold correspond to mutated codons), following manufacturer’s instructions. Briefly, a PCR product was generated with Pfu Turbo DNA polymerase (Stratagene) and treated with DpnI endonuclease before transformation into XL1-Blue supercompetent cells (Stratagene) to obtain pcDNA3-C1C5 RANTES. CHO cells were cultured in DMEM medium (Lonza BioWhittaker) supplemented with 10% fetal bovine serum (Lonza BioWhittaker). 1.5 × 106 CHO cells were transfected separately with 5 µg each of the two pcDNA3.1+ constructs, using an AMAXA electroporator (AMAXA Biosystems), according to manufacturer’s instructions. Two days after electroporation, cells were selected with 800 µg/ml of G-418 (Sigma) and stable CHO transfectant clones, obtained by limited dilution, were screened for secretion of the two RANTES variants using a commercial ELISA kit (RANTES/CCL5 DuoSet, R&D Systems). RANTES-expressing CHO clones were cultured for 72 hrs in Optimem medium (Lonza BioWhittaker) and, after centrifugation at 1200 rpm in a Heraeus 1.0 Megafuge to eliminate cell debris, supernatants were concentrated in Amicon filter devices (Amicon Centriplus YM-3) and analyzed by Western blot.
The E. coli/Lactobacillus shuttle p1063 expression vector for L. jensenii, a modified version of pOsel175 [15], was used for construction of wild type (wt) and C1C5 RANTES plasmids. The wt RANTES sequence (aa 1–68) was PCR amplified from pcDNA3-RANTES using specific primers (RANTES-forward 5’-CTAGCTAGCCCATATTCCTCGGACACC-3’ and RANTES-reverse 5’-CTAGCTAGCCTAACTCATCTCCAAAGAGT-3’, bases in bold correspond to Nhel restriction sites) and the PCR product cloned into the p1063 vector to obtain p1063-RANTES. pl063-C1C5 RANTES was obtained by the QuikChange site-directed mutagenesis kit (Stratagene) from pl063-RANTES using specific primers (C1C5-forward 5’-GTTTCTACTGTTTCAGCTTGTCCATATTCCTGCGACACCACACCCTG-3’ and C1C5-reverse 5’-CAGGGTGTGGTGTCGCAGGAATATGGACAAGCTGAAACAGTAGAAAC-3’, bases in bold correspond to mutated codons), following manufacturer’s instructions. Human vaginal isolate of Lactobacillus jensenii 1153 [11] was routinely cultivated at 37 °C (5% CO2) in MRS or Rogosa SL broth (Difco). Plasmids were introduced by electroporation into E. coli XL1-Blue (Stratagene). For plasmid maintenance, transformed E. coli XL1-Blue clones were grown in LB broth (Difco) at 37 °C, supplemented with erythromycin (300 µg/ml). Plasmids were transformed by electroporation into L. jensenii 1153 essentially as described [11]. Transformed L. jensenii clones were routinely propagated in liquid media containing 20 µg/ml erythromycin. To verify protein secretion, 30 µl of an overnight culture supernatant were analyzed by Western blot.
HIV-1 infection assay
CHO cells expressing wt RANTES were cultured in Optimem for 72 hrs. Then, 40 ml of cell supernatant were concentrated in Amicon filter devices to 1 ml, diluted with RPMI medium to 10 ml and concentrated again to 640 ng/ml. RANTES quantification was assayed by specific ELISA. Different amounts of RANTES ranging from 100 ng/ml to 0.1 ng/ml were used in the HIV-1 infection assay.
Acute HIV-1 infection was obtained by adding the HIV-1BaL stock (50 TCID50/well) to PM1 cells (2 × 104/well) in complete RPMI medium in the presence or absence of inhibitors. PM1 is a unique CD4+ T-cell clone susceptible to a wide variety of primary HIV-1 isolates, including those exclusively using CCR5 as a coreceptor [25]. Experiments were performed in triplicate in 96-well round-bottom microtiter plates. After incubation at 37 °C for 16 hrs, the wells were washed twice and complete medium, with or without the inhibitors, was added. Virus replication was assayed at day 4 post-infection by the p24 antigen ELISA. Supernatants were diluted in 1% Empigen BB detergent (Calbiochem) to disrupt virions and added to a 96-well ELISA plate coated with anti-HIV-1 p24 polyclonal antibodies (Abs) (Aalto Bio Reagents Ltd.) and incubated 2 hrs at room temperature. The plate was then washed three times in TBS buffer (1.5 M NaCl, 250 mM Tris pH 7.5) and an alkaline phosphatase-conjugated anti-HIV-1 p24 monoclonal antibody (Aalto Bio Reagents Ltd.) was added for 1 h at room temperature. After washing three times with TROPIX buffer (10 mM MgCl2, 200 mM Tris pH 9.8), p24 was detected adding the luminescence substrate CSPD TROPIX (Applied Biosystems) and the signal was analyzed using a Mithras LB 940 luminometer (Berthold Technologies). Levels of p24 were calculated generating a standard curve with HIV-1 p24 antigen standards. Human recombinant wt RANTES expressed in E. coli and refolded from inclusion bodies was donated by Dr. Proudfoot (Merck Serono Geneva Research Centre, Geneva, Switzerland) and used as standard in this assay and throughout all this work. Dose-response curves and histograms were fit using GraphPad Prism (GraphPad Software), also to calculate ID50 concentrations.
Intracellular analysis and Western blot
Stable CHO clones were cultured for 72 hrs in Optimem medium, washed in PBS (Lonza BioWhittaker) and then 1 × 106 CHO cells were lysed in 50 µl sample buffer 2X (Laemmli buffer: 4% SDS, 20% glycerol, 125 mM Tris pH 6.8, 0.004% bromophenol blue and, when necessary, 10% β-mercaptoethanol) and analyzed in Western blot for the intracellular content of wt and C1C5 RANTES. For the intracellular detection in L. jensenii, bacterial pellet derived from 1.5 ml cell suspension of overnight Rogosa culture was washed in 1 ml of PBS and resuspended in 150 µl of miniprep solution 1 (Qiagen). Lysozyme (Sigma) was added at a final concentration of 4 mg/ml and samples incubated at 37 °C for 30 min. Then, 30 µl of Laemmli sample buffer 6X were added and 60 µl samples used for Western blot analysis.
Samples for Western blot analysis were separated by reducing or non-reducing 13% SDS-PAGE, blotted onto Protran-83 nitrocellulose membranes (Schleicher & Schuell) and incubated overnight with rabbit anti-human RANTES Abs 1:1000 (PeproTech). Chemiluminescent signals were developed with horseradish peroxidase-conjugated goat anti-rabbit Abs 1:5000 (Sigma) and the ECL reagent (Amersham).
Chemical cross-linking
Five ml of RANTES-containing CHO cell supernatants (Optimem) were concentrated to 200 µl in Amicon filter devices and diluted in PBS to 2 ml. For the Lactobacillus-secreted proteins, 100 ng of purified RANTES variants were diluted in PBS to 2 ml. Samples were treated with 1 mM of 3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP, Pierce), with spacer arm length of 1.2 nm, for 2 hrs at 4 °C. The cross-linker modifies freely exposed amines on the N-terminus and lysine residues. Then, the reactions were blocked with 20 mM Tris pH 7.5 for 15 min at room temperature and the reacted material precipitated with 20% trichloroacetic acid (Sigma). Samples were resuspended in 100 µl of Laemmli sample buffer 2X, equally divided in two parts and analyzed in Western blot under non-reducing conditions or reducing conditions by adding 5% β-mercaptoethanol. As control, 100 ng of E. coli-produced recombinant RANTES standard diluted to 2 ml in PBS were treated with DTSSP in the same conditions.
Protein purification
In the screening system based on anti-HIV-1 activity, wt or C1C5 RANTES were partially purified from 10 ml of transformed L. jensenii overnight culture. Rogosa supernatants were clarified by centrifugation at 4000 rpm for 15 min, filtered with 0.22 µm filter membrane (Millipore) and incubated in a 15 ml tube with 0.5 ml Q-Sepharose Fast Flow (FF) resin (Amersham) for 1 h at 4 °C on a rotary shaker. Flow through was incubated with 0.5 ml SP-Sepharose FF resin (Amersham) as described for the previous step. Both resins were equilibrated with 20 mM Tris pH 6.0 (buffer A) before use. Then, SP-Sepharose FF resin was extensively washed with buffer A and RANTES variants eluted in batch in a two step gradient with 2 ml buffer A containing 300 mM NaCl and 2 ml buffer A containing 1 M NaCl (each elution step was collected in two fractions of 1 ml). RANTES concentration was evaluated by human RANTES ELISA (R&D Systems). The first 1 M NaCl fractions contained 1.45 µg/ml wt RANTES and 1.75 µg/ml C1C5 RANTES, respectively (about a half of the total supernatant content), and were used in the previously described p24-based assay. Non-transformed L. jensenii supernatant was treated identically as a control for the system and identical volumes of the first 1 M NaCl fraction were added to the HIV-1 infection assay, resulting in a negligible effect, yet subtracted to the final anti-HIV-1 activity values of RANTES variants.
To purify RANTES variants, transformed L. jensenii clones were cultured in Rogosa broth overnight. Two liters of the resulting broth were centrifuged at 6000 rpm for 30 min to discard cell debris and filtered with 0.22 µm stericup filters (Millipore). Clarified supernatant was diluted 1:2 with water to decrease ionic strength and loaded at a 4 ml/min flow on a column (20 cm length, 1.5 cm diameter, Biorad) packed with 15 ml Q-Sepharose FF resin (Amersham), previously equilibrated in buffer B (buffer A containing 50 mM NaCl). Flow through was directly loaded in the same buffer conditions on an identical column packed with 15 ml SP-Sepharose FF resin (Amersham) and, after extensive wash in buffer B, proteins were eluted using a sodium chloride multi-step gradient (from 250 mM to 2 M) in buffer B. Fractions were analyzed by ELISA (RANTES/CCL5 DuoSet, R&D Systems), 13% SDS-PAGE stained with Coomassie brillant Blue R-250 (Applichem) and Western blot. The best fractions were pooled, dialyzed against 20 mM Tris pH 8.0, 50 mM NaCl (buffer C) and loaded at 4 ml/min on a column packed with 5 ml Q-Sepharose FF resin, previously equilibrated in buffer C. As in previous step, most RANTES content was in the flow through and this was loaded onto a column packed with 5 ml SP-Sepharose FF resin, previously equilibrated in buffer C. After extensive wash in buffer B, proteins were eluted using a sodium chloride multi-step gradient (from 250 mM to 2 M) in buffer C. Purified RANTES variants were analyzed by 13% SDS-PAGE stained with Coomassie brillant Blue R-250 (Applichem) and by Western blot.
Results and discussion
Wild type and C1C5 RANTES expression in CHO cells and L. jensenii
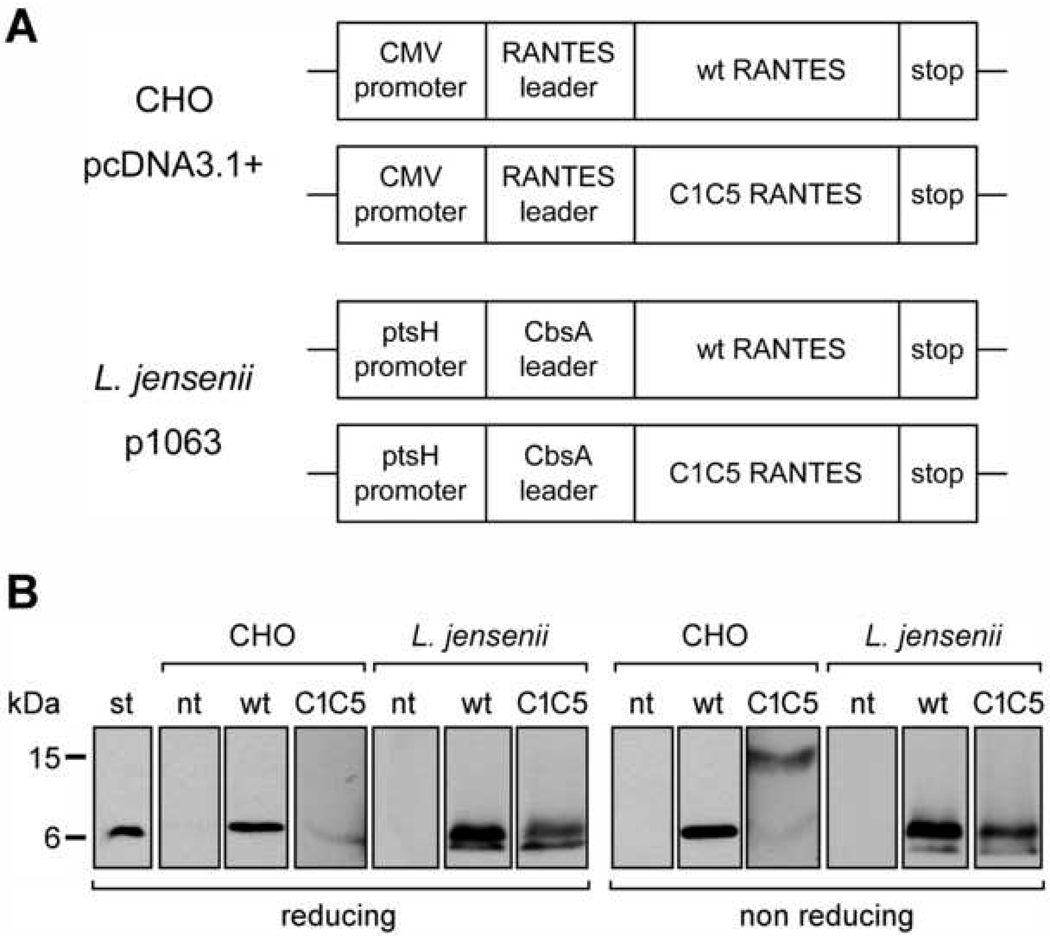
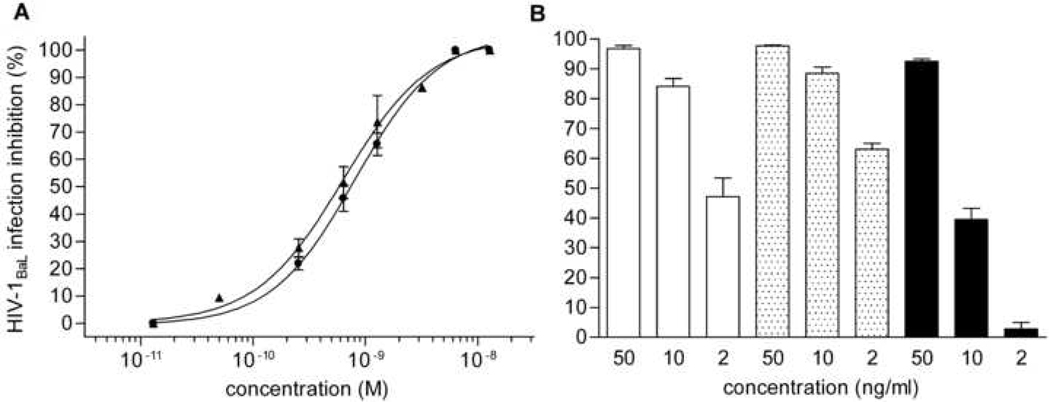
In an effort to establish a source for RANTES variants secreted as correctly folded proteins, we first evaluated the mammalian cell line CHO. Although the level of protein production was expected to be lower, as compared to E. coli, this limitation should be balanced by the release of properly folded material directly into the culture supernatant, thus making CHO cells a faster and superior screening system. Wt human RANTES cDNA, including the natural leader sequence for endoplasmic reticulum (ER) translocation, was cloned into the mammalian expression vector pcDNA3, downstream of the human cytomegalovirus (CMV) promoter (Fig. 1A). CHO cells were then transfected and, by means of a human RANTES-specific ELISA assay, stable clones were selected that secreted RANTES into the cell supernatant. A first hurdle encountered in this system was the very low expression level, reaching a maximum of 50 µg/L in the highest-producing CHO clones. However, wt RANTES could be detected in Western blot as a single secretory form migrating as a monomer in both reducing and non-reducing SDS-PAGE (Fig. 1B). Encouragingly, wt RANTES from CHO cells showed an HIV-1 inhibitory activity comparable to that of E. coli-produced wt RANTES, which is used in our laboratory as a standard (Fig. 2A). This result prompted us to continue with the CHO cell system and two new RANTES mutants have recently been produced providing further validation to this mammalian cell system (unpublished results).
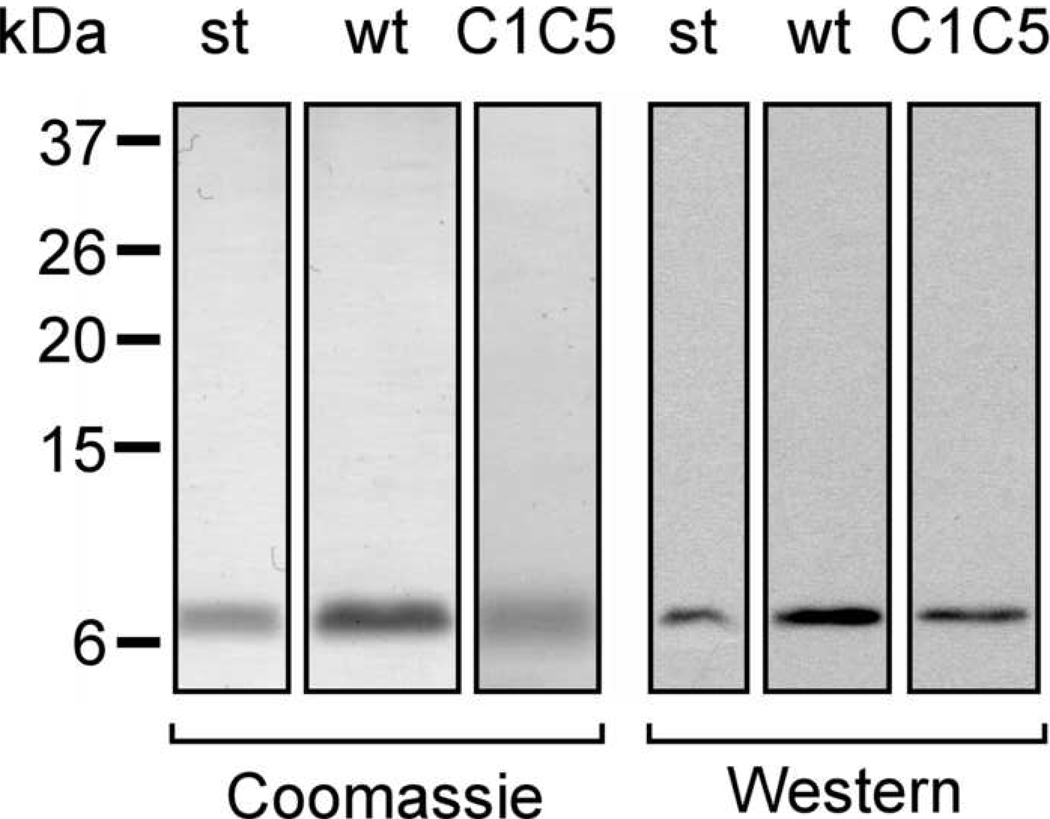
Fig. 1.
Wild type and C1C5 RANTES construct design and protein secretion. (A) Schematic representation of pcDNA3.1+ and p1063 constructs for wt and C1C5 RANTES expression in CHO cells and L. jensenii, respectively. (B) Reducing and non-reducing Western blot analysis of secreted RANTES variants using anti-human RANTES Abs. st, RANTES standard from E. coli; nt, supernatants from non-transfected CHO cells or non-transformed L. jensenii; and wt and C1C5, wt and C1C5 RANTES-containing supernatants, respectively.
Fig. 2.
Anti-HIV-1 activity of secreted wt and C1C5 RANTES. Inhibition of HIV-1BaL infection in PM1 cells was measured by the p24-based assay after 4 days of infection. (A) (circles) E. coli-expressed RANTES standard (ID50% = 0.8 nM) and (triangles) CHO cell-secreted wt RANTES (ID50% =0.63 nM). Values are the mean ± SD of two independent experiments performed in triplicate. (B) (white columns) E. coli-expressed RANTES standard; (dotted columns) partially purified wt RANTES from L. jensenii; and (black columns) partially purified C1C5 RANTES from L. jensenii. Values are the mean ± SEM of one experiment performed in triplicate, representative of two experiments yielding similar results.
Next, C1C5 RANTES, a RANTES mutant in which the serine residues at position 1 and 5 were mutated to cysteine [23], was selected as a second probe to validate CHO cells as a suitable platform to screen new RANTES mutants. The choice of C1C5 RANTES was dictated by two major features: i) its binding to CCR5 as antagonist, where CCR5 antagonism is a fundamental requisite to avoid pro-inflammatory conditions induced by anti-HIV-1 blockers; and ii) its peculiar need for an N-terminal intramolecular disulphide bridge, possibly representing a folding challenge to the expression system. C1C5 RANTES cloning, CHO cell transfection and stable clone selection was carried out using the same protocol as for wt RANTES (Fig. 1A). Unfortunately, the already low level of protein secretion obtained for wt RANTES was further reduced in the case of C1C5 RANTES, with a mere 1 µg/L reached for the highest-expressing clones, accounting for a dramatic 50-fold reduction. Somewhat unexpectedly, when analyzed in non-reducing SDS-PAGE, C1C5 RANTES was revealed as a covalent dimer with only an extremely faint monomeric form detectable (Fig. 1B). Dimerization occurred via intermolecular disulphide bridging, since, upon reduction, the secretory form migrated as a single band with a molecular mass corresponding to the monomer. The biological activity of C1C5 RANTES could not be investigated given the very low quantity of protein secreted. Despite the fact that the intramolecular C1C5 disulphide bridge is a non-natural feature, secretion of a covalent dimer might be interpreted as a lack of folding competence or, paradoxically, as the intervention of a too-specialized folding machinery. The generally low RANTES secretion level and impairment of C1C5 RANTES folding led us to abandon mammalian cells as a secretion system for new RANTES mutants acting as anti-HIV-1 CCR5 antagonists.
Interestingly, a second secretion system that could satisfy the needs described above came from a completely different approach, i.e., the development of a RANTES-producing anti-HIV-1 live microbicide (manuscript submitted). A human vaginal isolate L. jensenii strain was engineered to secrete wt and C1C5 RANTES. The chemokine cDNA was fused to the secretion leader peptide of the L. crispatus S-layer gene (CbsA) under the L. jensenii ptsH promoter (Fig. 1A). RANTES variants were cloned in the Lactobacillus-dedicated p1063 vector. In order to verify protein secretion, Rogosa medium derived from an overnight culture of transformed L. jensenii was harvested and analyzed by the RANTES-specific ELISA mentioned above. The expression level of wt and C1C5 RANTES was estimated to be in the 0.3–0.6 mg/L range, with a ~2 fold higher expression level for the wt protein. Western blot analysis under reducing and non-reducing conditions confirmed the presence of the proteins in the supernatants (Fig. 1B). Both RANTES variants appeared to be secreted as the expected monomer, indicating that in C1C5 RANTES the N-terminal intramolecular disulphide bond was correctly formed. A second form of lower molecular mass could be detected that has been identified by mass spectrometry as a proteolytic cleavage product (manuscript submitted). However, recent evidence from L. jensenii chromosomal integration of C1C5 RANTES indicates that the protein is secreted exclusively in the native form (unpublished results), a fundamental issue in view of the potential clinical application of this product.
In view of the need for a rapid protocol to screen several RANTES mutants and select the most promising ones for further characterization, we developed a system coupling partial purification to anti-HIV-1 activity testing. In order to proof this system, 10 ml of lactobacilli supernatants containing wt and C1C5 RANTES were applied to small aliquots of Q-Sepharose resin (in batch) and the unbound fractions were incubated in batch with small aliquots of SP-Sepharose. Bound proteins were eluted in two steps and RANTES variants were found mostly in the 1 M NaCl fraction. This procedure derives from our purification protocol (described below) and allows both the elimination of broth contaminants and the concentration of secreted RANTES. RANTES variants anti-HIV-1 activity was tested in the p24-based acute infection assay, using E. coli-produced wt RANTES as control (Fig. 2B). Clearly, this rapid protocol allows an initial evaluation of the quantity and quality of lactobacilli-produced RANTES mutants. As an alternative approach to screen anti-HIV-1 activity, supernatant concentration could be attempted, as described by other authors [16]. However, we abandoned this approach because we observed a strong HIV-1 inhibition with the RANTES-free lactobacilli supernatants (not shown), likely deriving from components of the lactobacilli culture broth.
This result implies a superior folding and secretory capability by a prokaryote (L. jensenii), compared to the highly coordinated quality control machinery present in the ER of mammalian cells, such as that of CHO cells. These lines of evidence, together with reports on other disulphide bond-containing (as well as multi-domain) proteins expressed by lactobacilli, strongly suggest that lactobacilli possess unexpectedly high proficiency in producing and secreting complex proteins from higher organisms. The biosynthesis stage where recombinant protein disulphide bonding occurs in lactic acid bacteria is still matter of a debate, firstly emerged on class IIa bacteriocins, disulphide bond-containing proteins secreted by Gram-positive bacteria [26]. Yet, Gram-positive disulphide oxidoreductases exist and are being characterized for their mechanism of action [27]. Hence, engineered lactobacilli proved to be an efficient and convenient expression system, as well as a promising source of anti-HIV-1 proteins as live microbicides. Taken together, these data indicate a superior capacity by lactobacilli to produce and secrete RANTES derivatives in comparison to mammalian cells.
Intracellular content of RANTES variants in CHO cells and L. jensenii
Considering the low amount of RANTES variants produced by CHO cells in comparison to L. jensenii and the differential folding exhibited for C1C5 RANTES, the intracellular content in the two systems was evaluated. The highly-specialized secretion system in CHO cells could induce the RANTES variants to undergo extensive ER retention and intracellular degradation. Analysis of the intracellular RANTES content in L. jensenii could further proof the efficiency of the system, considering also the fact that inclusion bodies formation in lactic acid bacteria has rarely been reported. In one such case, distribution of a difficult-to-express protein in soluble and inclusion bodies fractions was reported as an example of expression improvement as compared to its total sequestration within inclusion bodies in E. coli [28].
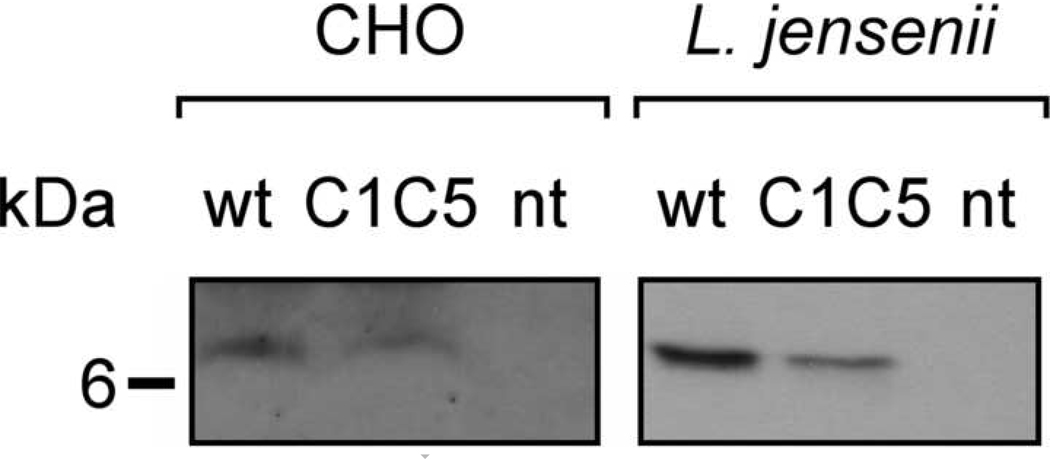
Wt and C1C5 RANTES-expressing CHO cell lysates were analyzed in Western blot under non-reducing conditions. A strong cross-reactivity background covered the 10–100 kDa molecular mass range, compromising the possible detection of intracellular disulphide-linked aggregates (not shown). Faint detection of the monomeric form was possible only when 1 × 106 cells/well were loaded on gel and autoradiography films were subjected to long exposure time (Fig. 3). Although the detection of a faint RANTES monomer band in the intracellular compartment might suggest an efficient secretion for wt RANTES by CHO cells, this could not be ascertained completely, given the lack of information on the presence or absence of higher order misfolded (disulphide-linked) oligomers.
Fig. 3.
Intracellular content of RANTES variants in CHO cells and L. jensenii. Cell lysates were analyzed in Western blot under non-reducing condition and RANTES variants revealed using anti-human RANTES Abs. nt, wt and C1C5 as in Fig. 1.
Since the intensity of the monomer band is only slightly lower compared to that of wt RANTES, a different assumption should be made for C1C5 RANTES. Thus, while the intracellular content of monomeric RANTES appears to be similar for both variants, this cannot account for the 50-fold reduction in the quantity of protein secreted. Therefore, it is likely that intracellular monomeric C1C5 RANTES is mostly retained in the ER and that only a small proportion of the biosynthesized protein is secreted and that this occurs only after a disulphide-linked dimer is formed.
Conversely, RANTES intracellular content in engineered L. jensenii was clearly identifiable as a single monomeric form of the expected molecular mass for both variants. Samples loaded on gel were cell pellets deriving from 0.5 ml of resuspended L. jensenii culture. Considering the fact that the intensity of the intracellular RANTES bands roughly corresponded to that of secreted RANTES obtained from 100 µl of cell supernatant, the intracellular protein detected is likely to belong to the pool of molecules that have just been synthesized and are being exported from the lactobacillus to the culture medium. In other words, an impaired or inefficient secretion for these RANTES variants does not appear to take place in L. jensenii. Interestingly, the RANTES intracellular pools do not appear to present the cleavage products observed in the secreted protein, suggesting that proteolysis may occur after secretion.
Oligomeric state analysis of recombinant RANTES variants by chemical cross-linking
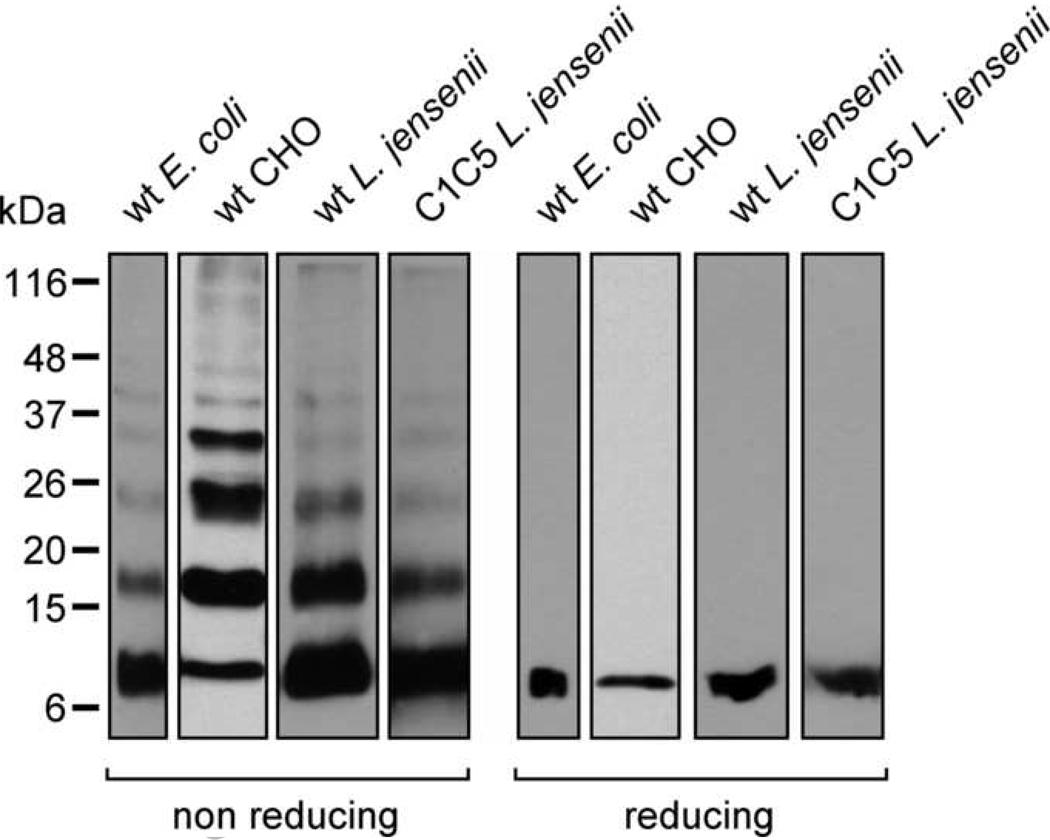
Another important issue when characterizing the expression and folding of CC chemokines is represented by their naturally-occurring non-covalent oligomerization [19]. CC chemokine dimerization is a major feature that has been extensively characterized and found to be determined by a dimer interface in which specific residues contribute to the monomer-monomer binding affinity [29,30]. Conversely, higher-order oligomer formation is governed by electrostatic interactions among monomers [31]. Therefore, analysis of the oligomeric state of recombinant RANTES variants may represent a further proof of appropriate folding. To this aim, proteins secreted by CHO cells (wt RANTES) and L. jensenii (wt and C1C5 RANTES) were chemically cross-linked with DTSSP, a cleavable homo-bifunctional cross-linker containing an internal disulphide bridge. E. coli-produced wt RANTES was subjected to chemical cross-linking and its oligomerization pattern was used as a reference. When analyzed in non-reducing SDS-PAGE, the pattern of RANTES oligomerization was similar in the four different samples, suggesting that all variants possessed a correct folding; under reducing conditions, the pattern resolved to a single form corresponding to the protein monomer (Fig. 4). The oligomerization pattern can clearly be distinguished into monomer, dimer, trimer, tetramer and less represented higher-order oligomers. C1C5 RANTES-containing supernatant from CHO cells was not included in this analysis for two reasons: i) its too scarce level of secretion hampered the analysis/detection of the protein; and ii) the protein is expressed predominantly as a disulphide-linked dimer, which is by itself an evidence for non-optimal folding. Yet, it would be interesting to see whether the disulphide-linked C1C5 RANTES dimer possesses the canonic RANTES fold except for an N-terminal domain swapping involving C1 and C5 residues that, upon intertwining, cross-bridge with the C1’ and C5’ residues from the second monomer. Similar or even deeper domain swapping events involving disulphide bond-mediated cross-bridging among monomers have been reported for other proteins, either as an alternative to monomer folding or as a natural solution to protein function [32,33]. Chemical cross-linking has recently been used also by another group as an approach to assess chemokine oligomerization [34]. This appears to be a simple and fast method that could be very useful in structural studies as well as in the generation of chemokine mutants with desired oligomeric content variation.
Fig. 4.
Chemical cross-linking of RANTES variants produced by E. coli, CHO cells and L. jensenii. RANTES variants were chemically-cross-linked using DTSSP, resolved by SDS-PAGE under non-reducing or reducing conditions and analyzed by Western blot using anti-human RANTES Abs. wt E. coli, inclusion bodies-refolded wt RANTES from E. coli (standard); wt CHO, CHO cell-secreted wt RANTES; wt L. jensenii, L. jensenii-secreted wt RANTES; and C1C5 L. jensenii, L. jensenii-secreted C1C5 RANTES.
Purification of L. jensenii-secreted RANTES variants
Wt and C1C5 RANTES were purified to homogeneity from L. jensenii culture supernatants following the same procedure for both variants, taking advantage of their similar calculated isoelectric point (9.3 and 8.9, respectively). A four-step ionic-exchange protocol was established that yielded purified proteins and allowed separation of the full-length protein from the cleaved form. In the first chromatography step (Q-Sepharose, pH 6.0), part of the supernatant proteins were retained in the column, allowing recovery of RANTES variants in the flow through. A sodium chloride gradient was applied to elute RANTES variants in the second step (SP-Sepharose, pH 6.0). The 1 M NaCl fraction contained most semi-purified RANTES, a feature exploited also in the fast screening system described above. This fraction was then dialyzed against a buffer at pH 8.0 and applied sequentially onto Q-Sepharose and SP-Sepharose columns, following the same procedure described for the first two steps. Purified full-length wt and C1C5 RANTES were detected in the 1 M NaCl fraction eluted from the last step (Fig. 5). Purified RANTES variants were successfully tested for anti-HIV-1 activity and the intramolecular disulphide bonds were properly formed, as confirmed by mass spectrometry (manuscript submitted). In addition to the purification protocol presented here, L. jensenii-secreted RANTES variants bind to heparin and the use of heparin columns, together with a gel filtration step on the semi-purified material, represent alternative or complementary purification steps. RANTES binding to heparin is well-known and is among the biological features of this chemokine, i.e., recognition of cell surface glycosaminoglycans [19].
Fig. 5.
Purification of L. jensenii-secreted wt and C1C5 RANTES. L. jensenii-secreted wt and C1C5 RANTES were purified to homogeneity using a four-step ionic exchange protocol. After SDS-PAGE in non-reducing conditions, proteins were stained by Coomassie blue or detected by Western blot using anti-human RANTES Abs. st, wt and C1C5 as in previous figures.
It is obvious that fusion of RANTES to a short sequence tag would speed up the system for protein purification. However, the present work stems from an initial approach in which special attention was given to ensuring native protein production. Now that the system has proved to be efficient and reliable, a tag fusion to screen new RANTES mutants could be implemented at the chemokine C-terminus. Conversely, the RANTES N-terminal portion is crucial for CCR5 recognition and the C1C5 mutant has revealed it as a hotspot for protein folding. Thus, it is advisable to maintain it in its native conformation.
Conclusions
RANTES is the most potent natural inhibitor of HIV-1 [18] and the most promising protein-based CCR5-binding HIV-1 inhibitor [19]. Yet, the absence of CCR5 activation is a fundamental requisite to exclude pro-inflammatory activity deriving from chronic activation of CCR5-expressing cells. In this context, C1C5 RANTES, a mutated RANTES antagonist for CCR5 and powerful HIV-1 inhibitor [23], represents an excellent scaffold for further search and discovery of new mutants with even higher anti-HIV-1 potency. The major issue with C1C5 RANTES production concerns the need for the intramolecular oxidation of the cysteine residues at positions 1 and 5. After an initial effort to develop a mammalian cell-based recombinant RANTES screening system (CHO cells), hampered by the extremely low quantity of protein secreted and an unwanted covalent dimerization event, we found that a prokaryotic system (L. jensenii) was able to secrete C1C5 RANTES as a folded, active monomer. RANTES-engineered L. jensenii was employed within the framework of a completely different approach, i.e., the development of vaginal anti-HIV-1 live microbicides, currently a very promising strategy to prevent HIV-1 infection. This approach involves the administration of commensal lactobacilli (i.e., bacteria naturally colonizing vaginal mucosa) engineered to deliver anti-HIV-1 molecules with the purpose to reduce heterosexual HIV-1 transmission [3,10]. The correct disulphide bonding of wt and C1C5 RANTES, the presence of the characteristic CC chemokine non-covalent dimerization and oligomerization, and the functional anti-HIV-1 activity altogether attest to the correct folding of the L. jensenii-secreted proteins. These lines of evidence satisfy the necessary requisites for the live microbicide strategy (for a news and views see a web-based comment at www.aidsmap.com Gus Cairns 4/3/2008 Microbicides 2008: Third-generation microbicides might act as ‘bacterial vaccine’) and represent a quality proof for lactobacilli as an ideal system to implement a recombinant RANTES expression/screening platform (see also highlights of a congress proceedings in reference 35). Indeed, chromosomal integration of C1C5 RANTES in L. jensenii has been established as an obvious development towards a safe live microbicide for women (unpublished results). Besides their impact in the fight against HIV-1, engineered lactic acid bacteria are emerging as a powerful tool and a possible resource in the protein production field. It is our intent to use these bacteria as a platform to test conceptually new RANTES modifications with the aim of creating even more powerful anti-HIV-1 CCR5 antagonists. Given its importance as anti-HIV-1 protein and the results presented here, C1C5 RANTES and analogue mutants will be used as a scaffold for further mutagenesis, taking advantage of lactobacilli as optimal expression system for protein production and testing.
Acknowledgements
We wish to thank Thomas P. Parks and Xiaowen Liu for invaluable help with plasmid construction and protein purification. This work was supported by NIH grant U19-AI060615.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used: spp., species; HIV-1, human immunodeficiency virus type 1; CHO, chinese hamster ovary; RANTES, regulated on activation normal T cell expressed and secreted; wt, wild type; Abs, antibodies; ER, endoplasmic reticulum; CMV, Cytomegalovirus; CbsA, S-layer protein of Lactobacillus crispatus.
References
- 1.Seegers JF. Lactobacilli as live vaccine delivery vectors: progress and prospects. Trends Biotechnol. 2002;20:508–515. doi: 10.1016/s0167-7799(02)02075-9. [DOI] [PubMed] [Google Scholar]
- 2.Wells JM, Mercenier A. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat. Rev. Microbiol. 2008;6:349–362. doi: 10.1038/nrmicro1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klasse PJ, Shattock R, Moore JP. Antiretroviral drug-based microbicides to prevent HIV-1 sexual transmission. Annu. Rev. Med. 2008;59:455–471. doi: 10.1146/annurev.med.59.061206.112737. [DOI] [PubMed] [Google Scholar]
- 4.Hladik F, Sakchalathorn P, Ballweber L, Lentz G, Fialkow M, Eschenbach D, McElrath MJ. Initial events in establishing vaginal entry and infection by human immunodeficiency virus type-1. Immunity. 2007;26:257–270. doi: 10.1016/j.immuni.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutler B, Justman J. Vaginal microbicides and the prevention of HIV transmission. Lancet Infect. Dis. 2008;8:685–697. doi: 10.1016/S1473-3099(08)70254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fauci AS. 25 years of HIV/AIDS science: reaching the poor with research advances. Cell. 2007;131:429–432. doi: 10.1016/j.cell.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 7.Moscicki AB. Vaginal microbicides: where are we and where are we going? J. Infect. Chemother. 2008;14:337–341. doi: 10.1007/s10156-008-0630-3. [DOI] [PubMed] [Google Scholar]
- 8.Balzarini J, Van Damme L. Microbicide drug candidates to prevent HIV infection. Lancet. 2007;369:787–797. doi: 10.1016/S0140-6736(07)60202-5. [DOI] [PubMed] [Google Scholar]
- 9.Grant RM, Hamer D, Hope T, Johnston R, Lange J, Lederman MM, Lieberman J, Miller CJ, Moore JP, Mosier DE, Richman DD, Schooley RT, Springer MS, Veazey RS, Wainberg MA. Whither or wither microbicides? Science. 2008;321:532–534. doi: 10.1126/science.1160355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolton M, van der Straten A, Cohen CR. Probiotics: potential to prevent HIV and sexually transmitted infections in women. Sex. Transm. Dis. 2008;35:214–225. doi: 10.1097/OLQ.0b013e31815b017a. [DOI] [PubMed] [Google Scholar]
- 11.Chang TL, Chang CH, Simpson DA, Xu Q, Martin PK, Lagenaur LA, Schoolnik GK, Ho DD, Hillier SL, Holodniy M, Lewicki JA, Lee PP. Inhibition of HIV infectivity by a natural human isolate of Lactobacillus jensenii engineered to express functional two-domain CD4. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11672–11677. doi: 10.1073/pnas.1934747100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pusch O, Boden D, Hannify S, Lee F, Tucker LD, Boyd MR, Wells JM, Ramratnam B. Bioengineering lactic acid bacteria to secrete the HIV-1 virucide cyanovirin. J. Acquir. Immune Defic. Syndr. 2005;40:512–520. doi: 10.1097/01.qai.0000187446.76579.d3. [DOI] [PubMed] [Google Scholar]
- 13.Chancey CJ, Khanna KV, Seegers JF, Zhang GW, Hildreth J, Langan A, Markham RB. Lactobacilli-expressed single-chain variable fragment (scFv) specific for intercellular adhesion molecule 1 (ICAM-1) blocks cell-associated HIV-1 transmission across a cervical epithelial monolayer. J. Immunol. 2006;176:5627–5636. doi: 10.4049/jimmunol.176.9.5627. [DOI] [PubMed] [Google Scholar]
- 14.Pusch O, Kalyanaraman R, Tucker LD, Wells JM, Ramratnam B, Boden D. An anti-HIV microbicide engineered in commensal bacteria: secretion of HIV-1 fusion inhibitors by lactobacilli. AIDS. 2006;20:1917–1922. doi: 10.1097/01.aids.0000247112.36091.f8. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Lagenaur LA, Simpson DA, Essenmacher KP, Frazier-Parker CL, Liu Y, Tsai D, Rao SS, Hamer DH, Parks TP, Lee PP, Xu Q. Engineered vaginal lactobacillus strain for mucosal delivery of the human immunodeficiency virus inhibitor cyanovirin-N. Antimicrob. Agents Chemother. 2006;50:3250–3259. doi: 10.1128/AAC.00493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu JJ, Reid G, Jiang Y, Turner MS, Tsai CC. Activity of HIV entry and fusion inhibitors expressed by the human vaginal colonizing probiotic Lactobacillus reuteri RC-14. Cell Microbiol. 2007;9:120–130. doi: 10.1111/j.1462-5822.2006.00772.x. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Lagenaur LA, Lee PP, Xu Q. Engineering of a human vaginal Lactobacillus strain for surface expression of two-domain CD4 molecules. Appl. Environ. Microbiol. 2008;74:4626–4635. doi: 10.1128/AEM.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 19.Vangelista L, Secchi M, Lusso P. Rational design of novel HIV-1 entry inhibitors by RANTES engineering. Vaccine. 2008;26:3008–3015. doi: 10.1016/j.vaccine.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lederman MM, Veazey RS, Offord R, Mosier DE, Dufour J, Mefford M, Piatak M, Jr, Lifson JD, Salkowitz JR, Rodriguez B, Blauvelt A, Hartley O. Prevention of vaginal SHIV transmission in rhesus macaques through inhibition of CCR5. Science. 2004;306:485–487. doi: 10.1126/science.1099288. [DOI] [PubMed] [Google Scholar]
- 21.Magistrelli G, Gueneau F, Muslmani M, Ravn U, Kosco-Vilbois M, Fischer N. Chemokines derived from soluble fusion proteins expressed in Escherichia coli are biologically active. Biochem. Biophys. Res. Commun. 2005;334:370–375. doi: 10.1016/j.bbrc.2005.06.091. [DOI] [PubMed] [Google Scholar]
- 22.Cho HJ, Lee Y, Chang RS, Hahm MS, Kim MK, Kim YB, Oh YK. Maltose binding protein facilitates high-level expression and functional purification of the chemokines RANTES and SDF-1alpha from Escherichia coli. Protein Expr. Purif. 2008;60:37–45. doi: 10.1016/j.pep.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Polo S, Nardese V, De Santis C, Arcelloni C, Paroni R, Sironi F, Verani A, Rizzi M, Bolognesi M, Lusso P. Enhancement of the HIV-1 inhibitory activity of RANTES by modification of the N-terminal region: dissociation from CCR5 activation. Eur. J. Immunol. 2000;30:3190–3198. doi: 10.1002/1521-4141(200011)30:11<3190::AID-IMMU3190>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 24.Comuzzi B, Arcelloni C, Polo S, Nardese V, Lusso P, Paroni R. Multi-step purification strategy for RANTES wild-type and mutated analogues expressed in a baculovirus system. J. Chromatogr. B. Biomed. Sci. Appl. 2000;737:47–54. doi: 10.1016/s0378-4347(99)00361-8. [DOI] [PubMed] [Google Scholar]
- 25.Lusso P, Cocchi F, Balotta C, Markham PD, Louie A, Farci P, Pal R, Gallo RC, Reitz MS., Jr Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drider D, Fimland G, Héchard Y, McMullen LM, Prévost H. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 2006;70:564–582. doi: 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heras B, Kurz M, Jarrott R, Shouldice SR, Frei P, Robin G, Cemazar M, Thöny-Meyer L, Glockshuber R, Martin JL. Staphylococcus aureus DsbA does not have a destabilizing disulfide. A new paradigm for bacterial oxidative folding. J. Biol. Chem. 2008;283:4261–4271. doi: 10.1074/jbc.M707838200. [DOI] [PubMed] [Google Scholar]
- 28.Straume D, Axelsson L, Nes IF, Diep DB. Improved expression and purification of the correctly folded response regulator PlnC from lactobacilli. J. Microbiol. Methods. 2006;67:193–201. doi: 10.1016/j.mimet.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 29.Skelton NJ, Aspiras F, Ogez J, Schall TJ. Proton NMR assignments and solution conformation of RANTES, a chemokine of the C-C type. Biochemistry. 1995;34:5329–5342. doi: 10.1021/bi00016a004. [DOI] [PubMed] [Google Scholar]
- 30.Chung CW, Cooke RM, Proudfoot AE, Wells TN. The threedimensional solution structure of RANTES. Biochemistry. 1995;34:9307–9314. doi: 10.1021/bi00029a005. [DOI] [PubMed] [Google Scholar]
- 31.Czaplewski LG, McKeating J, Craven CJ, Higgins LD, Appay V, Brown A, Dudgeon T, Howard LA, Meyers T, Owen J, Palan SR, Tan P, Wilson G, Woods NR, Heyworth CM, Lord BI, Brotherton D, Christison R, Craig S, Cribbes S, Edwards RM, Evans SJ, Gilbert R, Morgan P, Randle E, Schofield N, Varley PG, Fisher J, Waltho JP, Hunter MG. Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants. J. Biol. Chem. 1999;274:16077–16084. doi: 10.1074/jbc.274.23.16077. [DOI] [PubMed] [Google Scholar]
- 32.Vangelista L, Cesco-Gaspere M, Lamba D, Burrone O. Efficient folding of the Fc epsilon RI alpha-chain membrane-proximal domain D2 depends on the presence of the N-terminal domain D1. J. Mol. Biol. 2002;322:815–825. doi: 10.1016/s0022-2836(02)00853-7. [DOI] [PubMed] [Google Scholar]
- 33.Maekawa A, Schmidt B, Fazekas de St Groth B, Sanejouand YH, Hogg PJ. Evidence for a domain-swapped CD4 dimer as the coreceptor for binding to class II MHC. J. Immunol. 2006;176:6873–6878. doi: 10.4049/jimmunol.176.11.6873. [DOI] [PubMed] [Google Scholar]
- 34.Brandner B, Rek A, Diedrichs-Möhring M, Wildner G, Kungl AJ. Engineering the glycosaminoglycan-binding affinity, kinetics and oligomerization behavior of RANTES: a tool for generating chemokine-based glycosaminoglycan antagonists. Protein Eng. Des. Sel. 2009;22:367–373. doi: 10.1093/protein/gzp013. [DOI] [PubMed] [Google Scholar]
- 35.Sevastsyanovich S, Alfasi S, Cole J. Recombinant Protein Production: a comparative view on host physiology. N. Biotechnol. 2009;25:175–180. doi: 10.1016/j.nbt.2009.01.006. [DOI] [PubMed] [Google Scholar]