Abstract
The evolutionarily conserved Eps15 homology domain (EHD)/receptor-mediated endocytosis (RME)-1 family of C-terminal EH domain proteins has recently come under intense scrutiny because of its importance in intracellular membrane transport, especially with regard to the recycling of receptors from endosomes to the plasma membrane. Recent studies have shed new light on the mode by which these adenosine triphosphatases function on endosomal membranes in mammals and Caenorhabditis elegans. This review highlights our current understanding of the physiological roles of these proteins in vivo, discussing conserved features as well as emerging functional differences between individual mammalian paralogs. In addition, these findings are discussed in light of the identification of novel EHD/RME-1 protein and lipid interactions and new structural data for proteins in this family, indicating intriguing similarities to the Dynamin superfamily of large guanosine triphosphatases.
Keywords: C. elegans, EH domain, EHD1, EHD2, EHD3, EHD4, endocytic trafficking, Rab effectors, recycling, RME-1
The C-terminal EHD/RME-1 proteins
The Eps15 homology domain (EHD)/receptor-mediated endocytosis (RME)-1 family of EH domain proteins is a novel group of endosomal scaffolding molecules required for receptor recycling. While much is known about the regulation of the early steps of receptor internalization, considerably less is known about how recycling from endosomes to the plasma membrane, and the sorting of recycling cargo within endosomes, is mediated. Analysis of EHD/RME-1 family function, including its interactions with important protein and lipid partners, is providing new insights into the mechanisms that promote recycling within the endosomal system.
Mammalian cells express four highly homologous C-terminal EHD paralogs, EHD1, EHD2, EHD3 and EHD4, each of which contains a single EH domain at the C-terminus. The genomes of many invertebrate organisms, including Caenorhabditis elegans and Drosophila melanogaster, contain a single EHD family gene, most closely resembling EHD1/EHD3, although many isoforms of the C. elegans EHD family protein RME-1 are known to be expressed because of alternative splicing (1). The mammalian paralogs themselves exhibit 70–86% overall identity, with EHD1 and EHD3 being the most closely related (2). Remarkably, even the C. elegans family member RME-1 is 67% identical to mammalian EHD1 over its entire length (1). The EH domains of human EHD1–EHD4, C. elegans RME-1 and Drosophila dEHD/PAST1 display a greater level of homology with one another than with EH domains of other proteins (Table 1). For example, the EH domains of EHD1, EHD2, EHD3 and EHD4 show between 53 and 81% amino acid identity compared with only 37% identity between each of the EH domains of the C-terminal EHDs and the most closely related N-terminal EH domain, the second of the three Eps15 EH domains (Eps15 EH-2).
Table 1.
Comparison of amino acid sequence identity between C-terminal EH domains and the second Eps15 EH domain
| Comparison of EH domains | Percent identity |
|---|---|
| EHD1 EH/EHD2 EH | 59 |
| EHD1 EH/EHD3 EH | 81 |
| EHD1 EH/EHD4 EH | 68 |
| EHD2 EH/EHD3 EH | 57 |
| EHD2 EH/EHD4 EH | 53 |
| EHD3 EH/EHD4 EH | 67 |
| Eps15 EH-2/EHD1 EH | 37 |
| Eps15 EH-2/EHD2 EH | 37 |
| Eps15 EH-2/EHD3 EH | 37 |
| Eps15 EH-2/EHD4 EH | 37 |
Recently, new light has been shed on the domain architecture and structure of C-terminal EHDs, including the first crystal structure of a full-length family member, mouse EHD2 (3) (Figure 1A,B). This work showed that EHD2 contains highly helical regions that produce a lipid-binding surface. Like all EHD/RME-1 proteins, EHD2 contains a nucleotide-binding domain. This domain in EHD2 is remarkably similar in three dimensional structure, but not in primary sequence, to the G-domain of Dynamin, another large nucleotide-binding protein that mediates endocytosis (4). Thus, the EHD2 nucleotide-binding domain retains the nomenclature ‘G-domain’, although it binds to ATP rather than GTP (see below). These regions of the protein connect to a disordered ‘linker region’, prior to the EH domain (Figure 1B). Further functional parallels of EHD2 to Dynamin were also identified, and the authors suggested that EHD proteins could be considered members of the Dynamin-like superfamily (4). Detailed implications of the recent structural developments are discussed below.
Figure 1. Domain architecture and structure of C-terminal EHD proteins.
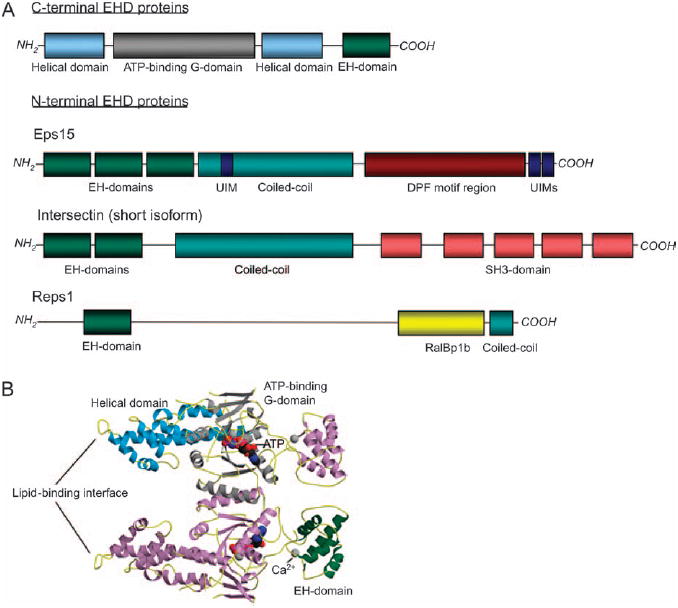
A) Comparison of the domain architecture of C-terminal/RME-1 proteins and N-terminal EHD proteins. UIM, ubiqutin interaction motif. B) The crystal structure of mouse EHD2 dimer. EHD2 is shown by a ribbon-type representation. The top monomer is colored to match the domain architecture depicted in (A).
The Structure and Function of EH Domains
The EH domain in general is a highly conserved 90–100 amino acid structure that was originally identified as a repeated domain in the endocytic adaptor protein known as the EGF receptor tyrosine kinase substrate (Eps15) (5,6). Over 500 proteins are now known to contain at least a single EH domain (SMART Architecture Analysis, http://smart.embl-heidelberg.de). Apparently a eukaryotic innovation, EH domains are present in diverse phyla ranging from the relatively simple budding yeast and paramecium to higher plants and metazoans such as nematodes, insects and mammals (reviewed in 7,8). Like other scaffolding proteins, EH domain proteins are generally modular, containing additional other well-characterized signaling domains such as SH3 regions, PH domains, C2 domains, proline-rich regions as well as aspartate-proline-phenylalanine (DPF) and ubiquitin interaction motifs (reviewed in 8,9). Despite the variety of functions implicated by these additional domains and motifs, regulation of endocytic trafficking appears to be a primary role for these proteins in the cell.
The first structure of an EH domain (Eps15 EH2) was solved by nuclear magnetic resonance (NMR) spectroscopy (10). Eps15 EH2 was found to contain a pair of EF-Hand type helix-loop-helix motifs that are linked by a short antiparallel β-sheet (10). Since then, several other EH domain structures have been solved, including those of C-terminal EHD proteins. The recent solution structure of the human EHD1 EH domain and the crystal structure of the complete mouse EHD2 protein have demonstrated the conservation of the basic EH domain fold and have provided new information on its function (3,11).
A variety of experimental methods, including phage display screens (12) and the screening of a human fibroblast expression library, have demonstrated that EH domains bind to peptides containing an asparagine-proline-phenylalanine (NPF) sequence (13). An explanation of the structural basis for EH-NPF interactions was first provided by the solution structure of Eps15 EH2 bound to NPFL or NPLR (10,14). These studies led to the conclusions that interacting NPF residues, in the conformation of a type I β-turn, fit into a hydrophobic pocket on the surface of the EH domain, between the αB; and the αC helices (14). Moreover, these studies precisely defined the conserved hydrophobic patch that accommodates the proline of the NPF, permitting the phenylalanine to insert deeply within the conserved pocket [(14) and reviewed in 15]. A recent study has also described an additional hydrophobic pocket within Eps15 EH2 (16) and raises the possibility that other EH domains may similarly be capable of additional modes of NPF binding.
Structural Insights into the EH Domain of EHD/RME-1 Proteins: NPF and Lipid Binding
An interesting question relates to how the specificity of individual EH domains, for particular NPF motifs in target proteins, is attained. For instance, a search of the predicted C. elegans proteome reveals over 800 proteins containing at least one NPF sequence (S. Pant and B. G., unpublished data). The answer, at least in part, appears to derive from differential preferences of specific EH domains for flanking residues surrounding the target NPF. Previous work showed that each EH domain has its own inherent preference for NPF-flanking residues (12). Indeed, the elegant work of Paoluzi et al. showed that the first and third EH domains of Eps15R prefer peptides with an arginine immediately following the NPF motif (12).
In contrast, our analysis, and that of Braun et al. (17), indicates that the EH domains of EHD proteins prefer acidic residues following the NPF (Figure 2). Although these data are not derived from a comprehensive phage display, the comparisons shown in this study in Figure 2 suggest that proteins ranked as strong binders to EHD proteins [i.e. Rabenosyn-5 NPFs 1 and 2 (18), Rab11-FIP2 (19), SNAP29 (20) and Syndapins (17,20)] have a strong acidic consensus in positions +1, +2 and +3. While not all NPFs implicated in EHD/RME-1 binding are surrounded by acidic residues in their primary sequence (Figure 2), those that lack this feature have only been found in proteins that contain multiple EHD/RME-1-binding sites (17-19,21). In fact, some interactors appear to require multiple intact NPFs for binding, suggesting a preference for interaction with assembled EHD oligomers (17-19).
Figure 2. Selectivity of C-terminal EH domains for NPF motifs flanked by acidic residues.

Alignment of NPF motifs and surrounding residues that exhibit weak or strong binding with EHD proteins. NPF motifs are indicated by red shading, whereas flanking acidic residues are denoted by light blue shading.
Recent structural advances have allowed new insights into the likely mechanisms by which the EHD EH domain produces binding specificity (3,11,22). The recent NMR solution structure of the EH domain of EHD1 has provided some important clues as to the reason that C-terminal EH domains preferentially bind to NPF motifs followed by a cluster of acidic residues (Figure 3). The electrostatic surface potential of the EH domain of EHD1 is highly positive, possibly facilitating interactions with the aspartate and glutamate residues that follow the NPF motifs of binding proteins (11). Indeed, upon comparison of the six residues of the EHD1 EH domain modeled into the NPF-binding pocket, these amino acids display a net electrostatic charge of +4, whereas the homologous residues in the Eps15 EH2 exhibit a net electrostatic charge of −3 (Figure 3). This analysis supports the hypothesis that positive electrostatic surface potential is an important mechanism to fine-tune the C-terminal EH domains and ensure that they interact with a select group of NPF-containing interaction partners, particularly those containing acidic clusters following their NPF motif.
Figure 3. C-terminal EH domains contain positively charged surface residues within the NPF-binding pocket.
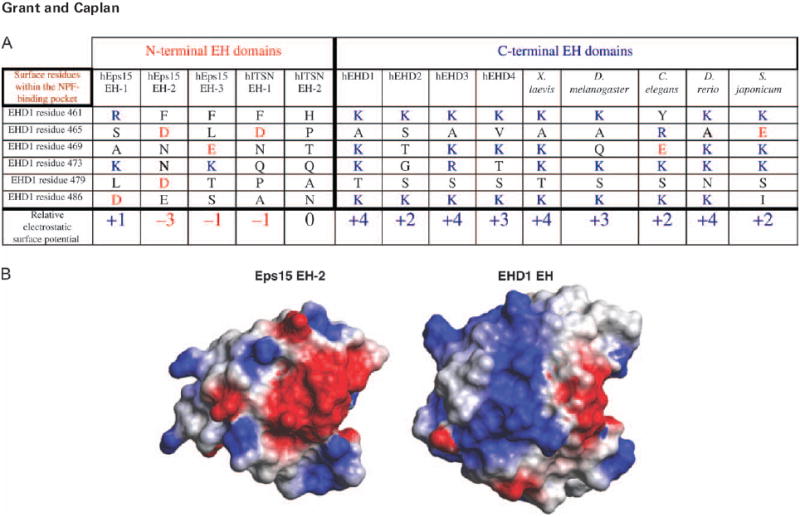
A) Comparison of the relative electrostatic surface potential of C-terminal and N-terminal EH domains. B) Comparison of the relative electrostatic surface potential of the human EHD1 EH domain and Eps15 EH-2. Blue represents residues with positively charged side chains, whereas red represents negatively charged residues.
One of the interesting features of EHD/RME-1 proteins is their distinctive localization to long tubular endosomal membranes (up to 10 μm), in addition to punctate endosomes (23). A recent study has also shed new light on this localization, suggesting that EH domain interaction with phosphoinositide lipids is important for interaction with, or formation of, such extended tubular endosomal domains (22). Chemical shift experiments, using two-dimensional NMR, detected perturbation of select residues of the EHD1 EH domain upon titration of phosphoinositides. A conserved, positively charged amino acid, lysine 483, was hypothesized to be involved in the EH domain/lipid binding (22). Indeed, mutation of lysine 483 to glutamic acid did not alter the overall structure of the EH domain but caused an altered subcellular distribution of the mutant EHD1, from tubulovesicular membranes to entirely punctate structures. This phenotype resembles truncation of the entire EH domain, indicating that this residue, and therefore EH phosphoinositide interactions, is critical for EHD-mediated tubulation (22).
Oligomerization, tubulation and nucleotide hydrolysis
It was recently demonstrated that all four full-length EHDs can bind to various phosphoinositides (24). However, because both the EH domain truncation and the lysine-483-to-glutamic acid mutant are nonetheless capable of interacting with punctate endosomal membranes, these data implicated the involvement of an additional region of the protein as the primary determinant of membrane association. The recent crystal structure of mouse EHD2 identified this binding site as a polybasic cluster of residues in close proximity to the tip of helix α9 (3). The dimerization of EHD2 allows the formation of a highly curved, scissor-like, membrane interface composed of the lipid-binding sites of the two helical domains (3). Further oligomerization of dimers to form rings around lipid tubules and the ability to tubulate negatively charged liposomes in isolation suggest that EHD2 and by extension the entire EHD family are functionally similar to Dynamin (3). Importantly, these results suggest that like Dynamin, EHD proteins may function to tubulate membranes and/or detach budding vesicles or tubules from endosomal membranes (promote fission). Further analysis will be required to determine how far the Dynamin analogy holds.
In addition to identifying the dimeric lipid-binding interface, the crystal structure of EHD2 also produced several somewhat surprising findings. For example, the authors identified a conserved GPF motif in the linker that separates the EH domain from the central helical region and were able to predict that in solution, the EH domains of the dimer would be adjoined, with each EH domain occupied by the conserved linker GPF motif (3). The authors further propose that in the course of oligomerization, the EH domains shift from their interaction with the linker GPF motif to interact with a disordered loop sequence XPFRKLNPF localized within the G-domain (3). Interestingly, while the XPFRKL motif is conserved among mammalian EHDs, the NPF motif itself is not found in EHD1, EHD3 or EHD4 (bold sequences indicate residues proposed to make contact with the EH domain binding pocket). Furthermore, the EHD homologs from some other species, such as isoform D of C. elegans RME-1, lack amino terminal XPF motifs, yet can fully rescue a null mutant lacking all RME-1 isoforms (21). These differences leave open the question of whether EHD1, EHD3, EHD4 and RME-1 are capable of the same proposed EH domain movement from the linker region to the side site of the G-domain upon higher order oligomerization. One possibility is that NPF-containing binding partners provide this function ‘in trans’ for these EHD proteins, promoting conformational changes in the EHD that in turn promote its polymerization around membrane tubules.
Based on sequence alignments, it was predicted that EHD proteins contain a nucleotide-binding site (1,23) most similar to the site found in guanosine triphosphatases (GTPases) such as Ras and Dynamin (25). The first evidence demonstrating the functional significance of this nucleotide-binding site came from in vivo endocytic assays showing that oocytes possessing a glycine-to-arginine mutation within the conserved nucleotide-binding site of the C. elegans RME-1 protein exhibited impaired recycling of yolk receptors and in fact behaved as dominant negatives (1). A somewhat surprising finding, which has been strongly supported by the recent crystal structure of EHD2 in the presence of a non-hydrolysable ATP analog (3), is that the G-domain of the EHD1 protein displays a strong preference for ATP binding over GTP binding, with a Km of about 80 μm for ATP (26). A structural explanation for this specificity was provided by the evidence that arginine 536 of EHD2 forms a hydrogen bond with aspartate 222 of the conserved NKxD motif in the G-domain (3), thus disrupting the typical function of such aspartates in GTP recognition and binding in GTPases (27). Furthermore, the EHD2 structure showed that a methionine at position 223 in EHD2 would likely sterically interfere with GTP binding (3).
An important feature of EHD proteins is their ability to oligomerize. EHD2 dimerization has been localized to a highly hydrophobic interface spanning about 2100 Å2 within the G-domain (3). EHD2 adenosine triphosphatase activity is stimulated by about 10-fold in the presence of liposomes and correlates with the apparent assembly of EHD2 rings around the membranes. This behavior is highly reminiscent of the assembly-stimulated GTPase activity characteristic of Dynamin (28-31). It remains to be determined if EHD/RME-1 family proteins contain an intrinsic assembly stimulated GTPase-activating protein (GAP) domain, as is known to be the case for Dynamin family proteins (31,32). There is no apparent homology between EHD2 and the proposed GAP domain (GED) of Dynamin, suggesting that the stimulation of nucleotide hydrolysis is achieved differently. Furthermore, the EHD2 structure indicates that the beta and gamma phosphate of the ATP are capped, preventing direct insertion of a catalytic residue to promote ATP hydrolysis (3).
While it has been demonstrated that EHD proteins are found in large complexes (23), the stoichiometry of the EHD paralogs interacting with one another has been difficult to resolve. Various laboratories have demonstrated in vitro with purified proteins, by two-hybrid analysis and by overexpression in cells that EHDs are capable of hetero-oligomerizing (19,26,33,34), but thus far, the only in vivo interaction documented has been between EHD1 and EHD4 (35). Such oligomerization has functional significance as depletion of either EHD1 or EHD4 dramatically alters the subcellular localization of its binding partner (35). Moreover, oligomerization may regulate the binding of EHDs to NPF-containing binding partners (19). The main exception to this is EHD2, which appears not to hetero-oligomerize with other EHDs in vivo, even when co-overexpressed in the same cells (34,35). Future studies are likely to provide exciting new information on the mode by which oligomerization and lipid binding affect the functional roles of EHD proteins.
Functional Roles of EHD Proteins in the Regulation of Endocytic Recycling
The first evidence for the role of an EHD family protein in controlling receptor recycling from endosomes to the plasma membrane, and its effects on the morphology of the endocytic recycling compartment (ERC), was provided by C. elegans genetics (1) and parallel studies utilizing mammalian Chinese hamster ovary cells (36) (Table 2). In C. elegans, the RME-1 protein is required for yolk receptor recycling, which in turn is required for efficient yolk uptake by oocytes (1). Many additional cell types show endocytic defects in rme-1 mutants, and the RME-1 protein is expressed in all cells, localizing to recycling endosomes and the plasma membrane (1). Interestingly, RME-1 displays a preference in the C. elegans intestine for basolateral recycling endosomes, and rme-1 mutants accumulate baso-laterally endocytosed fluid in grossly enlarged endosomes. Likewise perturbation or loss of EHD1 in mammalian cells interferes with the recycling of the transferrin receptor, major histocompatibility complex class I, Cystic fibrosis transmembrane conductance receptor, long-term potentiation AMPA receptors and β1 integrins to name a few known EHD/RME-1-dependent cargo proteins [(23, 36-40) and Table 2]. Notably, EHD1 is important for the recycling of transmembrane cargo that is internalized by clathrin-dependent and clathrin-independent mechanisms (23). Recent evidence suggests that this dual requirement for recycling both types of cargo is also true in C. elegans (21,41).
Table 2.
Functional roles for EHD1/RME-1
| Function | Model | Reference |
|---|---|---|
| Yolk receptor recycling | C. elegans | (1) |
| Basolateral fluid recycling | C. elegans | (1) |
| Transferrin receptor recycling | mammalian cells | (36) |
| Major histocompatibility complex class I recycling | mammalian cells | (23) |
| Transferrin receptor recycling | Ehd1−/− MEF cells | (40) |
| Cystic fibrosis transmembrane conductance regulator recycling | Mammalian cells | (39) |
| Insulin-regulated GLUT4 glucose transporter recycling | Mammalian cells | (60) |
| Long-term potentiation AMPA type glutamate receptor recycling | Mammalian cells | (38) |
| β1 integrin recycling | Mammalian cells | (37) |
| Low-density lipoprotein receptor endocytosis/cholesterol and lipid droplet homeostasis | Mammalian cells | (64) |
| Endosome-to-Golgi retrieval | Mammalian cells | (65) |
While the role of RME-1/EHD1 appears to be well conserved between C. elegans and mammalian cells, the roles of the other three mammalian EHD paralogs, EHD2, EHD3 and EHD4, have not been as well characterized, and there are some partially conflicting reports with regards to their function. To some extent, such studies have been hampered by the high level of identity between these proteins and difficulties in generating specific antibodies directed against each paralog. However, recent studies have begun to unravel the functional differences between the EHD family proteins (Figure 4).
Figure 4. Schematic diagram showing the proposed roles for EHD proteins in the regulation of endocytic transport.
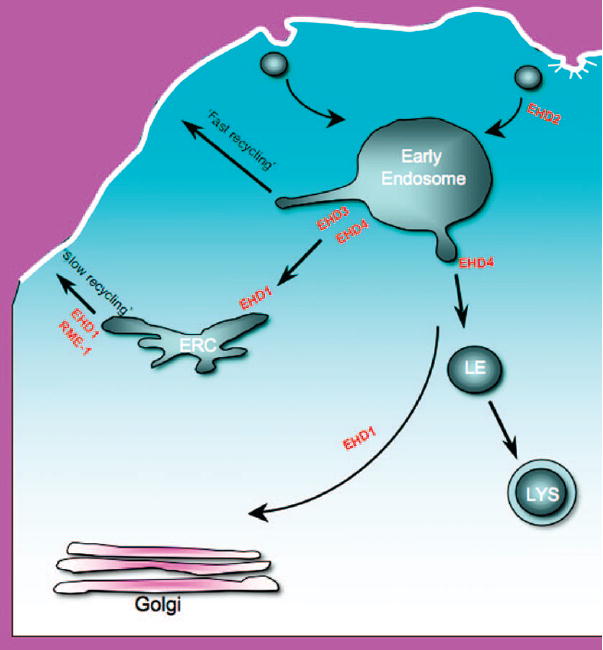
EHD2 has also been reported to function at the ERC (34), while EHD4/pincher controls the internalization of nerve growth factor receptor (45). LE, late endosome; LYS, lysosome.
EHD2 is the least homologous paralog, displaying about 70% amino acid identity with EHD1. While the crystal structure of EHD2 has led to some very interesting observations, the precise function of this protein remains vague. Studies by Guilherme et al. demonstrated that small interfering RNA (siRNA)-based depletion of EHD2 (or its calponin homology domain interaction partner, EHBP1) inhibits the internalization of transferrin receptor and GLUT4 in adipocytes (42). Consistent with these findings, Park et al. have identified interactions between EHD2 and μ1 and μ2 subunits of the clathrin adaptor complexes AP-1 and AP-2, respectively (43). However, EHD2 has also been depicted as a regulator controlling the exit of vesicular cargo from the ERC, similar to the function described for EHD1 (34). Additional experiments will be needed to definitively assess whether EHD2 is primarily involved in the internalization of receptors or their recycling.
EHD3 is the most homologous paralog to EHD1, showing 85% residue identity. Along with this high level of sequence identity, studies have shown that overexpressed EHD3 displays a strong colocalization with EHD1 and a similar tubulovesicular distribution pattern (19,24,33,34). However, unlike EHD1, depletion of EHD3 from HeLa cells causes internalized cargo to accumulate in enlarged peripheral early endosomes rather than at the centralized ERC (19,34). This function in the early endosome may be a vertebrate innovation because rme-1 null mutants in C. elegans do not appear to have any early endosomal defects (1).
Perhaps, the most conflicting reports have surfaced with regards to the function of EHD4. Despite displaying 74% identity to EHD1, and the lack of a secretory signal sequence, EHD4 was first described as a component of the extracellular matrix (44). Subsequently, EHD4 was characterized as an intracellular regulator of nerve growth factor receptor (TrkA) internalization (45). Intriguingly, TrkA does contain an NPF motif, but a direct interaction between these proteins has not been reported. One recent study has proposed that EHD4 is not involved in trafficking through endosomes but rather suggests that EHD4 functions in other highly specialized transport processes (24). A second study provides evidence that EHD4 does play a role in transport at the early endosome (34). In support of this, endogenous EHD4 has now been shown to colocalize with markers of the early endosome (35). This study also demonstrates the formation of enlarged early endosomal structures upon EHD4 depletion and the accumulation of internalized cargo in these compartments, which are enriched in active, GTP-bound, Rab5 (35). Moreover, the interaction described between endogenous EHD4 and EHD1, as well as the dramatic effects of depleting one EHD protein on the localization of the other, indicates a functional role for the interaction between these two paralogs (35).
Co-ordination of EHDs and Rab Proteins in Membrane Trafficking
Over 60 different mammalian Rab proteins have been described that participate in various aspects of regulating subcellular trafficking (46). One of the important questions arising from studying the function of EHD proteins is how they are able to co-ordinately regulate endocytic trafficking events with the Rab family of small GTP-binding proteins. For example, both EHD1 and Rab11 have been ascribed a role in regulating the exit of recycling cargo from the perinuclear ERC to the plasma membrane (1,23,36,47), and both EHD4 and Rab4 have been localized to peripheral early endosomes and implicated in controlling the recycling of receptors from this organelle (35,48). These findings have led to the hypothesis that EHD proteins and Rab proteins may interact with one another to co-ordinately regulate endocytic trafficking events. However, despite extensive colocalization between some EHDs and Rab proteins, to date, no direct interactions between these proteins have been reported.
One potential link between Rab proteins and EHD proteins is through Rab effectors. Each Rab protein is known to mediate its functional effects in the cell through binding to a series of ‘Rab effector proteins’. With few exceptions, these effectors bind exclusively to the GTP-bound Rab proteins, and each effector is thought to provide specific transport promoting functions. Two recent studies have linked Rab effectors with EHD proteins. First, the divalent Rab4/5 effector, Rabenosyn-5, was identified as an EHD interaction partner (18). Indeed, siRNA-mediated depletion of Rabenosyn-5 led to a delay in receptor recycling similar to that observed upon depletion of EHD1. However, whereas receptors affected by EHD1 depletion accumulated in the cell at the perinuclear ERC, knockdown of Rabenosyn-5 caused their accumulation in enlarged peripheral endosomes (18). These findings led to the proposition that Rabenosyn-5 interactions with EHD1 facilitate the transport of internalized receptors from peripheral sorting endosomes to the ERC. It is not yet known if Rabenosyn-5 interacts with early endosomal EHD3 or EHD4. A second study has identified the Rab11 effector, Rab11-FIP2, as an interaction partner for EHD proteins, suggesting a possible mechanism by which Rab11 and EHD1 might regulate the exit of receptors from the ERC to the plasma membrane (19). Additionally, Rab8a and Myosin Vb have been identified on EHD1/3 tubules (49), although no physical interaction between EHDs and Rab8a has been identified thus far. Similarly, C. elegans RME-1 acts downstream of RAB-10 in the basolateral recycling pathway of the intestine, but no direct link has been identified between the two proteins (41). Recent work suggests that Rab8 mediates Golgi-to-recycling endosome transport in polarized Madin-Darby Canine Kidney cells, while Rab10 mediates early endosomes-to-recycling endosome transport in such cells (50-52). Thus, Rab8 and Rab10 may regulate entry of distinct cargos into the EHD-regulated ERC.
In addition to the direct interactions between EHD proteins and Rab effectors, EHD proteins may be capable of regulating Rab activity. For example, depletion of EHD4 leads to the activation of Rab5, producing enhanced levels of GTP-bound Rab5 associated with endosomal membranes (35). Although the precise manner by which this interplay between Rab5 and EHD4 occurs has not yet been deciphered, it is clear that Rab and EHD proteins have evolved a co-ordinated strategy for the regulation of endocytic events.
Interaction Partners of EHD Proteins
The mode of interaction of Rabenosyn-5, Rab11-FIP2 and another endocytic regulator, Syndapin/Pacsin, with EHD proteins was found to be through EH domain/NPF motifs, and other EHD partners interact through similar mechanisms (Table 3). Rabenosyn-5, Rab11-FIP2 and Syndapin II all contain multiple NPF motifs (human Rabenosyn-5 carries five NPFs, mouse Rabenosyn-5 has six NPFs, mammalian Rab11-FIP2 has three NPF motifs and Syndapin II contains three NPF motifs). Other interaction motifs have also been observed aside from the typical NPF-EH interface. One such atypical interactor is the C. elegans protein ALX-1/ALIX, the V domain of which binds to a YPXL motif in the extreme C-terminus of RME-1. This strong V-domain–YPXL interaction requires augmentation in vivo, provided by an NPF-EH interface (21). This work uncovered a novel role for ALX-1/ALIX in the regulation of receptor recycling in worms and mammalian cells, with specificity for cargo taken up independently of clathrin (21). Bro1p/ALIX was already well known for its role in membrane protein degradation through interaction with endosomal sorting complex required for transport-III (ESCRT-III) on multivesicular endosomes in yeast and mammals, a function that is also conserved in C. elegans (21,53-56). One exciting possibility is that this dual role of ALX-1/ALIX, in recycling and degradation, indicates co-ordinate regulation of the decision to degrade or recycle cargo after endocytosis.
Table 3.
Interactions between EHD proteins and binding partners
| EHD protein | Interaction partner | Mode of EHD interaction | Reference |
|---|---|---|---|
| Rme-1 (C. elegans) | Reticulon-C protein | C-terminal region | (66) |
| Rme-1 (C. elegans) | Alix/ALX-1 | 1) Rme-1 EH domain binds ALX-1 NPF motif 2) Rme-1 YPSL motif binds ALX-1 V-domain |
(21) |
| EHD1 (D. melanogaster) | Numb | EH domain | (62) |
| EHD1 | Insulin-like growth factor 1 receptor | Not characterized | (67) |
| EHD1 | SNAP29/GS32 | EH domain | (20) |
| EHD1 | AP-2 α-adaptin | Not characterized | (67) |
| EHD1 | Clathrin heavy chain | Not characterized | (67) |
| EHD1 | Rabenosyn-5 | EH domain | (18) |
| EHD1 | Syndapin I | EH domain | (17,20) |
| EHD1 | Syndapin II | EH domain | (17) |
| EHD1 | Rab11-FIP2 | EH domain | (19) |
| EHD1 | Retromer subunits | Not through EH domain | (65) |
| EHD1 | Epsin1 | EH domain | (our unpublished data) |
| EHD1 | Stonin2 | EH domain | (our unpublished data) |
| EHD2 | GLUT4 | Not characterized | (43) |
| EHD2 | AP-1 μ1 | Not characterized | (43) |
| EHD2 | AP-2 μ2 | Not characterized | (43) |
| EHD2 | CALM | Not characterized | (43) |
| EHD2 | EHBP1 | EH domain | (42) |
| EHD2 | Arp2/3 | Acidic region prior to EH domain | (42) |
| EHD2 | Rabenosyn-5 | EH domain | (18) |
| EHD3 | Syndapin I and II | EH domain | (17) |
| EHD3 | Rabenosyn-5 | EH domain | (18) |
| EHD3 | Rab11-FIP2 | EH domain | (19) |
| EHD4 | Numb | EH domain | (62) |
| EHD4 | Syndapin I and II | EH domain | (17) |
| EHD4 | Type VI collagen | Not characterized | (44) |
Much remains to be learned regarding the role for interaction of EHD/RME-1 proteins with diverse partners and how they contribute to endocytic traffic. As alluded to above, some of these partners may regulate the assembly and/or nucleotide state of EHD oligomers. In addition, these proteins may act to link EHD/RME-1 proteins to the actin cytoskeleton. The Syndapin/Pacsin interaction with EHDs on recycling endosomes may be a particularly relevant to such discussions (17) because Syndapin also interacts with Dynamin during receptor internalization and activates actin polymerization through N-Wasp (57). Thus, association with Syndapin further extends the EHD/Dynamin parallel and suggests that like Dynamin, EHD proteins may help link membrane remodeling to actin dynamics, a process suggested to generate membrane tension during scission events (58,59). Likewise, EHD/RME-1-interacting proteins EHBP1, Numb and ALIX are also associated with actin binding or remodeling (21,42,60-62), and EHD/RME-1 colocalizes with the known actin regulators Arf6 and Cdc42 on endosomal tubules (23,63).
Conclusions/Perspectives
Since the turn of the millennium, rapid advances have been made in elucidating the function of EHD/RME-1 proteins. Despite the high levels of homology between the four mammalian paralogs, it is clear that in addition to partially overlapping functions, each of the EHDs also carries out distinct endocytic regulatory roles. The identification of novel EHD/RME-1 interactors through genetic and biochemical approaches, and the recent structural advances, have continued to highlight the pivotal role of EHDs in endocytic traffic and promise to further the understanding of the molecular mechanisms by which these proteins act.
Acknowledgments
The authors thank S. Pant and N. Naslavsky and O. Daumke for critical readings and O. Daumke, F. Kieken and P. Sorgen for kindly contributing figures. Support for this work was provided by National Institutes of Health grants GM074876 (S. C.) and GM067237 (B. D. G.).
References
- 1.Grant B, Zhang Y, Paupard MC, Lin SX, Hall DH, Hirsh D. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- 2.Naslavsky N, Caplan S. C-terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J Cell Sci. 2005;118:4093–4101. doi: 10.1242/jcs.02595. [DOI] [PubMed] [Google Scholar]
- 3.Daumke O, Lundmark R, Vallis Y, Martens S, Butler PJ, McMahon HT. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449:923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- 4.van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 5.Fazioli F, Minichiello L, Matoskova B, Wong WT, Di Fiore PP. eps15, a novel tyrosine kinase substrate, exhibits transforming activity. Mol Cell Biol. 1993;13:5814–5828. doi: 10.1128/mcb.13.9.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong WT, Schumacher C, Salcini AE, Romano A, Castagnino P, Pelicci PG, Di Fiore P. A protein-binding domain, EH, identified in the receptor tyrosine kinase substrate Eps15 and conserved in evolution. Proc Natl Acad Sci U S A. 1995;92:9530–9534. doi: 10.1073/pnas.92.21.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miliaras NB, Wendland B. EH proteins: multivalent regulators of endocytosis (and other pathways) Cell Biochem Biophys. 2004;41:295–318. doi: 10.1385/CBB:41:2:295. [DOI] [PubMed] [Google Scholar]
- 8.Santolini E, Salcini AE, Kay BK, Yamabhai M, Di Fiore PP. The EH network. Exp Cell Res. 1999;253:186–209. doi: 10.1006/excr.1999.4694. [DOI] [PubMed] [Google Scholar]
- 9.Polo S, Confalonieri S, Salcini AE, Di Fiore PP. EH and UIM: endocytosis and more. Sci STKE. 2003;2003:re17. doi: 10.1126/stke.2132003re17. [DOI] [PubMed] [Google Scholar]
- 10.de Beer T, Carter RE, Lobel-Rice KE, Sorkin A, Overduin M. Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain. Science. 1998;281:1357–1360. doi: 10.1126/science.281.5381.1357. [DOI] [PubMed] [Google Scholar]
- 11.Kieken F, Jovic M, Naslavsky N, Caplan S, Sorgen PL. EH domain of EHD1. J Biomol NMR. 2007;39:323–329. doi: 10.1007/s10858-007-9196-0. [DOI] [PubMed] [Google Scholar]
- 12.Paoluzi S, Castagnoli L, Lauro I, Salcini AE, Coda L, Fre S, Confalonieri S, Pelicci PG, Di Fiore PP, Cesareni G. Recognition specificity of individual EH domains of mammals and yeast. EMBO J. 1998;17:6541–6550. doi: 10.1093/emboj/17.22.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salcini AE, Confalonieri S, Doria M, Santolini E, Tassi E, Minenkova O, Cesareni G, Pelicci PG, Di Fiore PP. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Beer T, Hoofnagle AN, Enmon JL, Bowers RC, Yamabhai M, Kay BK, Overduin M. Molecular mechanism of NPF recognition by EH domains. Nat Struct Biol. 2000;7:1018–1022. doi: 10.1038/80924. [DOI] [PubMed] [Google Scholar]
- 15.Confalonieri S, Di Fiore PP. The Eps15 homology (EH) domain. FEBS Lett. 2002;513:24–29. doi: 10.1016/s0014-5793(01)03241-0. [DOI] [PubMed] [Google Scholar]
- 16.Rumpf J, Simon B, Jung N, Maritzen T, Haucke V, Sattler M, Groemping Y. Structure of the Eps15-stonin2 complex provides a molecular explanation for EH-domain ligand specificity. EMBO J. 2008;27:558–569. doi: 10.1038/sj.emboj.7601980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun A, Pinyol R, Dahlhaus R, Koch D, Fonarev P, Grant BD, Kessels MM, Qualmann B. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol Biol Cell. 2005;16:3642–3658. doi: 10.1091/mbc.E05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naslavsky N, Boehm M, Backlund PS, Jr, Caplan S. Rabenosyn-5 and EHD1 interact and sequentially regulate protein recycling to the plasma membrane. Mol Biol Cell. 2004;15:2410–2422. doi: 10.1091/mbc.E03-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Mol Biol Cell. 2006;17:163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y, Shi H, Wei S, Wong SH, Hong W. Mutually exclusive interactions of EHD1 with GS32 and Syndapin II. Mol Membr Biol. 2004;21:269–277. doi: 10.1080/09687680410001716871. [DOI] [PubMed] [Google Scholar]
- 21.Shi A, Pant S, Balklava Z, Chen CC, Figueroa V, Grant BD. A novel requirement for C elegans Alix/ALX-1 in RME-1-mediated membrane transport. Curr Biol. 2007;17:1913–1924. doi: 10.1016/j.cub.2007.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naslavsky N, Rahajeng J, Chenavas S, Sorgen PL, Caplan S. EHD1 and Eps15 interact with phosphatidylinositols via their Eps15 homology domains. J Biol Chem. 2007;282:16612–16622. doi: 10.1074/jbc.M609493200. [DOI] [PubMed] [Google Scholar]
- 23.Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blume JJ, Halbach A, Behrendt D, Paulsson M, Plomann M. EHD proteins are associated with tubular and vesicular compartments and interact with specific phospholipids. Exp Cell Res. 2007;313:219–231. doi: 10.1016/j.yexcr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Saraste M, Sibbald PR, Wittinghofer A. The P-loop – a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 26.Lee DW, Zhao X, Scarselletta S, Schweinsberg PJ, Eisenberg E, Grant BD, Greene LE. ATP binding regulates oligomerization and endosome association of RME-1 family proteins. J Biol Chem. 2005;280:280–290. doi: 10.1074/jbc.M412751200. [DOI] [PubMed] [Google Scholar]
- 27.Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 28.Shpetner HS, Vallee RB. Dynamin is a GTPase stimulated to high levels of activity by microtubules. Nature. 1992;355:733–735. doi: 10.1038/355733a0. [DOI] [PubMed] [Google Scholar]
- 29.Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 30.Warnock DE, Schmid SL. Dynamin GTPase, a force-generating molecular switch. Bioessays. 1996;18:885–893. doi: 10.1002/bies.950181107. [DOI] [PubMed] [Google Scholar]
- 31.Sever S, Muhlberg AB, Schmid SL. Impairment of dynamin’s GAP domain stimulates receptor-mediated endocytosis. Nature. 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- 32.Warnock DE, Hinshaw JE, Schmid SL. Dynamin self-assembly stimulates its GTPase activity. J Biol Chem. 1996;271:22310–22314. doi: 10.1074/jbc.271.37.22310. [DOI] [PubMed] [Google Scholar]
- 33.Galperin E, Benjamin S, Rapaport D, Rotem-Yehudar R, Tolchinsky S, Horowitz M. EHD3: a protein that resides in recycling tubular and vesicular membrane structures and interacts with EHD1. Traffic. 2002;3:575–589. doi: 10.1034/j.1600-0854.2002.30807.x. [DOI] [PubMed] [Google Scholar]
- 34.George M, Ying G, Rainey MA, Solomon A, Parikh PT, Gao Q, Band V, Band H. Shared as well as distinct roles of EHD proteins revealed by biochemical and functional comparisons in mammalian cells and C. elegans. BMC Cell Biol. 2007;8:3. doi: 10.1186/1471-2121-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma M, Naslavsky N, Caplan S. A role for EHD4 in the regulation of early endosomal transport. Traffic. 2008;9:995–1018. doi: 10.1111/j.1600-0854.2008.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin SX, Grant B, Hirsh D, Maxfield FR. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol. 2001;3:567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- 37.Jovic M, Naslavsky N, Rapaport D, Horowitz M, Caplan S. EHD1 regulates beta1 integrin endosomal transport: effects on focal adhesions, cell spreading and migration. J Cell Sci. 2007;120:802–814. doi: 10.1242/jcs.03383. [DOI] [PubMed] [Google Scholar]
- 38.Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305:1972–1975. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- 39.Picciano JA, Ameen N, Grant BD, Bradbury NA. Rme-1 regulates the recycling of the cystic fibrosis transmembrane conductance regulator. Am J Physiol Cell Physiol. 2003;285:C1009–C1018. doi: 10.1152/ajpcell.00140.2003. [DOI] [PubMed] [Google Scholar]
- 40.Rapaport D, Auerbach W, Naslavsky N, Pasmanik-Chor M, Galperin E, Fein A, Caplan S, Joyner AL, Horowitz M. Recycling to the plasma membrane is delayed in EHD1 knockout mice. Traffic. 2006;7:52–60. doi: 10.1111/j.1600-0854.2005.00359.x. [DOI] [PubMed] [Google Scholar]
- 41.Chen CC, Schweinsberg PJ, Vashist S, Mareiniss DP, Lambie EJ, Grant BD. RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol Biol Cell. 2006;17:1286–1297. doi: 10.1091/mbc.E05-08-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guilherme A, Soriano NA, Bose S, Holik J, Bose A, Pomerleau DP, Furcinitti P, Leszyk J, Corvera S, Czech MP. EHD2 and the novel EH domain binding protein EHBP1 couple endocytosis to the actin cytoskeleton. J Biol Chem. 2004;279:10593–10605. doi: 10.1074/jbc.M307702200. [DOI] [PubMed] [Google Scholar]
- 43.Park SY, Ha BG, Choi GH, Ryu J, Kim B, Jung CY, Lee W. EHD2 interacts with the insulin-responsive glucose transporter (GLUT4) in rat adipocytes and may participate in insulin-induced GLUT4 recruitment. Biochemistry. 2004;43:7552–7562. doi: 10.1021/bi049970f. [DOI] [PubMed] [Google Scholar]
- 44.Kuo HJ, Tran NT, Clary SA, Morris NP, Glanville RW. Characterization of EHD4, an EH domain-containing protein expressed in the extracellular matrix. J Biol Chem. 2001;276:43103–43110. doi: 10.1074/jbc.M106128200. [DOI] [PubMed] [Google Scholar]
- 45.Shao Y, Akmentin W, Toledo-Aral JJ, Rosenbaum J, Valdez G, Cabot JB, Hilbush BS, Halegoua S. Pincher, a pinocytic chaperone for nerve growth factor/TrkA signaling endosomes. J Cell Biol. 2002;157:679–691. doi: 10.1083/jcb.200201063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 47.Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Sluijs P, Hull M, Webster P, Male P, Goud B, Mellman I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell. 1992;70:729–740. doi: 10.1016/0092-8674(92)90307-x. [DOI] [PubMed] [Google Scholar]
- 49.Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell. 2007;18:2828–2837. doi: 10.1091/mbc.E07-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babbey CM, Ahktar N, Wang E, Chen CC, Grant BD, Dunn KW. Rab10 regulates membrane transport through early endosomes of polarized Madin-Darby canine kidney cells. Mol Biol Cell. 2006;17:3156–3175. doi: 10.1091/mbc.E05-08-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henry L, Sheff DR. Rab8 regulates basolateral secretory, but not recycling, traffic at the recycling endosome. Mol Biol Cell. 2008;19:2059–2068. doi: 10.1091/mbc.E07-09-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuck S, Gerl MJ, Ang A, Manninen A, Keller P, Mellman I, Simons K. Rab10 is involved in basolateral transport in polarized Madin-Darby canine kidney cells. Traffic. 2007;8:47–60. doi: 10.1111/j.1600-0854.2006.00506.x. [DOI] [PubMed] [Google Scholar]
- 53.Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 54.Katoh K, Shibata H, Suzuki H, Nara A, Ishidoh K, Kominami E, Yoshimori T, Maki M. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J Biol Chem. 2003;278:39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- 55.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci. 2003;116:1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- 57.Qualmann B, Roos J, DiGregorio PJ, Kelly RB. Syndapin I, a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol Biol Cell. 1999;10:501–513. doi: 10.1091/mbc.10.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 59.Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- 60.Guilherme A, Soriano NA, Furcinitti PS, Czech MP. Role of EHD1 and EHBP1 in perinuclear sorting and insulin-regulated GLUT4 recycling in 3T3-L1 adipocytes. J Biol Chem. 2004;279:40062–40075. doi: 10.1074/jbc.M401918200. [DOI] [PubMed] [Google Scholar]
- 61.Pan S, Wang R, Zhou X, He G, Koomen J, Kobayashi R, Sun L, Corvera J, Gallick GE, Kuang J. Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. J Biol Chem. 2006;281:34640–34650. doi: 10.1074/jbc.M602263200. [DOI] [PubMed] [Google Scholar]
- 62.Smith CA, Dho SE, Donaldson J, Tepass U, McGlade CJ. The cell fate determinant Numb interacts with EHD/Rme-1 family proteins and Has a role in endocytic recycling. Mol Biol Cell. 2004;15:3698–3708. doi: 10.1091/mbc.E04-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol. 2007;9:1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 64.Naslavsky N, Rahajeng J, Rapaport D, Horowitz M, Caplan S. EHD1 regulates cholesterol homeostasis and lipid droplet storage. Biochem Biophys Res Commun. 2007;357:792–799. doi: 10.1016/j.bbrc.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gokool S, Tattersall D, Seaman MN. EHD1 interacts with retromer to stabilise SNX1-tubules and facilitate endosome-to-Golgi retrieval. Traffic. 2007;8:1873–1886. doi: 10.1111/j.1600-0854.2007.00652.x. [DOI] [PubMed] [Google Scholar]
- 66.Iwahashi J, Kawasaki I, Kohara Y, Gengyo-Ando K, Mitani S, Ohshima Y, Hamada N, Hara K, Kashiwagi T, Toyoda T. Caenorhabditis elegans reticulon interacts with RME-1 during embryogenesis. Biochem Biophys Res Commun. 2002;293:698–704. doi: 10.1016/S0006-291X(02)00282-6. [DOI] [PubMed] [Google Scholar]
- 67.Rotem-Yehudar R, Galperin E, Horowitz M. Association of insulin-like growth factor 1 receptor with EHD1 and SNAP29. J Biol Chem. 2001;276:33054–33060. doi: 10.1074/jbc.M009913200. [DOI] [PubMed] [Google Scholar]


