Abstract
Bacterial sensing circuits may be triggered by molecules originating from the environment (e.g., nutrients, chemoattractants). Bacteria also actively probe the environment for information, by releasing molecular probes to measure conditions beyond the cell surface -- telesensing. Perceiving the environment beyond is achieved by sensing environmentally induced changes in those probes, such as occurs when a siderophore chelates an iron atom, or a quorum sensing signal is inactivated by a specific enzyme or adsorbent. This information, captured by chemical and physical changes induced in specifically produced molecules transiting through the environment, enable bacteria to mount a contextually appropriate response.
Introduction
We now understand that bacteria are highly sentient organisms with a sophisticated and diverse array of systems for sensing their surroundings. Rapidly accumulating genomic information shows that microbes in fluctuating environments are rich in two-component signaling and other sensing systems. For example, Pseudomonas aeruginosa strains possess as many as 53 two-component systems (Kiil et al., 2005). Contact sensing systems function when a molecule originating from the environment directly collides with a cell surface or periplasmic receptor, as in the binding of nutrients or chemoattractants. Other mechanisms for analyzing the environment, such as quorum sensing, require the microbe to generate a probe to query conditions beyond the cell surface. Local versus distance sensing systems are not unique to bacteria – it is at the heart of subjects as diverse as the debate over the need for manned space flight, where concepts of “touch sensing” and “telesensing” are well established (Genta and Perino, 2005).
Quorum sensing is a specific type of telesensing, and new variations on the theme are continually discovered (Svenningsen et al., 2009; Swem et al., 2008). There are several outstanding recent reviews of quorum sensing (Camilli and Bassler, 2006; Williams et al., 2007). The goal of quorum sensing is to count siblings or close relatives, by measuring a molecule that has diffused from a distance. Other telesensing systems probe the environment for specific external conditions, and when that condition is found, trigger a contextually appropriate response.
As one of the most thoroughly studies types of telesensing, it is of value to briefly examine quorum sensing so that it can be contrasted and compared to other telesensing systems. Telesensing involves the production, release and diffusion of a signal, followed by subsequent recognition of an environmentally imposed change in that signaling molecule. In the case of quorum sensing, the environment dictates the fate of the quorum molecule, including the rate of its accumulation to a threshold concentration, which then triggers a contextually appropriate genetic program. In its simplest form, quorum sensing enables bacteria to coordinate gene expression according to local population density. As a bacterial population increases, the concentration of the quorum molecule, or autoinducer, in the external environment increases proportionally. Thus, by controlling gene expression in response to a chemical that is synthesized, released, and either exchanged with a sibling or returns to the producer, bacteria query the environment beyond the cell surface for information on the population of siblings, and alter their behavior accordingly. Logically, the behaviors that are triggered are social involving community structure in biofilms, group expression of bioluminescence, sporulation, conjugation, motility, competence, and secretion of virulence factors (Miller and Bassler, 2001).
Quorum sensing mechanisms appear to have evolved along three tracks – acylhomoserine lactone-based (AHL) signaling systems of Gram-negative bacteria, peptide-based systems in Gram-positive bacteria, and a furanone-based system common to both (Hardie and Heurlier, 2008). AHL circuits are typically homologous to the LuxI/LuxR system of Vibrio fischeri (Engebrecht et al., 1983). LuxI catalyzes the formation of specific fatty acylhomoserine lactones (AHLs) that diffuse through the membrane. Once a threshold level is reached, AHLs bind to a LuxR-type regulator, which then induces or represses expression of target genes (Lazdunski et al., 2004; Miller and Bassler, 2001). In contrast, Gram-positive bacteria typically use oligopeptides, such as the Agr system of Staphylococcus aureus (Lyon and Novick, 2004), to actively probe the environment. These are generically referred to as autoinducing peptides (AIPs). Once exported, these AIPs interact with the environment, then are sensed in most cases by the external domains of membrane bound sensor kinase proteins. This interaction induces a phosphorylation cascade that leads to the activation of target genes. Autoinducers enable specific intraspecies communication. More universal communication is achieved between microbes of various species through the autoinducer 2 (AI2) system, which is shared by both Gram-positive and Gram-negative bacteria (Xavier and Bassler, 2003).
Common to all quorum sensing systems is the release of specific, microbially synthesized molecules into the environment. In an inert environment, the accumulation of a monotonically produced signal becomes a direct reflection of a) the number of microbes present, b) the length of time a closed environment has been occupied by the signal producing cell, or a combination of a) and b). Some highly co-evolved systems, such as the Vibrio fischeri/sepiolid squid light organ, approximate a closed environment with reproducible diffusion characteristics. Other environments with consistent, reproducible diffusion characteristics include the internal milieu of a monospecies bacterial colony. In a bacterial colony, a telesensing system may function just as a morphogen does in the development of higher organisms: A signaling molecule is produced by one cell in the population, which then diffuses outward. Rapid dilution into the environment from the surface of a colony creates a zone of reduced concentration at the colony/environment interface, whereas limited solvent movement between cells leads to locally high accumulation internally, overall resulting in a concentration gradient across the colony. Cells along that gradient express varying developmental programs, which in higher eukaryotes, results in the creation of complex tissue and organ architectures (Kicheva et al., 2007). The discovery of a quorum-regulated Streptococcus pneumoniae system that causes lysis of neighboring sibling cells in a colony along a concentration gradient (Guiral et al., 2005), led to a proposed mechanism and role for autolysis in the release of DNA and formation of higher order architecture in stable microbial communities, such as biofilms (Gilmore and Haas, 2005). In a variation on signal-induced fratricide, the soil bacterium Paenibacillus dendritiformis secretes a lethal factor that diffuses out from the colony. If another colony of the same strain is in close proximity, the concentration of the lethal factor can exceed the inhibitory threshold, producing a distinct zone of inhibition and cell death where the colonies approach each other (Be'er et al., 2009).
However, not all environments into which quorum or other sensing molecules are released have consistent diffusion or chemical characteristics, and the importance of the of external environment in altering telesensing signals is beginning to be appreciated (Horswill et al., 2007). Telesensing processes are now known to be influenced by environmental cues including temperature, ligand concentration, pH and water and oxygen availability (Bollinger et al., 2001; Bose et al., 2007; Dulla and Lindow, 2008; Hasegawa et al., 2005; Jensen et al., 2006; Latour et al., 2007; McGowan et al., 2005; Palmer et al., 2007; Pessi and Haas, 2000; Shrout et al., 2006; Surette and Bassler, 1998; Wagner et al., 2006). These observations highlight the importance of telesensing systems being contextually sensitive. Here, we review several well characterized telesensing systems that have evolved contextual sensitivity. Common to all of these systems is the production and release of a telesensing molecule by organisms that may inhabit diverse niches. If the molecule is released in one biologically relevant environment, the molecule is specifically affected by conditions within that environment, and this dictates the decision algorithm of the microbe. In another environment where the molecule may not be affected and returns unaltered, or simply increased in concentration behaving purely as a quorum sensor, another behavioral decision is made by the microbe. In short, we review here telesensing systems that have the characteristic of being environmentally responsive.
Environmentally responsive telesensing systems
Telesensing may be used by microbes to sense conditions in animal or plant niches, or occurring at large in the environment. Telesensing may regulate traits that dictate the relationship in a host niche, leading the microbe to break out of a stable commensal relationship and cause disease. Alternatively, telesensing may be used by a microbe to sense when it has crossed a line from stable colonization to being in direct conflict with the host, and down-regulate potentially pathogenic traits to promote a return to a stable host-commensal relationship. There is increasing evidence in some cases to support this more nuanced latter view.
A. Telesensing signals that are modified by host factors
1-The Enterococcus faecalis cytolysin signal senses target cells, and produces a toxin in response
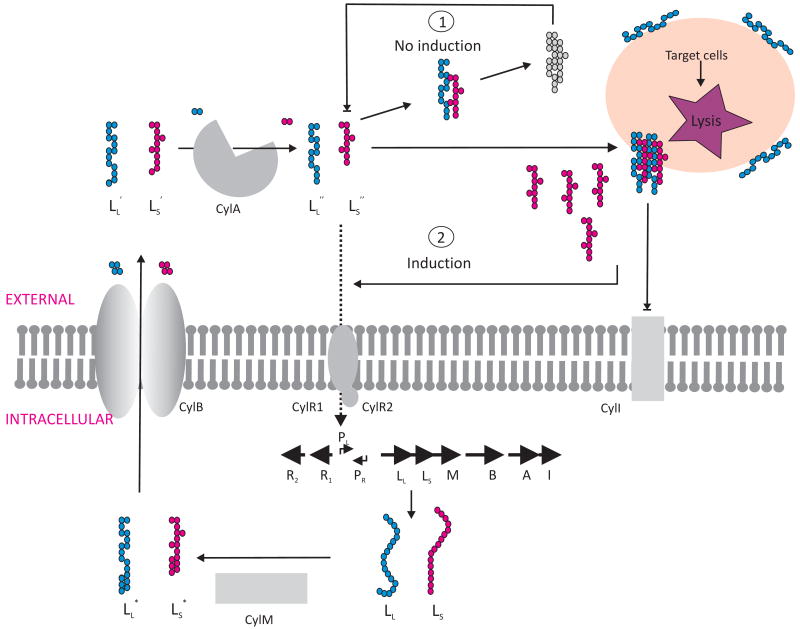
Enterococcus faecalis is a common resident of the gastrointestinal tract of a wide variety of hosts, ranging from mammals to insects. Moreover, it is a leading cause of hospital-acquired infection, and also the main cause of sub-acute endocarditis in the community ((McCormick et al., 2001). Highly virulent strains of E. faecalis produce a two subunit toxin termed the cytolysin. This toxin has broad activity against both eukaryotic and prokaryotic cells, and contributes to the severity of infection in all models tested (Coburn and Gilmore, 2003; Garsin et al., 2001). Cytolysin is a variable trait of E. faecalis, and is usually encoded within pheromone-responsive plasmids (plasmids that encode the ability to recognize molecules released into the environment by a potential recipient bacterium, and in response upregulate the conjugation machinery that transfers a plasmid copy into that recipient), or on a pathogenicity island (Shankar et al., 2002). The cytolysin consists of two small peptides, CylLL and CylLs. Both are posttranslationally modified and secreted into the extracellular environment by accessory proteins encoded within the cytolysin operon. Once activated by a final proteolytic step outside of the cell, the mature toxin subunits, termed CylLL″ and CylLS″, form a complex in eukaryotic or prokaryotic target cell membranes that results in membrane rupture, and in the case of erythrocytes which are often used as targets in vitro, hemoglobin release. Eight genes are involved in expression and regulation of cytolysin production. Six of these genes are transcribed from the promoter, PLys, and are required for toxin production, maturation, secretion, and immunity. Two additional divergently transcribed genes, expressed from the promoter PReg, encode regulatory proteins CylR1 and CylR2.
Cytolysin expression is regulated by an environmentally influenced telesensing system (Coburn et al., 2004). The smaller of the two toxin subunits, CylLS″, autoinduces expression of the toxin operon through a quorum sensing-like mechanism (Haas et al., 2002). Local accumulation of mature CylLS″ leads to autoinduction. Enigmatically, little cytolysin is expressed in liquid E. faecalis cultures, even though it is readily produced on blood agar leading to clear zones of hemolysis. Because of this, the cytolysin was originally termed a “pseudohemolysin” (Todd, 1934). More recently it was observed that when erythrocytes are added to broth culture, cytolysin activity is produced at high levels (Coburn et al., 2004). It was therefore of interest to determine the mechanism by which E. faecalis was able to sense the presence of target cells in the environment, and to express the cytolysin when target cells were sensed.
It was found that in the absence of target cells, the larger cytolysin subunit, CylLL″, interacts with the smaller subunit/autoinducer, CylLS″, to form an oligomeric complex. This very stable complex is devoid of cytolytic activity, and sequesters CylLS″ in a way that prevents its induction of the cytolysin operon (Coburn et al., 2004). In other words, CylLL″ titrates back the level of free CylLS″ in solution, and holds it below the threshold necessary to trigger high-level cytolysin expression. However, in the presence of target cells it was found that CylLL″ binds to lipid membranes with a 6 – 7 fold greater affinity than CylLS″. CylLL″ binds membranes in way that prevents its sequestration of CylLS″, allowing a pool of free CylLS″ to accumulate, at least transiently, and induce high-level cytolysin expression (Figure 1). Stated another way, when target cells are absent, the two cytolysin subunits CylLS″ and CylLL″ form inactive aggregates, but in the presence of target cells, CylLL″ rapidly adsorbs to the target cell surface leaving behind a pool of free CylLS″, which is capable of autoinduction. Using this mechanism, E. faecalis is able to sense cytolysin target cells and produce cytolysin only when target cells are present. Whether this regulatory system evolved to sense competing Gram-positive bacterial cells in the competitive GI tract (in addition to being a toxin, it is also a bacteriocin specific for Gram-positive bacteria), or perhaps single-celled eukaryotic organisms in the external environment, or cells of more complex life forms such as mammals, where it invariably contributes to virulence (Coburn and Gilmore, 2003), is currently unknown.
Figure 1. The Enterococcus faecalis cytolysin senses and destroys target cells.
The two subunits of the toxin, CylLL and CylLS are modified and secreted in the extracellular environment. Cytolysin expression is regulated by CylLS″ through a quorum sensing-like mechanism. In absence of target cells, the mature subunit CylLL″, interacts with CylLS″ to form an inactive oligomeric complex (1) that is not able to induce expression of the cytolysin operon. In the presence of target cells, CylLL″ binds them preferentially, creating a transient pool of free CylLS″ (2) to induce high-level cytolysin expression, prior to the slow reaction of CylLS″ joining the pore complex.
2-Neutralization of a pheromone inhibitor enables pheromone accumulation, leading to binding to a potential plasmid recipient, or to bacterial accumulation on host tissues in infection
Pheromone-responsive conjugative plasmids are widely distributed among enterococci where they contribute to the proliferation and distribution of antimicrobial resistance and other traits. Well characterized pheromone-responsive conjugation systems have been identified on plasmids pAD1 and pCF10. Expression of the large set of gene products required for conjugative transfer is controlled by communication between plasmid-free recipients and plasmid-carrying donor cells. Potential recipient cells release 7 – 8 amino acid oligopeptides, which are generally encoded within and processed from the C-terminal signal sequence of lipoprotein precursors, that act as pheromone attractants (Clewell et al., 2000). The pheromone peptide from a potential recipient cell is internalized by the potential donor E. faecalis cell (Dunny, 2007), where it induces expression of a plasmid-encoded cell surface protein. This protein, named aggregation substance, mediates stable mating pair-formation between donor and recipient, and if not carefully controlled, can also lead to donor clumping or autoaggregation. (Donor cells also produce the pheromone. However, donor cells additionally produce a plasmid-encoded surface protein as well as a plasmid-encoded inhibitor peptide that prevent donor-produced pheromone uptake by a molecular mechanism yet to be precisely defined). The plasmid-encoded pheromone inhibitor is a small peptide, which, like the pheromone, is synthesized as a precursor. Blockage of the pheromone by these donor cell activities prevents autoaggregation and futile activation of the transfer pathway. Expression of additional pheromone by the potential recipient, however, exceeds the capacity of the inhibitory activities, resulting in local pheromone concentrations that tip the balance in favor of mounting a response and expression of aggregation substance. Disturbing the balance of pheromone to pheromone antagonizing activities, even in a pure culture of plasmid-containing donor cells, can lead to autoaggregation.
The surface protein that mediates autoaggregation, originally termed aggregation substance prior to determination of its molecular character, in addition to mediating effective mating pair formation leading to plasmid transfer, also contributes to virulence in most infection models (Wirth, 2000). This suggested that aggregation substance is somehow expressed in the absence of recipient cells in sites of infection. Using pCF10, it was found that a host factor, most likely an albumin/lipid complex, selectively sequesters or degrades the pheromone inhibitor, resulting in a local pool of free pheromone even in the absence of recipient cells. This leads to expression of aggregation substance on the surface of E. faecalis in an infection (Chandler et al., 2005), where it results in larger vegetations on heart valves, and likely also to clumping making phagocytic clearance more difficult. Whether or not this behavior at the site of infection is the result of selective pressure in this setting is an interesting question. By far, enterococcal existence is mainly as a commensal in the GI tract in nearly all animals, despite its occurrence as a leading cause of multiple antibiotic resistant hospital acquired infection. Nevertheless, this mechanism of producing an aggregation effector along with an inhibitor, allows enterococci to sense the presence of either bacterial partners or its occurrence at the site of infection, and in response induce the expression of aggregation substance leading to attachment to recipient bacterial cells or accumulation on host tissue (Figure 2).
Figure 2. Pheromone inducible plasmids enable Enterococcus faecalis to sense either recipients or conditions in the bloodstream.
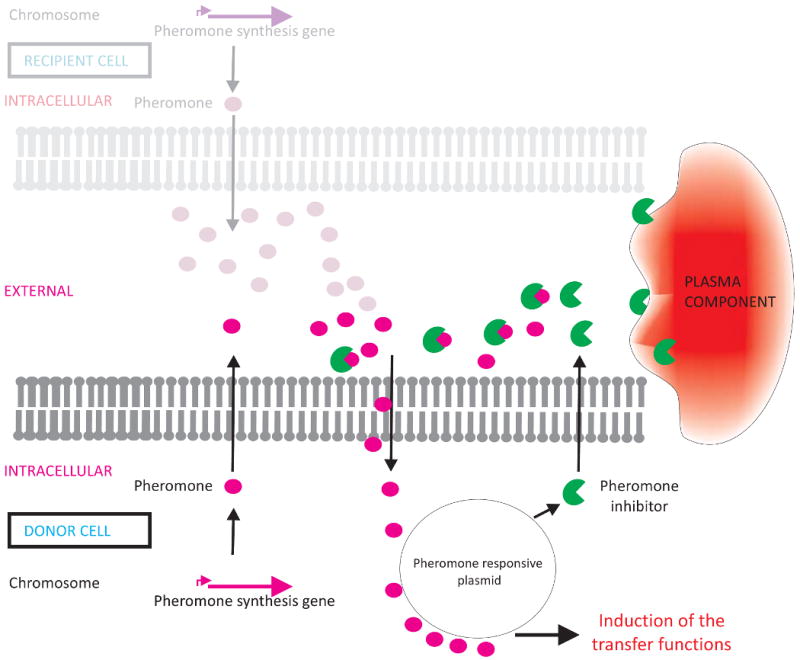
Chromosomally encoded pheromones activate plasmid encoded factors that promote transfer of pheromone-responsive plasmids (e.g., pAD1 and pCF10) by conjugation from donor to recipient. Both donor and recipient cells produce pheromones. In donor cells, however, pheromone activity is held in check via a plasmid encoded cell wall protein (not shown) and a pheromone inhibitor also encoded by the plasmid. An increase in external pheromone concentration can be driven by additional pheromone production by a potential recipient (grayed image), or by the presence of substances that selectively neutralize the inhibitor, as occurs in blood.
3-Evidence for détente between Staphylococcus aureus and man, mediated in part by environmentally responsive telesensing
Nearly all humans asymptomatically carry Staphylococcus aureus as a member of the commensal flora, yet it is the leading bacterial cause of invasive infection in the developed world, causing more deaths annually in the US than HIV/AIDS (Bancroft, 2007; Fridkin et al., 2005; Klevens et al., 2007). S. aureus virulence is centrally regulated by the agr system and other regulators (Novick and Geisinger, 2008). The agr locus is transcribed from two promoters, P2 and P3. Promoter P2 initiates transcription of a 4-gene operon, agrB, D, C and A. AgrD encodes the precursor of the signal molecule, which is trimmed and secreted by AgrB to generate the autoinducer signal peptide, a 7-9 mer peptide containing a 5 membered thiolactone ring. The secreted, cyclized autoinducer then binds the sensor kinase, AgrC, on the surface of S. aureus, which then activates the AgrA response regulator via a cascade of phosphorylation. Once phosphorylated, AgrA upregulates expression from the P2 promoter, as well as the P3 promoter, which initiates transcription of a regulatory RNA molecule, termed RNAIII. RNAIII is the effector of the agr regulon.
RNAIII down regulates expression of genes encoding surface adhesins while it up regulates capsule, toxins and protease production (Novick and Geisinger, 2008). This phenotypic conversion from an adhesive and colonizing behavior to a tissue damaging/invasive form, is central to the current understanding of the pathogenesis of systemic infection (Novick, 2003). Suggesting that there may be important complexities to this understanding, Rothfork and colleagues (Rothfork et al., 2004) made the surprising observation that phagocyte NADPH oxidase, myeloperoxidase or inducible nitric oxide synthetase inactivate the type I Agr signaling thiolactone. This results in inhibition of bacterial communication, and limits the upregulation of the virulence traits that constitute the virulent form. Oxidation of the C-terminal methionine of the Agr thiolactone was primarily responsible for the loss of activity. The use of oxygen radical generating defense and predation systems is common among most eukaryotes, and clearly predates the evolution of modern primates. Moreover, other amino acids are known to be functional at this position. Nevertheless, this microbe still employs a virulence signaling system that is inactive in the oxidative intracellular milieu of a phagocytic cell. Therefore, in interacting with the human host, a quorum of S. aureus behaves in a virulent way in some human niches, but in an avirulent way when it is exposed to host produced reactive oxygen and nitrogen species. Although this oxidative inactivation was only shown for Agr type I, the oxidized methionine is conserved in Arg type IV, and also in the E. faecalis gelatinase-regulating system, fsr (Lyon and Novick, 2004), which would be predicted to be similarly inactivated at sites of reactive oxygen and nitrogen species generation. Interestingly, the Agr homologs of coagulase-negative staphylococci, as well as Agr types II and III of S. aureus, lack this C-terminal methionine and have other terminal amino acids instead.
The Agr telesensing system is not the first evidence suggesting that S. aureus has struck a careful balance between virulence and host colonization. The Skaar laboratory (Torres et al., 2007) initially observed, perhaps counterintuitively, that when S. aureus detected the presence of heme (presumably a signal of its presence in an invasive infection) through the HssRS two component system, virulence was down-regulated. Inactivation of this two component system led to increased virulence (Torres et al., 2007). This indicated that in the bloodstream or other site of invasive infection where heme is abundant, S. aureus down regulates the factors necessary for the translation to the virulent form.
Supporting the concept of managed virulence by S. aureus, organisms that much more often colonize than infect, others observed that in conditions where the codY system of was likely to be active, such as in the bloodstream, S. aureus virulence was again attenuated (Camargo and Gilmore, 2008; Majerczyk et al., 2008). The codY global regulatory system of low G+C Gram-positive bacteria connects nutritional status, through sensing extracellular concentrations of branched chain amino acids and intracellular GTP levels, with regulation of metabolic and virulence genes. Further support for the management of S. aureus virulence in the bloodstream derives from the following: Whereas components of the bloodstream induce an imbalance in E. faecalis donor pheromone homeostasis, resulting in the autoaggregation of donor cells and exacerbation of virulence; VLDL and LDL lipoproteins antagonize Agr signaling and induction of virulence, very likely by a direct adsorption mechanism (Peterson et al., 2008).
All of these observations are consistent with an evolutionary model (Camargo and Gilmore, 2008) that proposes that S. aureus down-regulates virulence in a well nourished, immunologically intact host (that is, one capable of oxidatively inactivating the Agr thiolactone within phagocytic cells and clearing S. aureus from the bloodstream before sheer bacterial numbers tip the balance away from the additional suppression of virulence mediated by LDL/VLDL adsorption of the autoinducer, or the self-suppression of virulence by the codY regulon and by the sensing of heme). The net effect is that S. aureus is of limited virulence to a healthy host, but may contribute to the demise of a weakened, malnourished, immune compromised host, such that the microbe is first to the table to benefit from host demise. By culling the herd of weakened individuals, such a model may be net positive in selecting for host fitness, as well as advantageous for the microbe that can proliferate on the culled individual. These observations argue strongly for a much more nuanced view of virulence and the dynamic between hosts and microbes near the fine line that separates commensal from pathogen. Moreover, introduction of strong selective pressures into the human ecology, such the use of antibiotics, may promote the destabilization of this finely balanced host-microbe homeostasis, by selecting for the introduction of mobile genetic elements into the S. aureus genome that have the potential to inadvertently disrupt highly co-evolved gene expression patterns.
Expressing a telesensing signal that is conditionally responsive within various host niches is certainly not unique to S. aureus. Chun et al. (Chun et al., 2004) demonstrated that differentiated human airway epithelia have the ability to inactivate the N-(3-oxododecanoyl)-L-homoserine lactone signal of P. aeruginosa. However, in this case the argument would have to be made that the vast majority of P. aeruginosa human association is in the form of asymptomatic commensal colonization, and that this colonization/carriage, and not pathogenic infection, was the main selective pressure in evolving the C12 lactone signal of P. aeruginosa. While this relationship is clearer for S. aureus, commensal colonization is currently an area of substantial controversy for P. aeruginosa and other microbes, as new microbiome technologies reveal previously unappreciated species associations with the healthy human host (Fierer et al., 2008). Whether inactivating a quorum molecule is a host response (in which case the point would have to be argued that the host is prevailing despite the ability of bacteria to rapidly out-replicate the host and evolve), or a compromise reached between host and microbe, is currently unknown. A conceptually similar question exists in microbial ecology where, for example, several Bacillus species produce a lactonase, named AiiA, that inactivates among other things, acyl homoserine lactones (Dong et al., 2002). Whether this activity nonspecifically inactivates signaling by Gram-negative microbes, or by inactivating signaling, promotes specific microbe-microbe associations between lactonase-producing Bacillus species and Gram-negative microbes that otherwise would not occur, is likely to vary on a case by case basis. Interestingly, it was shown that 3-oxo-N-acylhomoserine lactones spontaneously hydrolyze to antimicrobial, antioxidant, iron-binding tetramic acid derivatives. The production of a lactonase by Bacillus and other organisms may simply represent a form of self defense (Kaufmann et al., 2005). Nevertheless, the tools for using telesensing to foster stable associations between microbe and host (or pathogenic relationships between microbes and host) likely derived from those used to establish cooperative and predatory relationships between microbes in the environment.
B. Telesensing probes that incorporate environmental molecules
1-“Ferrimones”: siderophores and hemophores detect and mediate responses to the presence of iron in the environment
Iron is an essential element for almost all organisms, but is generally not soluble in aerobic environments at neutral pH. Thus, despite its abundance on earth, there is little free iron to satisfy microbial requirements. Iron availability is further limited in mammalian hosts by being complexed to protein carriers, which protect cells against iron toxicity due to Fenton reactions. Pathogens must be able to compete for these limited supplies of available iron to colonize and cause disease.
In response to iron deprivation, many bacteria synthesize and release into the environment compounds that bind ferric iron (siderophores) or heme (hemophores) with high affinity (Wandersman and Delepelaire, 2004). The complexes diffuse back and bind to specific receptors on the cell surface, and the ferri-siderophore or heme is transported into the cell. Thus they serve as both iron acquisition and iron sensing molecules. Regulation of expression of the synthesis and transport of these compounds is accomplished by an iron binding repressor, Fur or DtxR, which recognizes a conserved sequence in the promoters of iron-regulated genes (Hantke, 2001). In the absence of iron, the repressor is inactive and the genes are expressed, but when iron is abundant, the repressor binds DNA and prevents expression of the iron acquisition genes, thus avoiding iron toxicity. Under iron starvation conditions in vitro, a pathogen may express all of its iron transport systems, and many pathogens have large numbers of systems. V. cholerae, for example, has genes for synthesis, secretion and transport of the siderophore vibriobactin (Wyckoff et al., 2007). It also has three transport systems for heme (Mey and Payne, 2001), at least three systems for transport of siderophores made by other bacteria (Wyckoff et al., 2007), and two ferrous and one ferric iron transporter (Wyckoff et al., 2006). The genes encoding these systems make up more than 1% of the V. cholerae genome and many more genes, including some encoding virulence factors, are regulated by the concentration of iron in the cell (Mey et al., 2005). Derepressing all these systems simultaneously is metabolically expensive, and forcing derepression by mutating fur inhibits growth or is lethal (Barton et al., 1996; Mey et al., 2005). The picture that is emerging is one in which the bacteria produce basal levels of the various chelators to probe the environment, and then up regulate or downregulate the various systems depending on which iron sources are available as revealed by which environmental probe is chemically altered. This has been termed ferrimone sensing (Brickman and Armstrong, 2009), and is another well studied class of telesensing.
Bordetella pertussis, the agent of whooping cough, has multiple iron transport systems. Three of these, production and transport of the siderophore alcaligen, transport of the catechol siderophore enterobactin, and heme transport, have been characterized at the genetic level (Brickman and Armstrong, 2009). Each of these is positively regulated by a transcription factor interacting with the cognate, iron-containing compound. This allows the bacterium to sense the iron sources available at a given site within the host and to monitor changes in iron sources over time. Temporal expression patterns of these systems, all of which are required for virulence, have been shown in vivo (Brickman et al., 2008). The siderophore biosynthesis and transport genes are induced early during infection while heme transport is induced late. Alcaligin is secreted, binds iron from the host and is transported into the bacterial cell, where it interacts with the AraC-type transcriptional regulator AlcR, which induces high levels of alcaligin gene expression (Brickman et al., 2001). A second siderophore receptor gene, bfeA, is also up-regulated early in infection. This receptor is induced by the enteric siderophore enterobactin, but its ligand in the respiratory tract appears to be neuroendocrine catecholamines including norepinephrine (Anderson and Armstrong, 2006). The catecholamines can bind iron and can be used as iron sources by B. pertussis. Thus, B. pertussis can monitor and respond to the presence of catecholamine released by the host as a result of damage to the respiratory epithelium. The heme transport system is induced late in infection. Heme binding to the heme receptor BhuR initiates signaling through an N-terminal extension of the receptor that is similar to extensions in proteins involved in iron-inducible extracytoplasmic function (ECF) sigma factor dependent regulators (Vanderpool and Armstrong, 2003). In the absence of heme, basal expression of the bhu operon results from read-through from an upstream promoter in low iron conditions. If heme is present in the environment, heme bound to BhuR results in binding of the anti-sigma membrane sensor HurR which allows release or activation of sigma factor HurI. HurI allows increased expression of BhuR from the bhu promoter.
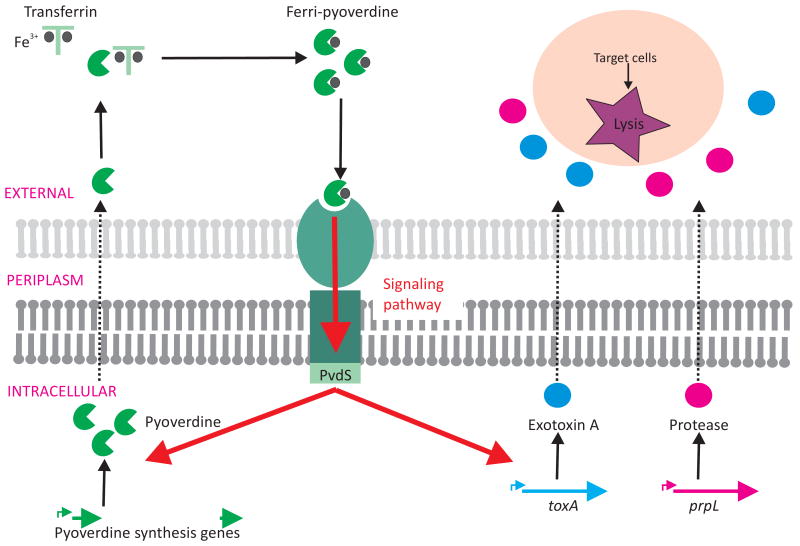
P. aeruginosa secretes two different siderophores – pyoverdine and pyochelin. Pyoverdine contributes to P. aeruginosa virulence in multiple animal models (Meyer et al., 1996; Takase et al., 2000), and has been isolated from sites of P. aeruginosa human infection (Haas et al., 1991). Lamont and co-workers (Beare et al., 2003; Lamont et al., 2002) have shown that pyoverdine can stimulate its own biosynthesis, and also influences production of at least two other P. aeruginosa virulence factors, exotoxin A and PrlP protease. Activation of the expression of these genes is mediated via the alternative sigma factor, PvdS, which recruits RNA polymerase to target promoters (Figure 3).
Figure 3. Pyoverdine “ferrimone” signaling.
Ferri-pyoverdine, having extracted a ferric ion from the environment, complexes with a specific outer-membrane receptor inducing a signaling pathway ultimately sensed by the PvdS alternative sigma factor. Activated PvdS recruits RNA polymerase to target promoters leading to activation of gene expression including genes involved in pyoverdine production, prpL encoding a protease and indirectly toxA coding for the exotoxin A.
Some Gram-negative pathogens secrete hemophores, heme- or hemopexin (a high affinity heme-binding protein in mammalian serum)-binding proteins that act as telesensing molecules for heme. Haemophilus influenzae HxuA is an example of a hemopexin-binding hemophore (Cope et al., 1995). Heme-binding hemophore systems have been identified in Serratia marcescens, P. aeruginosa, and Yersinia pestis, among others. Synthesis of the hemophore, HasA, by S. marcescens is mediated via an autoinduction control mechanism (Cescau et al., 2007; Wandersman and Delepelaire, 2004). The HasA outer membrane receptor, HasR, can bind heme-free (Apo-HasA) and heme-loaded (Holo-HasA) hemophores with similar affinity. However, only Holo-HasA activates has gene transcription (Cescau et al., 2007; Rossi et al., 2003). Since siderophores and hemophores not only deliver nutrients to the microbe, but also differentially induce specific genetic response programs when ligand is bound, they fulfill the criteria for environmentally responsive telesensing molecules.
2-Rhodopseudomonas palustris incorporates potentially host produced molecules into a telesensing signal
The fatty acyl group of fatty acyl-homoserine lactones provides signal specificity, but the variety of signal structures is limited. R. palustris, a Gram-negative plant saprophyte, possesses a LuxRI-like quorum sensing circuit consisting of RpaI and RpaR. Schaefer and co-workers (Schaefer et al., 2008) discovered that rpaI expression increased specifically in the presence of p-coumarate, a phenolic component of plant lignin. Further investigation showed that the acyl-homoserine lactone signal incorporated a p-coumaroyl group into its structure, generating a novel p-coumaroyl-homoserine lactone. RpaI was shown to catalyze the synthesis of p-coumaroyl-homoserine lactone from S-adenosylmethionine and p-coumaryol coenzyme A. Transcriptome analyses demonstrated that expression of 17 genes, including rapI, were significantly altered when bacteria where grown in the presence of either this modified autoinducer or p-coumarate (Cooley et al., 2008). Rather than sending out a telesensing molecule and responding differentially to it after it is altered by the environment, this system first takes up a specific plant metabolite and uses it to generate a telesensing quorum signal, which could thereby mediate specific interactions with a plant host. Thus, the use of p-coumarate results in a signal that perceives both cell-density and an environmental cue. Production of p-coumaroyl derivatives of homoserine lactones is not unique to R. palustris, in that it was found to occur in at least two other species of soil bacteria, Bradyrhizobium s.p and Silicibacter pomeroyi (Schaefer et al., 2008).
C. Telesensing system integration with host signalling
P. aeruginosa: The host responds to microbial signals and the microbe responds to host signals
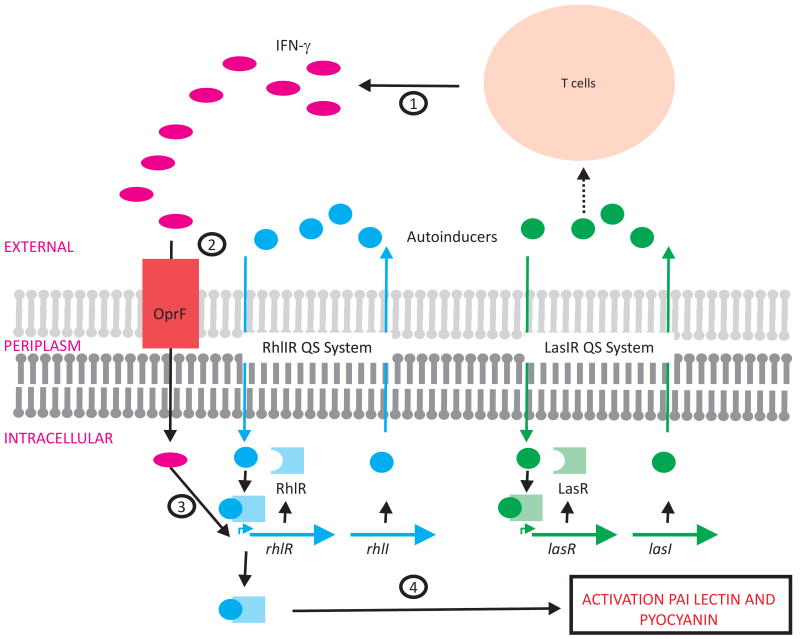
The opportunistic pathogen P. aeruginosa possesses two AHL-type quorum sensing systems, las and rhl, consisting of the LuxRI homologues, LasRI and RhlRI (Whiteley and Greenberg, 2001). Both systems contribute to P. aeruginosa pathogenicity in different animal models (Smith and Iglewski, 2003) by regulating expression of virulence determinants, including the galactophilic P. aeruginosa lectin (LecA) and pyocyanin (de Kievit and Iglewski, 2000).
Human host cells express a variety of pattern recognition receptors and other mechanisms for detecting bacteria. Wu et al. (Wu et al., 2005) hypothesized that bacteria themselves might possess specialized receptors that in turn recognize and respond to the host immune activation. They observed that the upregulation of lecA was substantially enhanced in the presence of exogenous interferon-γ (IFN-γ). Moreover, this effect was specific for this particular cytokine, and was abrogated in rhl mutant strains. IFN-γ was found to bind specifically to OprF, an outer membrane protein, which in turn activated the rhl quorum sensing machinery by an unknown mechanism. The result, however, was increased expression of virulence factors including PAI lectin and pyocyanin (Wu et al., 2005). In addition to its signaling function, the autoinducer produced by the lasRI quorum sensing system was shown to act on host signaling pathways, including stimulating IFN-γ production by T-cells establishing a potential feedback loop (Smith and Iglewski, 2003) (Figure 4). In this case, the microbe induces a host response, to which the microbe responds in a way that induces further host response.
Figure 4. Pseudomonas aeruginosa recognizes the host immune response.
Interferon γ (IFN-γ) activates expression of P. aeruginosa rhlIR QS genes. (1) Cells of the innate immunity system sense bacteria and secrete IFN-γ. Autoinducers produced via the LasIR QS system can induce IFN-γ release. (2) INF-γ binds to the OprF membrane protein leading to (3) activation of the rhl genes by an unknown mechanism. (4) This leads to RhlIR QS system induced activation of virulence factors, such as PAI lectin and pyocyanin.
Conclusions and perspectives
It is increasingly clear that many systems have evolved to be specifically responsive to particular variations in the environment. Bacterial telesensing systems enable a microbe to actively probe the environment beyond the cell surface, and based on changes induced in the environmental probe by environmental factors, respond in a manner that is contextually appropriate. Main subcategories of telesensing include 1) quorum sensing in its many forms, 2) iron or “ferrimone” signaling, and 3) variations on these themes often related to virulence factor expression by opportunists. Contextually sensitive telesensing systems are diverse and occur in both Gram-positive and Gram-negative bacteria. These systems are central to bacterial decision making, promoting host/microbe stability in some cases, and promoting parasitism in others, especially in situations where the microbe senses stress or host compromise. Telesensing inputs are integrated into complex multilayered signal transduction networks that overlap with additional pathways involved in environmental sensing and response. Examples of multilayered systems include Vibrio harveyi bioluminescence, which is not only cell density dependent, but is also influenced by MetR and CRP nutritional sensors (Chatterjee et al., 2002). Other organisms, such as Xanthomonas campestris, regulate virulence by both quorum sensing and the RavS/RavR two-component regulatory system (He et al., 2009). It is clear that tremendous complexity and sophistication has evolved in the mechanisms microbes possess for sensing the environment, even at a distance, and making contextually appropriate decisions. More research will be required to fully understand the algorithms used by microbes to integrate sensory inputs and make the contextually appropriate decisions that lead to perpetuation of the species.
Acknowledgments
Portions of the work described in this manuscript were supported by NIH grants AI072360, EY017381, and EY08289 (MSG), and AI016935 and AI050669 (SMP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson MT, Armstrong SK. The Bordetella bfe system: growth and transcriptional response to siderophores, catechols, and neuroendocrine catecholamines. J Bacteriol. 2006;188:5731–5740. doi: 10.1128/JB.00495-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft EA. Antimicrobial resistance: it's not just for hospitals. Jama. 2007;298:1803–1804. doi: 10.1001/jama.298.15.1803. [DOI] [PubMed] [Google Scholar]
- Barton HA, Johnson Z, Cox CD, Vasil AI, Vasil ML. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol. 1996;21:1001–1017. doi: 10.1046/j.1365-2958.1996.381426.x. [DOI] [PubMed] [Google Scholar]
- Be'er A, Zhang HP, Florin EL, Payne SM, Ben-Jacob E, Swinney HL. Deadly competition between sibling bacterial colonies. Proc Natl Acad Sci U S A. 2009;106:428–433. doi: 10.1073/pnas.0811816106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare PA, For RJ, Martin LW, Lamont IL. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol Microbiol. 2003;47:195–207. doi: 10.1046/j.1365-2958.2003.03288.x. [DOI] [PubMed] [Google Scholar]
- Bollinger N, Hassett DJ, Iglewski BH, Costerton JW, McDermott TR. Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J Bacteriol. 2001;183:1990–1996. doi: 10.1128/JB.183.6.1990-1996.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Kim U, Bartkowski W, Gunsalus RP, Overley AM, Lyell NL, Visick KL, Stabb EV. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol Microbiol. 2007;65:538–553. doi: 10.1111/j.1365-2958.2007.05809.x. [DOI] [PubMed] [Google Scholar]
- Brickman TJ, Armstrong SK. Temporal signaling and differential expression of Bordetella iron transport systems: the role of ferrimones and positive regulators. Biometals. 2009;22:33–41. doi: 10.1007/s10534-008-9189-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Hanawa T, Anderson MT, Suhadolc RJ, Armstrong SK. Differential expression of Bordetella pertussis iron transport system genes during infection. Mol Microbiol. 2008;70:3–14. doi: 10.1111/j.1365-2958.2008.06333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman TJ, Kang HY, Armstrong SK. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J Bacteriol. 2001;183:483–489. doi: 10.1128/JB.183.2.483-489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo IL, Gilmore MS. Staphylococcus aureus--probing for host weakness? J Bacteriol. 2008;190:2253–2256. doi: 10.1128/JB.00043-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cescau S, Cwerman H, Letoffe S, Delepelaire P, Wandersman C, Biville F. Heme acquisition by hemophores. Biometals. 2007;20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- Chandler JR, Hirt H, Dunny GM. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc Natl Acad Sci U S A. 2005;102:15617–15622. doi: 10.1073/pnas.0505545102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee J, Miyamoto CM, Zouzoulas A, Lang BF, Skouris N, Meighen EA. MetR and CRP bind to the Vibrio harveyi lux promoters and regulate luminescence. Mol Microbiol. 2002;46:101–111. doi: 10.1046/j.1365-2958.2002.03128.x. [DOI] [PubMed] [Google Scholar]
- Chun CK, Ozer EA, Welsh MJ, Zabner J, Greenberg EP. Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc Natl Acad Sci U S A. 2004;101:3587–3590. doi: 10.1073/pnas.0308750101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell DB, An FY, Flannagan SE, Antiporta M, Dunny GM. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol Microbiol. 2000;35:246–247. doi: 10.1046/j.1365-2958.2000.01687.x. [DOI] [PubMed] [Google Scholar]
- Coburn PS, Gilmore MS. The Enterococcus faecalis cytolysin: a novel toxin active against eukaryotic and prokaryotic cells. Cell Microbiol. 2003;5:661–669. doi: 10.1046/j.1462-5822.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- Coburn PS, Pillar CM, Jett BD, Haas W, Gilmore MS. Enterococcus faecalis senses target cells and in response expresses cytolysin. Science. 2004;306:2270–2272. doi: 10.1126/science.1103996. [DOI] [PubMed] [Google Scholar]
- Cooley M, Chhabra SR, Williams P. N-Acylhomoserine lactone-mediated quorum sensing: a twist in the tail and a blow for host immunity. Chem Biol. 2008;15:1141–1147. doi: 10.1016/j.chembiol.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Cope LD, Yogev R, Muller-Eberhard U, Hansen EJ. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kievit TR, Iglewski BH. Bacterial quorum sensing in pathogenic relationships. Infect Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Gusti AR, Zhang Q, Xu JL, Zhang LH. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl Environ Microbiol. 2002;68:1754–1759. doi: 10.1128/AEM.68.4.1754-1759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulla G, Lindow SE. Quorum size of Pseudomonas syringae is small and dictated by water availability on the leaf surface. Proc Natl Acad Sci U S A. 2008;105:3082–3087. doi: 10.1073/pnas.0711723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution. Philos Trans R Soc Lond B Biol Sci. 2007;362:1185–1193. doi: 10.1098/rstb.2007.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkin SK, Hageman JC, Morrison M, Sanza LT, Como-Sabetti K, Jernigan JA, Harriman K, Harrison LH, Lynfield R, Farley MM. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005;352:1436–1444. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
- Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, Calderwood SB, Ausubel FM. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A. 2001;98:10892–10897. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genta G, Perino MA. Teleoperation support for early human planetary missions. Ann N Y Acad Sci. 2005;1065:271–284. doi: 10.1196/annals.1370.008. [DOI] [PubMed] [Google Scholar]
- Gilmore MS, Haas W. The selective advantage of microbial fratricide. Proc Natl Acad Sci U S A. 2005;102:8401–8402. doi: 10.1073/pnas.0503828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiral S, Mitchell TJ, Martin B, Claverys JP. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A. 2005;102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B, Kraut J, Marks J, Zanker SC, Castignetti D. Siderophore presence in sputa of cystic fibrosis patients. Infect Immun. 1991;59:3997–4000. doi: 10.1128/iai.59.11.3997-4000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas W, Shepard BD, Gilmore MS. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature. 2002;415:84–87. doi: 10.1038/415084a. [DOI] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr Opin Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Hardie KR, Heurlier K. Establishing bacterial communities by ‘word of mouth’: LuxS and autoinducer 2 in biofilm development. Nat Rev Microbiol. 2008;6:635–643. doi: 10.1038/nrmicro1916. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Chatterjee A, Cui Y, Chatterjee AK. Elevated temperature enhances virulence of Erwinia carotovora subsp. carotovora strain EC153 to plants and stimulates production of the quorum sensing signal, N-acyl homoserine lactone, and extracellular proteins. Appl Environ Microbiol. 2005;71:4655–4663. doi: 10.1128/AEM.71.8.4655-4663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YW, Boon C, Zhou L, Zhang LH. Co-regulation of Xanthomonas campestris virulence by quorum sensing and a novel two-component regulatory system RavS/RavR. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06617.x. [DOI] [PubMed] [Google Scholar]
- Horswill AR, Stoodley P, Stewart PS, Parsek MR. The effect of the chemical, biological, and physical environment on quorum sensing in structured microbial communities. Anal Bioanal Chem. 2007;387:371–380. doi: 10.1007/s00216-006-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen V, Lons D, Zaoui C, Bredenbruch F, Meissner A, Dieterich G, Munch R, Haussler S. RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J Bacteriol. 2006;188:8601–8606. doi: 10.1128/JB.01378-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann GF, Sartorio R, Lee SH, Rogers CJ, Meijler MM, Moss JA, Clapham B, Brogan AP, Dickerson TJ, Janda KD. Revisiting quorum sensing: Discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc Natl Acad Sci U S A. 2005;102:309–314. doi: 10.1073/pnas.0408639102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kicheva A, Pantazis P, Bollenbach T, Kalaidzidis Y, Bittig T, Julicher F, Gonzalez-Gaitan M. Kinetics of morphogen gradient formation. Science. 2007;315:521–525. doi: 10.1126/science.1135774. [DOI] [PubMed] [Google Scholar]
- Kiil K, Ferchaud JB, David C, Binnewies TT, Wu H, Sicheritz-Ponten T, Willenbrock H, Ussery DW. Genome update: distribution of two-component transduction systems in 250 bacterial genomes. Microbiology. 2005;151:3447–3452. doi: 10.1099/mic.0.28423-0. [DOI] [PubMed] [Google Scholar]
- Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. Siderophore-mediated signaling regulates virulence factor production in Pseudomonasaeruginosa. Proc Natl Acad Sci U S A. 2002;99:7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latour X, Diallo S, Chevalier S, Morin D, Smadja B, Burini JF, Haras D, Orange N. Thermoregulation of N-acyl homoserine lactone-based quorum sensing in the soft rot bacterium Pectobacterium atrosepticum. Appl Environ Microbiol. 2007;73:4078–4081. doi: 10.1128/AEM.02681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazdunski AM, Ventre I, Sturgis JN. Regulatory circuits and communication in Gram-negative bacteria. Nat Rev Microbiol. 2004;2:581–592. doi: 10.1038/nrmicro924. [DOI] [PubMed] [Google Scholar]
- Lyon GJ, Novick RP. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides. 2004;25:1389–1403. doi: 10.1016/j.peptides.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol. 2008;190:2257–2265. doi: 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JK, Hirt H, Waters CM, Tripp TJ, Dunny GM, Schlievert PM. Antibodies to a surface-exposed, N-terminal domain of aggregation substance are not protective in the rabbit model of Enterococcus faecalis infective endocarditis. Infect Immun. 2001;69:3305–3314. doi: 10.1128/IAI.69.5.3305-3314.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan SJ, Barnard AM, Bosgelmez G, Sebaihia M, Simpson NJ, Thomson NR, Todd DE, Welch M, Whitehead NA, Salmond GP. Carbapenem antibiotic biosynthesis in Erwinia carotovora is regulated by physiological and genetic factors modulating the quorum sensing-dependent control pathway. Mol Microbiol. 2005;55:526–545. doi: 10.1111/j.1365-2958.2004.04397.x. [DOI] [PubMed] [Google Scholar]
- Mey AR, Payne SM. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol Microbiol. 2001;42:835–849. doi: 10.1046/j.1365-2958.2001.02683.x. [DOI] [PubMed] [Google Scholar]
- Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect Immun. 2005;73:8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G, Haas D. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J Bacteriol. 2000;182:6940–6949. doi: 10.1128/jb.182.24.6940-6949.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MM, Mack JL, Hall PR, Alsup AA, Alexander SM, Sully EK, Sawires YS, Cheung AL, Otto M, Gresham HD. Apolipoprotein B Is an innate barrier against invasive Staphylococcus aureus infection. Cell Host Microbe. 2008;4:555–566. doi: 10.1016/j.chom.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi MS, Paquelin A, Ghigo JM, Wandersman C. Haemophore-mediated signal transduction across the bacterial cell envelope in Serratia marcescens: the inducer and the transported substrate are different molecules. Mol Microbiol. 2003;48:1467–1480. doi: 10.1046/j.1365-2958.2003.03516.x. [DOI] [PubMed] [Google Scholar]
- Rothfork JM, Timmins GS, Harris MN, Chen X, Lusis AJ, Otto M, Cheung AL, Gresham HD. Inactivation of a bacterial virulence pheromone by phagocyte-derived oxidants: new role for the NADPH oxidase in host defense. Proc Natl Acad Sci U S A. 2004;101:13867–13872. doi: 10.1073/pnas.0402996101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, Harwood CS. A new class of homoserine lactone quorum-sensing signals. Nature. 2008;454:595–599. doi: 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- Shankar N, Baghdayan AS, Gilmore MS. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature. 2002;417:746–750. doi: 10.1038/nature00802. [DOI] [PubMed] [Google Scholar]
- Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- Surette MG, Bassler BL. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci U S A. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen SL, Tu KC, Bassler BL. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. Embo J. 2009;28:429–439. doi: 10.1038/emboj.2008.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Swem DL, Wingreen NS, Bassler BL. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell. 2008;134:461–473. doi: 10.1016/j.cell.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase H, Nitanai H, Hoshino K, Otani T. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect Immun. 2000;68:1834–1839. doi: 10.1128/iai.68.4.1834-1839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd EW. A comparative serological study of streptolusins derived from human and from animal infections, with notes on pneumococcal hemolyson, telanolysin and staphylococcus toxin. J Pathol Bacteriol. 1934;39:299–321. [Google Scholar]
- Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, Anderson KL, Dunman PM, Joyce S, Skaar EP. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe. 2007;1:109–119. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK, Armstrong SK. Heme-responsive transcriptional activation of Bordetella bhu genes. J Bacteriol. 2003;185:909–917. doi: 10.1128/JB.185.3.909-917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner VE, Frelinger JG, Barth RK, Iglewski BH. Quorum sensing: dynamic response of Pseudomonas aeruginosa to external signals. Trends Microbiol. 2006;14:55–58. doi: 10.1016/j.tim.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Greenberg EP. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J Bacteriol. 2001;183:5529–5534. doi: 10.1128/JB.183.19.5529-5534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P, Winzer K, Chan WC, Camara M. Look who's talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond B Biol Sci. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth R. Sex pheromones and gene transfer in Enterococcus faecalis. Res Microbiol. 2000;151:493–496. doi: 10.1016/s0923-2508(00)00163-7. [DOI] [PubMed] [Google Scholar]
- Wu L, Estrada O, Zaborina O, Bains M, Shen L, Kohler JE, Patel N, Musch MW, Chang EB, Fu YX, et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005;309:774–777. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
- Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J Bacteriol. 2006;188:6515–6523. doi: 10.1128/JB.00626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff EE, Mey AR, Payne SM. Iron acquisition in Vibrio cholerae. Biometals. 2007;20:405–416. doi: 10.1007/s10534-006-9073-4. [DOI] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol. 2003;6:191–197. doi: 10.1016/s1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]