Abstract
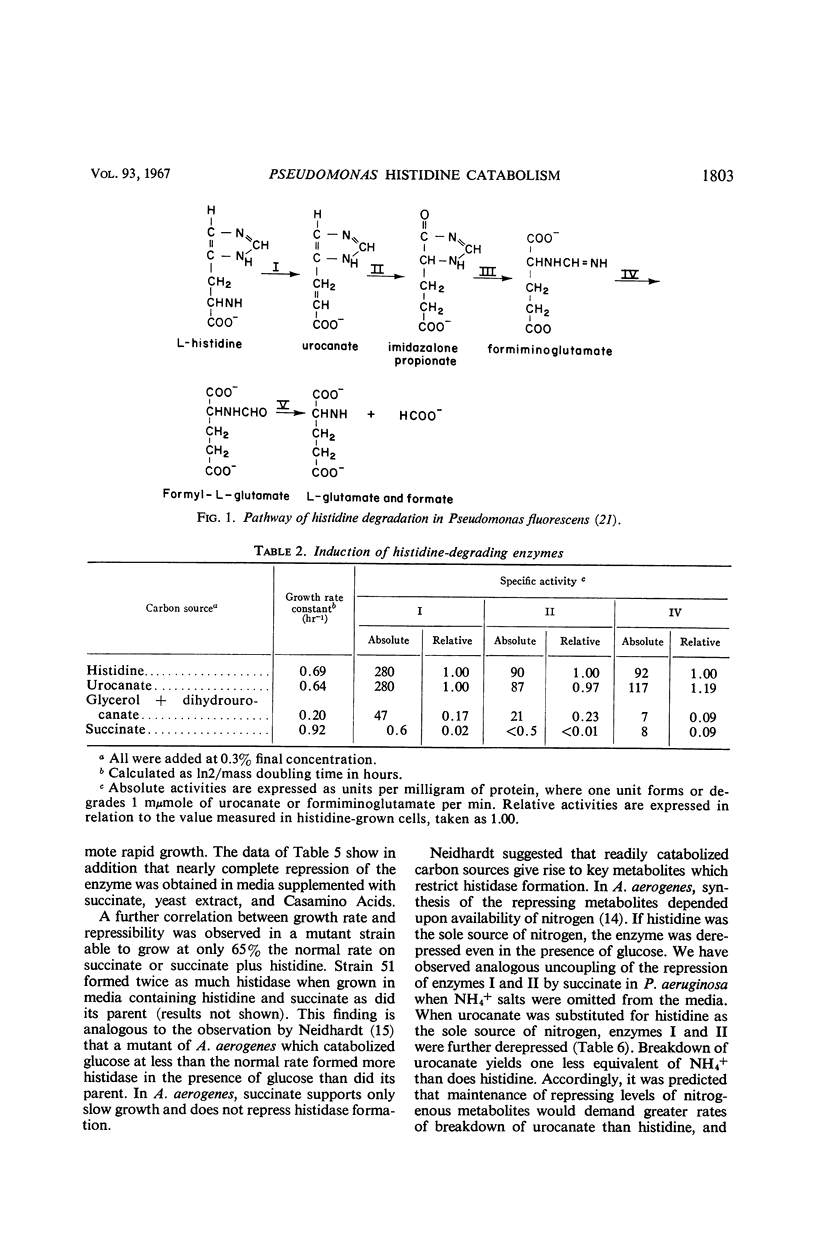
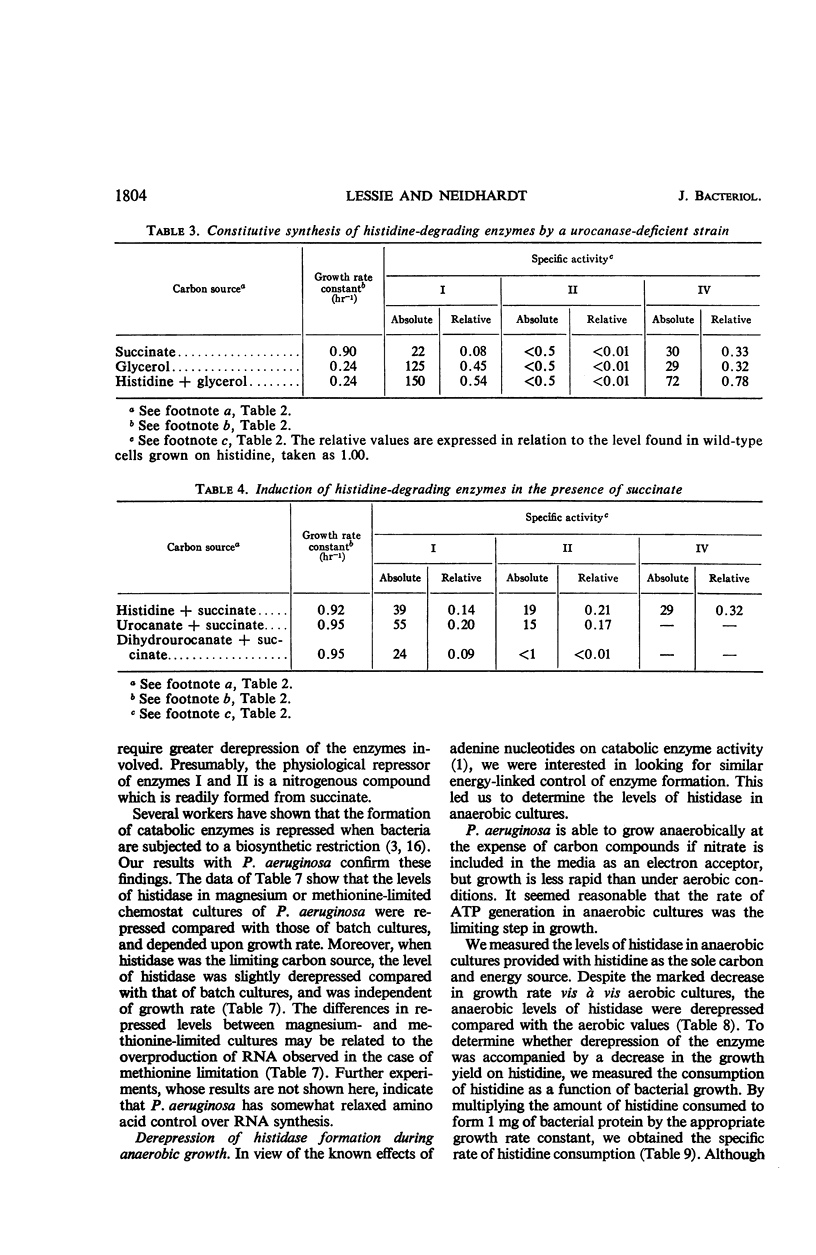
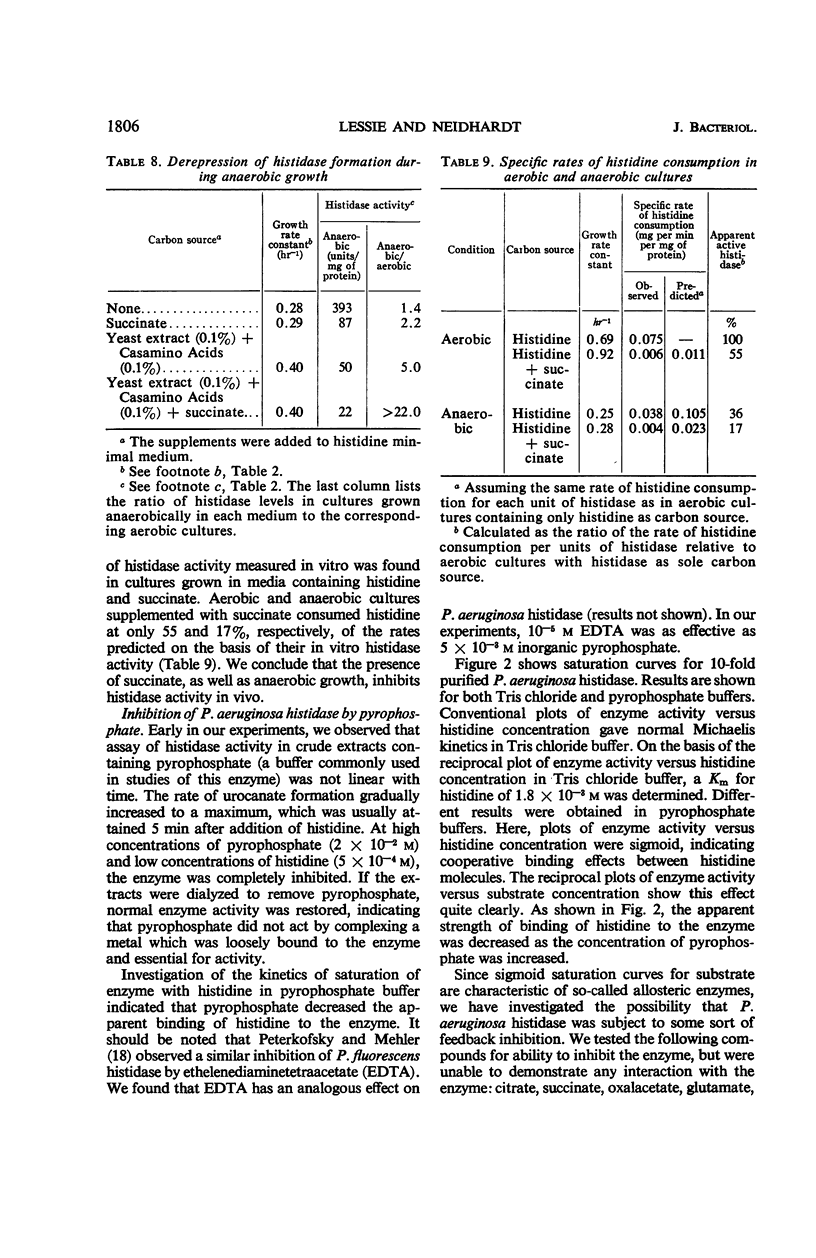
Histidine ammonia lyase (histidase), urocanase, and the capacity to degrade formiminoglutamate, which are respectively involved in steps I, II, and IV in the catabolism of histidine, were induced during growth of Pseudomonas aeruginosa on histidine or urocanate, and were formed gratuitously in the presence of dihydro-urocanate. Urocanase-deficient bacteria formed enzymes I and IV constitutively; presumably they accumulate enough urocanate from the breakdown of endogenous histidine to induce formation of the pathway. Urocanate did not satisfy the histidine requirement of a histidine auxotroph, indicating that it probably acted as an inducer without being converted to histidine. The results imply that urocanate is the physiological inducer of the histidine-degrading enzymes in P. aeruginosa. Enzymes of the pathway were extremely sensitive to catabolite repression; enzymes I and II, but not IV, were coordinately repressed. Our results suggest a specific involvement of nitrogenous metabolites in the repression. Mutant bacteria with altered sensitivity to repression were obtained. The molecular weight of partially purified histidase was estimated at 210,000 by sucrose gradient centrifugation. Its Km for histidine was 2 × 10−3 m in tris(hydroxymethyl)aminomethane chloride buffer. Sigmoid saturation curves were obtained in pyrophosphate buffer, indicating that the enzyme might have multiple binding sites for histidine. Under certain conditions, histidase appeared to be partially inactive in vivo. These findings suggest that some sort of allosteric interaction involving histidase may play a role in governing the operation of the pathway of histidine catabolism.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- HARTWELL L. H., MAGASANIK B. THE MOLECULAR BASIS OF HISTIDASE INDUCTION IN BACILLUS SUBTILIS. J Mol Biol. 1963 Oct;7:401–420. doi: 10.1016/s0022-2836(63)80033-9. [DOI] [PubMed] [Google Scholar]
- Jorpes E. The colorimetric determination of histidine. Biochem J. 1932;26(5):1507–1511. doi: 10.1042/bj0261507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessie T. G. RNA metabolism of Rhodopseudomonas spheroides during preferential photopigment synthesis. J Gen Microbiol. 1965 Oct;41(1):37–45. doi: 10.1099/00221287-41-1-37. [DOI] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Magasanik B., Lund P., Neidhardt F. C., Schwartz D. T. Induction and repression of the histidine-degrading enzymes in Aerobacter aerogenes. J Biol Chem. 1965 Nov;240(11):4320–4324. [PubMed] [Google Scholar]
- NEIDHARDT F. C., MAGASANIK B. Reversal of the glucose inhibition of histidase biosynthesis in Aerobacter aerogenes. J Bacteriol. 1957 Feb;73(2):253–259. doi: 10.1128/jb.73.2.253-259.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]


