Abstract
Changes in gene expression may represent an important mode of human adaptation. However, to date, there are relatively few known examples in which selection has been shown to act directly on levels or patterns of gene expression. In order to test whether single nucleotide polymorphisms (SNPs) that affect gene expression in cis are frequently targets of positive natural selection in humans, we analyzed genome-wide SNP and expression data from cell lines associated with the International HapMap Project. Using a haplotype-based test for selection that was designed to detect incomplete selective sweeps, we found that SNPs showing signals of selection are more likely than random SNPs to be associated with gene expression levels in cis. This signal is significant in the Yoruba (which is the population that shows the strongest signals of selection overall) and shows a trend in the same direction in the other HapMap populations. Our results argue that selection on gene expression levels is an important type of human adaptation. Finally, our work provides an analytical framework for tackling a more general problem that will become increasingly important: namely, testing whether selection signals overlap significantly with SNPs that are associated with phenotypes of interest.
Keywords: population genetics, recent positive selection, eQTL mapping, humans, iHS
Introduction
Mutations in cis-regulatory regions can produce precise changes in gene function by changing the expression, timing, or location of gene expression. Therefore, it seems likely that changes in cis-regulation might be an important mode of adaptive evolution, and indeed, there are now several known examples of this kind of adaptation (Wray [2007]; but see also Hoekstra and Coyne [2007]). Examples in humans include mutations upstream of the lactase gene that cause lactase production in the intestine to persist into adulthood (Bersaglieri et al. 2004; Enattah et al. 2004; Tishkoff et al. 2007) and the Duffy-null mutation that stops expression of the DARC receptor in erythrocytes as a defense against Plasmodium vivax (Hamblin and Di Rienzo 2000). Other examples include selection on cis regulation of human prodynorphin (Rockman et al. 2005) and selection on regulatory variation at the human factor VII locus (Hahn et al. 2004). However, genome-scale studies of the evolution of expression in humans have been hindered by our limited knowledge of which sequences are actually regulatory. Recent studies have reported instances of rapid evolution of either conserved noncoding sequences or promoter regions, but in most cases, it is difficult to connect sequence changes to biological function (Pollard et al. 2006; Prabhakar et al. 2006; Haygood et al. 2007; Kim and Pritchard 2007).
As an alternative way forward, we used recent advances in expression quantitative trait locus (eQTL) mapping that allow identification of large numbers of single nucleotide polymorphisms (SNPs) that are strongly associated with gene expression levels, using data from the HapMap lymphoblast cell lines (Morley et al. 2004; Dixon et al. 2007; Stranger, Forrest, et al. 2007; Stranger, Nica, et al. 2007; Gilad et al. 2008; Veyrieras et al. 2008). We set out to test whether such eQTL signals are frequent targets of positive selection. Our essential logic was that if eQTLs are rarely targets of positive selection, then eQTLs should be independent of selection signals. In contrast, if expression changes that can be detected in lymphoblast cell lines are frequently adaptive, then there should be an enrichment for eQTLs among SNPs that show evidence of positive selection.
More broadly, this type of approach can potentially enable us to link selection signals to particular genes and molecular phenotypes. One of the important next steps in genome-wide studies of selection is to understand how selected alleles affect molecular or organism-level phenotypes. By overlaying large-scale data sets of QTLs for gene expression or other phenotypes (including disease studies), it may be possible to link more signals of selection with function. In this paper, we lay out an analytical framework for testing for an enrichment of signals of selection among a particular class of SNPs (e.g., eQTLs or disease-associated SNPs), while controlling for important confounders.
Detecting Recent Selection
To identify signals of recent or ongoing natural selection on variants that are currently polymorphic within populations (and hence can be eQTLs), we calculated the “integrated haplotype score” (iHS) statistic (Voight et al. 2006) for SNPs in the HapMap Phase II data. The iHS is one of a number of measures introduced to detect signatures of very recent selection on variants that have not yet reached fixation (Hudson et al. 1994; Sabeti et al. 2002, 2007; Wang et al. 2006; Tang et al. 2007).
The iHS statistic aims to identify SNPs for which one allele has changed frequency very rapidly, a hallmark of strong selection. It does this by comparing the extent of haplotype homozygosity on haplotypes carrying the ancestral and derived alleles, respectively, at a SNP. The presence of an unusual difference in homozygosity between the two alleles can be an indicator of selection (Hudson et al. 1994; Sabeti et al. 2002). Several lines of evidence indicate that iHS is effective at identifying instances of strong, recent selection in which the selected allele has not yet reached fixation (Voight et al. 2006). Our previous analysis found that the iHS signals are most reliable in the Yoruba population, from Ibadan, Nigeria (YRI; Voight et al. 2006). This is to be expected because bottlenecks—as experienced by the non-African populations—reduce the power of this type of approach (Teshima et al. 2006; Sabeti et al. 2007).
The iHS statistic is constructed to have an approximately standard normal distribution for all allele frequencies. A value of iHS ≪ 0 at an SNP indicates that the derived allele is on an unusually long haplotype; iHS ≫ 0 indicates that the ancestral allele is on an unusually long haplotype. Therefore, for a standard sweep model in which the derived allele increases in frequency rapidly, we expect the selected site to have a negative iHS value. In simulations, we find that selected sites are often surrounded by numerous SNPs with either strongly positive or negative iHS values and that the selected site generally does not have the most negative iHS value (Voight et al. [2006] and Kudaravalli S, unpublished data). Therefore, most of our analysis below focuses on SNPs that have an extreme iHS value, either positive or negative and that lie within a cluster of other SNPs that also have |iHS| > 2. We consider that such SNPs are likely to lie on the same haplotype as the selected site.
Note that our approach does not provide formal P values for candidate selection signals because it is difficult to simulate a fully accurate null model (Voight et al. 2006). Instead, we focus on SNPs that lie in the tail of the overall genome-wide distribution of iHS, with the view that these are likely to be enriched for true selection signals (Teshima et al. 2006). We will evaluate whether such SNPs are more likely than random SNPs to be eQTLs.
Methods
HapMap Genotype Data
All analyses were based on the HapMap Project Phase II/rel#21 datafiles (http://www.hapmap.org) (International HapMap Consortium 2007). For the CEPH European (CEU) and YRI samples, we analyzed the data from the 60 unrelated parents. Due to their close genetic similarity and in order to have a single larger sample, we pooled the Han Chinese from Beijing and Japanese from Tokyo samples to form a single analysis panel of 90 unrelated Asian individuals, denoted here as the “ASN” sample. Haplotype phase estimation for all the data was performed by the HapMap consortium using Phase 2.0 (Stephens and Scheet 2005; International HapMap Consortium 2007). The phasing procedure imputed all missing genotypes at SNPs with < 20% missing data. Our analyses were restricted to the autosomes. In total, we analyzed 2,419,983, 2,557,252, and 2,856,346 SNPs for the ASN, CEU, and YRI populations, respectively.
SNP and Gene Annotation Information
Gene annotation information was obtained from the RefSeq database (Pruitt et al. 2007). This information was primarily used for obtaining the gene start and gene end coordinates. Where required, genome coordinates were converted from NCBI build 36 (hg18) to build 35 (hg17) using the Batch Coordinate Conversion tool available at the University of California-Santa Cruz (UCSC) Web browser (Karolchik et al. 2008).
Estimating Ancestral States of SNPs
Ancestral states for all SNPs were estimated using whole-genome human–chimpanzee alignments from the UCSC database (Karolchik et al. 2008). Based on the physical position of the SNP in the human genome (Build hg17), the allele at the corresponding position in the chimp genome (Build pantro2) was obtained. If the human SNP position aligned to missing data in the chimpanzee genome or if the chimpanzee allele did not match either human allele, then the corresponding SNP was excluded from further analysis.
Calculation of |iHS|
|iHS| values were calculated separately in each population using methods described previously (Voight et al. 2006). We estimated recombination rates separately for each HapMap population as described previously (Voight et al. 2006). We did not compute |iHS| for SNPs without an estimated ancestral state, or whose population minor allele frequency is <5%, or for some SNPs that are close to chromosome ends or large regions without SNPs (Voight et al. 2006). The total numbers of autosomal SNPs with valid |iHS| scores were 2,143,201 for CEU, 2,383,208 for YRI, and 1,966,892 for ASN. The locations of iHS peaks agreed very closely with the earlier Phase I results (supplementary information, Supplementary Material online).
Criteria for Identifying SNPs as Lying in Selection Signals
For all analyses in the main text, we require that SNPs have |iHS| > 2 to be considered as possessing candidate selection signals. Additionally, for the logistic regression and hierarchical model analyses (and the data in red in fig. 2), we required that candidate selected SNPs also lie within “clusters” of other high iHS SNPs. Specifically, for each SNP, we considered a window of 151 consecutive SNPs centered on the SNP of interest (75 SNPs on either side). We counted the proportion of SNPs within this window for which |iHS| > 2 and considered the window to be of interest if this proportion lies within the top 5% of all windows genome-wide for that population. (For other thresholds in the Supplementary Material online, see results.) About 1.5% of all the SNPs meet these criteria. For SNPs that lie close to chromosome ends, the proportion of SNPs with |iHS| > 2 was calculated based on the maximum possible window size, from a minimum of 75 SNPs up to 151 SNPs.
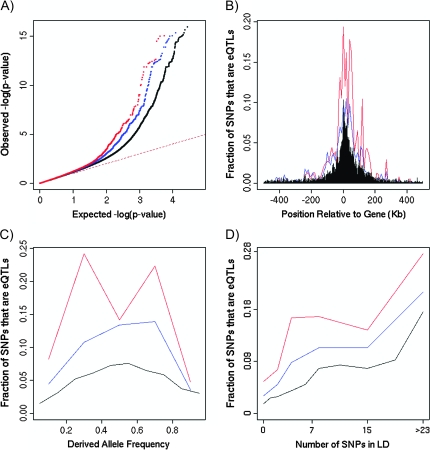
FIG. 2.—
The abundance of eQTL signals in SNPs with and without evidence for selection (YRI data). In each plot, the red data correspond to SNPs with strong evidence for selection (|iHS| > 2 and surrounded by an unusual cluster of other high |iHS| SNPs); blue data are for SNPs with |iHS| > 2; and black data are for all SNPs. (A) Quantile–quantile plots of the distributions of −log10(P values) obtained from testing the expression levels at each gene for association with nearby SNPs. The dashed line indicates the expected distribution of P values if there were no true associations between SNPs and gene expression levels. Notice that SNPs with high |iHS| (red and blue data) show a higher rate of significant P values compared with SNPs without a signal of selection. (B) SNPs with high |iHS| show an enrichment for eQTLs at various distances from the transcription start site. (C) SNPs with high iHS tend to be enriched for eQTLs after controlling for allele frequency. The enrichment may be highest in the frequency ranges where iHS has the greatest power (roughly 50–80%; Voight et al. 2006). (D) SNPs with high iHS show generally higher rates of eQTLs after controlling for LD levels, as measured by the number of SNPs in high LD with the SNP in question (r2 > 0.8). For analogous plots of the other two populations, see the Supplementary Material online.
Gene Expression Data
We used gene expression levels that were measured previously in lymphoblastoid cell lines from all 210 unrelated HapMap individuals, using Illumina's human whole-genome expression array (WG-6 version 1) (Stranger, Forrest, et al. 2007). We downloaded the data that were normalized first by quantile normalization within replicates and then median normalized across all HapMap individuals (Stranger, Forrest, et al. 2007). The expression data for each probe were further modified by quantile normalization within each population to bring the data to a standard normal distribution for each probe in each population.
Mapping Illumina Probe Sequences onto the Human Genome
To determine the genes associated with the probe sequences on the Illumina chip, each probe sequence was aligned against whole-genome RNA sequences using BLAT (Kent 2002). RNA sequences were downloaded from RefSeq (Pruitt et al. 2007). We aligned to RNA sequences instead of the human reference sequence to account for situations where a probe crosses exon boundaries. Each probe was 50 bp long. Probes that aligned to multiple genes with >80% sequence matching were dropped from our analysis. In total, this procedure led to a total of 19,536 probes in 16,155 unique autosomal genes for analysis.
Testing for Association between SNPs and Gene Expression Levels
For each gene with expression data, we tested all SNPs that are either inside the gene or within 100 kb of the gene's transcription start or end site and with minor allele frequency >5% in the relevant population, for association with the measured expression level (separately at each probe). The analysis was performed separately in each population.
We also performed the analysis using larger windows around the target gene, such as including all SNPs within 500 kb of the gene (for other cutoffs, see the supplementary information, Supplementary Material online). However, the false discovery rate (FDR) for eQTLs detected outside 100 kb is very high and so we focus on this smaller window size. Given that these eQTLs are tightly clustered around the corresponding genes, we will refer to these as cis-eQTLs, though we do not have direct evidence of a true cis-acting mechanism.
We tested for association between each SNP and gene expression level using a standard linear regression model with the genotypes being the predictor variable and with the quantile normalized gene expression level being the dependent variable. The genotypes were coded as 0, 1, and 2 (corresponding to the number of copies of the minor allele), which means that we assume that the average quantile normalized expression levels of heterozygotes are halfway between the expression levels of each homozygote. As described above, we used the phased HapMap data with all missing genotypes imputed.
Any SNP that was significantly associated with the expression profile at P < 10−4 was considered an eQTL in that population. The gene-level FDR associated with this significance threshold is low (FDR < 18%) in all three population groups (see supplementary information, Supplementary Material online). Simulations indicate that our P values have the correct distribution under the null hypothesis for minor allele frequencies ≥10% and are slightly conservative at lower allele frequencies (supplementary information, Supplementary Material online). When multiple probes mapped to a single gene (<10% of genes), we tested for association separately with each probe. For our analysis, we then used only the probe that had the largest number of associated SNPs (at P < 10−4). We also performed separate analyses based on selecting the probe with the single most “significant” association, and the overall results were very similar (data not shown).
Odds Ratio
We used the logistic regression and hierarchical models to estimate an odds ratio that measures the relative enrichment of eQTLs among SNPs with selection signals, compared with those without. The odds ratio is defined as
| (1) |
where in a slight abuse of notation, “eQTL,” and “not eQTL,” are used as short hand to indicate that an SNP is, or is not, significantly associated with expression of a prespecified gene (at P < 10−4) and where “iHS,” and “not iHS,” indicate that an SNP does, or does not, have a significant selection signal as defined above.
Logistic Regression Model
To test whether there is an enrichment of eQTLs among iHS signals, we implemented a logistic regression model, as follows. The model is used to predict, independently for each gene, population, and for every SNP within 100 kb of that gene, whether or not that SNP is an eQTL, as a function of the selection information and other potential explanatory variables:
| (2) |
Here, I(eQTL = 1) is an indicator function that is 1 if an SNP is significantly associated with expression for the gene in question (at P < 10−4) and is otherwise 0; I(iHSsig = 1) is an indicator function that is 1 if the SNP shows signals of selection (as defined above) and is otherwise 0; LD is a measure of the extent of LD around the SNP in question; distTSS and distTES are the distances to the gene's transcription start and end sites, respectively, measured in base pairs; MAF is the minor allele frequency of the SNP in the relevant population; and Logit[x] is the function log[Pr(x)/Pr(1 − x)]. The β variables are the coefficients of the logistic regression. If β1 > 0, this implies an enrichment effect for eQTLs among SNPs with selection signals. Note that exp(β1) estimates the odds ratio for the effect of selection (defined above), while controlling for the other explanatory variables.
The explanatory variables LD, distTSS, distTES, and MAF were included because we observed that the locations of eQTLs are significantly associated with each of these variables. Among these, LD is the most plausible confounder with selection because the other measures show only minor correlation with the probability that an SNP shows signals of selection. For this reason, we investigated various measures of the extent of LD. Broadly speaking, we used two types of approaches to measure LD. One type of measure, used for the main paper, assesses how many other SNPs are in strong LD with the target SNP. (To be precise, we counted how many HapMap SNPs have r2 > 0.8 with the target SNP, calculated in a 500-kb window around the target SNP.) The rationale is that an SNP in strong LD with a large number of other SNPs has an increased probability of being associated with gene expression because it has a greater probability of being in LD with a functional variant (see fig. 2D, black line). We used the LD measure r2 because r2 quantifies the strength of association signal captured by a tag SNP (Pritchard and Przeworski 2001). Our results are robust to the choice of r2 cutoff (supplementary information, Supplementary Material online). We also considered two measures that describe the overall extent of LD in a region (the total number of eQTL SNPs—which gives some sense of the breadth of association signals—and the population recombination rate ρ per unit physical distance). Results for the latter measures are given in the Supplementary Material online and generally yield larger odds ratio estimates.
For each population, we restricted our logistic regression analysis to the set of genes that have at least one significant eQTL SNP (i.e., P < 10−4) in that population. This was done for two reasons. First, this would be more robust to any systematic differences in iHS between genes with and without eQTLs, as might happen if there are differences in average SNP density or recombination rate between the two sets. Second, it is likely that many of the genes for which we did not find eQTLs are simply not expressed in lymphoblast cell lines and hence including these genes simply adds noise to the overall data. Hence, the estimated odds ratio can be interpreted as the relative enrichment within genes with an eQTL.
Logistic Regression Confidence Intervals
To estimate confidence intervals (CIs) for the logistic regression model, we used a bootstrap approach to account for the fact that clumps of nearby SNPs may all be eQTLs for the same gene. In effect, our bootstrap analysis resamples across the set of genes with a detected eQTL in each population (638, 1,060, and 1,289 genes in CEU, YRI, and ASN, respectively). In YRI, for example, random sets of genes were drawn with replacement from the full set of 1,060 genes. We reran the logistic regression analysis using the new set of 1,060 genes (including multiples of some genes) and output the estimated odds ratio. This was done for 5,000 independent replicates in order to approximate the sampling variation of the observed odds ratio. We used the central 95% of the odds ratio estimates as estimates of the 95% CIs for each population.
Hierarchical Model
In a separate project, we have developed a hierarchical model for fine mapping the functional SNPs that underlie eQTLs and for identifying covariates that are predictive of the locations and identities of these SNPs (Veyrieras et al. 2008). In brief, the method starts from the Bayesian regression framework developed by Servin and Stephens (2007). For each SNP in the cis-candidate region around a gene, we compute a Bayes factor that is the ratio of the probability of the expression data assuming that the genotype at this SNP affects expression levels to the probability of the expression data assuming that the genotype does not affect expression levels. (The expression data are generated either as a mixture of three normal distributions corresponding to the three genotypes or, under the null hypothesis, as a single normal distribution. We assume that dominance effects are usually small so that the heterozygote mean is approximately the average of the two homozygotes' means Servin and Stephens [2007].) Then, if we assume that there is exactly one eQTL for a gene, the Bayes factors allow us to compute the posterior probability that each SNP is the causal SNP. (When the causal SNP is not in HapMap, simulations show that its signal is usually absorbed by a nearby SNP in strong LD.) We then use these Bayes factors in a hierarchical model that allows us to estimate the relative contributions of different types of covariates to predicting whether an SNP will be an eQTL. Specifically, conditional on there being an eQTL for a particular gene, we consider that the jth SNP has a prior probability πj that it is the functional site, where
| (3) |
where M is the number of SNPs in the cis-candidate region and where
| (4) |
Here is a vector of annotation effect parameters and δjl is an indicator function that is 1 if SNP j has the lth annotation and is 0 otherwise. In this framework, exp(λl) is the odds ratio of the effect of the lth annotation. For this analysis, each SNP was assigned to a single location bin that indicates the position of the SNP relative to the gene in question (setting δjl = 1 for one location bin and to 0 for all other locations). Additionally, there was a λ for the effect of an SNP having a signal of selection or not. The analysis was performed separately in each population group. For the hierarchical model analysis, we used a subset of the full gene set of 16,155 genes used in the logistic regression analysis. For the hierarchical model analysis, we considered 11,446 genes with a single known transcript (Veyrieras et al. 2008).
Results
We analyzed expression data for 16,155 autosomal genes generated by the Illumina Sentrix Human-6 Expression BeadChip for all HapMap individuals (Stranger, Forrest, et al. 2007). For each gene with expression data, we used linear regression to test all HapMap SNPs within 100 kb of the gene for association with expression levels (Methods). We defined the region of interest in this way because most of the strong eQTLs in the data set lie within these boundaries (Stranger, Nica, et al. 2007) and the gene-level FDR for eQTLs beyond 100 kb is extremely high (supplementary information, Supplementary Material online).
We identified SNPs that are significant at P < 10−4 for association with expression levels of 638, 1,060, and 1,289 genes for the CEPH (CEU), Yoruba (YRI), and east Asian (ASN) samples, respectively. This significance threshold yields a gene-level FDR ≤18% in all three populations (supplementary information, Supplementary Material online). The lower number of genes with eQTLs in CEU may reflect some anomalous patterns of gene expression evident in the older CEU cell lines (supplementary information, Supplementary Material online; Stranger, Nica, et al. [2007]). The numbers of genes that we consider significant here is larger than in the previous analysis of these data by Stranger, Nica, et al. (2007) because we use a less stringent significance threshold in order to increase the number of observed signals. Nonetheless, the FDR implies that the large majority (>80%) of the eQTLs identified are true positives.
Overlap of iHS and eQTL Signals
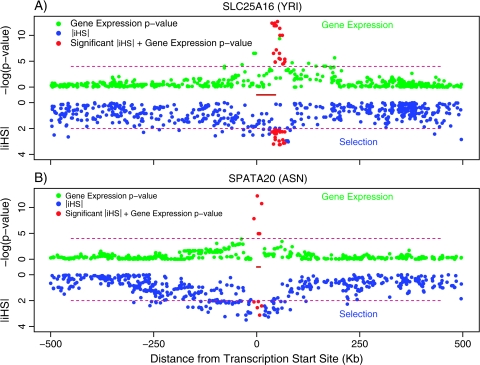
Visual inspection of the data reveals examples in which there is a strong overlap between the eQTL and selection signals. Data for several of these genes are illustrated in figure 1 and in the supplementary information (Supplementary Material online). In these examples, almost all the SNPs that are correlated with expression level also have high |iHS|, suggesting that in each case the eQTL itself may be the actual target of selection. However, these examples also illustrate that it is usually unclear which site is the actual target of selection, and whether this coincides with the functional site in the eQTL.
FIG. 1.—
Two examples in which an eQTL is centered on a strong signal of selection. The upper half of each plot (green and red points) shows the strength of association between SNPs and gene expression levels (plotted as −log10(P values) of the indicated gene). The lower half of each plot (blue and red points) indicates −|iHS| scores for the same set of SNPs. Red points indicate SNPs that are both strongly associated with expression (P < 10−4) and have |iHS| > 2. The positions of the genes of interest are indicated by the red bars at the center of each plot. (A) Data from SCL25A16 (YRI). (B) Data from SPATA20 (ASN). According to the sliding-window analysis, the clusters of high |iHS| signals are in the 2.5% and 1% tails of the empirical Yoruba (A) and Asian (B) distributions, respectively. The favored haplotypes are at 60% and 89% frequency, respectively.
To look more quantitatively at the overlap of selection and eQTL signals, we next examined genome wide whether SNPs with high |iHS| are more likely to be eQTLs compared with SNPs with low |iHS|. Figure 2A shows, for the YRI population, that SNPs with |iHS| > 2 (roughly 5% of SNPs) are considerably more likely than random SNPs to be associated with expression of nearby genes. When these SNPs lie within clusters of high-|iHS| SNPs (shown to be a more reliable indicator of selection; Voight et al. 2006), the abundance of eQTLs is further enriched (fig. 2A, red data). Overall, the signal of enrichment is strongest in YRI, the population that has the clearest signals of selection according to iHS (Voight et al. 2006), but enrichment is also seen in the other HapMap groups (supplementary information, Supplementary Material online).
Another view of this effect is provided by figure 2B. As expected, most SNPs that are significantly associated with expression (at P < 10−4) lie close to the transcription start site of the relevant gene (Stranger, Nica, et al. 2007). Among SNPs with high |iHS|, the pattern is the same, but the abundance of eQTLs is considerably higher. This also shows that iHS does not appear to create a bias in the location of the eQTLs. Similarly, figure 2C plots eQTL frequency as a function of derived allele frequency and shows a general excess of eQTLs among SNPs with selection signals. Both these factors are important because we find that SNPs with a high |iHS| are enriched in genic regions and most eQTLs also occur close to the relevant genes; the power to detect a partial sweep using iHS is highest when the selected allele is in the 50–85% frequency range. This frequency range overlaps with range of frequencies over which we have maximum power to detect an eQTL (fig. 2C).
Although the results in figure 2 certainly seem to suggest a genome-wide enrichment of selection signals on SNPs that are associated with gene expression, these analyses raise two important caveats. First, SNPs with high iHS are often in strong LD with many other SNPs. Hence, they may simply be better than random SNPs at tagging nearby eQTLs (cf. Pe'er et al. [2006]). However, when we control for local LD, the enrichment remains (fig. 2D). Second, the results might plausibly be driven by chance overlap of iHS and eQTL signals in a small number of regions.
To address these concerns more systematically, we implemented a logistic regression model that estimates the probability that an SNP is associated with expression of a nearby gene as a function of 1) presence of a selection signal, 2) the number of other SNPs tagged by that SNP, 3) the distance from the transcription start and end sites of the gene whose expression data we are considering, and 4) the minor allele frequency of the SNP (see Methods). In the results reported below, we consider that an SNP shows a signal of selection if and only if |iHS| > 2 and it lies within a cluster of other high |iHS| SNPs (see Methods). To account for the fact that clusters of SNPs within a selection signal may all be eQTLs for the same gene, we obtained CIs on the effect sizes by bootstrapping over genes (Methods). The logistic regression model estimates an odds ratio for the odds that an SNP with a signal of selection is an eQTL, compared with the odds that an SNP without a signal of selection is an eQTL, while controlling for the confounders above (Methods). Values of the odds ratio >1 imply an enrichment of eQTLs among SNPs with signals of selection.
The results in table 1 show that indeed the enrichment of eQTLs among SNPs with signals of selection is significant in the Yoruba, where the estimated odds ratio ( ) is 2.41 and where the 95% CI [1.23–4.27] does not overlap 1. We find that the results suggest a similar effect in the CEPH (
) is 2.41 and where the 95% CI [1.23–4.27] does not overlap 1. We find that the results suggest a similar effect in the CEPH ( R =2.29, CI = [0.83–4.35]) and east Asians (
R =2.29, CI = [0.83–4.35]) and east Asians ( R, CI = [0.82–2.38]); however in these populations, the confidence regions overlap 1.
R, CI = [0.82–2.38]); however in these populations, the confidence regions overlap 1.
Table 1.
Enrichment of eQTLs among SNPs with Signals of Selection, as Estimated by Two Different Methods
| Population | Analysis | Odds Ratio | 95% CI | Number of Genes |
| YRI | LR | 2.41 | 1.23–4.27 | 35 |
| HM | 5.43 | 2.17–13.87 | ||
| CEU | LR | 2.29 | 0.83–4.35 | 16 |
| HM | 2.26 | 0.45–8.03 | ||
| ASN | LR | 1.41 | 0.82–2.38 | 47 |
| HM | 1.52 | 0.58–3.43 |
NOTE.—For each population separately, we used logistic regression (LR) and a hierarchical model (HM) to estimate the odds ratio that an SNP with a selection signal (|iHS| > 2 and a cluster-based signal in the top 5%) is an eQTL, compared with a comparable SNP without a selection signal. The 95% CIs were estimated as described in the Methods. “Number of genes” indicates the number of genes for which at least one SNP is both associated with gene expression and has a selection signal.
We hypothesize that the lack of a significant result in the CEPH and east Asians is a reflection of the lower power and higher false-positive rates of our selection scans in the non-Africans. Compared with the Yoruba, both non-African groups have undergone substantial bottlenecks (Keinan et al. 2007). Bottlenecks are expected to reduce the power of selection scans in general (Teshima et al. 2006) and for iHS in particular (Pickrell J, personal communication). Moreover, by most measures, the iHS signals detected by Voight et al. (2006) were more reliable in the Yoruba than in the CEPH and east Asians. Consequently, it may be that the nonsignificant correlation with gene expression in the non-Africans is due to a reduced fraction of true positives within the tail of the iHS distributions in those populations. In addition to this, perhaps due to the aberrant expression patterns in the CEPH, we find fewer genes with an eQTL signal. So even though the odds ratio for the CEU and the YRI are in the same range (table 1), the CI for the CEPH overlaps with 1 possibly because of the smaller number of genes with SNPs that have an eQTL and an iHS signal.
It should be noted that correcting for the strength of local LD in the logistic regression model reduces the power to detect an association between iHS and eQTL. This is because this variable is correlated with the iHS signal and hence decreases the effect size of iHS in the model. If instead of the number of SNPs in LD, the point estimate of the local recombination rate (ρ) at an SNP, based on LD (International HapMap Consortium 2007), is used as a surrogate for local LD, the estimated effect size (odds ratio) of iHS on eQTL in the logistic regression model increases for all three populations. In that analysis, the 95% CIs of the odds ratios for YRI, CEU, and ASN do not overlap with 1 (supplementary information, Supplementary Material online). We further verified that the effect size estimates are fairly robust to various aspects of the analysis, including the measure of local LD and the size of the region analyzed around each gene (supplementary information, Supplementary Material online).
We also used a second, quite different analytical approach to assess the strength of the observed signals, applying a recently developed Bayesian hierarchical model for detecting eQTLs (Veyrieras et al. 2008); see Methods for further explanation. The hierarchical model assumes at most one eQTL per gene and, by sharing information across all genes, it assesses whether external information such as SNP location or iHS signal affects the odds that a particular SNP generates an observed eQTL. Because the model assumes at most one eQTL per gene, the method is robust to varying levels of LD (and hence variable numbers of associated SNPs); however, there may also be a small cost in power if some genes in fact have multiple eQTLs. The Yoruba odds ratio is again significantly >1 (estimated OR = 5.4, CI = [2.2–13.9]). As before, the 95% CIs for CEPH and east Asians both overlap 1.
Finally, large allele frequency differences between populations (high FST) can also be an indicator of potential selection. Consistent with our finding that regulation of gene expression is a target of natural selection, we also find that SNPs with large frequency differences between populations show a trend to be enriched for eQTLs compared with SNPs with low-frequency differences (supplementary information, Supplementary Material online).
Expression Differences That May Be Targets of Selection
In the Yoruba, there are ∼ 30 genes in which SNPs with selection signals are also eQTLs. Given that the odds ratio of the enrichment is estimated to be ∼2.5 by one method and ∼3.5 by our other method (table 1), this would suggest that the majority of these overlaps are in fact due to selection on gene expression. (e.g., if the true odds ratio is 3:1, then we would anticipate only about 1/4 of the overlapping signals to be coincidental.) Hence, these results suggest that perhaps 20 or more of the expression QTLs detected in Yoruba are targets of natural selection. The odds ratio point estimates suggest that a smaller number of the CEPH and Asian signals may also represent meaningful overlaps (≈ 0–25 depending on the analysis).
We find that the set of genes identified by the overlap between iHS and eQTLs includes several that are worth highlighting (a full list is contained in the Supplementary Material online). For example, HLA-C has eQTL SNPs with signals of selection in both the CEPH and east Asians. An SNP upstream of this gene (rs9264942) has recently been associated with HIV-1 viral load (Fellay et al. 2007); furthermore that study observed that the protective allele at this SNP is associated with higher HLA-C gene expression in the Stranger, Forrest, et al. (2007) data. Further inspection of the data shows that rs9264942 lies within a clump of SNPs that are associated with gene expression, and many of which also have significant iHS. The SNP rs9264942 does not itself have a significant iHS but it is not clear which SNP in the region is actually functional, and there is at least one other nearby SNP with a much stronger association with HLA-C expression (that SNP, rs2249741 has iHS = −1.9). The selected haplotype is associated with lower expression of HLA-C. Given that HIV is a relatively novel human pathogen, we speculate that this selection signal is a response to a different pathogen. It is interesting to note that the set of genes with eQTL and selection SNPs is enriched overall for genes that interact with HIV proteins (12 in total, P = 0.07 compared with eQTL genes overall). These include MAN2C1, PAWR, and USF1 in YRI, TUBB2A, TUBB2B, and MAN1A2 in CEU, and B3GALNT1, RNGTT, and HLA-DRB5 in ASN.
We also identified several genes that are involved in susceptibility to diseases. We find that alleles associated with lower expression of PPARG show signals of positive selection. A common nonsynonymous variant in PPARG contributes to risk for type 2 diabetes (Altshuler et al. 2000). Genes involved in rare diseases with such signals in YRI include USF1 (hyperlidemia), NF1 (neurofibromatosis), and RNF135 (overgrowth and learning disabilities; Maglott et al. 2007; OMIM 2008). The observation that RNF135 is involved with overgrowth may be interesting in light of a separate observation that an expression variant in GDF5 with a signal of selection is also associated with height (see below).
Discussion
The overlap of signals of selection and eQTLs around a single gene does not automatically imply that expression change is the target of selection at that locus. However, a genome-wide enrichment of this observation is strongly suggestive of the fact that cis regulation of gene expression, as a specific class of phenotype, is an important target of recent positive selection in the human genome. In this paper, we report that in the Yoruba HapMap, there is a significant correlation between iHS-based signals of selection and gene eQTLs detected in lymphoblastoid cell lines. This observation argues that levels of gene expression are an important target of positive selection. We also see a trend toward a similar effect in the two non-African populations, however in both cases, this is not statistically significant. As discussed above, these nonsignificant results may reflect a higher false-positive rate for scans of recent positive selection in the non-African populations.
It should be pointed out that our approach likely underestimates the importance of positive selection acting on gene expression levels. First, we have incomplete power to detect both targets of selection and eQTLs. However, fundamentally, there will surely be many additional eQTLs that would be detected in other tissues but that cannot be detected in lymphoblast cells. Two examples epitomize how this could affect our results. The best documented case of a partial sweep on a human cis-regulatory change is for lactase (Bersaglieri et al. 2004; Enattah et al. 2004; Tishkoff et al. 2007). As might be expected, we did not detect an eQTL for lactase in these cell lines, and so this well-known example does not contribute to our overall signal of enrichment (despite a strong selection signal around lactase). Similarly, an SNP in the 5′ untranslated regions of GDF5 has been shown to affect expression levels of GDF5 in chondrogenic cells and to contribute to osteoarthritis in east Asians (Miyamoto et al. 2007). The allele that increases the risk of osteoarthritis is at the center of a strong iHS signal in the HapMap Asians; however, the expression change is also not observed in our data. This allele has also been associated with decreased height in samples of Europeans and African Americans (although in the latter study, the identity of the functional SNP is unclear; Sanna et al. 2008). Clearly, this region is associated with important human phenotypes, and the selected haplotype appears to be linked to these phenotypes. However, our analysis of the association between selection and gene expression of GDF5 is limited by the fact that we do not detect eQTLs for GDF5 in our data, possibly because it is not expressed in this tissue. Therefore, it will be important to repeat this type of analysis across a broad range of tissues and at a range of developmental stages, in order to obtain a more complete view of the importance of cis-regulation as a target of recent selection. It is also worth noting that these data were collected from transformed lymphoblasts and the expression patterns in these cells are likely to differ from those in the untransformed tissue. However, although this may complicate the interpretation of individual eQTL signals, it is hard to see how this could create a false overall association between eQTLs and selection signals.
This study also exemplifies the future role of overlaying phenotypic or functional information onto selection signals. Typically, haplotype-based signals span tens or hundreds of kilobases, including many genes. One of the greatest challenges facing studies of genomic selection scans is to interpret the selection signals that we find: which variants or genes are the actual targets of a selection signal and how do the variants affect phenotype? In a small subset of cases, broad signals include very strong candidate genes (lactase and the skin pigmentation gene SLC24A5 are two such examples Bersaglieri et al. [2004]; Lamason et al. [2005]). However, for most selection signals, there is little to guide us as to the targets of selection. By intersecting selection data with external information such as eQTL data or phenotype associations, we anticipate that it will become possible to link many more of the selection signals to genes and phenotypes. From there, we will begin to gain a better understanding of the role of selection in modifying the human phenotype.
Electronic Resources
An online browser providing haplotype plots and iHS scores for all HapMap SNPs is at http://haplotter.uchicago.edu.
Supplementary Material
Supplementary information is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank the Pritchard, Przeworski, and Stephens groups, Graham Coop, Anna Di Rienzo, Yoav Gilad, Charalampos Papachristou, and Molly Przeworski, and the anonymous reviewers for helpful conversations and comments on this work. Funding was provided by US National Institutes of Health grant HG002772 to J.K.P. E.T.D. and B.E.S. are funded by the Wellcome Trust. J.K.P. is an investigator of the Howard Hughes Medical Institute.
References
- Altshuler D, Hirschhorn J, Klannemark M, et al. (16 co-authors) The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- Bersaglieri T, Sabeti P, Patterson N, Vanderploeg T, Schaffner S, Drake J, Rhodes M, Reich D, Hirschhorn J. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon A, Liang L, Moffatt M, et al. (13 co-authors) A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Enattah N, Forsblom C, Rasinper H, Tuomi T, Groop P, Jrvel I. The genetic variant of lactase persistence C (-13910) T as a risk factor for type I and II diabetes in the Finnish population. Eur J Clin Nutr. 2004;58:1319–1322. doi: 10.1038/sj.ejcn.1601971. [DOI] [PubMed] [Google Scholar]
- Fellay J, Shianna K, Ge D, et al. (27 co-authors) A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Rifkin S, Pritchard J. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M, Rockman M, Soranzo N, Goldstein D, Wray G. Population genetic and phylogenetic evidence for positive selection on regulatory mutations at the factor VII locus in humans. Genetics. 2004;167:867–877. doi: 10.1534/genetics.103.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin M, Di Rienzo A. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am J Hum Genet. 2000;66:1669–1679. doi: 10.1086/302879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haygood R, Fedrigo O, Hanson B, Yokoyama K, Wray G. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat Genet. 2007;39:1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- Hoekstra H, Coyne J. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hudson R, Bailey K, Skarecky D, Kwiatowski J, Ayala F. Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics. 1994;136:1329–1340. doi: 10.1093/genetics/136.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Kuhn R, Baertsch R, et al. (25 co-authors) The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Mullikin J, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in east Asians than in Europeans. Nat Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Pritchard J. Adaptive evolution of conserved noncoding elements in mammals. PLoS Genet. 2007;3:1572–1586. doi: 10.1371/journal.pgen.0030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason R, Mohideen M, Mest J, et al. (25 co-authors) SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt K, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:26–31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Mabuchi A, Shi D, et al. (16 co-authors) A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007;39:529–533. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- Morley M, Molony C, Weber T, Devlin J, Ewens K, Spielman R, Cheung V. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OMIM. Online mendelian inheritance in man, omim (tm). McKusick-Nathans Institute of Genetic Medicine. Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information. Bethesda (MD): National Library of Medicine; 2008. Available from: http://www.ncbi.nlm.nih.gov/omim/ [Google Scholar]
- Pe'er I, Chretien Y, de Bakker P, Barrett J, Daly M, Altshuler D. Biases and reconciliation in estimates of linkage disequilibrium in the human genome. Am J Hum Genet. 2006;78:588–603. doi: 10.1086/502803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K, Salama S, King B, et al. (13 co-authors) Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar S, Noonan J, Pbo S, Rubin E. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K, Tatusova T, Maglott D. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M, Hahn M, Soranzo N, Zimprich F, Goldstein D, Wray G. Ancient and recent positive selection transformed opioid cis-regulation in humans. PLoS Biol. 2005;3:e387. doi: 10.1371/journal.pbio.0030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P, Reich D, Higgins J, et al. (17 co-authors) Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sabeti P, Varilly P, Fry B, et al. (13 co-authors) Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna S, Jackson A, Nagaraja R, et al. (38 co-authors) Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B, Forrest M, Dunning M, et al. (17 co-authors) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B, Nica A, Forrest M, et al. (14 co-authors) Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Thornton K, Stoneking M. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 2007;5:e171. doi: 10.1371/journal.pbio.0050171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima K, Coop G, Przeworski M. How reliable are empirical genomic scans for selective sweeps? Genome Research. 2006;16:702–712. doi: 10.1101/gr.5105206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S, Reed F, Ranciaro A, et al. (19 co-authors) Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras J, Kudaravalli S, Dermitzakis E, Gilad Y, Stephens M, Pritchard J. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4(10):e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B, Kudaravalli S, Wen X, Pritchard J. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Kodama G, Baldi P, Moyzis R. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.