Abstract
We performed a genome-wide screening for T cell epitopes using synthetic peptides that encompass all of the influenza A viral proteins, including subtype variants for hemagglutinin (HA) (H1, H3 and H5) and neuraminidase (NA) (human and avian N1 and N2) proteins, based on the sequence information of recently circulating strains. We found a total of 83 peptides, 54 of them novel, to which specific T cells were detectable in IFN-γ ELISPOT assays using peripheral blood mononuclear cells from four healthy adult donors. The surface glycoproteins, HA and NA, major components of vaccines, had many T cell epitopes. HA and matrix protein 1 had more T cell epitopes than other viral proteins, most of which were recognized by CD4+ T cells. We established several cytotoxic CD4+ T cell lines from these donors. We also analyzed H1 and H3 HA-specific T cell responses using the peripheral blood mononuclear cells of 30 hospital workers. 53% of donors gave a positive response to H3 HA peptides, while 17% gave a positive response to H1 HA peptides. Our genome-wide screening is useful in identifying T cell epitopes and complementary to the approach based on the predicted binding peptides to well-studied HLA-A, B and DR alleles.
Keywords: Human T cell epitopes, influenza A virus, CD4+ T cells, T cell epitope screening, hemagglutinin
INTRODUCTION
Human influenza is a contagious respiratory disease that results in substantial morbidity and mortality worldwide. With the recent cases of avian influenza infection in humans [1] and the heightened concern for an influenza pandemic [2], it is essential to understand host responses that would confer protective immunity to influenza. It is also important to generate influenza vaccines that can induce heterosubtypic immunity, which may in part be mediated by CD8+ and CD4+ T cells. A re-analysis of the archival records from the Cleveland Family Study, which was conducted before and during the 1957 pandemic when a shift from subtype H1N1 to H2N2 occurred, suggested an impact of accumulated heterosubtypic immunity in adults [3]. In addition, measures of the ex vivo cellular immune response to influenza in vaccinated older subjects correlated with protection against influenza while serum antibody responses had a limitation as a sole measure of vaccine efficacy [4]. Thus, understanding the roles of cell-mediated immune responses to influenza virus and evaluating the efficacy of influenza vaccines in humans require insight into the specificity and diversity of T cell epitopes elicited upon infection and upon vaccination.
In humans, the CD8+ and CD4+ T cell responses to influenza have not been fully elucidated. Earlier studies to identify T cell epitopes to influenza have found nucleoprotein (NP) and matrix protein 1 (M1) as the major antigens for CD8+ T cells [5-7] and the hemagglutinin (HA) for CD4+ T cells [7]. We [8, 9] and others [10-13] showed that human memory cytotoxic T lymphocyte (CTL) responses to influenza A virus are broadly directed to epitopes on a wide variety of viral proteins. We previously established influenza A-specific CTL lines from healthy volunteers and identified epitopes using recombinant vaccinia viruses that expressed influenza A virus proteins. We then used synthetic peptides based on the amino acid sequences of the influenza viral proteins that were recognized by the CTL lines. Other groups synthesized peptides based on epitope prediction algorithms to several common human leukocyte antigen (HLA) molecules and screened them by peptide binding assays, and the peptides which bound with high enough affinity to a given HLA molecule were screened in HLA-transgenic mice splenocytes or with human peripheral blood mononuclear cells (PBMCs) [11-13]. Wang et al. [12] screened for influenza epitopes by focusing on well-conserved peptide sequences among influenza A H1N1 strains and, as a result, most of HA peptides were excluded. Utilization of epitope prediction algorithms may bias the screening since it precludes atypical but potentially important epitopes because of their poor binding scores. Additionally, we [8, 9] and Gianfrani et al. [11] both utilized the sequences of the A/Puerto Rico/8/34 (H1N1) strain (A/PR/8), which was isolated more than 70 years ago and is not a circulating strain. Therefore, all of the above approaches were designed to identify highly conserved epitopes in viral internal proteins. As a result, we may have underestimated the CTL responses to the HA as well as the neuraminidase (NA), another surface glycoprotein which also undergoes antigenic drift.
In the present study, we performed an epitope screening approach using synthetic peptides that encompass all of the influenza A viral proteins based on sequence information from recently circulating influenza strains. Our peptide arrays include subtype variants for HA (H1, H3 and H5) and NA (human and avian N1 and N2) proteins. We screened these peptides using PBMCs from healthy adult volunteers without in vitro amplification of influenza A-specific T cells. Our genome-wide epitope screening confirmed a broad T cell response to influenza that was directed to several viral proteins. HA and M1 had more T cell epitopes than other viral proteins. We also detected cross-reactive T cell responses to H5 HA peptides in healthy humans who were unlikely to have been exposed to H5N1 viruses.
MATERIAL AND METHODS
Influenza A peptides and control peptides
17-mer peptides overlapping by 11-12 amino acids encompassing the entire sequence of all influenza viral proteins were obtained from the National Institutes of Health (NIH) Biodefense and Emerging Infections Research Resources Repository (BEI Resources) (Table 1). The length of the peptides was a debated compromise in an effort to detect most CD4+ and CD8+ T cell epitopes at a reasonable cost. The amino acid sequences of these peptides were based on the recent vaccine strains of influenza viruses A (H1N1) and (H3N2) (or an antigenically indistinguishable strain from the vaccine strain, when the amino acid sequences of the proteins were not available for the vaccine strain) and recent isolates of influenza virus A (H5N1). Because of considerable difference between avian and human N1 NA amino acid sequences, peptides sets for both neuraminidases were synthesized. Polymerase B1-F2 peptides of both strains were also synthesized because of differences between H1N1 and H3N2 strains. Peptide arrays of control peptides for known major histocompatibility complex (MHC) Class I and II epitopes of Influenza A viruses were also provided by BEI Resources. Lyophilized peptides were reconstituted by dissolving in 100% dimethyl sulfoxide (DMSO) at a concentration of 10 mg/ml per peptide. The CEF peptide pool was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, the National Institute of Allergy and Infectious Diseases (NIAID), NIH [14] and was used as a positive control. It contains MHC class I-restricted human T cell epitope peptides of Cytomegalovirus, Epstein-Barr virus and Influenza A virus at 2 μg/ml per peptide. Other peptides described in this paper were synthesized by AnaSpec, Inc. (San Jose, CA).
Table 1.
Influenza A peptides used in this paper
| Influenza Protein | Abbreviation | Strain | No. of Peptides | BEI Catalog No. |
|---|---|---|---|---|
| Hemagglutinin | H1 HA | A/New Caledonia/20/1999 H1N1 | 94 | NR-2602 |
| Hemagglutinin | H3 HA | A/New York/384/2005 H3N2 | 94 | NR-2603 |
| Hemagglutinin | H5 HA | A/Thailand/4(SP-528)/2004 H5N1 | 94 | NR-2604 |
| Neuraminidase | aN1 NA | A/Thailand/4(SP-528)/2004 H5N1 | 74 | NR-2607 |
| Neuraminidase | hN1 NA | A/New Caledonia/20/1999 H1N1 | 78 | NR-2606 |
| Neuraminidase | N2 NA | A/New York/384/2005 H3N2 | 78 | NR-2608 |
| Matrix Protein 1 | M1 | A/New York/348/2003 H1N1 | 41 | NR-2613 |
| Matrix Protein 2 | M2 | A/New York/348/2003 H1N1 | 15 | NR-2614 |
| Nucleoprotein | NP | A/New York/348/2003 H1N1 | 82 | NR-2611 |
| Nonstructural Protein 1 | NS1 | A/New York/444/2001 H1N1 | 37 | NR-2612 |
| Nonstructural Protein 2 | NS2 | A/New York/348/2003 H1N1 | 19 | NR-2615 |
| Polymerase A | PA | A/New York/348/2003 H1N1 | 119 | NR-2618 |
| Polymerase B1 | PB1 | A/New York/348/2003 H1N1 | 126 | NR-2617 |
| Polymerase B1-F2 | PB1-F2 (H1N1) | A/New York/348/2003 H1N1 | 8 | NR-2685 |
| Polymerase B1-F2 | PB1-F2 (H3N2) | A/New York/504/1998 H3N2 | 16 | NR-2685 |
| Polymerase B2 | PB2 | A/New York/348/2003 H1N1 | 126 | NR-2616 |
Study subjects
Blood samples were obtained from four healthy adults and PBMC were purified by Ficoll-Hypaque density gradient centrifugation as previously described [15]. The donors were chosen based on the availability of PBMC for large scale screening and their reactivity to the influenza peptides in the CEF peptide pool in enzyme-linked immunosorbent spot (ELISPOT) assays that had been previously performed. Donors 1, 3 and 4 received influenza vaccine almost every year. Donor 2 never received any influenza vaccine. None of these four donors had a history of laboratory-confirmed influenza infection. The HLA alleles of donor 1 are A2, A24, B7, B62, Cw3, DP2, DQw5, DQw6, DRB1*0103, and DRB1*1501; Donor 2 has the HLA alleles A1, B8, B44, Cw5, DQw1, DQw2, DR2, DR3, and DRw52; Donor 3 has A1, A24, B35, Cw04, DRB1*11 and DRB1*13; Donor 4 has A30, A31, B13, B51, DRB1*07 and DRB1*13. Additional screening of the HA (H1 and H3) was done using PBMC from 30 healthy hospital workers, whose HLA-A, -B, and -DR typing were described previously [16]. HLA-typing for donors 1 and 2 was determined by the HLA Typing Laboratory at the University of Massachusetts Medical Center, and the HLA-typing for donor 3 and 4, and the 30 vaccinated donors was determined by the HLA Core Facility of the Center for Infectious Diseases and Vaccine Research at the University of Massachusetts Medical School.
Peptide screening
In our preliminary experiments, we detected IFN-γ-producing cells to the M158-66 in a pool with 15 non-overlapping peptides using donor 1 PBMC by ELISPOT assays. The number of IFN-γ-producing cells did not differ significantly if we used 5, 10 or 15 non-overlapping peptides in a pool, but decreased if we used peptides that overlapped with the peptide M158-66 (data not shown). Therefore, we made peptide pools that contained 15 non-overlapping peptides and made two sets of peptide pools to facilitate the peptide screening. The first set of peptide pools contains all peptides of the surface glycoproteins of H1, H3 and H5 HA, and avian N1 (aN1) and human N1 (hN1) and N2 NA. This set of peptide pools consists of 33 pools; Pool 1-6 contain H1 HA peptides, Pool 7-12 contain H3 HA peptides, Pool 13-18 contain H5 HA peptides, Pool 19-23 contain hN1 NA peptides, Pool 24-28 contain aN1 NA peptides, and Pool 29-33 contain N2 NA peptides. The second set of peptide pools consists of 38 pools and included all peptides of the internal viral proteins: NP and nonstructural protein 1 (NS1) (Pool 1-8), M1 and nonstructural protein 2 (NS2) (Pool 9-12), polymerase A (PA) and matrix protein 2 (M2) (Pool 13-21), and polymerase B1 (PB1) and polymerase B2 (PB2) (Pool 22-38). Three additional peptide pools were also made. These contained PB1-F2 peptides and H5 HA peptides representative of regions of amino acid sequence diversity among different strains of H5N1 viruses. Donor PBMC was tested against the influenza A peptide pools in ELISPOT. The cut-off value for a positive response in ELISPOT was 20 SFC per 106 cells for donors 1, 2 and 4. This value was determined by the average spot forming cells (SFC) per 106 cells greater than 3 standard deviations of the negative control wells. The PBMC of donor 3 had higher media background than did the other PBMC and the cut-off value for a positive response in this donor was 37 SFC/106. Peptide pools that had SFC values equal to or greater than the determined cut-off for each donor were then deconvoluted to identify the individual peptide/s eliciting the IFN-γ response. A final peptide concentration of 2μg/ml for each peptide was used in all ELISPOT assays.
IFN-γ ELISPOT assay
ELISPOT assays were done as previously described [8]. Briefly, cryopreserved PBMC (2-2.5×106 cells per well) were seeded onto polyvinylidene difluoride membrane 96-well plates (Millipore, Bedford, MA) precoated with 5 μg/ml anti-IFN-γ mAb (clone D1K; Mabtech, Cincinnati, OH) in the presence or absence of peptide or peptide pools. Phytohemagglutinin (PHA) (Sigma-Aldrich, St. Louis, MO) (1:100), CEF peptide pool and/or virus were used as positive controls. After 18-24 hr incubation, cells were removed by washing with PBS plus 0.05% Tween 20. Secondary biotinylated anti- IFN-γ mAb (clone 7-B6-1; Mabtech) was added at 2 μg/ml and the plates were incubated for 2 hours at room temperature. Plates were washed again and IFN-γ was detected with avidin-peroxidase (3420-2H, Mabtech) and substrate kit (NovaRed, Vector Laboratories, Burlingame, CA). The frequency of IFN-γ producing cells was determined by using the ImmunoSpot® S4 Pro Analyzer and the ImmunoSpot® Academic V.4 Software (Cellular Technologies Ltd.). Experiments were performed in triplicate wells.
CD4+ or CD8+ T cells depletion of PBMC
CD4 or CD8 expressing cell populations were depleted from PBMC by negative selection using anti-CD4 or anti-CD8 antibody-coated magnetic beads from the MACS purification system (Miltenyi Biotec, Bergisch Gladbach, Germany) and were processed according to the manufacturer’s protocol. Depleted PBMC were used in ELISPOT to determine the cell population producing IFN-γ.
Preparation of antigen presenting cells (APCs) for 51Cr release assays and ICS
For virus-infected targets, autologous B lymphoblastoid cell lines (BLCL) were established by culturing with Epstein-Barr virus in 24 well plates as previously described previously [17]. BLCL target cells were infected with either of the following egg-adapted virus strains - A/New Caledonia/20/1999 IVR-66 or A/Wisconsin/67/2005X-161B. These virus strains were a gift from Dr. Michel DeWilde and Dr. Robert Ryall of Sanofi Pasteur. The optimal concentrations of the two strains were determined in preliminary experiments. Infected cells were incubated for 18 hours at 37°C. Virus-infected target cells were then radiolabeled for use in 51Cr release assays or used as APCs in ICS. Peptide-pulsed targets were prepared using autologous BLCLs that were either radiolabeled for one hour in CTL assays or used directly in ICS assays. Peptide is then added to the cells at a final concentration of 10 μg/ml per peptide, unless indicated otherwise.
51Cr release assay
T cell lines or bulk culture effector cells were added to 1.5 × 103 51Cr-labeled target cells at various effector to target (E:T) ratios [8]. After incubating for 4-6 hours at 37°C, supernatants were harvested (Skatron Instruments, Sterling, VA), and specific lysis was calculated as [(experimental release - spontaneous release)/(maximum release - spontaneous release)] × 100. All assays were performed in triplicates. Unpulsed target cells were used as negative control. Spontaneous lysis was <30% in all assays.
ICS and flow cytometry analysis
Bulk culture effector cells were washed and resuspended at 5×105 cells in RPMI-1640 medium supplemented with 10% FBS (RPMI-10). Autologous BLCLs were used as APCs at an E:T ratio of 10 and were added to the effector cells, together with the peptide of interest at 10μg/ml. These were incubated for 1 h at 37°C in a 5% CO2 incubator, followed by an additional 5 h in the presence of Golgi Plug (BD Biosciences, San Jose, CA). The cells were then washed with FACS buffer (2% FBS and 0.1% sodium azide in PBS) and stained using the Live/Dead Aqua Fixable Dead Cell Stain Kit (Invitrogen, Eugene, OR) to identify live and dead cells. Cells were then stained for surface markers such as CD3-PerCPCy5.5, CD8-FITC, CD4-APC or -Pacific Blue (BD Biosciences) for 30 min at 4°C. After washing with FACS buffer, the cells were fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences), and stained for the intracellular cytokine IFN-γ (PE-conjugate; BD Biosciences) for 30 min at 4°C. Cells were then washed with Permwash buffer (BD Biosciences) and resuspended in Cytofix (BD Biosciences) for flow cytometric analysis. Multiparameter flow cytometric analyses were performed using a FACSCalibur or ARIA flow cytometer. The number of events collected per donor varied from 150,000 to 300,000. List-mode data files were analyzed using FlowJo (Version 6.3, TreeStar 6 Inc., Ashland, CA). Graphs were plotted as dot plots of CD4+ vs. CD8+ T cells in the gated live, CD3+ IFN-γ+ cell population.
Generation of bulk culture T cell lines and clones
To generate peptide-specific bulk culture lines, PBMC (3 to 5 × 106 cells) were washed and resuspended in 2 ml of AIM/V-10% FBS supplemented with 1:100 sodium pyruvate (Gibco) and 1:1000 2-Mercaptoethanol (Gibco). The corresponding influenza peptide was added at a final concentration of 10 μg/ml. Human rIL-7 (Peprotech, Inc., Rockyhill, NJ) was also added to the culture (5ng/ml) and incubated at 37°C. On Day 3, human rIL-2 (BD Discovery Labware, Bedford, MA) (25-50 U/ml) was applied, and the medium was replenished with AIM/V-10% FBS and rIL-2 every three to four days. Bulk culture 51Cr release assays were done between days 10 and 13 of culture. The cultures were restimulated once with autologous PBMC on day 14 to reduce non-specific background lysis and to generate enough cells for the assays.
To establish influenza A-specific T cell clones, a limiting dilution assay (LDA) was done as previously described [8]. Briefly, PBMC which had been stimulated in bulk culture for 14 days were plated at a concentration of 1, 3, 10, or 30 cells per well in 96-well round-bottom microtiter plates in 50 μl of AIM-V medium containing 10% FBS, 25 U of IL-2, a 1:1,000 dilution of anti-CD3 monoclonal antibody 12F6 (gift from Dr. Johnson Wong), and 1×105 gamma-irradiated (3500 rads) allogeneic PBMC/well. On day 7, 50 μl of fresh AIM-V medium with FBS and IL-2 were added, and on day 14, fresh medium with 1×105 gamma-irradiated allogeneic PBMC/well and a 1:1,000 dilution of the anti-CD3 12F6 were added. The cells were assayed for cytolytic activity using 51Cr assays between days 21-28. Cells from wells with influenza A peptide-specific cytolytic activity (specific killing of 15% and above at a peptide concentration of 10 μg/ml) were expanded to 48-well plates.
RESULTS
The PBMC of donor 1 had a broad IFN-γ response to peptide-epitopes on several influenza A proteins
We used ELISPOT assays to quantitate the number of IFN-γ-producing cells in donor PBMC that are specific for the influenza A viral proteins. This method of screening for T cell epitopes directed to viral proteins had been used with much success (reviewed in [18]), and in particular with vaccinia virus [19]. Donor 1 is HLA-A2-positive and was known to have T cells specific to M158-66, which is considered an immunodominant epitope [20], when responses to the CEF peptide pool were analyzed (data not shown).
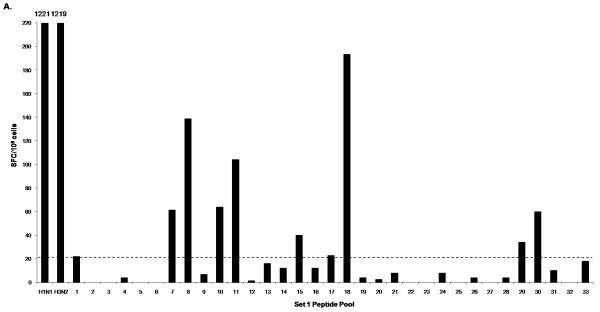
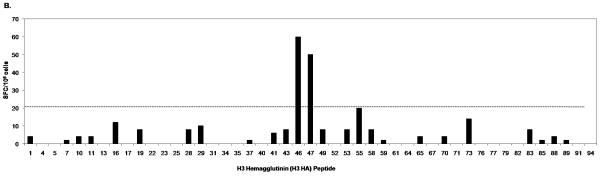
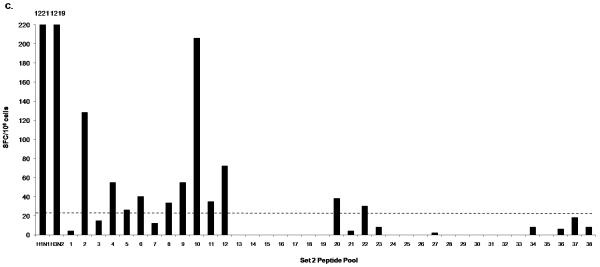
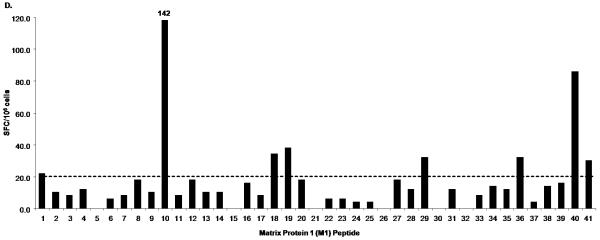
The PBMC of donor 1 were broadly reactive to peptide pools from several influenza proteins as shown by the IFN-γ responses to peptide pools containing HA peptides (Fig. 1A; Pool 1, 7, 8, 10, 11, 15, 18), NA peptides (Fig. 1A; Pool 29, 30), NP peptides and NS1 peptides (Fig. 1C; Pool 2, 4, 5, 6, 8), M1 and NS2 peptides (Fig. 1C; Pool 9, 10, 11, 12), PA peptides (Fig. 1C: Pool 20) and PB1 and PB2 peptides (Fig. 1C; Pool 22). IFN-γ responses to individual peptides from positive pools containing H3 HA and M1 peptides are shown in Fig. 1B and 1D respectively. In addition, donor 1 had positive responses to two pools containing PB1-F2 and H5 HA variant peptides (data not shown). We also detected several IFN-γ responses to H3 but not to H1. For the internal proteins, the majority of responses were seen in pools containing M1, NP, NS1 and NS2 peptides. The pool that contained the peptide with the HLA-A2-restricted M1 epitope M158-66 (Fig. 1C; Pool 10) had the highest SFC value among all the peptide pools. The sum of the number of IFN-γ-producing cells responding to these positive peptides was 1108, which is 90.7 % of the number of IFN-γ-producing cells responding to live influenza virus stimulation (1221 SFC/106 for H1N1 and 1219 SFC/106 for H3N2) (Table 2). We also performed intracellular cytokine staining (ICS) to determine the cells producing IFN-γ in response to live influenza virus-stimulation for both H1N1 and H3N2 using donor 1 PBMC. In this donor’s PBMC, ~96% of IFN-γ-producing cells responding to influenza virus were either CD4+ or CD8+ (with values ranging from 10-30 % CD4+ and 67-88 % CD8+ depending on the virus strain used to stimulate, data not shown).
Figure 1.
IFN-γ response of donor 1 PBMC to peptides from all influenza proteins. PBMC was first tested in ELISPOT against Set 1 peptide pools (1-33), which include the hemagglutinin (H1 HA, H3 HA and H5 HA) and neuraminidase (aN1 NA, hN1 NA and N2 NA) peptides (A) and Set 2 peptide pools (1-38), which include peptides spanning all the viral internal proteins (C). Positive pools were deconvoluted to test individual peptides. An example of such screening is shown in B and D. ELISPOT assays were performed using individual peptides in Pools 7, 10, and 11 consisting of hemagglutinin H3 HA peptides (B); and individual peptides in Pools 9, 10, 11, and 12 consisting of matrix protein 1 (M1) peptides (D). A/Wisconsin/67/2005X-161B H3N2 virus and A/New Caledonia/20/1999 IVR-66 H1N1 virus were used as positive controls for live influenza A virus infection. Peptide pools and individual peptides were tested in three replicate wells with 200,000 to 250,000 cells per well. The final concentration of peptide used in all experiments was 2μg/ml per each peptide. The dotted line indicates the cut-off SFC value for this donor (20 SFC/106).
Table 2.
Total SFC values. The sum of the number of IFN-γ producing cells that responded to influenza peptide stimulation in ELISPOT was determined for each donor and compared to live virus infection
| Total SFC valuesa |
||||||
|---|---|---|---|---|---|---|
| H1 and hN1 | H3 and N2 | H5 and aN1 | Internal Protein | H1N1 Virus | H3N2 Virus | |
| Peptides | Peptides | Peptides | Peptides | Infection | Infection | |
| Donor 1 | 202 | 264 | 180 | 620 | 1221 | 1219 |
| Donor 2 | 0 | 52.5 | 0 | 52.5 | N.D.b | 420 |
| Donor 3 | 0 | 985 | 0 | 1825 | 1182 | 1478 |
| Donor 4 | 0 | 273 | 193 | 714 | 1033 | 1135 |
Total SFC values are representative of one ELISPOT experiment, with triplicate wells per peptide or virus stimulation.
N.D., not determined
IFN-γ responses to influenza A proteins in three other healthy adults
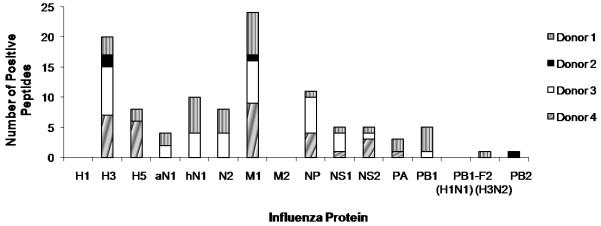
We selected three additional donors based on the availability of PBMC for large scale screening and their previous reactivity to the influenza peptides in the CEF peptide pool in ELISPOT assays (data not shown). All three of these donors responded to peptides from several influenza A viral proteins (Fig. 2). The PBMC of Donor 2 had less reactivity to influenza peptides with responses to only 4 peptides (Fig. 2, black box).
Figure 2.
Broad T cell responses to influenza viral proteins in PBMC from healthy adults. PBMC from four healthy adults were screened for IFN-γ responses to all influenza proteins in ELISPOT assays.
On average, the four donors’ PBMC responded to 21 different peptides from 9 different viral proteins, including H3 and H5 HA. Responses to H3 and H5 HA were comparable, with SFC values ranging from 20 to 60 SFC per 106 cells. Table 2 lists the sum of the number of IFN-γ-producing cells responding to these positive peptides for donors 2, 3 and 4 as well as the SFC values for live virus stimulation of these donors’ PBMC. The sum of the responses after stimulation with a complete set of the viral peptides is comparable to that of live virus stimulation in ELISPOT assays. Results of our ELISPOT screening using PBMC from the four donors are summarized in Table 3. Most of these peptides do not contain known epitope sequence (underlined in Table 3) based on our search using the Immune Epitope Database (IEDB) (www.immuneepitope.org) [21, 22]. We considered a peptide sequence to be a potential novel epitope if there was no record that the peptide sequence associated with positive T cell data in the IEDB. When we searched the IEDB, we considered both the amino acid sequence of the 17-mer peptide and the corresponding HLA allele of the donor’s PBMC that responded. Some of the peptides we identified have a record of some positive T cell data in IEDB, but were not fully characterized as to their HLA restriction or minimal epitopes (indicated in Table 3). About 60% of these peptides gave “moderate” IFN-γ responses, compared to the number of IFN-γ-producing cells responding to the “immunodominant” HLA-A2-restricted M158-66 epitope (Fig. 3).
Table 3.
Influenza peptides that gave a positive IFN-γ response in our ELISPOT screening using PBMC from donors 1, 2, 3 and 4
| Surface Glycoproteins | ||||||
|---|---|---|---|---|---|---|
| Protein | AA Position | Peptide Sequencea | Donor | SFC/106b | Commentsc | CD4/CD8d |
| H3 HA | 71-87 | PHQILDGENCTLIDALL | 3 | 37.5 | novel | ND |
| H3 HA | 179-195 | ALNVTMPNNEKFDKLYI | 4 | 22.5 | novel | ND |
| H3 HA | 209-225 | SLYAQASGRITVSTKRS | 4 | 42.5 | novel | ND |
| H3 HA | 215-231 | SGRITVSTKRSQQTVIP | 3 | 195 | novel | CD4 |
| H3 HAe | 243-259 | PSRISIYWTIVKPGDIL | 2, 4 | 27.5, 80 | novel | CD4 |
| H3 HAe | 249-265 | YWTIVKPGDILLINSTG | 3, 4 | 127.5, 47.5 | CD4 T cell epitope restriction not known | CD4 |
| H3 HA | 255-271 | PGDILLINSTGNLIAPR | 3 | 67.5 | CD4 T cell epitope restriction not known | CD4 |
| H3 HA | 267-283 | LIAPRGYFKIRSGKSSI | 1 | 60 | DRB1*1001; DQA1*0102; DQB1*0602 | CD4 |
| H3 HA | 273-289 | YFKIRSGKSSIMRSDAP | 1 | 50 | DRB1*1001; DQA1*0102; DQB1*0602 | CD4 |
| H3 HA | 279-295 | GKSSIMRSDAPIGKCNS | 4 | 20 | novel | ND |
| H3 HA | 285-301 | RSDAPIGKCNSECITPN | 3 | 75 | novel | ND |
| H3 HA | 321-337 | CPRYVKQNTLKLATGMR | 1, 3 | 20, 52.5 | DRB1*0101; DRB1*0401; DRB5 | CD4 |
| H3 HA | 350-366 | AIAGFIENGWEGMVDGW | 2 | 25 | novel | CD4 |
| H3 HA | 391-407 | NQINGKLNRLIGKTNEK | 3, 4 | 75, 20 | novel | ND |
| H3 HA | 505-521 | DVYRDEALNNRFQIKGV | 3 | 47.5 | CD4 T cell epitope restriction not known | CD4 |
| H3 HA | 528-543 | KDWILWISFAISCFLL | 4 | 40 | CD4 T cell epitope restriction not known | CD4 |
| H5 HA | 30-46 | EQVDTIMEKNVTVTHAQ | 4 | 35 | novel | ND |
| H5 HA | 136-152 | SSWSSHEASLGVSSACP | 4 | 22.5 | novel | ND |
| H5 HA | 151-167 | SSACPYQRKSSFFRNVV | 1 | 80 | novel | ND |
| H5 HA | 207-223 | YQNPTTYISVGTSTLNQ | 4 | 27.5 | novel | ND |
| H5 HA | 243-259 | EFFWTILKPNDAINFES | 4 | 55 | novel | CD4 |
| H5 HA | 351-367 | AIAGFIEFFWQGMVDGW | 4 | 20 | novel | ND |
| H5 HA | 387-403 | TQKAIDGVTNKVNSIID | 4 | 32.5 | novel | ND |
| H5 HA | 470-486 | LQLKDNAKELGNGCFEF | 1 | 34 | novel | ND |
| hN1 NA | 13-29 | ISIAIGIISLMLQIGNI | 1 | 44 | A*0201 | CD8 |
| hN1 NA | 42-58 | SQNHTGVCNQRIITYEN | 1 | 38 | novel | ND |
| hN1 NA | 309-325 | NLDYQIGYICSGVFGDN | 1 | 34 | novel | ND |
| hN1 NA | 434-450 | NTTIWTSGSSISFCGVN | 1 | 40 | novel | ND |
| hN1 NAe | 452-468 | DTANWSWPDGAELPFTI | 1 | 32 | novel | ND |
| hN1 NAe | 458-470 | WPDGAELPFTIDK | 1 | 28 | novel | ND |
| aN1 NA | 1-17 | MNPNKKIITIGSICMVT | 1 | 38 | novel | ND |
| aN1 NA | 24-40 | LQIGNLISIWVSHSIHT | 1 | 42 | novel | ND |
| N2 NA | 13-29 | VSLTISTICFFMQIAIL | 1 | 44 | novel | ND |
| N2 NAe | 176-192 | IAWSSSSCHDGKAWLHV | 1, 3 | 50, 120 | novel | CD4 |
| N2 NAe | 188-204 | AWLHVCVTGDDKNATAS | 1 | 22 | novel | ND |
| N2 NA | 206-222 | IYNGRLVDSIVSWSKEI | 3 | 87.5 | novel | ND |
| N2 NA | 236-252 | TCTVVMTDGSASGKADT | 3 | 47.5 | novel | ND |
| N2 NA | 266-282 | STLSGSAQHVEECSCYP | 1, 3 | 24, 52.5 | novel | ND |
| Internal Proteins | ||||||
|---|---|---|---|---|---|---|
| Protein | AA Position | Peptide Sequencea | Donor | SFC/106b | Commentsc | CD4/CD8d |
| M1e | 1-17 | MSLLTEVETYVLSIVPS | 1, 4 | 34, 45 | novel | ND |
| M1e | 7-23 | VETYVLSIVPSGPLKAE | 1 | 22 | A*1101 | CD8 |
| M1e | 13-29 | SIVPSGPLKAEIAQRLE | 1 | 26 | DRB1*0101; DRB1*1501 | CD4 |
| M1 | 37-53 | TDLEALMEWLKTRPILS | 1 | 50 | CD4 T cell epitope restriction not known | CD4 |
| M1 | 43-59 | MEWLKTRPILSPLTKGI | 4 | 47.5 | CD4 T cell epitope restriction not known; CW*0102 | CD4 |
| M1 | 55-71 | LTKGILGFVFTLTVPSE | 1 | 142 | A*0201 | CD8 |
| M1e | 91-107 | NNMDRAVKLYRKLKREI | 3, 4 | 157.5, 22.5 | DRB1*0103 | CD4 |
| M1e | 97-113 | VKLYRKLKREITFHGAK | 3 | 212.5 | DRB1*0103 | CD4 |
| M1 | 121-137 | AGALASCMGLIYNRMGA | 3 | 42.5 | B*3501 | CD8 |
| M1 | 139-155 | TTESAFGLICATCEQIA | 4 | 45 | novel | ND |
| M1 | 163-179 | RQMVTTTNPLIRHENRM | 4 | 20 | DRB1*0103 | CD4 |
| M1 | 169-185 | TNPLIRHENRMVLASTT | 1 | 42 | DRB1*0103; DRB1*1501; DRB5 | CD4 |
| M1e | 187-203 | KAMEQMAGSSEQAAEAM | 3, 4 | 47.5, 37.5 | DRB5 | CD4 |
| M1e | 193-209 | AGSSEQAAEAMEVASQA | 4 | 37.5 | novel | ND |
| M1e | 205-221 | VASQARQMVQAMRAIGT | 3 | 210 | novel | CD4 |
| M1e | 210-226 | RQMVQAMRAIGTHPSSS | 3, 4 | 142.5, 65 | novel | CD4 |
| M1e | 216-232 | MRAIGTHPSSSTGLKND | 3 | 115 | novel | ND |
| M1 | 228-244 | GLKNDLLENLQAYAKRM | 2 | 22.5 | DRB5 | CD4 |
| M1 | 234-250 | LENLQAYQKRMGVQMQR | 1 | 40 | DRB1*0103; DQw3, DQw1 | CD4 |
| M1 | 240-252 | YQKRMGVQMQRFK | 4 | 35 | DQw1 | CD4 |
| NS1 | 13-29 | CFLWHVRKQVADQDKGD | 4 | 25 | novel | ND |
| NS1 | 120-136 | DQAIMDKNIILKANFSV | 1 | 24 | A*0201 | CD8 |
| NS2e | 13-29 | LMRMSKMQLGSSSGDLN | 4 | 35 | novel | ND |
| NS2e | 19-35 | MQLGSSSGDLNGMITQF | 4 | 30 | novel | ND |
| NS2 | 37-53 | SLKLYRDSLGEAVMRLG | 1, 4 | 50, 35 | novel | ND |
| NS2 | 85-101 | HKLKTTENSFEQITFMQ | 4 | 37.5 | novel | ND |
| NP | 55-71 | RLIQNSLTIERMVLSAF | 4 | 47.5 | novel | ND |
| NPe | 109-125 | VLYDKEEIRRIWRQANN | 3 | 165 | novel | CD4 |
| NPe | 115-131 | EIRRIWRQANNGDDATA | 3 | 70 | Novel | CD4 |
| NPe | 175-191 | RSGAAGAAVKGVGTMVL | 3 | 95 | B*2705f | CD4 |
| NPe | 187-203 | GTMVLELIRMIKRGIND | 3 | 75 | A*1101f | CD4 |
| NPe | 193-209 | LIRMIKRGINDRNFWRG | 3 | 102 | novel | CD4 |
| NP | 246-262 | RNPGNAEIEDLTFLARS | 4 | 22.5 | novel | ND |
| NP | 258--274 | FLARSALILRGSVAHKS | 1 | 40 | A3 | CD8 |
| NP | 294-310 | EGYSLVGVDPFKLLQTS | 4 | 22 | novel | ND |
| PA | 220-236 | PPNFSCIENFRAYVDGF | 1 | 22 | A*0201 | CD8 |
| PA | 310-326 | CMRTFFGWKEPTVVKPH | 4 | 32 | novel | ND |
| PA | 516-532 | DVVNFVSMEFSLTDPRL | 1 | 20 | novel | ND |
| PA | 576-592 | KWGMEMRRCLLQSLQQI | 4 | 36 | novel | ND |
| PB1 | 406-422 | GMMMGMFNMLSTVLGVS | 1 | 26 | A*0201 | CD8 |
| PB1 | 447-463 | FALIVNAPNYAGIQAGV | 1 | 44 | novel | ND |
| PB1e | 537-553 | NDLGPATAQMALQLFIK | 1 | 32 | B7 | CD8 |
| PB1e | 548-564 | LQLFIKDYRYTYRCHRG | 1 | 36 | novel | ND |
| PB1-F2 (H3N2) | 65-81 | KNPTQGSLRTHALKQWK | 1 | 48 | novel | ND |
| PB2 | 203-219 | VAYMLERELVRKTRFLP | 2 | 30 | novel | ND |
Underlined amino acid sequences are previously published T cell epitopes (also see comments). Amino acid/s highlighted in bold reflects an amino acid change in our peptide sequence compared to the previously published epitope.
SFC values are representative of one ELISPOT experiment with three replicate wells per donor.
The HLA class I and class II alleles described in the comments are the restricting MHC allele of the underlined amino acid sequence.
T-cell phenotype was determined by either IFN-γ ELISPOT using CD4 or CD8 depleted donor PBMC or IFN-γ ICS using peptide-stimulated bulk culture cells as effectors. ND (not determined).
This peptide contains amino acid residues that overlaps with another peptide and is identified by the same donor.
This peptide contains a known CD8 T-cell epitope, but the peptide-stimulated bulk culture we generated has a CD4 T-cell phenotype.
Figure 3.
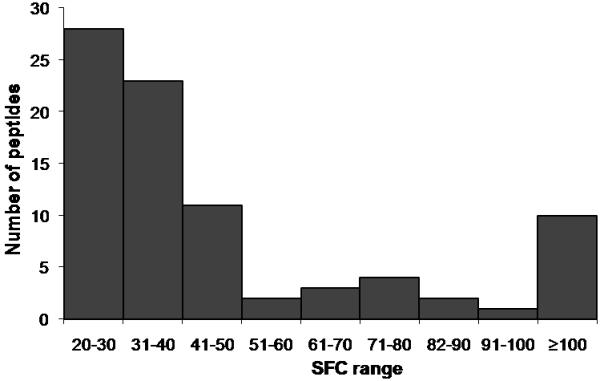
Distribution of SFC values of positive peptides in ELISPOT assays. The SFC values for each peptide that gave a positive IFN-γ response were plotted in a histogram chart to determine the frequency of IFN-γ-producing cells among PBMC of the four donors.
Determining phenotype of IFN-γ producing cells by depletion
To determine the T cell population that produced IFN-γ after peptide stimulation, we depleted CD4 or CD8 cells from the donors’ PBMC. We then performed ELISPOT using the peptide of interest to stimulate the CD4 or CD8 depleted cells. Peptides that had SFC values greater than 60 in Table 3 were first analyzed to ensure that there are enough peptide-specific precursor cells. Majority of the IFN-γ producing cells that responded to the peptides in ELISPOT were CD4+ cells, given by a drastic decrease in SFC values when CD4-expressing cells are depleted from whole PBMC (Table 4). For the other candidate peptides, bulk culture cell lines using donor PBMC were set-up and ICS was performed on days 12-14 to determine the phenotype of the IFN- γ producing cells. Figure 4 shows representative FACS plots determining the phenotype of IFN-γ producing cells from peptide-stimulated bulk culture using donor 3 PBMC. More than 80% of IFN-γ producing T cells expressed the CD4 surface marker upon cognate peptide stimulation in ICS (Figure 4). A complete list of candidate peptides tested by either ELISPOT or ICS is given in Table 3.
Table 4.
Phenotype of IFN-γ-producing cells after CD4+ or CD8+ cell depletion
| SFC/106 |
|||
|---|---|---|---|
| Peptide | Whole PBMC | CD4-depleted PBMC | CD8-depleted PBMC |
| DONOR 1 | |||
| aM158-66 | 401.7 | 281.7 | 0 |
| bH3 HA322-334 | 8.3 | 6.7 | 25 |
| H3 HA273-289 | 46.7 | 8.3 | 65 |
| M1234-250 | 53.3 | 1.3 | 57.3 |
| N2176-192 | 34.7 | 0 | 68 |
| DONOR 3 | |||
| M191-107 | 90.7 | 0.0 | 164.2 |
| M197-113 | 124 | 27.5 | 152.5 |
| M1205-221 | 68.0 | 7.5 | 174.2 |
| M1210-226 | 97.3 | 7.5 | 235 |
| M1216-232 | 22.7 | 0.8 | 50 |
| N2176-192 | 44 | 0.8 | 135 |
| NP109-125 | 74.7 | 7.5 | 95 |
| NP115-131 | 30 | 0.8 | 45 |
| NP175-191 | 22 | 0.8 | 20 |
| NP187-203 | 37.3 | 2.5 | 42.5 |
| NP193-209 | 52 | 4.2 | 102.5 |
| H3 HA249-265 | 25 | 2.5 | 25 |
M158-66 is an HLA-A2-restricted immunodominant epitope. It is used here as a control peptide for positive CD8 T cell responses in HLA-A2 donors.
H3 HA322-334 is a HLA-DR1-restricted epitope. It is used here as a control peptide for positive CD4 T cell responses in HLA-DR1 donors.
Figure 4.
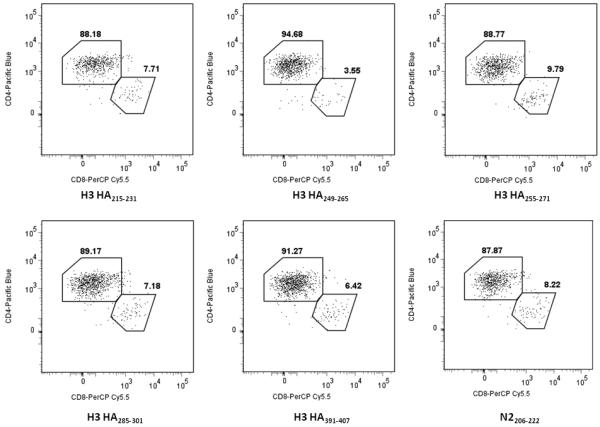
Flow cytometry to determine the phenotypes of IFN-γ producing cells in donor 3 PBMC. Peptide-stimulated bulk culture cells using donor 3 PBMC were used as effectors in an ICS. The peptides shown here were used to initially stimulate whole PBMC and establish the bulk culture. The final peptide concentration was 10μg/ml in all experiments. Events were gated for the live, CD3+ IFN-γ+ cells.
IFN-γ responses to HA proteins using additional source of PBMC
Since we detected HA-specific T cell responses in the first four donors and there was no well-characterized CD8+ T cells epitope, we decided to analyze HA-specific T cell responses using the PBMC of 30 hospital workers. The same ELISPOT strategy was used to screen these donors’ PBMC. Only H1 HA and H3 HA peptides were screened because of the limited number of PBMC available from these donors (Table 5). Sixteen of 30 donors (53.3%) gave a positive response to H3 HA peptides, and five of 30 donors (16.7%) gave a positive response to H1 HA peptides.
Table 5.
H1 and H3 HA peptides that gave a positive IFN-γ response in ELISPOT screening using PBMC from an additional 30 donors
| Protein | AA Position | Sequencea | No. of Positive Donors | SFC/106b |
|---|---|---|---|---|
| H3 HA | 13-29 | LVFAQKLPGNDNSTATL | 1 | 62.5 |
| H3 HA | 37-53 | PNGTIVKTITNDGIEVT | 2 | 50, 107.5 |
| H3 HA | 43-59 | KTITNDQIEVTNATELV | 2 | 30, 37.5 |
| H3 HA | 59-75 | VQSSSTGGICDSPHQIL | 1 | 57.5 |
| H3 HA | 83-99 | IDALLGDPQCDGFQNKK | 1 | 52.5 |
| H3 HA | 107-123 | SKAYSNCYPYDVPDYAS | 2 | 40, 50 |
| H3 HA | 119-135 | PDYASLRSLVASSGTLE | 1 | 22.5 |
| H3 HA | 131-147 | SGTLEFNNESFNWTGVT | 1 | 37.5 |
| H3 HA | 155-171 | CKRRSNNSFFSRLNWLT | 1 | 20 |
| H3 HAc | 209-225 | SLYAQASGRITVSTKRS | 4 | 20, 25, 330, 167.5 |
| H3 HAc | 215-231 | SGRITVSTKRSQQTVIP | 2 | 37.5 |
| H3 HAc | 243-259 | PSRISIYWTIVKPGDIL | 5 | 33.3, 37.5, 40, 42.5, 75 |
| H3 HAc | 249-265 | YWTIVKPGDILLINSTG | 4 | 27.5, 27.5, 32.5, 32.5 |
| H3 HAc | 255-271 | PGDILLINSTGNLIAPR | 4 | 30, 22.5, 35, 50 |
| H3 HA | 267-283 | LIAPRGYFKIRSGKSSI | 1 | 20 |
| H3 HA | 291-307 | GKCNSECITPNGSIPND | 1 | 22.5 |
| H3 HA | 321-337 | CPRYVKQNTLKLATGMR | 3 | 50, 35, 67.5 |
| H3 HA | 344-360 | TRGIFGAIAGFIENGWE | 1 | 50 |
| H3 HA | 368-384 | GFRHQNSEGIGQAADLK | 1 | 27.5 |
| H3 HA | 386-402 | TQAAINQINGKLNRLIG | 2 | 27.5, 32.5 |
| H3 HA | 397-413 | LNRLIGKTNEKFHQIEK | 1 | 22.5 |
| H3 HA | 403-419 | KTNEKFHQIEKEFSEVE | 1 | 57.5 |
| H3 HA | 409-425 | HQIEKEFSEVEGRIQDL | 1 | 57.5 |
| H3 HA | 421-437 | RIQDLEKYVEDTKIDLW | 1 | 90 |
| H3 HA | 433-449 | KIDLWSYNAELLVALEN | 3 | 35, 27.5, 57.5 |
| H3 HA | 439-455 | YNAELLVALENQHTIDL | 2 | 22.5, 35 |
| H3 HA | 457-473 | DSEMNKLFERTKKQLRE | 1 | 55 |
| H3 HA | 463-479 | LFERTKKQLRENAEDMG | 1 | 20 |
| H3 HA | 481-497 | GCFKIYHKCDNACIGSI | 3 | 62.5, 22.5, 80 |
| H3 HA | 505-521 | DVYRDEALNNRFQIKGV | 1 | 85 |
| H3 HA | 528-543 | KDWILWISFAISCFLL | 1 | 50 |
| H3 HA | 550-566 | FIMWACQKGNIRCNICI | 1 | 47.5 |
| H1 HA | 37-53 | LEKNVTVTHSVNLLEDS | 1 | 27.5 |
| H1 HA | 156-172 | GKSSFYRNLLWLTGKNG | 1 | 35 |
| H1 HA | 262-278 | GNLIAPWYAFALSRGFG | 1 | 60 |
| H1 HA | 416-432 | LERRMENLNKKVDDGFL | 1 | 40 |
| H1 HA | 434-450 | IWTYNAELLVLLENERT | 1 | 25 |
| H1 HA | 458-474 | VKNLYEKVKSQLKNNAK | 1 | 37.5 |
| H1 HA | 464-479 | KVKSQLKNNAKEIGNG | 2 | 65, 75 |
| H1 HA | 480-496 | CFEFYHKCNNECMESVK | 1 | 45 |
| H1 HA | 510-526 | KLNREKIDGVKLESMGV | 1 | 35 |
| H1 HA | 527-543 | YQILAIYSTVASSLVLL | 1 | 20 |
| H1 HA | 545-560 | SLGAISFWMCSNGSLQ | 1 | 42.5 |
| H1 HA | 550-565 | SFWMCSNGSLQCRICI | 1 | 20 |
Underlined sequences are known influenza HA epitopes. Amino acids highlighted in bold reflect an amino acid change in our peptide sequence compared to the previously published epitope.
SFC values are representative of one ELISPOT experiment with three replicate wells for each donor positive for the peptide.
This peptide contains amino acid residues that overlaps with another peptide and tested positive for IFN-γ response using PBMC from the same donor.
Peptide-specific CD4+ T cell lines produce IFN-γ upon cognate peptide stimulation
We generated bulk culture lines by stimulating donor PBMC with peptides of interest to further characterize the peptide-specific responses we identified in our ELISPOT screening. We selected peptides that potentially contain novel epitopes by searching MHC class I and class II binding motifs within the peptides by two prediction algorithms, HLA Peptide Binding Predictions (http://www-bimas.cit.nih.gov/molbio/hla_bind/) [23] and SYFPEITHI (http://www.syfpeithi.de) [24]. After two weeks in culture, peptide-stimulated bulk culture lines that had specific CTL killing of ≥15% in 51Cr release assays (E:T ratios of 10, 30 and 90 were tested) were used in a limiting dilution set-up to generate peptide-specific T cell lines. We generated T cell lines that are specific to peptides H3 HA267-283, H3 HA350-366, M1205-221, and M191-107, as well as to known T-cell epitopes contained in peptides H3 HA321-337 (PRYVKQNTLKLAT, HA322-334) restricted by HLA-DR1 and M155-71 (GILGFVFTL, M158-66) restricted by HLA-A2. A 51Cr release assay was performed to determine the ability of the T cell lines to kill targets pulsed with decreasing doses of cognate peptide. All T cell lines were able to kill peptide-pulsed targets at a peptide concentration of 10 μg/ml (> 15% specific lysis, Table 6). We also determined the surface expression of CD4 or CD8 of these T cell lines by flow cytometry. The T cell lines that were specific to H3 HA267-283 (1-3E2), H3 HA350-366 (2-10D8), and M1205-221 (3-1C9) were CD4+ (Table 6, >95% purity), while the M1-10-specific T cell line was CD8+ (data not shown). We also performed ICS for IFN-γ by pulsing autologous BLCLs with the cognate peptide and adding the appropriate T cell line prior to Golgi plug application (Table 6). T cell lines were able to produce IFN-γ upon cognate peptide stimulation of autologous APCs, comparable to the virus-infected control, although only a low percentage of the 2-10D8 T cell line produced IFN-γ after stimulation with either live influenza A virus or the peptide.
Table 6.
Recognition of influenza virus peptides by CTL lines generated from donor PBMC
| % Specific 51Cr releasea |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cell Lineb |
Protein | Epitope (AA position) |
CD4/CD8c | % CD4 T-cells producing IFN-γd |
HLA restrictione |
10μg/ml peptide |
1 μg/ml peptide |
0.1 μg/ml peptide |
0.001 μg/ml peptide |
| 1-3E2 | H3 HA | 267-283 | CD4 | 84.5 | DRB1*01 | 59.2 | 38.0 | 31.7 | 31.5 |
| 2-10D8 | H3 HA | 350-366 | CD4 | 0.73f | N.D. | 30.2 | 8.5 | 12.9 | 8.4 |
| 3-1C9 | M1 | 205-221 | CD4 | 27.6 | N.D. | 55.7 | 35.5 | 37.9 | 7.4 |
E:T ratio was 10:1; values are representative of one experiment. Specific lysis was determined by subtracting the percentage of lysis of unpulsed targets by effector cells from that of peptide-pulsed targets. Lysis of unpulsed targets were <5%.
Cell lines were tested in CTL assay at least twice in triplicate wells.
T cell phenotype was determined by flow cytometry. Purity of T cell population is >98%.
IFN-γ producing cells were identified by ICS. Cell lines were added to either virus or peptide-stimulated autologous BLCLs. PMA/ionomycin was used as a positive control.
HLA restriction was determined by CTL assay against various BLCLs expressing different combinations of MHC class II molecules and by pre-treating target cells with anti-HLA-DR, -DQ, and -DP antibodies.
For this T-cell line, 62.4% of CD4 T-cells produced IFN-γ upon PMA/ionomycin stimulation, but only 0.22% of CD4 T-cells produced IFN-γ upon H3N2 virus infection.
DISCUSSION
Characterization of epitope-specific T cell responses is critical in the understanding of immune responses to infection or vaccination. In this study, we performed a less biased and comprehensive screening of peptides covering all influenza A virus proteins. We confirmed previous reports by us [8, 9] and others [10-13] that the T cell responses to influenza are broadly directed to several viral proteins. We found that surface glycoproteins, HA and NA, which are major components of inactivated vaccines, had many T cell epitopes. Recently we [16, 25] and others [26] reported that trivalent inactivated influenza vaccine can induce T cell responses and part of these T cell responses may be targeting epitopes on HA, NA and M1. Overall, HA and M1 had more T cell epitopes than other viral proteins, most of which were recognized by CD4+ T cells. One limitation of this study is that for some of the peptides identified in our screening, we were not able to determine whether T cells responding to a given peptide were CD4+ or CD8+ T cells. We did not use in vitro amplification of influenza A-specific T cells to avoid the skewing the relative frequency of the T cells specific to each epitope. As a result, the frequency of most peptide-specific T cells in PBMC was not high enough to determine the phenotype by performing ICS or depletion experiments. Based on the ICS of PBMC stimulated with live influenza A viruses it is estimated that approximately 80% of T cells responding to the viruses are CD8+ T cells (data not shown). However, for peptides that we were able to do further studies, they were all recognized by CD4+ T cells, with the exception of the CD8+ T cell epitope, M158-66. The peptides we used for the screening are 17mers, which may have stimulated CD4+ T cells better than CD8+ T cells and as a result the sensitivity of detecting CD4+ T cells may have been relatively higher. Ideally we should have included influenza-naïve donors as a negative control to show that T cells responding to influenza peptides have been generated against influenza virus. However, it is practically not possible to find influenza-naïve adults (we cannot obtain large volume of blood for genome-wide screening experiments from very young influenza-naïve children).
Assarsson et al. recently found that PB1 was the major target for both CD4+ and CD8+ T cell responses [13]. In our screening, HA (especially H3) and M1 were major targets of T cell responses. The peptides they screened were predicted to have high-affinity binding to HLA class I or class II molecules and to be highly conserved. Our peptides were 17mers overlapping by 11 amino acids covering all influenza A viral proteins. Using minimal epitope peptides in the assays is likely to increase the sensitivity of detecting responding T cells, especially in the case of CD8+ T cells. Using longer peptides covering all viral proteins is probably less sensitive in detecting specific CD8+ T cells, but can detect T cells recognizing atypical T cell epitopes or epitopes restricted by MHC class I or class II molecules whose binding motif predictions are not available (for example HLA-DP and DQ alleles and rare HLA-A, B and C alleles). These differences in the peptide sets may explain why the two screenings by Assarsson et al. and the present study produced different results. Because HLA-restriction of the epitope candidate peptides identified in this screening has not been determined, we were not able to test if these epitopes could be identified by the computer predictions. We think that our results are complementary to those of Assarsson et al. and that both approaches may be used, if possible, to identify T cell epitopes on a virus.
The abundance of T-cell responses against HA was confirmed by analyzing the PBMC of 30 more donors. In screening using the PBMC of 30 hospital workers, more responses to H3 HA were seen than to H1 HA (53.3% to H3 and 16.7% to H1). These may reflect the epidemiology of currently circulating influenza A strains, prior infections with H3N2 versus H1N1 viruses, or higher virulence of H3N2 strain than H1N1 strain. Additionally, only 17% of the hospital workers had responses to H1, thus it is probably not surprising that none of the four healthy donors we previously screened had responses to H1. The amino acid sequence identity between the HA and NA of A/New Caledonia/20/99 (H1N1) and A/Wisconsin/67/2005 (H3N2), is 40% for the HA (AAP34324 and ABW80978) and 42% for the NA (CAD57252 and ABP52004) respectively. This suggests a low probability of identifying subtype cross-reactive T cell epitopes in these proteins. However, in two of the four healthy adult donors whose PBMC were screened, we detected T cells responding to the peptides encoded by the H5 HA gene in IFN-γ ELISPOT assays. Although the frequencies of these T cells were not high, they were comparable to those responding to the peptides encoded by H3 HA gene. Recently Roti and colleagues [27] reported the presence of CD4+ T cells recognizing epitopes encoded by H5 HA gene in healthy individuals, who were unlikely to have been exposed to the H5N1 virus, although in vitro amplification of specific T cells were needed to detect them, suggesting a low frequency of these H5 HA cross-reactive T cells. They found that none of the H5 HA epitopes identified were uniquely cross-reactive to H2 HA, which is the closest subtype to H5 and suggested that exposure to H2N2 viruses is not essential for cross-reactivity to H5 HA. Except for two peptides (H5151-167 and H5243-259 in Table 3) which have a four- to eight-amino acid overlap with the H5 HA epitopes identified by them, the H5 HA peptides that our donors’ PBMCs responded to are different. We did not observe responses to the H1 and H3 HA peptides corresponding to these eight H5 HA peptides in these donors. Other groups have also identified cross-reactive memory T cell responses to avian H5N1 proteins in healthy individuals who were previously infected or exposed to seasonal influenza [28, 29], as well as in individuals who were recently vaccinated for influenza [30]. Most of the cross-reactive responses were towards the internal proteins M1 and NP [28, 29], which is expected since the internal proteins are highly conserved even among the different subtypes. They were also able to identify cross-reactive responses to the HA [28] and NA [30] proteins.
Many of the novel H3 HA T cell epitopes we detected and those previously defined are situated at conserved segments of the H3 protein sequence, with the majority of them clustering at the C-terminus. This confirms a previous study that correlated the H3 three-dimensional structure and the epitopes that had been identified in mice and humans and found that dominant epitopes to HA are primarily located in conformationally stable segments of the C-terminal region [31]. In addition, a recent study using HLA-DR1 transgenic mice [32] identified a diverse HA-specific, HLA-DR1-restricted CD4+ T cell response, with the majority of epitopes located in conserved HA regions. Our data also suggest that the T cell responses are directed to influenza HA regions that are structurally and functionally conserved. Repeated infection or immunization by different virus strains may selectively stimulate T cells specific to the epitopes located in conserved regions.
CD4+ T cell effector and memory responses to influenza infection have been studied to some extent in mice (reviewed in [33, 34]), however, our knowledge of CD4+ T cell responses to influenza in humans is quite limited. A few HA CD4+ T cell epitopes have been identified in humans [35-38], and from recent study done using HLA-DR1 transgenic mice [32]. These influenza-specific CD4+ T cell responses may have important roles during influenza infection. An earlier study done in athymic mice reported a differential ability of influenza T helper clones to afford help to B-cells, depending on whether the B cells presented either viral surface proteins or internal viral components on their cell surface [39]. In Balb/c mice, CD4+ effector T cells mediated protection against a lethal influenza infection by perforin-mediated cytotoxicity [40]. Moreover, higher neutralizing antibody titers were attributed to CD4+ T cell help [40]. In another study, murine CD4+ T cells were able to traffic to the lungs during influenza infection, but needed antigen to be present to proliferate [41]. By stimulating PBMC with peptides of interest, we established CD4+ T cell lines specific to three peptides. These three lines were able to lyse target cells pulsed with the peptides. One CD4+ T cell line, 2-10D8, produced little IFN-γ after stimulation with autologous BLCL pulsed with the peptide or infected with influenza A virus, although the peptide recognized by the line was initially identified in IFN-γ ELISPOT assays, suggesting that measuring IFN-γ production alone is likely to underestimate the T cell responses to influenza A virus.
In summary, our screening data showed the strength and breadth of T cell responses against influenza A virus at baseline levels of healthy adults. These T cell responses target both subtype specific (most of the epitopes localized with HA and NA peptides) and subtype cross-reactive (internal protein peptides) peptides. Genome-wide screening using overlapping peptides covering all viral proteins is useful for identifying T cell epitopes and complementary to the approach based on the predicted binding peptides to well-studied HLA-A, B and DR alleles. It is important to analyze the effect of influenza vaccination on T cell responses, and the information reported here will be useful for that purpose.
ACKNOWLEDGEMENT
We would like to thank Marcia Woda for assistance with our ICS experiments, Christine Turcotte and Denise Marengo for assistance with HLA typing, Dr. Jeffrey S. Kennedy who helped obtain some blood samples, and Kim West and Dr. Alan L. Rothman for discussion. We also thank Dr. Shibani Mitra-Kaushik for her critical reading of the manuscript, and Dr. Michel DeWilde and Dr. Robert Ryall of Sanofi Pasteur for the influenza virus strains used in this study. This work was supported by NIH / NIAID grant U19 AI-057319. The reagents listed in Table 1 were obtained through the NIH BEI Resources Repository. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID/NIH.
Abbreviations used
- NP
nucleoprotein
- M1
matrix protein 1
- HA
hemagglutinin
- CTL
cytotoxic T lymphocyte
- HLA
human leukocyte antigen
- A/PR/8
A/Puerto Rico/8/34 (H1N1) strain
- NA
neuraminidase
- PBMCs
peripheral blood mononuclear cells
- NIH
National Institutes of Health
- BEI Resources
Biodefense and Emerging Infections Research Resources Repository
- MHC
major histocompatibility complex
- DMSO
dimethyl sulfoxide
- NIAID
the National Institute of Allergy and Infectious Diseases
- ELISPOT
enzyme-linked immunosorbent spot
- IFN
interferon
- aN1
avian N1
- hN1
human N1
- M2
matrix protein 2
- NS1
nonstructural protein 1
- NS2
nonstructural protein 2
- PB1
polymerase B1
- PB2
polymerase B2
- PA
polymerase A
- SFC
spot forming cells
- PHA
Phytohemagglutinin
- APCs
antigen presenting cells
- ICS
intracellular cytokine staining
- BLCL
B lymphoblastoid cell lines
- FBS
fetal bovine serum
- PBS
phosphate buffered saline
- rIL
recombinant interleukin
- IEDB
Immune Epitope Database
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.The Writing Committee of the World Health Organization Consultation on Human Influenza AH Avian Influenza A (H5N1) Infection in Humans. N Engl J Med. 2005 September 29;353(13):1374–85. doi: 10.1056/NEJMra052211. 2005. [DOI] [PubMed] [Google Scholar]
- 2.Doherty PC, Turner SJ, Webby RG, Thomas PG. Influenza and the challenge for immunology. Nat Immunol. 2006;7(5):449–55. doi: 10.1038/ni1343. [DOI] [PubMed] [Google Scholar]
- 3.Epstein S. Prior H1N1 Influenza Infection and Susceptibility of Cleveland Family Study Participants during the H2N2 Pandemic of 1957: An Experiment of Nature. The Journal of Infectious Diseases. 2006;193(1):49–53. doi: 10.1086/498980. [DOI] [PubMed] [Google Scholar]
- 4.McElhaney JE, Xie D, Hager WD, Barry MB, Wang Y, Kleppinger A, et al. T Cell Responses Are Better Correlates of Vaccine Protection in the Elderly. J Immunol. 2006 May 15;176(10):6333–9. doi: 10.4049/jimmunol.176.10.6333. 2006. [DOI] [PubMed] [Google Scholar]
- 5.Gotch F, Rothbard J, Howland K, Townsend A, McMichael A. Cytotoxic T lymphocytes recognize a fragment of influenza virus matrix protein in association with HLA-A2. Nature. 1987;326(6116):881–2. doi: 10.1038/326881a0. [DOI] [PubMed] [Google Scholar]
- 6.Yewdell JW, Bennink JR, Smith GL, Moss B. Influenza A virus nucleoprotein is a major target for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proceedings of the National Academy of Science. 1985;82:1785–9. doi: 10.1073/pnas.82.6.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamb JR, Green N. Analysis of the antigen specificity of influenza haemagglutinin-immune human T lymphocyte clones: identification of an immunodominant region for T cells. Immunology. 1983;50:659–66. [PMC free article] [PubMed] [Google Scholar]
- 8.Jameson J, Cruz J, Ennis FA. Human Cytotoxic T-Lymphocyte Repertoire to Influenza A Viruses. J Virol. 1998 November 1;72(11):8682–9. doi: 10.1128/jvi.72.11.8682-8689.1998. 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jameson J, Cruz J, Terajima M, Ennis FA. Human CD8+ and CD4+ T Lymphocyte Memory to Influenza A Viruses of Swine and Avian Species. J Immunol. 1999 June 15;162(12):7578–83. 1999. [PubMed] [Google Scholar]
- 10.Boon ACM, de Mutsert G, Graus YMF, Fouchier RAM, Sintnicolaas K, Osterhaus ADME, et al. The Magnitude and Specificity of Influenza A Virus-Specific Cytotoxic T-Lymphocyte Responses in Humans Is Related to HLA-A and -B Phenotype. J Virol. 2002 January 15;76(2):582–90. doi: 10.1128/JVI.76.2.582-590.2002. 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gianfrani C, Oseroff C, Sidney J, Chesnut RW, Sette A. Human memory CTL response specific for influenza A virus is broad and multispecific. Human Immunology. 2000;61(5):438–52. doi: 10.1016/s0198-8859(00)00105-1. [DOI] [PubMed] [Google Scholar]
- 12.Wang M, Lamberth K, Harndahl M, Roder G, Stryhn A, Larsen MV, et al. CTL epitopes for influenza A including the H5N1 bird flu; genome-, pathogen-, and HLA-wide screening. Vaccine. 2007;25(15):2823–31. doi: 10.1016/j.vaccine.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 13.Assarsson E, Bui H-H, Sidney J, Zhang Q, Glenn J, Oseroff C, et al. Immunomic Analysis of the Repertoire of T-Cell Specificities for Influenza A Virus in Humans. J Virol. 2008 December 15;82(24):12241–51. doi: 10.1128/JVI.01563-08. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Currier JR, Kuta EG, Turk E, Earhart LB, Loomis-Price L, Janetzki S, et al. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. Journal of Immunological Methods. 2002;260(12):157–72. doi: 10.1016/s0022-1759(01)00535-x. [DOI] [PubMed] [Google Scholar]
- 15.Boyam A. Isolation of mononuclear cells and granulocytes from human blood. Scand J Clin Lab Invest. 1968;21:77. [PubMed] [Google Scholar]
- 16.Co MDT, Orphin L, Cruz J, Pazoles P, Rothman AL, Ennis FA, et al. Discordance between antibody and T cell responses in recipients of trivalent inactivated influenza vaccine. Vaccine. 2008;26(16):1990–8. doi: 10.1016/j.vaccine.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green S, Kurane I, Edelman R, Tacket CO, Eckels KH, Vaughn DW, et al. Dengue virus-specific human CD4+ T-lymphocyte responses in a recipient of an experimental live-attenuated dengue virus type 1 vaccine: bulk culture proliferation, clonal analysis, and precursor frequency determination. J Virol. 1993 October 1;67(10):5962–7. doi: 10.1128/jvi.67.10.5962-5967.1993. 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anthony DD, Lehmann PV. T-cell epitope mapping using the ELISPOT approach. Methods. 2003;29(3):260–9. doi: 10.1016/s1046-2023(02)00348-1. [DOI] [PubMed] [Google Scholar]
- 19.Oseroff C, Kos F, Bui H-H, Peters B, Pasquetto V, Glenn J, et al. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proceedings of the National Academy of Sciences of the United States of America. 2005 September 27;102(39):13980–5. doi: 10.1073/pnas.0506768102. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMichael AJ, Gotch FM, Santos-Aguado J, Strominger JL. Effect of mutations and variations of HLA-A2 on recognition of a virus peptide epitope by cytotoxic T lymphocytes. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9194–8. doi: 10.1073/pnas.85.23.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bui H-H, Peters B, Assarsson E, Mbawuike I, Sette A. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proceedings of the National Academy of Sciences. 2007 January 2;104(1):246–51. doi: 10.1073/pnas.0609330104. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters B, Sidney J, Bourne P, Bui H-H, Buus S, Doh G, et al. The Immune Epitope Database and Analysis Resource: From Vision to Blueprint. PLoS Biology. 2005;3(3):e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994 January 1;152(1):163–75. 1994. [PubMed] [Google Scholar]
- 24.Rammensee HG, Bachmann J, Emmerich NPN, Bachor OA, Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3):213–9. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 25.Terajima M, Cruz J, Leporati AM, Orphin L, Babon JAB, Co MDT, et al. Influenza A Virus Matrix Protein 1-Specific Human CD8+ T-Cell Response Induced in Trivalent Inactivated Vaccine Recipients. J Virol. 2008 September 15;82(18):9283–7. doi: 10.1128/JVI.01047-08. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X-S, Holmes TH, Zhang C, Mahmood K, Kemble GW, Lewis DB, et al. Cellular Immune Responses in Children and Adults Receiving Inactivated or Live Attenuated Influenza Vaccines. J Virol. 2006 December 1;80(23):11756–66. doi: 10.1128/JVI.01460-06. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roti M, Yang J, Berger D, Huston L, James EA, Kwok WW. Healthy Human Subjects Have CD4+ T Cells Directed against H5N1 Influenza Virus. J Immunol. 2008 February 1;180(3):1758–68. doi: 10.4049/jimmunol.180.3.1758. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee LY, Ha do LA, Simmons C, de Jong MD, Chau NV, Schumacher R, et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest. 2008 Oct;118(10):3478–90. doi: 10.1172/JCI32460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreijtz JHCM, de Mutsert G, van Baalen CA, Fouchier RAM, Osterhaus ADME, Rimmelzwaan GF. Cross-Recognition of Avian H5N1 Influenza Virus by Human Cytotoxic T-Lymphocyte Populations Directed to Human Influenza A Virus. J Virol. 2008 June 1;82(11):5161–6. doi: 10.1128/JVI.02694-07. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gioia C, Castilletti C, Tempestilli M, Piacentini P, Bordi L, Chiappini R, et al. Cross-subtype immunity against avian influenza in persons recently vaccinated for influenza. Emerg Infect Dis. 2008 Jan;14(1):121–8. doi: 10.3201/eid1401.061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landry SJ. Three-Dimensional Structure Determines the Pattern of CD4+ T-Cell Epitope Dominance in Influenza Virus Hemagglutinin. J Virol. 2008 February 1;82(3):1238–48. doi: 10.1128/JVI.02026-07. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richards KA, Chaves FA, Krafcik FR, Topham DJ, Lazarski CA, Sant AJ. Direct Ex Vivo Analyses of HLA-DR1 Transgenic Mice Reveal an Exceptionally Broad Pattern of Immunodominance in the Primary HLA-DR1-Restricted CD4 T-Cell Response to Influenza Virus Hemagglutinin. J Virol. 2007 July 15;81(14):7608–19. doi: 10.1128/JVI.02834-06. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown DM, Roman E, Swain SL. CD4 T cell responses to influenza infection. Seminars in Immunology. 2004;16(3):171–7. doi: 10.1016/j.smim.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Swain SL, Agrewala JN, Brown DM, Jelley-Gibbs DM, Golech S, Huston G, et al. CD4+ T-cell memory: generation and multi-faceted roles for CD4+ T cells in protective immunity to influenza. Immunological Reviews. 2006;211(1):8–22. doi: 10.1111/j.0105-2896.2006.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gelder C, Davenport M, Barnardo M, Bourne T, Lamb J, Askonas B, et al. Six unrelated HLA-DR-matched adults recognize identical CD4+ T cell epitopes from influenza A haemagglutinin that are not simply peptides with high HLA-DR binding affinities. Int Immunol. 1998 February 1;10(2):211–22. doi: 10.1093/intimm/10.2.211. 1998. [DOI] [PubMed] [Google Scholar]
- 36.Gelder CM, Lamb JR, Askonas BA. Human CD4+ T-cell recognition of influenza A virus hemagglutinin after subunit vaccination. J Virol. 1996 July 1;70(7):4787–90. doi: 10.1128/jvi.70.7.4787-4790.1996. 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gelder CM, Welsh KI, Faith A, Lamb JR, Askonas BA. Human CD4+ T-cell repertoire of responses to influenza A virus hemagglutinin after recent natural infection. J Virol. 1995 December 1;69(12):7497–506. doi: 10.1128/jvi.69.12.7497-7506.1995. 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, James EA, Huston L, Danke NA, Liu AW, Kwok WW. Multiplex mapping of CD4 T cell epitopes using class II tetramers. Clinical Immunology. 2006;120(1):21–32. doi: 10.1016/j.clim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Scherle PA, Gerhard W. Differential Ability of B Cells Specific for External Vs. Internal Influenza Virus Proteins to Respond to Help from Influenza Virus-Specific T-Cell Clones in vivo. Proceedings of the National Academy of Sciences. 1988 June 15;85(12):4446–50. doi: 10.1073/pnas.85.12.4446. 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown DM, Dilzer AM, Meents DL, Swain SL. CD4 T Cell-Mediated Protection from Lethal Influenza: Perforin and Antibody-Mediated Mechanisms Give a One-Two Punch. J Immunol. 2006 September 1;177(5):2888–98. doi: 10.4049/jimmunol.177.5.2888. 2006. [DOI] [PubMed] [Google Scholar]
- 41.Chapman TJ, Castrucci MR, Padrick RC, Bradley LM, Topham DJ. Antigen-specific and non-specific CD4+ T cell recruitment and proliferation during influenza infection. Virology. 2005;340(2):296–306. doi: 10.1016/j.virol.2005.06.023. [DOI] [PubMed] [Google Scholar]