Abstract
In Parkinson’s disease (PD), oxidative stress is implicated in protein misfolding and aggregation, which may activate the unfolded protein response (UPR) by the endoplasmic reticulum (ER). Dopamine (DA) can initiate oxidative stress via H2O2 formation by DA metabolism and by oxidation into DA quinone (DAQ). We have previously shown that DAQ induces oxidative protein modification, mitochondrial dysfunction in vitro, and dopaminergic cell toxicity in vivo and in vitro. In this study, we used cysteine- and lysine-reactive fluorescent dyes with 2-D difference in-gel electrophoresis (2D-DIGE), mass spectrometry, and peptide mass fingerprint analysis to identify proteins in PC12 cell mitochondrial-enriched fractions that were altered in abundance following DA exposure (150μM,16h). Quantitative changes in proteins labeled with fluorescent dyes indicated increases in a subset of proteins after DA exposure: calreticulin, ERp29, ERp99, Grp58, Grp78, Grp94, and Orp150 (149-260%), and decreased levels of aldolase A (39-42%). Changes in levels of several proteins detected by 2D-DIGE were confirmed by Western blot. Using this unbiased proteomics approach, our findings demonstrated that in PC12 cells, DA exposure leads to a cellular response indicative of ER stress prior to the onset of cell death, providing a potential link between DA and the UPR in the pathogenesis of PD.
Keywords: proteomics, 2D-DIGE, mitochondria, PC12, dopamine, endoplasmic reticulum stress
In Parkinson’s disease (PD), the dopaminergic neurons of the substantia nigra (SN) degenerate, leading to movement dysfunction. Oxidative stress appears to play a role in the loss of these dopaminergic neurons, since increased levels of oxidized lipids, DNA, and protein, as well as decreased levels of the major cellular antioxidant, glutathione (GSH), have been measured in PD SN (for review, see Dawson and Dawson, 2003; Ischiropoulos and Beckman, 2003; Jenner, 2003).
Oxidative stress can contribute to cellular damage commonly observed in neurodegenerative disorders like PD by inducing mitochondrial dysfunction, endoplasmic reticulum (ER) stress, and protein accumulation (Fahn and Sulzer, 2004; Gandhi and Wood, 2005; Halliwell, 2006a, b; Lin and Beal, 2006; Calabrese et al., 2007). Mitochondrial dysfunction is well-documented in PD, with decreased mitochondrial complex I activity observed directly in the SN and systemically in PD patients (for review, see Lin and Beal, 2006; Mancuso et al., 2006). Mitochondrial complex I forms superoxide anion during normal respiration, and reactive oxygen species (ROS) can, in turn, inhibit mitochondrial complexes (Fiskum et al., 2003; Turrens, 2003). Partial inhibition of complex I has been shown to generate even more ROS, suggesting that ROS-induced mitochondrial dysfunction, similar to that observed in PD, can lead to further ROS production and increased oxidative protein damage (Pitkanen and Robinson, 1996; Votyakova and Reynolds, 2001).
Mitochondria are linked to the ER both by proximity and through common signaling mechanisms, including calcium homeostasis and pro- and anti-apoptotic pathways (Breckenridge et al., 2003; Rao et al., 2004). Thus mitochondria can influence ER stress, a cellular response to the accumulation of misfolded and oxidized proteins (Breckenridge et al., 2003; Rao et al., 2004). Oxidative stress can also contribute to ER stress via protein oxidation, by affecting calcium signaling, and by altering the redox balance necessary to maintain ER function (Chakravarthi et al., 2006; Gorlach et al., 2006). Over time, the accumulation of misfolded proteins, which is a characteristic of many neurodegenerative diseases, activates the unfolded protein response (UPR) in the ER, and may ultimately lead to cell death (Lindholm et al., 2006).
Dopaminergic neurons may be especially susceptible to oxidative stress due to the potentially reactive nature of dopamine (DA) and its metabolite 3,4-dihydroxyphenylacetic acid (DOPAC), which readily oxidize to produce reactive quinones and ROS (Tse et al., 1976; Graham, 1978). Dopamine exposure has been shown to cause oxidative damage to protein through direct modification of cysteinyl residues by DA quinone (DAQ) and to selectively damage DA neurons in vivo (Hastings and Zigmond, 1994; Hastings et al., 1996; Rabinovic et al., 2000). Therefore, DA exposure is an apt model to observe the effects of toxin-induced oxidative stress in dopaminergic cells.
Because DA exposure has previously been shown to initiate cell death in PC12 cells (Walkinshaw and Waters, 1995; Cantuti-Castelvetri and Joseph, 1999; Jones et al., 2000; Koshimura et al., 2000; Xiao-Qing et al., 2005), we used this model combined with proteomic techniques to examine changes in proteins following DA exposure in PC12 cells. As examining whole-cell proteomes by gel electrophoresis techniques can be potentially complex, we chose to examine cellular subfractions individually. In this study, we focused on mitochondrial-enriched fractions due to the role mitochondrial dysfunction plays in PD, and because DAQ has been shown to inhibit the activity of mitochondrial protein complexes (including complex I), to uncouple mitochondrial respiration, and to open the permeability transition pore (Ben-Shachar et al., 1995; Berman and Hastings, 1999; Li and Dryhurst, 2001; Khan et al., 2005; Schapira, 2007). We used cysteine reactive maleimide-CyDyes or lysine reactive NHS-ester-CyDyes to label proteins, followed by 2-D difference in-gel electrophoresis, mass spectrometry (MS), and peptide mass fingerprint analysis for identification of proteins. Using this unbiased proteomics approach, we identified a subset of proteins in the mitochondrial-enriched fraction whose relative levels were altered following DA exposure. The only identified protein that was decreased in the mitochondrial-enriched fraction following DA was the glycolytic protein aldolase A. Surprisingly, most of the proteins which were increased after DA were ER chaperone proteins, and a subset of these proteins were confirmed to be changed by Western blot analyses of PC12 whole-cell lysates. The relative increase in the levels of several ER stress proteins suggests that ER stress occurs early and prior to cell death in a DA-induced toxicity model.
Materials and Methods
Chemicals and Reagents
Dulbecco’s Modified Eagle Medium (DMEM, Gibco brand), fetal bovine serum (HyClone brand), and horse serum (HyClone brand) were purchased from Invitrogen (Carlsbad, CA). Nerve growth factor (NGF) was purchased from BD Bioscience (San Diego, CA). Immobiline Drystrips pH 3-10, maleimide Cy3/Cy5 dyes, N-hydroxysuccinimidyl (NHS)-ester Cy3/Cy5 dyes, and IPG buffer pH 3-10 were purchased from GE Healthcare (Piscataway, NJ). Trypsin for MS analysis was obtained from Promega (Madison, WI). Protease inhibitor cocktail (PIC) included in mitochondrial isolation was obtained from Sigma (P2714; St. Louis, MO), and PIC added to PC12 cell whole cell lysate for Western blot analysis was obtained from Roche. All other non-specified reagents were purchased from Sigma. All solutions were made in distilled water purified with a Milli-Q system (Millipore Corp., Bedford, MA) unless otherwise noted.
Cell culture
PC12 cells, a dopaminergic cell line, were grown in media containing 7% horse serum (HS) and 7% fetal bovine serum (FBS). For differentiation, PC12 cells were plated at a density of 19,000 cells/cm2 in differentiation media (DMEM, 1% HS, 1% FBS, and 0.1 μg/ml NGF) for 6 d. The differentiated PC12 cells were then exposed to control media or 150 μM DA in media for 2-24 h for Western blot analysis of cell lysates and 16 h for isolated mitochondria experiments. Control cultures were treated at the same time as DA-exposed cultures.
Mitochondrial-enriched Fraction Preparation
PC12 cell mitochondrial-enriched fractions were prepared by a modified version of a previously described mitochondria isolation protocol (Berman and Hastings, 1999). Following exposure to control or DA-containing media, cells from 10-12 plates (100mm) were pooled for each group, collected in PBS, and combined with the treatment media prior to gentle centrifugation (800 × g, 3min). The supernatant was discarded, and the cell pellet was resuspended in isolation buffer containing 225 mM mannitol, 75 mM sucrose, 5 mM HEPES, 1 mg/mL BSA, PIC (2 μL/mL), pH 7.4, and the cells were lysed in a 2 mL Dounce homogenizer. The cell homogenate was then centrifuged (12,000 × g) three times at 3°C to isolate mitochondria. Both control and DA exposed mitochondrial-enriched pellets were lysed in buffer containing 9M urea, 2% w/v CHAPS, 30 mM Tris-base, and PIC (2.5 μL/mg protein), pH 8.0. The amount of protein in the lysed control and DA exposed mitochondrial-enriched fraction was measured by Bradford assay (Bradford, 1976). The mitochondria isolation procedure utilized with the DA-treated and control PC12 cells was identical. The mitochondrial-enriched samples from the two treatment groups remained separate until after the reaction with the fluorescent dyes, as described below.
2D Gel Electrophoresis
For first dimension separation of proteins, Immobiline DryStrips (linear pH 3-10; GE Healthcare) were rehydrated overnight at RT, according to the manufacturer’s protocol for cup loading of samples. Mitochondrial-enriched lysate from control and DA-exposed PC12 cells were reacted separately with either charge-matched fluorescent Cy5 or Cy3 maleimide dyes or Cy5 or Cy3 NHS ester dyes (GE Healthcare) prior to reduction by DTT. To confirm that the dyes exhibited no preferential chemistries, control and DA-exposed samples were reciprocally labeled with either Cy5 and Cy3 dyes, and direct comparison showed no evidence of any differential labeling affinities. Maleimides react with reduced sulfhydryl groups, and thus label reduced cysteines. The optimal dye:protein ratio was chosen so that protein spots visualized using the Typhoon 9400 scanner with PMT=600 were clearly visible, not overexposed, and with no apparent dye-shifting of spots. Preliminary trials using variable dye to protein ratios indicated that a maleimide reaction ratio of 1 pmol dye to 2 μg protein for 45 min at room temperature was optimal. The maleimide reaction was quenched by the addition of an equal volume of sample loading buffer containing 130 mM dithiothreitol (DTT), 9 M urea, 2% w/v CHAPS, 2% v/v IPG buffer (pH 3-10; GE Healthcare), and trace amounts of bromophenol blue.
The NHS-ester dye reaction ratio of 2 pmol dye to 1 μg protein was determined to be sufficient for lysine labeling. After the addition of the NHS-ester dye to the mitochondrial-enriched protein (prior to DTT reduction), the reaction was incubated on ice for 30 min and quenched by the addition of 10 nmol lysine (in 1 μL) at 4°C. Sample loading buffer was then added in a 1:1 ratio.
Following the addition of sample loading buffer for either maleimide or NHS-ester reacted samples, 125 μg protein from the control mitochondrial-enriched fraction were combined with 125 μg protein from DA exposed PC12 cell mitochondrial-enriched fraction, and the total 250 μg protein were loaded onto the first dimension strip using sample cup loading. To separate proteins by isoelectric point, the first dimension was run on a Multiphor II system according to the manufacturer’s protocol (GE Healthcare). The strips were prepared for the second dimension by washing with equilibration buffer (75 mM Tris-HCl, pH 6.8, 6M urea, 30% glycerol, 1% w/v SDS) containing 30 mM DTT followed by 240 mM iodoacetamide. The strip was trimmed to 13.5 cm, the molecular weight standards were loaded, and the second dimension was run on a 12% SDS polyacrylamide gel on a Hoeffer SE600 Ruby Electrophoresis Unit (GE Healthcare).
Visualization of Difference Gels and Spot Picking
Gels were rinsed with Milli-Q H2O and fixed in 40% methanol, 1% acetic acid. The gel was then scanned into the automated spot picker in the Genetics and Proteomics Core Laboratories at the University of Pittsburgh (developed by Dr. Jonathan Minden of Carnegie Mellon University). Protein spots that showed differences in relative fluorescence, plus several spots that appeared not to change, were picked.
Trypsin Digest and Mass Spectrometry of Target Proteins
Gel plugs were washed twice in 1:1 solution of methanol and 50 mM NH4HCO3, dehydrated in acetonitrile, and dried by speed-vacuum. In-gel trypsin digest was done at 42°C for 4 h, using Promega Gold trypsin reconstituted in 50% acetonitrile, 0.3% trifluoroacetic acid, and 1 mM ammonium citrate (0.2 μg/sample). The gel plug was then rinsed twice in extraction buffer (1% trifluoroacetic acid, in 50% acetonitrile and 50 % H2O). The trypsinized protein solution and extraction washes were dried by speed-vacuum.
The dried trypsinized protein was reconstituted in extraction buffer containing 100 μM ammonium citrate and mixed with a saturated solution of CHCA (α-cyano-4-hydroxycinnamic acid) and spotted on a metal MALDI plate along with a trypsin blank and mass standards (Applied Biosystems). The identification of the protein was determined using a 4700 MALDI-TOF-TOF (Applied Biosystems) mass spectrometer. The resulting ion peak spectra were used for peptide mass fingerprinting, searched against the NCBI database by GPS Data Explorer™ MS analysis software ver. 2 or 3 (Applied Biosystems), with the peptide mass tolerance set to 50 ppm and allowing a maximum of 1 missed trypsin cleavage. A positive protein identification was accepted when a confidence interval of >90% was obtained in coordination with a probability-based MOWSE (MOlecular Weight SEarch) protein score using the GPS Data Explorer™ MS analysis software coupled to the MASCOT database engine. Every protein identification was confirmed in two or more experiments. In all experiments, each protein spot was identified as only one protein and its related isoforms, so each protein spot appears to correspond to a single protein. No limitations other than species designation (All species and Rattus) were used in the database search, with no bias towards cell type or organelle.
Decyder Analysis of Fold Change
Spots with protein identifications were analyzed using the DeCyder Difference In-gel Analysis (DIA) software (GE Healthcare). Gel images from the Typhoon 9400 laser scanner (GE Healthcare) using equal PMT levels (typically 600 V) between the Cy5 and Cy3 scans, were loaded on the software. Each individual spot was identified and mapped using the DeCyder DIA software. Previously MS-identified spots were located on each gel image, and the normalized volume ratio calculated by the DeCyder DIA software was recorded for each spot. The normalized volume ratios were determined by the DeCyder DIA software by first calculating the direct volume ratio of the DA-exposed /control volume densities for every spot on the gel. All of the ratios were then normalized so that the modal peak of all the ratios equals 1, because most protein levels should be similar. Percent change of DA-exposed PC12 cell mitochondrial protein from control mitochondrial protein were determined from the normalized volume ratios, and the percent change within each gel was averaged over all gels in which the volumes for identified proteins could be determined. Limitations in the DeCyder software led to inaccurate spot mapping, and prevented analysis of one protein. This protein is designated as “UN” (undetermined) in Supplementary Data. Proteins with corresponding spots in both cysteine- and lysine-labeled gels which were not positively identified by our strict MS criteria in both sets of gels, are listed as “NI” (no identification) in Figure 3B and Supplementary Data. Proteins that did not have corresponding spots in both cysteine- and lysine-labeling experiments are listed as “ND” (no spot detected) in Figure 3B and Supplementary Data. Eight (8) maleimide (cysteine) dye gels from 6 separate experiments and 5 NHS-ester (lysine) dye gels from 4 separate experiments were used in the DeCyder analysis.
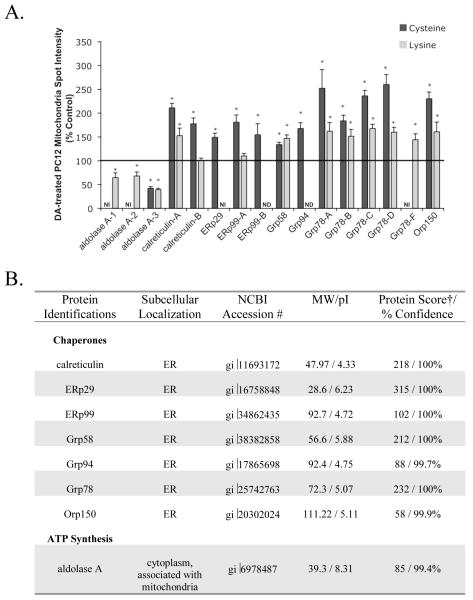
Figure 3. Changes in protein spot intensity of PC12 cell in the mitochondrial fraction following DA-exposure.
Mitochondrial-enriched fractions from control and DA-exposed (150 μM, 16 h) differentiated PC12 cells were reacted with Cy3 or Cy5 maleimide or NHS-ester dyes. Proteins listed above were identified by MALDI peptide mass fingerprint. Multiple spots with the same protein identification are denoted by letters (−A, −B, −C, etc) or numbers (−1, −2, and −3). (A.) The density of protein spots was analyzed using Decyder DIA software, and changes in DA exposed compared to control protein were determined in both maleimide (cysteine) and NHS-ester (lysine) reacted protein for most spots shown. Proteins spots that appear to correspond between cysteine- and lysine-labeling experiments, but were not identified according to our MS criteria, are labeled as NI (no identification). Proteins that do not have corresponding spots in both labeling experiments, are labeled with ND (no spot detected). DA treated PC12 cell protein intensity is measured as average % control ± SEM, n=5-8. *, significance p<0.05. (B.) Proteins identified that were significantly changed from control as quantified by DeCyder analysis and that were changed at least ± 1.2-fold from control in cysteine (maleimide) and/or lysine (NHS-ester) DIGE experiments are grouped by function. Actual molecular weight (MW in kDa) and isoelectric point (pI), best protein score and % confidence for each identified protein are also listed. † MASCOT probability-based protein MOWSE scores provided represent the highest protein score obtained across all experiments.
Collection of PC12 Whole-Cell Lysate for Western Blotting
PC12 cells were cultured and treated with 150 μM DA as described above. Following 2, 4, 8, 16, and 24 h of DA exposure or treatment with control media, the media was removed and PC12 cells were collected in PBS via force-pipetting. Cells from each plate were combined with the media before pelleting, to collect any floating cells. Cells were rinsed in PBS and re-pelleted prior to lysis in a buffer containing: 20mM Tris (pH 7.5), 150mM NaCl, 1mM EDTA, 1mM EGTA, 1% Triton X-100, 2.5mM sodium pyrophosphate, 1mM DTT, 1mM sodium orthovanadate, and 1X PIC (Roche).
Western Blot Analysis of MS-identified spots
Protein (20 μg) from control and DA-exposed PC12 whole cell and mitochondria-enriched lysates, were separated by 10, 12, or 15% SDS-PAGE and transferred to nitrocellulose membranes using a Trans-blot SD Semi-Dry Electrophoretic Transfer Cell (Biorad). Following transfer, the blots were washed in Tris buffered saline (50 mM Tris, pH 7.5, 150 mM NaCl; TBS), blocked with 0.2% w/v dry milk in TBS-T (TBS + 0.1% Tween 20), then incubated overnight at 4°C with primary antibody in TBS-T with 0.2% w/v dry milk. The blots were then washed in TBS-T and incubated at RT with the appropriate alkaline phosphatase conjugated secondary antibody (Biorad). Blots were then washed again in TBS-T prior to the application of chemiluminescent substrate (Biorad), and exposure to Biomax MR Film (Kodak) for visualization of bands. Antibodies used were Grp78 (1: 2000; Stressgen), Grp58 (1:2000; Stressgen), ERp29 (1:15,000; Abcam), Grp94 (1:5000; Abcam), aldolase-A (1:1000; Abnova), actin (1:50,000; Chemicon), and tubulin (1:12,500; Sigma). The densities of immunoreactive bands were quantified using UN-SCAN-IT software (Silk Scientific; Orem, UT). Actin and tubulin were used as loading controls. Data from both the DeCyder analysis and Western blots indicated that actin and tubulin levels were similar between control and DA-exposed samples.
Statistical Analysis
Statistical analyses of DA-exposed cysteine and lysine Cy-dye volume densities from isolated mitochondrial MS-identified proteins were performed using a 1-sample two-tailed Z-test on the DA-treated mitochondrial protein spot volume intensities expressed as percent of control. The Z-test was chosen because the DeCyder DIA software calculates changes between control and treated samples as a normalized volume ratio of the two groups, generating one value comparing both groups and normalizing the entire constellation of labeled spots. Significance for each DA-exposed protein from control (valued at 100% control) was determined when p<0.05 and the change was greater than ± 1.2-fold from control (<83.3% or >120%). The percent control values were directly calculated from the normalized DeCyder volume ratios.
Differences among group means for Western blot data analysis were determined by ANOVA followed by post-hoc student’s t-test, with significance determined at p<0.05.
Results
2-D DIGE using Cysteine-Reactive And Lysine-Reactive CyDyes
To determine which proteins were changed in abundance following a 16 h exposure to 150 μM DA, control and DA-exposed PC12 cell mitochondrial-enriched protein fractions were analyzed together by 2D-DIGE. The advantages of using 2D-DIGE methods are that two different samples (control and treated) can be analyzed and compared within one gel. The 16 h timepoint was chosen because it was prior to the onset of significant cell death observed at 24 h and 48h, and because we observed, by HPLC analysis, increased levels of cysteinyl-catechol residues in whole-cell protein following 16h of 150 μM DA exposure (unpublished observations).
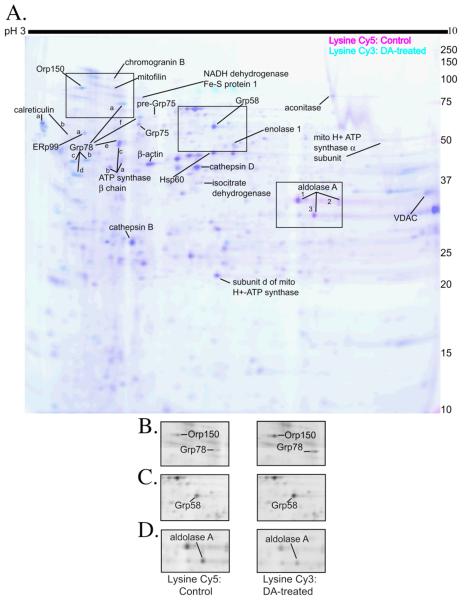
Mitochondrial-enriched protein fractions isolated from either control or DA-exposed cells were incubated separately with the cysteine-reactive maleimide Cy3- and Cy5-conjugated dyes (GE Healthcare). The control and DA-treated fluorescently labeled samples were then pooled and proteins were separated in 2-dimensions, first by isoelectric point, and then by apparent molecular weight. In Figure 1A, the fluorescent scan of a representative DIGE gel is displayed with inverted colors for clarity (control: Cy3, cyan and DA treated: Cy5, pink). Protein spots that were more strongly labeled in either the control or the DA-treated samples and a few unchanged test spots were excised from the gel, trypsinized, and subjected to MS analysis for identification. All protein identifications were confirmed in multiple cysteine-labeled gels, and are indicated in Figure 1A.
Figure 1. 2D-DIGE of PC12 cell mitochondrial-enriched fractions using cysteine-reactive dyes with insets of sample proteins.
(A.) Mitochondrial-enriched fractions from control and 16h, 150 μM DA-exposed PC12 cells were isolated by differential centrifugation, and equal protein amounts were reacted with either Cy3 (cyan scan; control) and Cy5 (pink scan; DA-exposed) maleimide dyes. Pink spots designate proteins in which more cysteine labeling occurred in the DA-exposed sample, indicating increased protein levels following DA exposure. Cyan spots designate proteins in which more cysteine labeling occurred in control, indicating cysteine modification or decreased protein levels following DA exposure. Dark blue spots indicate proteins in which equal cysteine labeling occurred in the control and DA-exposed mitochondrial-enriched fraction. MS-identified proteins are indicated on the gel and listed in Figure 3B. In cases where more than one spot with the same protein identification was determined by MS, letter (−A, −B, −C, etc) or numerical (−1, −2, and −3) notations were used. All spots identified corresponded to only one protein and related isoforms. Identification of each spot was replicated in 2 or more experiments. The gel is representative of n=8 cysteine 2D-DIGE gels. Boxes outline inset pictures in B-D. (B.) Black and white representation of the control protein, Cy3 fluorescent scan and the DA-treated protein, Cy5 fluorescent scan with Orp150 and Grp78 spots indicated. (C.) Black and white representation of the control protein, Cy3 fluorescent scan and the DA-treated protein, Cy5 fluorescent scan with the Grp58 spot indicated. (D.) Black and white representation of the control protein, Cy3 fluorescent scan and the DA-treated protein, Cy5 fluorescent scan with the aldolase A-3 spot indicated.
Differences between Cy3 and Cy5 spot intensities were quantified using DeCyder DIA software (GE Healthcare). The identified proteins that were significantly changed in the DA-treated fraction compared to control are graphed and listed in Figure 3. Proteins identified and determined to be unchanged are listed in Supplemental Data. Our results showed a subset of proteins, mainly ER stress proteins, that displayed increased cysteine dye binding in the DA-treated mitochondrial fraction, including calreticulin, ER protein 29 (ERp29), ER protein 99 (ERp99), Grp58, Grp 78, Grp94, and oxygen regulated protein (Orp150) (Figure 1A, 1B, 1C). Only one protein, aldolase A, was identified with reduced cysteine dye labeling in the DA-treated fraction (Figure 1A, D). Increases in cysteine dye-labeling following DA treatment may be attributed to increased protein levels (either due to up-regulation of the protein or a reduced rate of degradation). Decreased labeling in the DA-treated pool is likely due to decreased protein levels. However, cysteine modification prior to dye-labeling may also contribute to decreased cysteine labeling, because DAQ or DA-induced ROS can oxidize cysteine residues on proteins and potentially lead to a reduced number of protein thiols available to react with the maleimide dye. Therefore, we confirmed the apparent changes in protein abundance observed by cysteine labeling with a second, lysine-specific dye.
Control or DA-exposed mitochondrial-enriched fractions were incubated separately with the lysine-reactive NHS-ester Cy3 and Cy5-conjugated dyes (GE Healthcare), then combined, and analyzed by 2D-DIGE. In Figure 2A, the fluorescent scan of a representative lysine gel is displayed with inverted colors (control: Cy5=pink and DA treated: Cy3=cyan) with all confirmed protein identifications indicated. In both cysteine and lysine labeled 2D-DIGE gels, the vast majority of proteins in both the control and DA treated groups exhibited similar levels of labeling, indicating no change in the amount of these proteins following DA exposure. Protein spots that appeared unchanged between the two groups were not generally selected for protein identification, with a few exceptions.
Figure 2. 2D-DIGE of PC12 cell mitochondrial-enriched fraction using lysine-reactive dyes with insets of sample proteins.
(A.) Mitochondrial-enriched fraction from control and 16h, 150 μM DA-exposed differentiated PC12 cells were isolated by differential centrifugation. Mitochondrial protein was reacted with either Cy5 (pink scan; control) and Cy3 (cyan scan; DA-exposed) NHS-ester dyes to label lysine residues. Cyan (blue) spots designate proteins in which more lysine labeling occurred in the DA-exposed sample, indicating increased protein levels. Pink spots designate proteins in which more lysine labeling occurred in the control sample, indicating decreased protein levels induced by DA exposure. Dark blue spots indicate proteins in which equal lysine labeling occurred in the control and DA-exposed samples. MS-identified proteins are indicated on the gel and listed in Figure 3B. When multiple spots were determined by MS with the same protein identification, notations with letters (−A, −B, −C, etc) or numbers (−1, −2, and −3) were used. All listed identified protein spots corresponded to only one protein and related isoforms, and was replicated in at least 2 experiments. The gel is representative of n=5 lysine 2D-DIGE gels. Boxes outline inset pictures in figures B-D. (B.) Black and white representation of the control protein (Cy5 fluorescent scan) and the DA-treated protein (Cy3 fluorescent scan) with Orp150 and Grp78 spots indicated. (C.) Black and white representation of the control protein (Cy5 fluorescent scan) and the DA-treated protein (Cy3 fluorescent scan) with the Grp58 spot indicated. (D.) Black and white representation of the control protein (Cy5 fluorescent scan) and the DA-treated protein (Cy3 fluorescent scan) with the aldolase A-3 spot indicated.
Comparing the two types of dyes, our results showed that most of the proteins that changed in the cysteine-labeling experiments also changed similarly in the lysine-labeling studies (Figure 3). In fact, most proteins that displayed increased cysteine dye binding in the DA-treated mitochondrial fraction, like calreticulin A, Orp150, Grp78, and Grp58 (Figure 1A, 1B, and 1C), also displayed increased lysine dye binding in the DA-treated mitochondrial fraction (Figure 2A, 2B, 2C). Decreases in cysteine and lysine labeling of aldolase A following DA exposure were also observed (Figure 1A, 1D, 2A, 2D). Equivalent increases in both cysteine and lysine dye-labeling following DA treatment indicate increased protein levels. Similarly, corresponding decreases in both cysteine and lysine dye-labeling following DA treatment, indicate decreased protein levels. Interestingly, we also found that the intensity of the labeling by the cysteine and lysine dyes were similar, even though we reacted less cysteine dye than lysine dye per μg protein. Utilizing both dyes allowed us to control for modifications on either cysteine or lysine residues that could have altered the labeling of proteins.
Levels of cysteine and lysine labeling did not correlate for two identified proteins following DA exposure. Calreticulin-B and ERp99-A had increased cysteine dye labeling, while lysine dye labeling remained unchanged following DA exposure (Figure 3). Both protein spots had lower dye labeling intensities compared to other protein spots in both cysteine and lysine dye experiments, suggesting they are low abundance proteins, close to the limit of detection by dye labeling. Since the cysteine dye has previously been shown to have an order of magnitude greater range of detection (Shaw et al., 2003), it is possible that the lysine dye labeling for these proteins were outside their quantitative range. Thus, we were unable to detect changes in protein levels with the lysine dye, but we were able to measure changes with the cysteine dye for Calreticulin-B and ERp99-A spots.
Most of the Identified Proteins Function as Chaperones
We successfully identified many proteins in the PC12 cell mitochondrial-enriched fraction that are changed following DA exposure (Figures 1, 2, and 3). Most of the changed proteins identified are typical ER localized chaperone proteins, with one identified protein involved in ATP synthesis (Figure 3B). Previously, ER proteins have been found associated with isolated mitochondrial preparations (Fountoulakis et al., 2002). A more recent study has shown that the ER and mitochondria are physically tethered to one another (Csordas et al., 2006), and thus it is not surprising that we observed ER proteins in our crude, detergent-free mitochondrial fraction. Other proteins including cytosolic structural proteins, proteins involved in secretion, proteases, and some mitochondrial proteins were also identified (Supplemental Data). Sixteen identified spots were significantly changed in the DA-treated mitochondrial fraction as compared to control (Figure 3).
Increased Levels of ER proteins Following DA Exposure
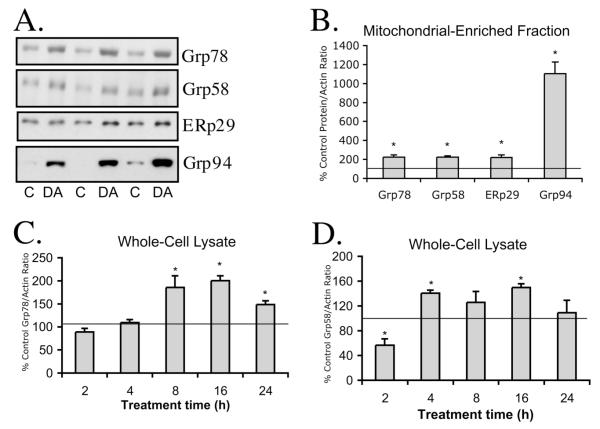
Since many of the proteins with the greatest increases in the DA-exposed samples were ER chaperone proteins (Figure 3), we were interested in whether the increases were due to an increased ER response to DA-induced toxicity. Because the ER may swell following oxidative stress (Atlante et al., 2001; Esrefoglu et al., 2003) and are closely associated with mitochondria (Levine and Rabouille, 2005; Csordas et al., 2006), it is possible that DA-exposure resulted in an increased amount of ER isolated with the crude mitochondrial preparation, rather than an overall increase in cellular levels of ER stress proteins. To confirm our 2-D DIGE findings and evaluate the total cellular ER response, we measured the levels of four ER-associated chaperone proteins Grp78, Grp58, ERp29, and Grp94 using Western blot analysis in PC12 cell mitochondrial-enriched fractions, in addition to analyses of Grp78 and Grp58 in whole cell lysate (Figure 4). These four ER chaperone proteins, which were increased following DA exposure in mitochondrial-enriched fractions on 2D-DIGE, also increased on Western blot analysis, although not surprisingly, the magnitude of the protein level increase varied somewhat between the two methods. Lanes from representative Western blots for Grp78, Grp58, ERp29, and Grp94 in 16 h control and DA-exposed mitochondrial fractions are shown in Figure 4A.
Figure 4. Western blot analysis of Grp78, Grp58, ERp29, and Grp94 in PC12 mitochondrial-enriched fraction and Grp78 and Grp58 in whole cell lysate following DA-exposure.
Mitochondria-enriched protein and PC12 whole-cell lysate from control and 150 μM DA-exposed PC12 cells were collected. (A.) Representative blots for proteins Grp78, Grp58, ERp29, and Grp94 are shown for PC12 mitochondrial fractions isolated from 16h control and 150 μM DA-treated cells. (B.) Grp78/actin, Grp58/actin, ERp29/actin, and Grp94/actin ratios for PC12 mitochondrial fraction DA treated cells were quantified and compared to control. Values for (C.) Grp78/actin and (D.) Grp58/actin ratios were measured in whole cell lysates at various time-points and expressed as % Control ± SEM, n=3 separate experiments, each measured in duplicate. *, significance p<0.05.
Western blot analysis of Grp78 in the mitochondrial fraction showed a 224% increase compared to control (Figure 4B), validating the results obtained in 2D-DIGE DeCyder analysis (cysteine: +184-260%; lysine: +152-168%; Figure 3). We also observed a 224% increase in Grp58 on Western blot in the DA exposed mitochondrial fraction as compared to control (Figure 4B), which was slightly above levels of Grp58 in the DA-exposed mitochondrial fraction measured by 2D-DIGE, DeCyder analysis (cysteine: +134%; lysine: +147%; Figure 3). Levels of ERp29 in mitochondrial fractions following DA exposure were elevated to 219% of control by Western blot, compared to 149% of control as determined by DeCyder analysis with the cysteine dyes (Figures 3 and 4B). Levels of Grp94 in mitochondrial fractions following DA exposure were elevated to 1100% of control by Western blot, compared to 169% of control as determined by DeCyder analysis with the cysteine dyes (Figures 3 and 4B). Both methods detected an increase in Grp94, however the reason for the large discrepancy in values between the two methods is not clear, but may be related to the very low levels of protein observed in control samples and the limits of detection by these two methods.
In addition, we utilized Western blot analysis of PC12 cell whole cell lysate to determine whether the DA-induced changes in Grp78 and Grp58 protein were due to overall increases in the levels of these proteins in the cell or increased association of ER with mitochondria. Differentiated PC12 cells were treated with control media or media containing 150 μM DA for 2, 4, 8, 16, or 24 h prior to collection of cell lysate for Western blotting. A time-course of Grp78 and Grp58 levels in whole cell lysate allows us to more accurately assess the timing of the ER induced response to DA. Levels of Grp78 were significantly increased from control following 8 h of DA exposure (+186%; Figure 4C). The levels of Grp78 remained elevated above control after 16 and 24 h of DA exposure (+200% and +148%, respectively; Figure 4C). Levels of Grp58 in whole cell lysates were significantly decreased from control after 2h of DA exposure (−44%; Figure 4D). However, levels of Grp58 were increased after 4h and 16h of DA exposure (+141% and +150%, respectively; Figure 4D).
The change in levels of Grp78 measured by Western blot analyses in the DA-exposed PC12 cell mitochondrial-enriched fraction (224% of control; Figure 4B) are comparable to the 16 h DA exposed whole-cell lysate levels (200% of control; Figure 4C), and consistent with the increases observed by DeCyder analysis (cysteine: +184-260%, lysine: +152-168%; Figure 3A). In addition, the change in levels of Grp58 measured by Western blot after 16 h DA exposure in the mitochondrial enriched fraction (224% of control; Figure 4B), whole cell lysate (150% of control; 4D), and DeCyder analysis (134-147%; Figure 3A) are similar. These results confirm the increases in ER protein and suggest that these increases occur throughout the cell in response to DA-induced toxicity.
Levels of Aldolase A Differ In Whole Cell Lysate Compared to the Mitochondrial Fraction Following DA Exposure
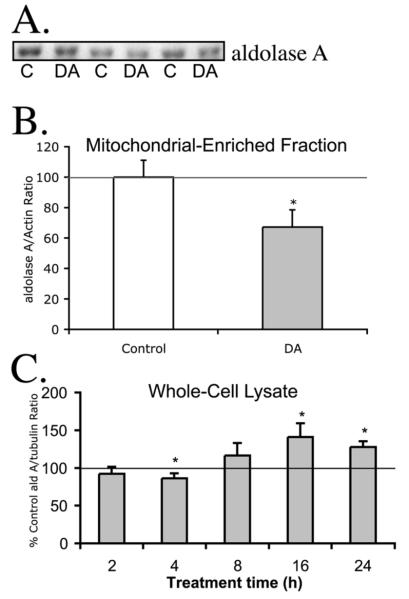
In addition to the ER chaperone proteins, we were also interested in confirming the levels of aldolase A, since it was the only identified protein significantly decreased following DA-exposure in the PC12-cell mitochondrial fraction, according to DeCyder analysis. Since aldolase A is found both in the cytoplasm (Pfleiderer et al., 1975; Wachsmuth, 1976) and associated with mitochondria (MacDonnell and Greengard, 1974), we were also interested in whether the aldolase A in the cytosolic fraction, the mitochondrial fraction, or both were lost following DA-exposure. We observed that the aldolase A levels in mitochondrial-enriched fraction measured by Western blot also followed a similar trend to levels measured by 2D-DIGE, DeCyder analysis. We observed a significant decrease (67% of control) in levels of aldolase A following 16 h DA exposure by Western blot (Figure 5A), consistent with the decreases observed in 2D-DIGE, DeCyder analysis (42% of control, cysteine; 39% of control, lysine; Figure 3A). Although Western blotting and 2D-DIGE DeCyder techniques are very different, we were able to confirm the decrease in aldolase A levels with both analyses. In whole cell lysates, a slight decrease in aldolase A was observed after 4 h of DA exposure (−14%; Figure 5C). However, after longer DA exposure times, the levels of whole cell aldolase A were increased above control (16 h, +141%; 24 h +128%; Figure 5C). Thus, the loss of aldolase A in the mitochondrial-enriched fraction appears to be specific to this fraction, the significance of which is not known, but may be related to changes in metabolic demands.
Figure 5. Western blot analysis of aldolase A in PC12 mitochondrial-enriched fraction and whole cell lysate following DA-exposure.
Mitochondria-enriched protein and PC12 whole-cell lysate from PC12 cells were collected after exposure to control media or media containing 150 μM DA. (A.) Representative blots for aldolase A are shown for PC12 mitochondrial-enriched fractions isolated from 16h control and 150 μM DA-treated cells. (B.) The quantification of control and DA exposed PC12 cell mitochondrial-enriched protein are reported as % control ± SEM of the average immunoblot intensity of aldolase A/actin. (C.) Aldolase A levels in PC12 whole cell lysate following DA exposure were examined at various time-points compared to control cells. Aldolase A/tubulin ratios were reported as % time-matched control ± SEM. n=3 separate experiments, each measured in duplicate. *, significance p<0.05.
Discussion
Proteins with Increased Levels Identified Using DIGE
Using an unbiased proteomics approach in this study, we found that DA exposure led to changes in specific proteins, consistent with an ER stress response in differentiated PC12 cells. We utilized cysteine dyes that label reduced cysteine residues from both the control and DA-exposed mitochondrial-enriched fractions to detect changes in protein levels and compared the results in parallel studies using lysine-labeling dyes. Using both methods, we observed increased levels of various ER chaperone proteins in response to DA exposure in PC12 cells, including: Grp78 (BiP), Grp58, Grp94, Orp150, calreticulin, ERp29 and ERp99. Some of these proteins, including Grp58, Grp78, Grp94, calreticulin, and Orp150 have previously been shown to be up-regulated following oxidative stress and ER stress (Kuwabara et al., 1996; Kaufman, 1999; Kaneda et al., 2000; Lee, 2001; Nunez et al., 2001; Berridge, 2002). ERp29 has been described as an escort chaperone, with possible involvement in secretion (Mkrtchian and Sandalova, 2006). ERp99 is speculated to be an ER membrane sorting chaperone, since the N-terminus has identical sequence homology to Grp94, an ATP-dependent chaperone (Mazzarella and Green, 1987).
ER Chaperone Proteins Grp78 and Grp58
Grp78 up-regulation is the classical marker for induction of the UPR, and serves to protect cells by binding to stress sensors, including PERK, IRE1, and ATF6 (Rao et al., 2004). Using 2D-DIGE and Western blot analysis of mitochondrial protein, we found that Grp78 was significantly increased above control, indicating that DA exposure led to Grp78 up-regulation. The time-course of Grp78 levels in whole cell lysate measured by Western blot revealed a significant increase in Grp78 as early as 8h. Levels of Grp78 remain increased above control at 16h and 24h DA exposure, indicating a sustained activation of the UPR. Prolonged UPR activation may contribute to cell death via multiple mechanisms, including calcium signaling, caspase-12 activation, ROS accumulation, the ASK1/JNK stress activated kinase apoptosis pathway, and the p53 apoptosis pathway (Gorlach et al., 2006; Szegezdi et al., 2006; Zhao and Ackerman, 2006).
Grp58 is an ER stress-inducible chaperone protein with thiol oxidoreductase activity that has been shown to be susceptible to oxidation by H2O2, and works in conjunction with calreticulin and calnexin to fold glycoproteins (Mazzarella et al., 1994; Murthy and Pande, 1994; High et al., 2000; van der Vlies et al., 2002; Antoniou and Powis, 2003). In the present study, we found that Grp58 levels were significantly elevated above control in 16h DA-exposed PC12 cell mitochondria using 2D-DIGE and Western blot analysis. Grp58 protein analyzed in whole cell lysate revealed a significant increase in Grp58 at 4 and 16 h of DA exposure. The increases in Grp58 and Grp78 levels as well as other ER stress proteins identified by 2D-DIGE and confirmed by Western blot, strongly indicate ER stress activation in PC12 cells exposed to DA.
Aldolase A
Thus far, only one identified protein, aldolase A, had significantly lower labeling using cysteine dyes and lysine dyes in DA-exposed mitochondria. Aldolase A catalyzes the breakdown of fructose-1,6-bisphosphate during glycolysis to glyceraldehyde 3-phosphate and dihydroxyacetone phosphate (Lorentzen et al., 2005) and is expressed in all cells throughout the body, including neurons (Buono et al., 2001). It is found both in the cytoplasm and associated with mitochondria (MacDonnell and Greengard, 1974; Pfleiderer et al., 1975). Recently, aldolase A was found to be associated with PD-linked proteins DJ-1 and α-synuclein in MES cells (Jin et al., 2007). This association was decreased following rotenone, suggesting a novel role for aldolase A in a PD model employing selective dopaminergic cell death (Jin et al., 2007).
Results from the Western blot analyses showed that aldolase A is decreased in the mitochondrial fraction, but not throughout the cell (Figure 5) following DA exposure. Aldolase A levels measured by Western blot in whole cell lysate showed an initial decrease following 4h of DA exposure, but at longer exposure times, aldolase A levels were increased above control. Differential aldolase A levels between the mitochondrial fraction and in whole cell lysate at 16 h of DA exposure could be due to various factors. Aldolase A has been shown to be part of multi-enzyme glycolytic complexes that are attached to mitochondria (Beeckmans et al., 1990; Minaschek et al., 1992). The release of mitochondrial-associated glycolytic complexes may occur following DA exposure, resulting in decreased levels of aldolase A in the mitochondrial-enriched fraction and increased levels in the whole cell lysate. Since ATP levels in PC12 cells decrease following 18 h DA exposure and rebound close to control levels after 24 h DA exposure (unpublished observations), cytosolic levels of glycolytic enzymes, such as aldolase A and GAPDH (unpublished observations), may be up-regulated to compensate for DA-induced energy deficits. Potential mechanisms and significance of the differential aldolase A levels are currently being investigated.
2D-DIGE Analyses of Cysteine Dye and Lysine Dye Labeling
Detection of differences in lysine dye labeling or cysteine dye labeling were used to measure relative changes in protein levels between control and DA-exposed PC12 cell mitochondrial fractions. Because reactive metabolites of DA may oxidize or covalently bind with cysteine residues, we recognized the possibility that cysteine labeling may decrease in a given protein without a similar decrease in lysine labeling. However, none of our observations in PC12 cells at 16 h showed this response in the mitochondrial-enriched fraction. Our observations with both PC12 cells and isolated brain mitochondria (Van Laar et al., 2008), suggested that bulk labeling of cysteines is not a sensitive enough method to detect redox modifications to cysteine residues. This conclusion was also reached by two other recent studies (Chan et al., 2005; Hurd et al., 2007). Spot migration due to shifts in pI or molecular weight caused by oxidative or posttranslational modifications could also affect the interpretation of 2D-DIGE data. We observed no pairs of increased Cy3- and Cy5-labeled spots with the same molecular weight, and thus no evidence for pI shifting (Figures 1 and 2). Although we have located multiple spots at different molecular weights with the same protein identification (e.g. Grp78), 2D-DIGE analyses of those spots revealed similarly increased cysteine labeling following DA-exposure, with no concomitant decreased labeled spot. Therefore, spot migration is unlikely to account for our observations. Our results utilizing 2D-DIGE consistently showed increased levels of ER chaperone proteins in the mitochondrial-enriched fractions following DA exposure, suggesting that the cells were mounting a response to the DA-induced oxidative stress.
ER Stress, Mitochondria, and PD
The ER plays a role in pro- and anti-apoptotic signaling in conjunction with mitochondria (Berridge, 2002; Breckenridge et al., 2003; Paschen, 2003; Rao et al., 2004). One way in which mitochondria and the ER communicate is via the calcium equilibrium, which is essential for the normal functions of both organelles (Berridge, 2002; Breckenridge et al., 2003; Paschen, 2003). Recently, a physical linkage between ER and mitochondria that plays a role in calcium signaling between the organelles has been described (Csordas et al., 2006). Mitochondria and ER communication is essential for the cellular response to stress and ultimately cell survival. Many of the Bcl-2 family of proteins, known to regulate mitochondrial-mediated apoptosis, also seem to influence ER-induced cell death and calcium signaling between the ER and mitochondria (Breckenridge et al., 2003; Rao et al., 2004; Gorlach et al., 2006; Szegezdi et al., 2006; Wu and Kaufman, 2006). Mitochondrial energy deficits have also been shown to initiate ER stress, including the up-regulation of Grp78 (Flores-Diaz et al., 2004; Xu et al., 2004). While short-term UPR activation serves to protect cells, sustained activation of ER stress can lead to activation of mitochondrial-dependent apoptosis involving cytochrome c release, in addition to a mitochondrial-independent caspase-12 pathway, leading to cell death (Breckenridge et al., 2003; Momoi, 2004; Rao et al., 2004; Zhao and Ackerman, 2006).
Using an unbiased proteomics approach, this study has shown that DA exposure leads to ER stress protein up-regulation in PC12 cells, which is likely involved in the mechanism of DA-induced cell death. Recent studies lend also showed that ER stress may be induced in vitro by dopaminergic toxins. Exposure to high levels of DA (500 μM), 6-OHDA, or MPP+ in SH-SY5Y cells, and expression of mutant A53T alpha synuclein in PC12 cells increased levels of Grp78 and Chop/GADD153, an ER stress inducible transcription factor (Chen et al., 2004; Gomez-Santos et al., 2005; Smith et al., 2005). Increases in the phosphorylation state of ER stress kinases were observed following 6-OHDA, MPP+, and rotenone treatment (Ryu et al., 2002; Holtz and O’Malley, 2003). In MN9D cells, 6-OHDA induced an ER stress response prior to cytochrome c release, indicating that ER stress activation occurred prior to apoptotic signaling pathways (Holtz et al., 2006). Others have also suggested ER stress leads to activation of apoptotic cell death pathways (Boyce and Yuan, 2006; Lindholm et al., 2006; Szegezdi et al., 2006; Wu and Kaufman, 2006). A recent study found that after rotenone exposure in MES cells, increased levels of Grp94 were associated with α-synuclein, linking the UPR to proteins thought to be involved in PD pathogenesis (Jin et al., 2007) .
The activation of ER stress has also been observed in in vivo models of PD and ischemia (DeGracia and Montie, 2004; Kitao et al., 2007), and is thought to be a common pathological pathway in many neurodegenerative disorders (Paschen and Mengesdorf, 2005; Xu et al., 2005; Lindholm et al., 2006). Many inherited forms of PD involve abnormalities in protein degradation or mitochondrial dysfunction (Lindholm et al., 2006). The relationships between inheritable PD, protein degradation, and inclusion formation suggest that the accumulation of unfolded proteins may contribute to neuronal death observed in PD (Lindholm et al., 2006). Recently, a study found activation of the PERK-eIF2α pathway of the UPR in the substantia nigra of PD patients, linking ER pathways directly to the disease (Hoozemans et al., 2007). Therefore, the relationship between oxidative stress, mitochondrial dysfunction, and abnormal protein degradation is connected to the activation of ER stress pathways in both DA-induced toxicity and the pathology of PD.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health, grants NS44076, AG20899, and DA09601. We thank Dr. Billy Day, Jiyan An, and Dr. Ashraf Elamin at the University of Pittsburgh Genomics and Proteomics Core Laboratories for providing their proteomics expertise and mass spectrometry facilities. We would also like to thank Michael Fischer at the University of Pittsburgh Department of Biostatistics Statistical Consulting Service.
Abbreviations Used
- 2D-DIGE
two-dimensional difference in-gel electrophoresis
- 6-OHDA
6-hydroxydopamine
- BiP
immunoglobulin heavy chain binding protein
- BSA
bovine serum albumin
- CHCA
α-cyano-4-hydroxycinnamic acid
- CHOP/Gadd153
C/EBP-homologous transcription factor
- DA
dopamine
- DAQ
dopamine quinone
- DMEM
Dulbecco’s modified Eagle Medium
- DOPAC
3,4 dihydroxyphenylacetic acid
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- ERp29
endoplasmic reticulum protein 29 kDa
- ERp99
endoplasmic reticulum protein 99 kDa
- FBS
fetal bovine serum
- Grp58
glucose regulated protein 58 kDa
- Grp78
glucose regulated protein 78 kDa
- Grp94
glucose regulated protein 94 kDa
- HS
horse serum
- MS
mass spectrometry
- NGF
nerve growth factor
- NHS
N-hydroxysuccinimidyl
- NO
nitric oxide
- Orp150
oxygen regulated protein 150 kDa
- PBS
phosphate buffered saline
- PD
Parkinson’s disease
- PIC
protease inhibitor cocktail
- ROS
reactive oxygen species
- SDS
sodium dodecyl sulfate
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SN
substantia nigra
- TBS
Tris buffered saline
- TBS-T
Tris buffered saline with 0.1% Tween20
- TFA
trifluoroacetic acid
- UPR
unfolded protein response
References
- Antoniou AN, Powis SJ. Characterization of the ERp57-Tapasin complex by rapid cellular acidification and thiol modification. Antioxid Redox Signal. 2003;5:375–379. doi: 10.1089/152308603768295104. [DOI] [PubMed] [Google Scholar]
- Atlante A, Calissano P, Bobba A, Giannattasio S, Marra E, Passarella S. Glutamate neurotoxicity, oxidative stress and mitochondria. FEBS Lett. 2001;497:1–5. doi: 10.1016/s0014-5793(01)02437-1. [DOI] [PubMed] [Google Scholar]
- Beeckmans S, Van Driessche E, Kanarek L. Clustering of sequential enzymes in the glycolytic pathway and the citric acid cycle. J Cell Biochem. 1990;43:297–306. doi: 10.1002/jcb.240430402. [DOI] [PubMed] [Google Scholar]
- Ben-Shachar D, Zuk R, Glinka Y. Dopamine neurotoxicity: inhibition of mitochondrial respiration. J Neurochem. 1995;64:718–723. doi: 10.1046/j.1471-4159.1995.64020718.x. [DOI] [PubMed] [Google Scholar]
- Berman SB, Hastings TG. Dopamine oxidation alters mitochondrial respiration and induces permeability transition in brain mitochondria: implications for Parkinson’s disease. J Neurochem. 1999;73:1127–1137. doi: 10.1046/j.1471-4159.1999.0731127.x. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. The endoplasmic reticulum: a multifunctional signaling organelle. Cell Calcium. 2002;32:235–249. doi: 10.1016/s0143416002001823. [DOI] [PubMed] [Google Scholar]
- Boyce M, Yuan J. Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death Differ. 2006;13:363–373. doi: 10.1038/sj.cdd.4401817. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Breckenridge DG, Germain M, Mathai JP, Nguyen M, Shore GC. Regulation of apoptosis by endoplasmic reticulum pathways. Oncogene. 2003;22:8608–8618. doi: 10.1038/sj.onc.1207108. [DOI] [PubMed] [Google Scholar]
- Buono P, D’Armiento FP, Terzi G, Alfieri A, Salvatore F. Differential distribution of aldolase A and C in the human central nervous system. J Neurocytol. 2001;30:957–965. doi: 10.1023/a:1021828421792. [DOI] [PubMed] [Google Scholar]
- Calabrese V, Guagliano E, Sapienza M, Panebianco M, Calafato S, Puleo E, Pennisi G, Mancuso C, Butterfield DA, Stella AG. Redox regulation of cellular stress response in aging and neurodegenerative disorders: role of vitagenes. Neurochem Res. 2007;32:757–773. doi: 10.1007/s11064-006-9203-y. [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri I, Joseph JA. Differential effect of dopamine catabolism and uptake inhibition on dopamine-induced calcium dysregulation and viability loss. Free Radic Biol Med. 1999;27:1393–1404. doi: 10.1016/s0891-5849(99)00188-4. [DOI] [PubMed] [Google Scholar]
- Chakravarthi S, Jessop CE, Bulleid NJ. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 2006;7:271–275. doi: 10.1038/sj.embor.7400645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HL, Gharbi S, Gaffney PR, Cramer R, Waterfield MD, Timms JF. Proteomic analysis of redox- and ErbB2-dependent changes in mammary luminal epithelial cells using cysteine- and lysine-labelling two-dimensional difference gel electrophoresis. Proteomics. 2005;5:2908–2926. doi: 10.1002/pmic.200401300. [DOI] [PubMed] [Google Scholar]
- Chen G, Bower KA, Ma C, Fang S, Thiele CJ, Luo J. Glycogen synthase kinase 3beta (GSK3beta) mediates 6-hydroxydopamine-induced neuronal death. Faseb J. 2004;18:1162–1164. doi: 10.1096/fj.04-1551fje. [DOI] [PubMed] [Google Scholar]
- Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- DeGracia DJ, Montie HL. Cerebral ischemia and the unfolded protein response. J Neurochem. 2004;91:1–8. doi: 10.1111/j.1471-4159.2004.02703.x. [DOI] [PubMed] [Google Scholar]
- Esrefoglu M, Gepdiremen A, Kurus M. Ultrastructural clues for glutamate-induced necrosis in parietal and cerebellar neurons. Fundam Clin Pharmacol. 2003;17:341–347. doi: 10.1046/j.1472-8206.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- Fahn S, Sulzer D. Neurodegeneration and neuroprotection in Parkinson disease. NeuroRx. 2004;1:139–154. doi: 10.1602/neurorx.1.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskum G, Starkov A, Polster BM, Chinopoulos C. Mitochondrial mechanisms of neural cell death and neuroprotective interventions in Parkinson’s disease. Ann N Y Acad Sci. 2003;991:111–119. doi: 10.1111/j.1749-6632.2003.tb07469.x. [DOI] [PubMed] [Google Scholar]
- Flores-Diaz M, Higuita JC, Florin I, Okada T, Pollesello P, Bergman T, Thelestam M, Mori K, Alape-Giron A. A cellular UDP-glucose deficiency causes overexpression of glucose/oxygen-regulated proteins independent of the endoplasmic reticulum stress elements. J Biol Chem. 2004;279:21724–21731. doi: 10.1074/jbc.M312791200. [DOI] [PubMed] [Google Scholar]
- Fountoulakis M, Berndt P, Langen H, Suter L. The rat liver mitochondrial proteins. Electrophoresis. 2002;23:311–328. doi: 10.1002/1522-2683(200202)23:2<311::AID-ELPS311>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Gandhi S, Wood NW. Molecular pathogenesis of Parkinson’s disease. Hum Mol Genet. 2005;14 Spec No. 2:2749–2755. [PubMed] [Google Scholar]
- Gomez-Santos C, Barrachina M, Gimenez-Xavier P, Dalfo E, Ferrer I, Ambrosio S. Induction of C/EBP beta and GADD153 expression by dopamine in human neuroblastoma cells. Relationship with alpha-synuclein increase and cell damage. Brain Res Bull. 2005;65:87–95. doi: 10.1016/j.brainresbull.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Gorlach A, Klappa P, Kietzmann T. The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control. Antioxid Redox Signal. 2006;8:1391–1418. doi: 10.1089/ars.2006.8.1391. [DOI] [PubMed] [Google Scholar]
- Graham DG. Oxidative pathways for catecholamines in the genesis of neuromelanin and cytotoxic quinones. Mol Pharmacol. 1978;14:633–643. [PubMed] [Google Scholar]
- Halliwell B. Oxidative stress and neurodegeneration: where are we now? J Neurochem. 2006a;97:1634–1658. doi: 10.1111/j.1471-4159.2006.03907.x. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Proteasomal dysfunction: a common feature of neurodegenerative diseases? Implications for the environmental origins of neurodegeneration. Antioxid Redox Signal. 2006b;8:2007–2019. doi: 10.1089/ars.2006.8.2007. [DOI] [PubMed] [Google Scholar]
- Hastings TG, Zigmond MJ. Identification of catechol-protein conjugates in neostriatal slices incubated with [3H]dopamine: impact of ascorbic acid and glutathione. J Neurochem. 1994;63:1126–1132. doi: 10.1046/j.1471-4159.1994.63031126.x. [DOI] [PubMed] [Google Scholar]
- Hastings TG, Lewis DA, Zigmond MJ. Role of oxidation in the neurotoxic effects of intrastriatal dopamine injections. Proc Natl Acad Sci U S A. 1996;93:1956–1961. doi: 10.1073/pnas.93.5.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High S, Lecomte FJ, Russell SJ, Abell BM, Oliver JD. Glycoprotein folding in the endoplasmic reticulum: a tale of three chaperones? FEBS Lett. 2000;476:38–41. doi: 10.1016/s0014-5793(00)01666-5. [DOI] [PubMed] [Google Scholar]
- Holtz WA, O’Malley KL. Parkinsonian mimetics induce aspects of unfolded protein response in death of dopaminergic neurons. J Biol Chem. 2003;278:19367–19377. doi: 10.1074/jbc.M211821200. [DOI] [PubMed] [Google Scholar]
- Holtz WA, Turetzky JM, Jong YJ, O’Malley KL. Oxidative stress-triggered unfolded protein response is upstream of intrinsic cell death evoked by parkinsonian mimetics. J Neurochem. 2006;99:54–69. doi: 10.1111/j.1471-4159.2006.04025.x. [DOI] [PubMed] [Google Scholar]
- Hoozemans JJ, van Haastert ES, Eikelenboom P, de Vos RA, Rozemuller JM, Scheper W. Activation of the unfolded protein response in Parkinson’s disease. Biochem Biophys Res Commun. 2007;354:707–711. doi: 10.1016/j.bbrc.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Hurd TR, Prime TA, Harbour ME, Lilley KS, Murphy MP. Detection of reactive oxygen species-sensitive thiol proteins by redox difference gel electrophoresis: implications for mitochondrial redox signaling. J Biol Chem. 2007;282:22040–22051. doi: 10.1074/jbc.M703591200. [DOI] [PubMed] [Google Scholar]
- Ischiropoulos H, Beckman JS. Oxidative stress and nitration in neurodegeneration: cause, effect, or association? J Clin Invest. 2003;111:163–169. doi: 10.1172/JCI17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner P. Oxidative stress in Parkinson’s disease. Ann Neurol. 2003;53(Suppl 3):S26–36. doi: 10.1002/ana.10483. discussion S36-28. [DOI] [PubMed] [Google Scholar]
- Jin J, Li GJ, Davis J, Zhu D, Wang Y, Pan C, Zhang J. Identification of novel proteins associated with both alpha-synuclein and DJ-1. Mol Cell Proteomics. 2007;6:845–859. doi: 10.1074/mcp.M600182-MCP200. [DOI] [PubMed] [Google Scholar]
- Jones DC, Gunasekar PG, Borowitz JL, Isom GE. Dopamine-induced apoptosis is mediated by oxidative stress and Is enhanced by cyanide in differentiated PC12 cells. J Neurochem. 2000;74:2296–2304. doi: 10.1046/j.1471-4159.2000.0742296.x. [DOI] [PubMed] [Google Scholar]
- Kaneda S, Yura T, Yanagi H. Production of three distinct mRNAs of 150 kDa oxygen-regulated protein (ORP150) by alternative promoters: preferential induction of one species under stress conditions. J Biochem (Tokyo) 2000;128:529–538. doi: 10.1093/oxfordjournals.jbchem.a022783. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Khan FH, Sen T, Maiti AK, Jana S, Chatterjee U, Chakrabarti S. Inhibition of rat brain mitochondrial electron transport chain activity by dopamine oxidation products during extended in vitro incubation: implications for Parkinson’s disease. Biochim Biophys Acta. 2005;1741:65–74. doi: 10.1016/j.bbadis.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Kitao Y, Imai Y, Ozawa K, Kataoka A, Ikeda T, Soda M, Nakimawa K, Kiyama H, Stern DM, Hori O, Wakamatsu K, Ito S, Itohara S, Takahashi R, Ogawa S. Pael receptor induces death of dopaminergic neurons in the substantia nigra via endoplasmic reticulum stress and dopamine toxicity, which is enhanced under condition of parkin inactivation. Hum Mol Genet. 2007;16:50–60. doi: 10.1093/hmg/ddl439. [DOI] [PubMed] [Google Scholar]
- Koshimura K, Tanaka J, Murakami Y, Kato Y. Effects of dopamine and L-DOPA on survival of PC12 cells. J Neurosci Res. 2000;62:112–119. doi: 10.1002/1097-4547(20001001)62:1<112::AID-JNR12>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Kuwabara K, Matsumoto M, Ikeda J, Hori O, Ogawa S, Maeda Y, Kitagawa K, Imuta N, Kinoshita T, Stern DM, Yanagi H, Kamada T. Purification and characterization of a novel stress protein, the 150-kDa oxygen-regulated protein (ORP150), from cultured rat astrocytes and its expression in ischemic mouse brain. J Biol Chem. 1996;271:5025–5032. doi: 10.1074/jbc.271.9.5025. [DOI] [PubMed] [Google Scholar]
- Lee AS. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci. 2001;26:504–510. doi: 10.1016/s0968-0004(01)01908-9. [DOI] [PubMed] [Google Scholar]
- Levine T, Rabouille C. Endoplasmic reticulum: one continuous network compartmentalized by extrinsic cues. Curr Opin Cell Biol. 2005;17:362–368. doi: 10.1016/j.ceb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Li H, Dryhurst G. Oxidative metabolites of 5-S-cysteinyldopamine inhibit the pyruvate dehydrogenase complex. J Neural Transm. 2001;108:1363–1374. doi: 10.1007/s007020100013. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13:385–392. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Siebers B, Hensel R, Pohl E. Mechanism of the Schiff base forming fructose-1,6-bisphosphate aldolase: structural analysis of reaction intermediates. Biochemistry. 2005;44:4222–4229. doi: 10.1021/bi048192o. [DOI] [PubMed] [Google Scholar]
- MacDonnell PC, Greengard O. Enzymes in intracellular organelles of adult and developing rat brain. Arch Biochem Biophys. 1974;163:644–655. doi: 10.1016/0003-9861(74)90525-6. [DOI] [PubMed] [Google Scholar]
- Mancuso M, Coppede F, Migliore L, Siciliano G, Murri L. Mitochondrial dysfunction, oxidative stress and neurodegeneration. J Alzheimers Dis. 2006;10:59–73. doi: 10.3233/jad-2006-10110. [DOI] [PubMed] [Google Scholar]
- Mazzarella RA, Green M. ERp99, an abundant, conserved glycoprotein of the endoplasmic reticulum, is homologous to the 90-kDa heat shock protein (hsp90) and the 94-kDa glucose regulated protein (GRP94) J Biol Chem. 1987;262:8875–8883. [PubMed] [Google Scholar]
- Mazzarella RA, Marcus N, Haugejorden SM, Balcarek JM, Baldassare JJ, Roy B, Li LJ, Lee AS, Green M. Erp61 is GRP58, a stress-inducible luminal endoplasmic reticulum protein, but is devoid of phosphatidylinositide-specific phospholipase C activity. Arch Biochem Biophys. 1994;308:454–460. doi: 10.1006/abbi.1994.1064. [DOI] [PubMed] [Google Scholar]
- Minaschek G, Groschel-Stewart U, Blum S, Bereiter-Hahn J. Microcompartmentation of glycolytic enzymes in cultured cells. Eur J Cell Biol. 1992;58:418–428. [PubMed] [Google Scholar]
- Mkrtchian S, Sandalova T. ERp29, an unusual redox-inactive member of the thioredoxin family. Antioxid Redox Signal. 2006;8:325–337. doi: 10.1089/ars.2006.8.325. [DOI] [PubMed] [Google Scholar]
- Momoi T. Caspases involved in ER stress-mediated cell death. J Chem Neuroanat. 2004;28:101–105. doi: 10.1016/j.jchemneu.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Murthy MS, Pande SV. A stress-regulated protein, GRP58, a member of thioredoxin superfamily, is a carnitine palmitoyltransferase isoenzyme. Biochem J. 1994;304(Pt 1):31–34. doi: 10.1042/bj3040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez MT, Osorio A, Tapia V, Vergara A, Mura CV. Iron-induced oxidative stress up-regulates calreticulin levels in intestinal epithelial (Caco-2) cells. J Cell Biochem. 2001;82:660–665. doi: 10.1002/jcb.1194. [DOI] [PubMed] [Google Scholar]
- Paschen W. Endoplasmic reticulum: a primary target in various acute disorders and degenerative diseases of the brain. Cell Calcium. 2003;34:365–383. doi: 10.1016/s0143-4160(03)00139-8. [DOI] [PubMed] [Google Scholar]
- Paschen W, Mengesdorf T. Endoplasmic reticulum stress response and neurodegeneration. Cell Calcium. 2005;38:409–415. doi: 10.1016/j.ceca.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Pfleiderer G, Thoner M, Wachsmuth ED. Histological examination of the aldolase monomer composition of cells from human kidney and hypernephroid carcinoma. Beitr Pathol. 1975;156:266–279. doi: 10.1016/s0005-8165(75)80166-1. [DOI] [PubMed] [Google Scholar]
- Pitkanen S, Robinson BH. Mitochondrial complex I deficiency leads to increased production of superoxide radicals and induction of superoxide dismutase. J Clin Invest. 1996;98:345–351. doi: 10.1172/JCI118798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovic AD, Lewis DA, Hastings TG. Role of oxidative changes in the degeneration of dopamine terminals after injection of neurotoxic levels of dopamine. Neuroscience. 2000;101:67–76. doi: 10.1016/s0306-4522(00)00293-1. [DOI] [PubMed] [Google Scholar]
- Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004;11:372–380. doi: 10.1038/sj.cdd.4401378. [DOI] [PubMed] [Google Scholar]
- Ryu EJ, Harding HP, Angelastro JM, Vitolo OV, Ron D, Greene LA. Endoplasmic reticulum stress and the unfolded protein response in cellular models of Parkinson’s disease. J Neurosci. 2002;22:10690–10698. doi: 10.1523/JNEUROSCI.22-24-10690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH. Mitochondrial dysfunction in Parkinson’s disease. Cell Death Differ. 2007;14:1261–1266. doi: 10.1038/sj.cdd.4402160. [DOI] [PubMed] [Google Scholar]
- Shaw J, Rowlinson R, Nickson J, Stone T, Sweet A, Williams K, Tonge R. Evaluation of saturation labelling two-dimensional difference gel electrophoresis fluorescent dyes. Proteomics. 2003;3:1181–1195. doi: 10.1002/pmic.200300439. [DOI] [PubMed] [Google Scholar]
- Smith WW, Jiang H, Pei Z, Tanaka Y, Morita H, Sawa A, Dawson VL, Dawson TM, Ross CA. Endoplasmic reticulum stress and mitochondrial cell death pathways mediate A53T mutant alpha-synuclein-induced toxicity. Hum Mol Genet. 2005;14:3801–3811. doi: 10.1093/hmg/ddi396. [DOI] [PubMed] [Google Scholar]
- Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse DC, McCreery RL, Adams RN. Potential oxidative pathways of brain catecholamines. J Med Chem. 1976;19:37–40. doi: 10.1021/jm00223a008. [DOI] [PubMed] [Google Scholar]
- Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlies D, Pap EH, Post JA, Celis JE, Wirtz KW. Endoplasmic reticulum resident proteins of normal human dermal fibroblasts are the major targets for oxidative stress induced by hydrogen peroxide. Biochem J. 2002;366:825–830. doi: 10.1042/BJ20020618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laar VS, Dukes AA, Cascio M, Hastings TG. Proteomic analysis of rat brain mitochondria following exposure to dopamine quinone: implications for Parkinson disease. Neurobiol Dis. 2008;29:477–489. doi: 10.1016/j.nbd.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votyakova TV, Reynolds IJ. DeltaPsi(m)-Dependent and -independent production of reactive oxygen species by rat brain mitochondria. J Neurochem. 2001;79:266–277. doi: 10.1046/j.1471-4159.2001.00548.x. [DOI] [PubMed] [Google Scholar]
- Wachsmuth ED. Differentiation of epithelial cells in human jejunum: localization and quantification of aminopeptidase, alkaline phosphatase and aldolase isozymes in tissue sections. Histochemistry. 1976;48:101–109. doi: 10.1007/BF00494548. [DOI] [PubMed] [Google Scholar]
- Walkinshaw G, Waters CM. Induction of apoptosis in catecholaminergic PC12 cells by L-DOPA. Implications for the treatment of Parkinson’s disease. J Clin Invest. 1995;95:2458–2464. doi: 10.1172/JCI117946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- Xiao-Qing T, Jun-Li Z, Yu C, Jian-Qiang F, Pei-Xi C. Hydrogen peroxide preconditioning protects PC12 cells against apoptosis induced by dopamine. Life Sci. 2005;78:61–66. doi: 10.1016/j.lfs.2005.04.048. [DOI] [PubMed] [Google Scholar]
- Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Liu L, Charles IG, Moncada S. Nitric oxide induces coupling of mitochondrial signalling with the endoplasmic reticulum stress response. Nat Cell Biol. 2004;6:1129–1134. doi: 10.1038/ncb1188. [DOI] [PubMed] [Google Scholar]
- Zhao L, Ackerman SL. Endoplasmic reticulum stress in health and disease. Curr Opin Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.