Abstract
The preparation and evaluation of a series of inhibitors of Myc—Max dimerization and Myc-induced cell transformation are described providing mycmycin-1 (3) and mycmycin-2 (4).
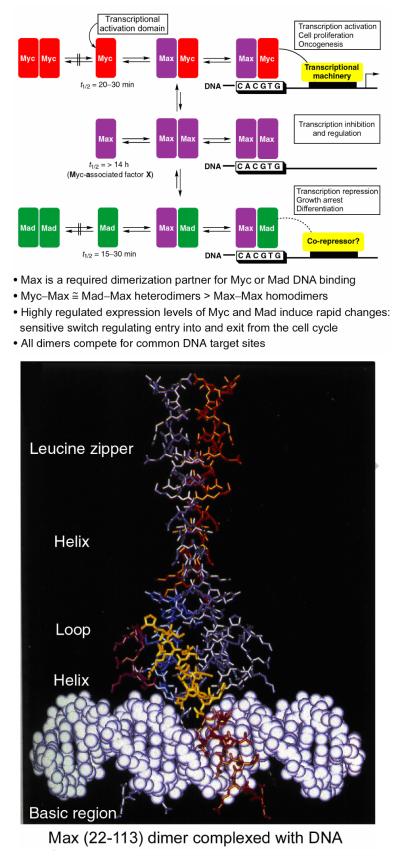
The c-myc proto-oncogene is involved in the progression of a wide range of human tumors.1-5 Proteins in the immediate Myc network are essential regulators of differentiation and proliferation. Oncogenic transformation occurs mainly through elevated expression of its gene product Myc, a short-lived nuclear protein and an important member of the basic helix-loop-helix leucine zipper (bHLHLZ) transcription factors (Figure 1).6 The oncogenic potential7,8 of c-myc has been demonstrated in transgenic animals and cell culture, and typically requires the action of at least one additional oncogene such as ras or bcl-2 (inhibits apoptosis). In normal cells, Myc is required for cell proliferation and prevents differentiation. It is transiently expressed in response to mitogenic stimuli and has a short half life (t1/2 = 20–30 min). Its aberrant expression in the absence of such growth factors drives cells into cell cycle but also induces apoptosis unless co-oncogenic mechanisms are present (i.e., inhibition by constitutive bcl-2 overexpression). All known activities of Myc require heterodimerization with Max,9-12 a constitutively expressed bHLHLZ protein that is stable (t1/2 >14 h). The Myc/Max heterodimer binds the DNA sequence CACGTG (E box element) and activates transcription through the transactivation domain of Myc. DNA binding occurs through the basic region while both the HLH and the leucine zipper form the dimerization interface.6 Max, but not Myc, forms homodimers that bind the same DNA site and inhibit transcription. In part, this results from the competition for Max and for the common DNA site. In addition, Max heterodimerizes with another family of bHLHLZ proteins that include Mad (Mad-1), Mxi-1, Mad-3, and Mad-4 that are also transiently expressed (t1/2 = 15–30 min) and signal growth arrest and differentiation.13 Their association behavior is similar to that of Myc in that they do not homodimerize or interact with Myc family members, but readily form heterodimers with Max that bind the CACGTG core consensus sequence. Mad and Myc compete for Max with approximately equal affinities and the heterodimers are formed in preference to Max homodimerization. Mad/Max heterodimers repress Myc/Max transcriptional activation. Thus, Myc/Max induces proliferation and apoptosis in nontransformed cells while Max/Max and Mad/Max are involved in growth arrest, differentiation, and cell survival. In transformed cells, when accompanied by oncogenic events inhibiting apoptosis, constitutive Myc expression maintains the cell proliferation state and prevents growth arrest and differentiation.
Figure 1.
Myc—Max—Mad network and structure of the Max dimer bound to DNA.
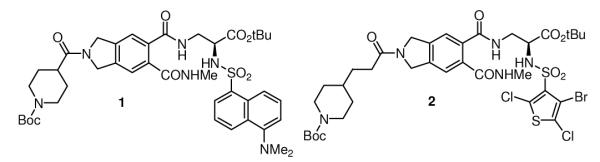
With the recognition that the aberrant expression of Myc leading to transformation may be addressed through inhibition of Myc/Max heterodimerization and its subsequent transcriptional activation, we screened our library of candidate modulators of protein—protein interactions for inhibitors of Myc/Max dimerization using a novel FRET assay.14 This led to the identificaton of the first prototypical inhibitors of Myc/Max dimerization whose behavior was not only confirmed in more classical ELISA and EMSA assays, but two (1 and 2) of which were also shown to inhibit Myc-induced oncogenic transformation in chicken embryo fibroblast cultures (CEF), providing the first validation of the approach for small molecule therapeutic intervention (Figure 2).14 Not only did this serve as the first example of inhibiting the (aberrant) function of a transcription factor by small molecule inhibition an integral protein—protein interaction, but it inspired our examination of a number of additional protein—protein interaction targets.15-24 Since this disclosure, additional inhibitors of Myc/Max dimerization have been reported and their therapeutic potential further validated.25-27 In addition, the examination of their interaction with Myc has revealed that the bHLHLZ interacting domain of Myc, which is disordered and undergoes coupled folding and binding upon heterodimerization, assumes a more rigid and defined conformation with the small molecule binding and limiting its ability to interact with Max.28
Figure 2.
Lead Compounds.
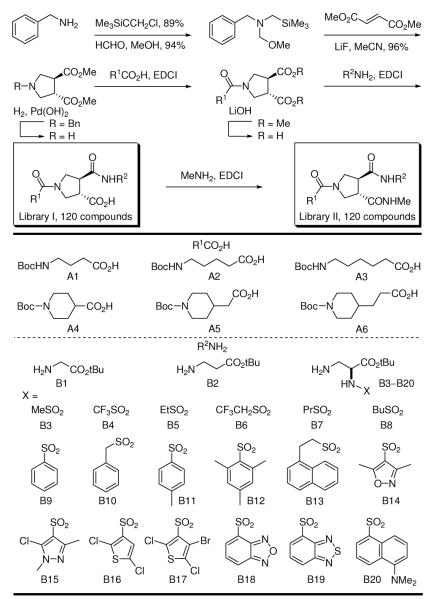
In a continuation of our studies and in efforts to improve on the modest potency of our original lead compounds, we have prepared two complementary 120-membered libraries to those that led to the discovery of 1 and 2.29 These were assembled using a symmetrical and racemic trans-3,4-dicarboxylic acid template enlisting acid—base liquid—liquid extractions30 for isolation of the pure intermediates or final products and employed the identical appended substituents (A1–A6, B1–B20, where B3–B20 are the single (S)-enantiomers)29 used in the libraries that provided 1 and 2, Scheme 1.
Scheme 1.
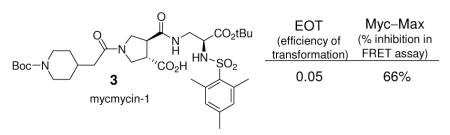
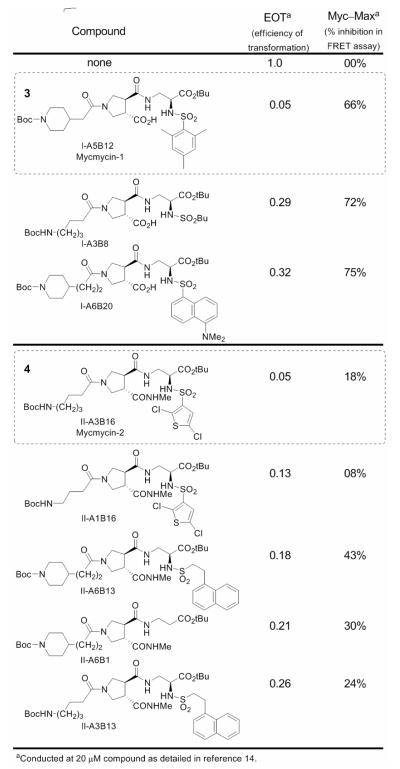
Since Myc induces the formation of oncogenically transformed cell foci in cultures of chicken embryo fibroblasts (CEF) and because this transforming activity of Myc depends on dimerization with Max, we examined all members of the candidate libraries directly for inhibition of this oncogenic transformation in this cell-based focus assay. Although laborious in requiring the maintenance of the cell culture over the three weeks of its conduct and a manual visual counting of the individual foci in each well, the use of the focus assay as the primary screen for activity ensured the identification of all members that display the desired functional activity in a cell-based assay.14 Thus, all 240 compounds produced were examined for inhibition of Myc-induced cell transfomation in the focus assay at 20 μM enlisting CEF infected with the RCAS viral vector expressing the transforming chicken cellular Myc.14 In these efforts, 40, 16, and finally 9 of the candidate compounds were examined more carefully in iterative rounds of assay displaying potent inhibition of the Myc-induced cell transformation in CEF. Two of the candidate inhibitors (3 and 4) proved especially potent exhibiting a complete inhibition of foci formation at 20 μM (IC95 = 20 μM). Additionally, both failed to exhibit inhibitory activity for foci formation induced by the oncogenic transcription factor Jun or the Src oncoprotein in CEF infected with the avian sarcoma virus 17 expressing the oncoprotein Jun31 or the Prague strain of Rous sarcoma virus encoding the Src oncoprotein.32 Thus, both 3 and 4 are selective for Myc-induced oncogenic transformation and neither possess an intrinsic cytotoxic acitivity for CEF. The top 8 candidates in the focus assay were also examined for inhibition of Myc—Max dimerization using the FRET assay disclosed in our original work,14 and the results are summarized in Figure 3 along with the tabulated results of the focus assay (EOT; efficiency of transformation, no inhibition = 1).
Figure 3.
Inhibitors of Myc-induced transformation in CEF and Myc/Max dimerization.
Of the final nine compounds examined in detail, eight exhibited significant inhibition of Myc-induced oncogenic transformation in CEF, two were found to completely inhibit foci formation at the tested concentration of 20 μM (EOT = 0.05), all had no effect on Jun- or Src-induced oncogenic transformation in CEF, and each displayed substantial inhibition of Myc/Max dimerization. The inhibitors fall into two classes being derived from Library I or II. Those derived from Library I including 3 exhibited the most effective inhibition of Myc/Max dimerization, but bear a free carboxylic acid that may impede cellular uptake limiting their inhibition of Myc-induced oncogenic transformation. Even with this potential limitation, compound 3 (mycmycin-1) displayed effective inhibition of the Myc-induced oncogenic transformation (EOT = 0.05) consistent with its inhibition of Myc/Max dimerization. The remaining five compounds including 4 (mycmycin-2) were derived from Library II and exhibited less effective inhibition of Myc/Max dimerization that, in part, correlated with their effectiveness for inhibiting the Myc-induced oncogenic transformation. The two exceptions, which include 4 (EOT = 0.05), displayed more potent inhibition of the oncogenic transformation than might be predicted simply from their relative inhibition of Myc/Max dimerization established in the FRET assay, yet bear structures that most closely resemble the original lead 2. Significantly, both mycmycin-1 and -2 (3 and 4) provide additional effective probes with which to continue examining this protein—protein interaction target.33
Acknowledgements
We gratefully acknowledge the financial support of the National Institutes of Health (CA78045). LRW is a Skaggs Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Oster SK, Ho CSW, Soucie EL, Penn LZ. Adv. Cancer Res. 2002;84:81. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 2.Henriksson M, Luscher B. Adv. Cancer Res. 1996;68:109. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 3.Calabretta B, Skorski T. Anti-Cancer Drug Design. 1997;12:373. [PubMed] [Google Scholar]
- 4.Amati B, Land H. Curr. Opin. Genet. Dev. 1994;4:102. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 5.Marcu KB, Bossone SA, Patel AJ. Annu. Rev. Biochem. 1992;61:809. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 6.Ferre—D’Amare AR, Prendergast GD, Ziff EB, Burley SK. Nature. 1993;363:38. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]; Brownlie P, Ceska TA, Lamers M, Romier C, Stier G, Teo H, Suck D. Structure. 1997;5:509. doi: 10.1016/s0969-2126(97)00207-4. [DOI] [PubMed] [Google Scholar]; Nair SK, Burley SK. Cell. 2003;112:193. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 7.Duesberg PH, Bister K, Vogt PK. Proc. Natl. Acad. Sci. USA. 1977;74:4320. doi: 10.1073/pnas.74.10.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duesberg PH, Vogt PK. Proc. Natl. Acad. Sci. USA. 1979;76:1633. doi: 10.1073/pnas.76.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackwood EM, Eisenman RN. Science. 1991;251:1211. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]; Prendergast GC, Lawe D, Ziff EB. Cell. 1991;65:395. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 10.Amati B, Dalton S, Brooks MW, Littlewood TD, Evan GI, Land H. Nature. 1992;359:423. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]; Blackwood EM, Luscher B, Eisenman RN. Gene Dev. 1992;6:71. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]; Kato GJ, Lee WMF, Chen L, Dang CV. Gene Dev. 1992;6:81. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]; Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Cell. 1993;72:233. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]; Tsuneoka M, Nakano F, Ohgusu H, Mekada E. Oncogene. 1997;14:2301. doi: 10.1038/sj.onc.1201067. [DOI] [PubMed] [Google Scholar]
- 11.Kretzner L, Blackwood EM, Elsenman RN. Nature. 1992;359:426. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- 12.Amati B, Littlewood TD, Evan GI, Land H. EMBO J. 1993;12:5083. doi: 10.1002/j.1460-2075.1993.tb06202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayer DE, Kretzner L, Eisenman RN. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]; Zervos AS, Gyuris J, Brent R. Cell. 1993;72:223. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 14.Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK. Proc. Natl. Acad. Sci. USA. 2002;99:3830. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lu X, Vogt PK, Boger DL, Lunec J. Oncol. Rep. 2008;19:825. [PubMed] [Google Scholar]
- 15.Review: Boger DL, Desharnais J, Capps K. Angew. Chem. Int. Ed. 2003;42:4138. doi: 10.1002/anie.200300574.
- 16.Silletti S, Kessler T, Goldberg J, Boger DL, Cheresh DA. Proc. Natl. Acad. Sci. USA. 2001;98:119. doi: 10.1073/pnas.011343298. [DOI] [PMC free article] [PubMed] [Google Scholar]; Boger DL, Goldberg J, Silletti S, Kessler T, Cheresh DA. J. Am. Chem. Soc. 2001;123:1280. doi: 10.1021/ja003579+. [DOI] [PubMed] [Google Scholar]
- 17.Goldberg J, Jin Q, Ambroise Y, Satoh S, Desharnais J, Capps K, Boger DL. J. Am. Chem. Soc. 2002;124:544. doi: 10.1021/ja0118789. [DOI] [PubMed] [Google Scholar]; Review: Boger DL, Goldberg J. Bioorg. Med. Chem. 2001;9:557. doi: 10.1016/s0968-0896(00)00276-5.
- 18.Boger DL, Goldberg J, Satoh S, Ambroise Y, Cohen SB, Vogt PK. Helv. Chim. Acta. 2000;83:1825. [Google Scholar]
- 19.Ambroise Y, Yuspan B, Ginsberg MH, Boger DL. Chem. Biol. 2002;9:1219. doi: 10.1016/s1074-5521(02)00246-6. [DOI] [PubMed] [Google Scholar]
- 20.Capps KJ, Humiston J, Dominique R, Hwang I, Boger DL. Bioorg. Med. Chem. Lett. 2005;15:2840. doi: 10.1016/j.bmcl.2005.03.094. [DOI] [PubMed] [Google Scholar]
- 21.Eubanks LM, Hixon MS, Jin W, Hong S, Clancy CM, Tepp WH, Malizio CJ, Goodnough MC, Barbieri JT, Johnson EA, Boger DL, Dickerson TJ, Janda KD. Proc. Natl. Acad. Sci. USA. 2007;104:2602. doi: 10.1073/pnas.0611213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee AM, Rojek JM, Spiropoulou CF, Gunderson A, Jin W, Shaginian A, York J, Nunberg JH, Boger DL, Oldstone MBA, Kunz S. J. Biol. Chem. 2008;283:18734. doi: 10.1074/jbc.M802089200. [DOI] [PMC free article] [PubMed] [Google Scholar]; Whitby L, Lee AM, Kunz S, Oldstone MBA, Boger DL. Bioorg. Med. Chem. Lett. 2009;19:3771. doi: 10.1016/j.bmcl.2009.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stover JS, Shi J, Jin W, Vogt PK, Boger DL. J. Am. Chem. Soc. 2009;131:3342. doi: 10.1021/ja809083d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaginian A, Whitby LR, Hong S, Hwang I, Faroogi B, Chen J, Vogt PK, Boger DL. J. Am. Chem. Soc. 2009;131:5564. doi: 10.1021/ja810025g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin XY, Glap C, Lazo JS, Prochownik EV. Oncogene. 2003;22:6151. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]; Wang HB, Hammoudeh DI, Follis AV, Reese BE, Lazo JS, Metallo SJ, Prochownik EV. Mol. Cancer Ther. 2007;6:2399. doi: 10.1158/1535-7163.MCT-07-0005. [DOI] [PubMed] [Google Scholar]; Mustata G, Follis AV, Hammoudeh DI, Metallo SJ, Wang H, Prochownik EV, Lazo JS, Bahar I. J. Med. Chem. 2009;52:1247. doi: 10.1021/jm801278g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiessling A, Weisinger R, Sperl B, Berg T. ChemMedChem. 2007;2:627. doi: 10.1002/cmdc.200600294. [DOI] [PubMed] [Google Scholar]; Kiessling A, Sperl B, Hollis A, Eick D, Berg T. Chem. Biol. 2006;13:745. doi: 10.1016/j.chembiol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y, Shi J, Yamamoto N, Mossa JA, Vogt PK, Janda KD. Bioorg. Med. Chem. 2006;14:2660. doi: 10.1016/j.bmc.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 28.Follis AV, Hammoudeh DI, Wang H, Prochownik EV, Metallo SJ. Chem. Biol. 2008;15:1149. doi: 10.1016/j.chembiol.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Boger DL, Lee JK, Goldberg J, Jin Q. J. Org. Chem. 2000;65:1467. doi: 10.1021/jo9916481. [DOI] [PubMed] [Google Scholar]
- 30.Cheng S, Comer DD, Williams JP, Boger DL. J. Am. Chem. Soc. 1996;118:2567. [Google Scholar]; Cheng S, Tarby CM, Comer DD, Williams JP, Caporale LH, Boger DL. Bioorg. Med. Chem. 1996;4:727. doi: 10.1016/0968-0896(96)00069-7. [DOI] [PubMed] [Google Scholar]; Boger DL, Tarby CM, Caporale LH. J. Am. Chem. Soc. 1996;118:2109. [Google Scholar]
- 31.Cavalieri F, Ruscio T, Tinoco R, Benedict S, Davis C, Vogt PK. Virology. 1985;143:680. doi: 10.1016/0042-6822(85)90412-x. [DOI] [PubMed] [Google Scholar]; Maki Y, Bos TJ, Davis C, Starbuck M, Vogt PK. Proc. Natl. Acad. Sci. USA. 1987;84:2848. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogt PK. Virology. 1971;46:939. doi: 10.1016/0042-6822(71)90092-4. [DOI] [PubMed] [Google Scholar]
- 33.3 (mixture of two diastereomers): 1H NMR (600 MHz, CDCl3, 25 °C) δ 9.03 (s, 1H), 7.44–7.26 (m, 1H), 6.90 (s, 2H), 6.21–6.05 (m, 1H), 4.21–3.17 (m, 12H), 2.68 (s, 1H), 2.57 (s, 6H), 2.24 (s, 3H), 2.22–2.13 (m, 2H), 1.98 (s, 1H), 1.69 (d, J = 11.2 Hz, 2H), 1.43 (s, 9H), 1.19 (d, J = 7.8 Hz, 9H), 1.10 (d, J = 11.8 Hz, 2H); 13C NMR δ 177.5, 176.05, 176.02, 175.81, 175.77, 174.9, 174.8, 173.20, 173.16, 172.3, 172.1, 171.7, 169.59, 169.57, 169.55, 169.4, 155.79, 155.78, 155.77, 143.54, 143.49, 143.48, 140.13, 140.12, 140.11, 140.08, 140.06, 140.05, 84.07, 84.05, 83.93, 83.91, 80.4, 80.3, 68.87, 68.86, 67.54, 67.49, 65.47, 65.42, 56.39, 56.36, 56.21, 56.16, 49.94, 49.90, 49.50, 49.47, 49.2, 49.1, 49.0, 47.5, 47.38, 47.35, 47.2, 46.8, 46.4, 46.2, 45.77, 45.74, 45.1, 44.3, 42.6, 42.5, 41.77, 41.75, 33.8, 33.7, 32.9, 30.6, 29.3, 28.4, 23.8, 21.7, 15.94, 15.92, 15.0; ESI-TOFMS (m/z) calcd for [C34H52N4O10S+H]+ 709.3477; found: 709.3473. For 4 (mixture of two diastereomers): 1H NMR (400 MHz, DMSO-d6, 25 °C) δ 8.64 (s, 1H), 8.24–8.14 (m, 1H), 7.96–7.88 (m, 1H), 7.27–7.25 (m, 1H), 6.78–6.70 (m, 1H), 4.04–3.88 (m, 1H), 3.74–3.60 (m, 2H), 3.54–3.44 (m, 1H), 3.42–3.34 (m, 1H), 3.30–3.14 (m, 3H), 3.14–3.02 (m, 1H), 2.88 (dd, J = 12.9, 6.6 Hz, 2H), 2.57 (dd, J = 4.5, 2.6 Hz, 3H), 2.20–2.14 (m, 2H), 1.65–1.56 (m, 1H), 1.50–1.41 (m, 2H), 1.40–1.32 (m, 11H), 1.31–1.27 (m, 9H), 1.26–1.17 (m, 2H); 13C NMR δ 171.79, 171.75, 171.71, 171.69, 171.24, 171.22, 170.7, 170.6, 168.88, 168.85, 168.84, 156.0, 154.3, 137.79, 137.75, 137.70, 137.66, 129.35, 129.33, 129.28, 129.26, 127.3, 126.5, 81.98, 81.93, 81.90, 79.08, 79.04, 77.8, 56.2, 56.0, 55.4, 49.44, 49.38, 49.35, 49.09, 49.03, 49.01, 46.93, 46.91, 46.86, 46.75, 46.72, 45.4, 45.3, 45.22, 45.17, 41.60, 41.58, 41.49, 41.47, 33.99, 33.91, 29.9, 28.8, 28.6, 28.1, 27.8, 26.57, 26.52, 26.18, 26.16, 24.48; ESI-TOFMS (m/z) calcd for [C29H45Cl2N5O9S2+H]+ 742.2108; found: 742.2088