Abstract
Family X polymerases like DNA polymerase λ (pol λ) are well suited for filling short gaps during DNA repair because they simultaneously bind both the 5′ and 3′ ends of short gaps. DNA binding and gap filling are well characterized for one nucleotide gaps, but the location of yet-to-be-copied template nucleotides in longer gaps is unknown. Here we present crystal structures revealing that when bound to a two-nucleotide gap, pol λ scrunches the template strand and binds the additional uncopied template base in an extrahelical position within a binding pocket comprised of three conserved amino acids. Replacing these amino acids with alanine results in less processive gap filling and less efficient NHEJ involving two nucleotide gaps. Thus, akin to scrunching by RNA polymerase during transcription initiation, scrunching occurs during gap filling DNA synthesis associated with DNA repair.
The DNA polymerases of family X are responsible for small scale DNA synthesis in the context of different DNA repair processes1. For instance, the best studied member of this family, pol β provides both a gap-filling and a rate-limiting dRP lyase activity to the Base Excision Repair (BER) pathway2. Pol λ is also capable of gap-filling3 and contains a dRP lyase activity4. However, while evidence exists suggesting its participation in BER 5,6, its main physiological role appears to be gap filling during repair of double strand breaks through the Non Homologous DNA End Joining (NHEJ) pathway 7,8,9. Like pol λ,the other two human members of the family, terminal deoxynucleotidyl transferase and pol μ, participate in double strand break repair1,7,9. Family X enzymes play a role both in the NHEJ repair process and in V(D)J recombination, a process that is critical for lymphocyte development10.
The availability of structural information for each of the mammalian members of the family and in particular the wealth of structural and biochemical data available for pols β and λ have resulted in a clear picture of the structural organization and catalytic mechanism of family X polymerases1. Important differences exist in the way that pol β and pol λ interact with DNA and in the nature of the conformational changes that take place throughout the catalytic cycle11. Nevertheless, there are many structural and biochemical similarities between both enzymes, including a similar behavior when filling in small gaps. Both polymerases processively fill short gaps in which the 5'-end of the gap contains a 5'-phosphate3,12. This behavior is explained by the presence of a family X polymerase-specific 8 kDa domain that interacts with the 5'-phosphate group2.
In pols β13, λ11 and μ14, the spatial relationship of the polymerase domain that binds the primer-template junction and the 8-kDa domain that binds to the 5′ end of a downstream DNA chain is well known for synthesis to fill a single nucleotide gap. Not only are the polymerase and 8-kDa domains tethered to each other within the same polypeptide chain, structure-function studies of pols β and λ reveal that these two domains also physically interact with each other during repair synthesis to fill gaps containing a single template base2. However, longer patch BER and NHEJ of certain broken ends involve filling gaps longer than a single nucleotide15,16. Consistent with this requirement, biochemical studies demonstrate that pol β and pol λ can fill gaps as long as 5 and 6 nucleotides in a processive manner, indicating simultaneous DNA binding by both the polymerase and 8 kDa domain3,12. Furthermore, recent studies have demonstrated that perturbing 5′ end binding by pol μ reduces the efficiency of NHEJ17 and that 5'-phosphate binding is also critical for bacterial NHEJ18. Yet, it is not evident from existing structures how simultaneous binding to both ends of a DNA gap could be achieved on gaps longer than one nucleotide. We decided to investigate how pol λ catalyzes gap-filling synthesis on longer DNA gaps. Here we present structural and biochemical evidence suggesting a mechanism by which human pol λ is capable of simultaneously engaging the two ends of a DNA gap containing more than one nucleotide in a catalytically relevant conformation.
Results
Structure of a gap-filling intermediate
The first crystal structure of pol λ captured the polymerase in an inactive binary complex in which the enzyme was bound to a two-nucleotide gap, but without a dNTP present19. In this structure, the 8 kDa domain was bound to the 5'-phosphate end of the gap, but the polymerase active site was not productively engaged at the 3' end of the gap. Although not representing a catalytically competent gap filling intermediate, this structure suggested that, in the absence of an incoming nucleotide, the polymerase might preferentially bind to the 5'-end of a gap, a feature that appeared consistent with a role in gap filling.
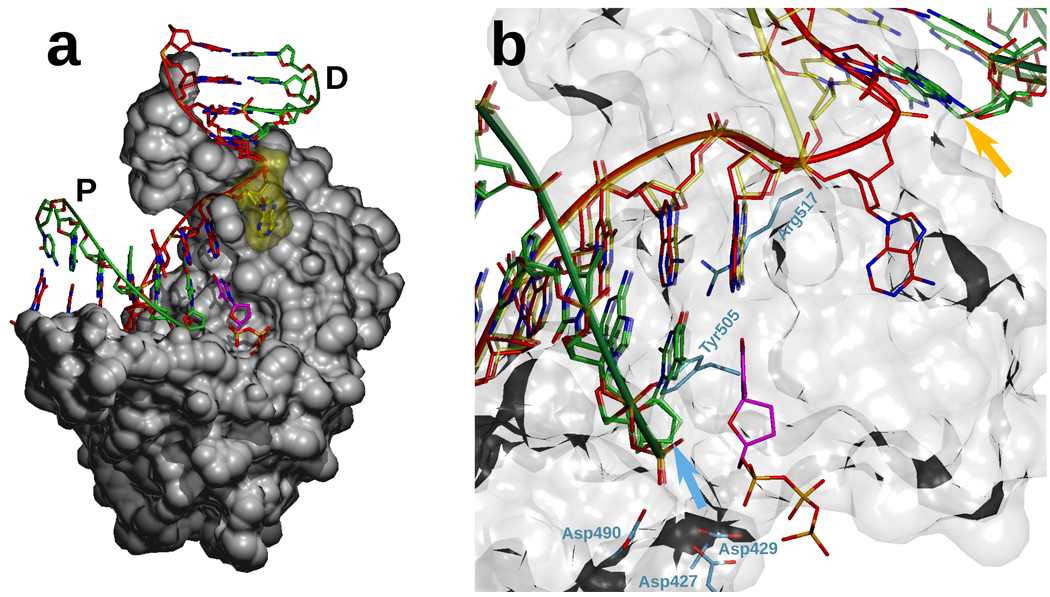
In order for the first step of gap-filling to occur, the active site of the polymerase must be bound in its catalytic conformation at the 3'-end of the primer. If, as suggested by the biochemical data, polymerization occurs while the 8kDa domain is engaging the 5'-phosphate, it is necessary to invoke a conformational change leading to an intermediate where, either through DNA or protein rearrangements, the polymerase active site would bind the 3' DNA end in a catalytic conformation while the 8 kDa domain would remain bound to the 5'-end of the gap. Because such a conformation is not seen in the 2-nucleotide gap binary complex (1RZT), we hypothesized that it could be dependent on dNTP binding. Thus, we attempted to determine the structure of a ternary complex of human pol λ bound to a 2-nucleotide gap with a correct dNTP bound in a pre-catalytic state. We grew crystals of pol λ after mixing the enzyme with a three-nucleotide gap in the presence of ddTTP, with the expectation that incorporation of ddTMP would result in a dideoxy-terminated two-nucleotide gap with ddTTP bound in the polymerase active site (see Methods). We obtained crystals that diffracted to a resolution of 1.95Å (Table 1) and we were able to solve the structure by molecular replacement (see Methods). The structure (Fig. 1) shows pol λ bound to DNA while attempting to incorporate a correct nucleotide opposite the first of the two single-stranded nucleotides in the gap. The polymerase binds in a remarkably similar manner to what was observed in previous ternary complex structures involving a single nucleotide gap11, despite the fact that the gap is one nucleotide longer (rmsd of 0.935 Å for 316 C-a atoms). All the polymerase active site residues are in conformations indistinguishable from those observed in a single-nucleotide gap structure, indicating that this structure represents a functional complex. Moreover, the 8 kDa domain is engaged in binding the 5'-phosphate, and does so in a similar conformation as is observed when the enzyme is in complex with a 1-nucleotide gap. The solution to this apparent paradox is that the template strand is scrunched, where one of the template nucleotides in the 2-nucleotide gap is extrahelical (Fig. 1), and bound in a manner that results in very little distortion of helix geometry (Fig. 1b). The largest difference is in the position of the phosphate 5'- to the extrahelical base (Fig. 1b). Importantly, the 3' primer terminus and the 5' phosphate of the downstream primer (arrows in Fig. 1b) remain in the same relative positions, and only a slight difference is observed in the conformation of the downstream DNA and the 8 kDa. By adopting this conformation, Pol λ engages both sides of the gap in the absence of functionally relevant protein conformational changes and without changing the relative locations of the polymerase and 8 kDa domains. For the latter reason, we refer to this as “scrunching”, by analogy to what was previously observed with RNA polymerases20,21,22.
Table 1.
Data collection and refinement statistics
| 3HWT | 3HW8 | 3HX0 | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | C2221 |
| Cell dimensions | |||
| a, b, c (Å) | 56.023, 63.328, 137.850 | 56.249, 68.386, 137.108 | 124.179, 131.912, 280.412 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50-1.95 | 50-1.95 | 50-3 |
| Rsym | 8.9 (39.9) | 9.2 (52.2) | 10.7 (74.5) |
| I / σI | 10.3 (2.9) | 17.5 (2.9) | 9.4 (2.6) |
| Completeness (%) | 96.0 (91.2) | 97.2 (89.5) | 99.9 (99.5) |
| Redundancy | 4.1 (2.8) | 6.1(3.7) | 9.3 (8.3) |
| Refinement | |||
| Resolution (Å) | 1.95 | 1.95 | 3 |
| No. reflections | 70757 | 38282 | 44203 |
| Rwork / Rfree | 22.8/26.1 | 21.2/23.3 | 22.3/26.8 |
| No. atoms | 3243 | 3321 | 11994 |
| Protein | 2410 | 2509 | 10027 |
| Ligand/ion | 480 | 486 | 1932 |
| Water | 350 | 331 | 35 |
| B-factors | |||
| Protein | 43.2 | 41.4 | 88.9 |
| Ligand/ion | 34.2 | 29.3 | 84.7 |
| Water | 48.9 | 42.7 | 64.3 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.003 | 0.005 | 0.003 |
| Bond angles (°) | 1.09 | 1.0 | 0.8 |
Values in parentheses are for highest-resolution shell.
Figure 1.
Structure of Pol λ bound to a two-nucleotide gap. a. Overview of the structure. The DNA polymerase was crystallized in a pre-catalytic state, prior to incorporation of the incoming dTTP (magenta). The primer (P) and downstream primer (D) strands (green) are bound in the same conformation as in a single-nucleotide gap. The template strand (red) contains an extrahelical single-stranded nucleotide (yellow surface). b. Overlay of the DNA strands in the one- and two-nucleotide gap structures. The only difference between the conformation of the template strand in the one-nucleotide (yellow) and the two-nucleotide (red) structure is the presence of the extrahelical nucleotide. The 3'-end of the primer (blue arrow) and the 5'-phosphate end of the downstream primer (orange arrow) are in similar positions in both complexes.
A binding pocket for purines and pyrimidines
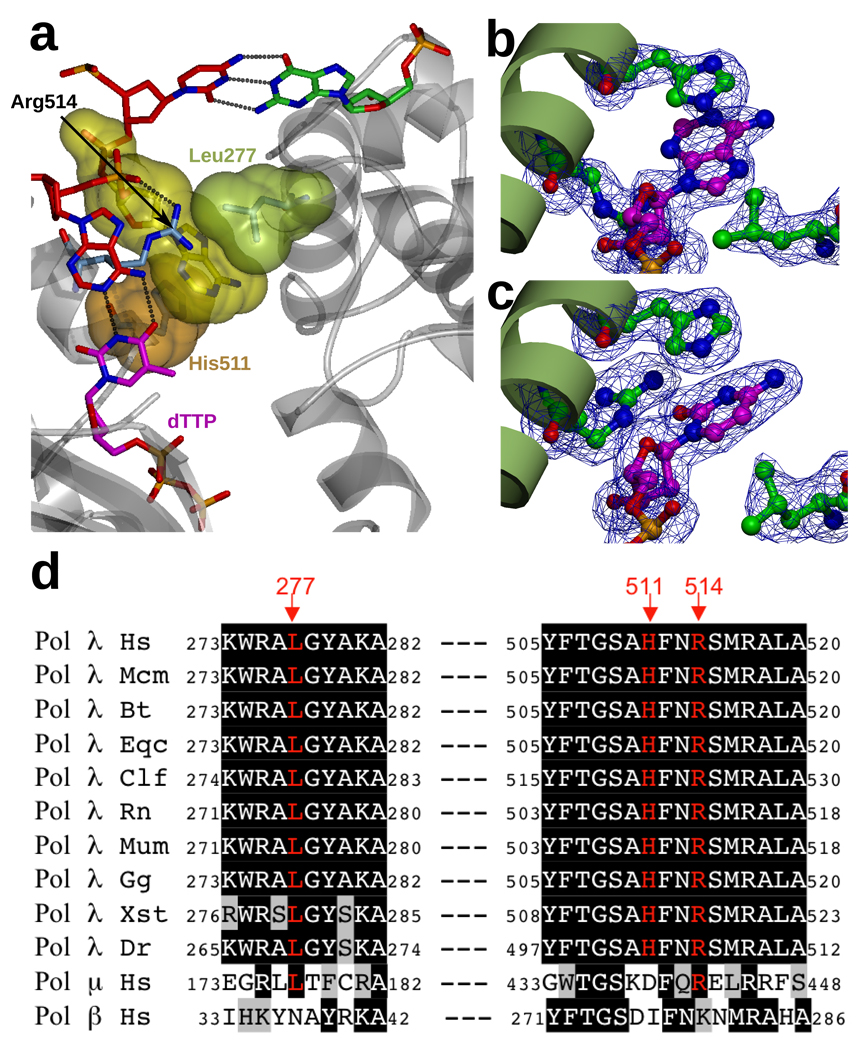
Inspection of the structure reveals that the extrahelical nucleotide is bound in a pocket formed by the side chains of three amino acids (Fig. 2a,b). When the nucleotide bound in the pocket is an adenine, Leu277 and His511 provide hydrophobic contacts with the extrahelical base, while Arg514 mainly interacts with the phosphate 3' to that nucleotide. To determine if this pocket also accommodates pyrimidines, we crystallized pol λ in complex with a 2-nucleotide gap and incoming dTTP as before, but altered the template sequence so that dCMP is present as the second single-stranded nucleotide in the gap. These crystals diffracted to a resolution of 1.95Å (see Table 1 and Methods), and the resulting electron density indicates cytosine is indeed bound in the pocket (Fig 2c). Binding is similar to that observed with adenine (Fig. 2b), except that the side chain of Arg514 adopts a different conformation that allows it to hydrogen bond with the O2 atom in the cytosine base, while Leu277 appears to undergo a slight rearrangement to maximize contact with the smaller pyrimidine base. These results suggest that the three residues comprising the binding pocket for the extrahelical base share an analogous role for all four bases normally present in DNA, and further suggest that the pocket may have a role in stabilizing the scrunched intermediate. The functional significance of this pocket is further implied by the fact that the three residues comprising the pocket are conserved in all chordate pol λ homologs (Fig. 2d). Interestingly, two of the three residues are conserved in human pol μ, while only the charge of one of the three residues (Arg514) has been conserved in human pol β, as Lys 280.
Figure 2.
Binding pocket for the scrunched nucleotide. a. Binding pocket for the scrunched nucleotide. The extrahelical nucleotide (yellow surface) is shown, together with the following and preceding template residues (red), the 5' residue in the downstream primer (green), the incoming dTTP (magenta), and the residues that form the binding pocket, His511 (orange surface), Leu277 (green surface) and Arg514 (blue). b/c. Detail of the binding pocket for the scrunched nucleotide. A simulated annealing Fo-Fc omit density map is shown, contoured at 3σ. The binding pocket adopts a similar conformation whether the base of the scrunched nucleotide is adenine (B) or cytosine (C). d. The scrunching pocket residues are conserved in pol λ from animal cells. Human (Hs) pol λ sequence is aligned with sequences of pol λ from: rhesus monkey, Macaca mulatta (Mcm); cattle, Bos taurus (Bt); horse, Equus caballus (Eqc); dog, Canis lupus familiaris (Clf); rat, Rattus norvegicus (Rn); mouse, Mus musculus (Mum); red jungle fowl, Gallus gallus (Gg); western clawed frog, Xenopus (Silurana) tropicalis (Xst); zebrafish, Danio rerio (Dr); and sequences of human pols μ and β. Invariant residues have black background. The residues of the scrunching pocket are presented in red and the red numbers correspond to the numbering of residues in the human sequence. Conserved residues have gray background.
A triple mutant polymerase has decreased processivity
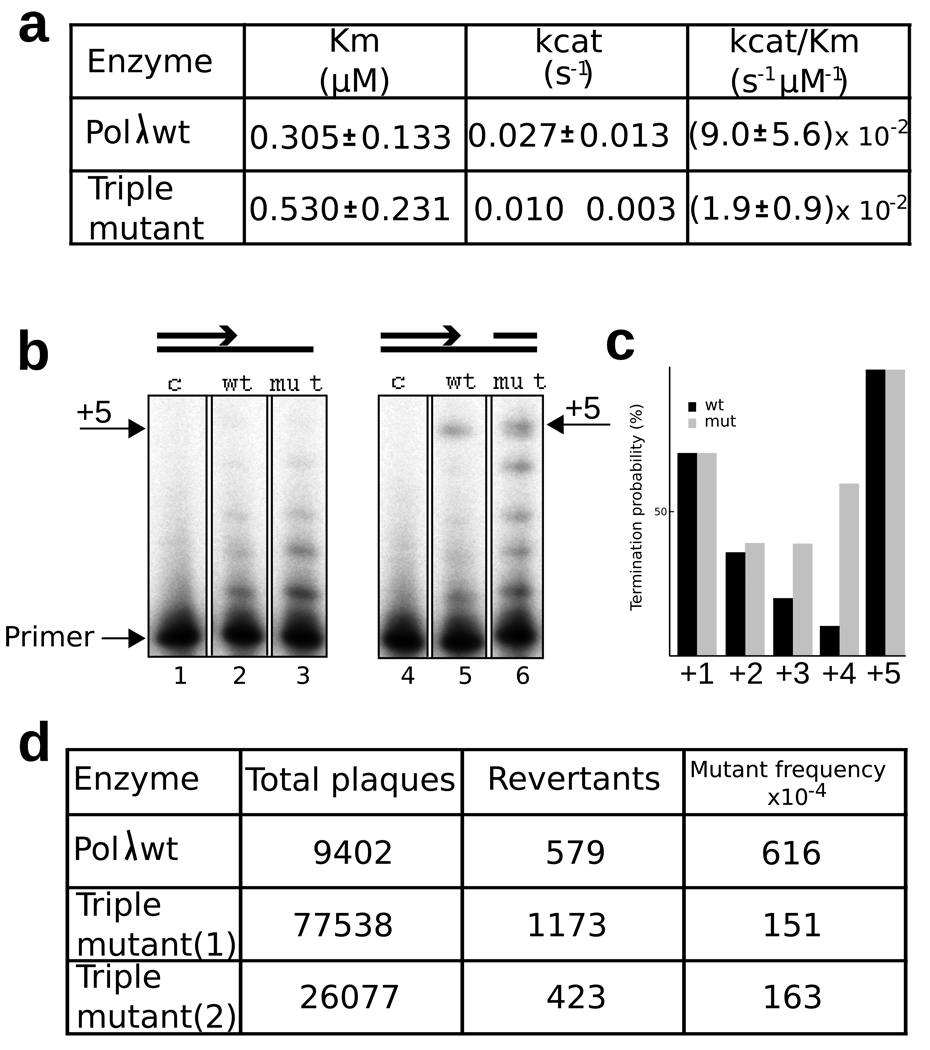
To test the functional significance of this template-binding pocket, we replaced the three conserved residues that contact the extrahelical nucleotide with alanine. Steady state kinetic analysis of single nucleotide gap filling demonstrated that, despite non-conservative replacement of three conserved residues, the catalytic efficiency of the purified triple mutant pol λ is only 5-fold lower than that of wild type pol λ(Fig. 3a). Moreover, the triple mutant enzyme behaves like wild-type pol λ when copying a non-gapped primer-template, in that both enzymes are largely distributive when no downstream primer with a 5′ phosphate is present (Fig. 3b, lanes 2 and 3). However, the results are different when filling a 5-nucleotide gap (Fig. 3b, lanes 5 and 6). The total amount of product generated is greater than for simple primer extension (e.g., compare product band intensities in lane 3 versus 6), suggesting that the triple mutant still benefits from binding the downstream 5'-phosphate. As expected, wild type pol λ fills the gap (lane 5) more processively than for simple primer extension (lane 2). In contrast, gap filling DNA synthesis by the triple mutant is less processive (lane 6) than for wild type pol λ (lane 5), demonstrating that it dissociates from the DNA more readily and/or translocates less efficiently than wild type pol λ. When termination probabilities were calculated (see Methods) for each position as the gap was being filled, wild type and mutant pol λ behaved similarly for the first and second incorporations (Fig. 3c, black versus gray bars at +1 and +2). In these instances, the numbers of uncopied template bases in the gap are four and three, respectively. However, at the +3 and +4 positions, the triple mutant enzyme terminated processive synthesis more often than did wild type pol λ. At the +4 position, which results from incorporation into a substrate where one uncopied base would need to be bound in the pocket, the difference in termination probability between the wild type and triple mutant is 5-fold. Thus, the effect of the pocket on processivity increases as the gap becomes shorter with the maximum effect seen on a two nucleotide gap with one nucleotide stabilized in the pocket.
Figure 3.
a. Steady state kinetic analysis of nucleotide incorporation. Reactions were performed as described in Materials and Methods. b. Gap-filling activity assay. The wild type and the triple alanine mutant polymerases have comparable activity when polymerizing on either an open template/primer (T/P) or a 5 nucleotide gap substrate (Note that relative to the wt enzyme, the triple mutant generated more total products, consistent with the fact that more enzyme was present in the reaction). However, only the wt polymerase fills in the 5 nucleotide gap processively. c. The bands in the right two lanes of panel a were quantified and termination probabilities were calculated after each incorporation during synthesis to fill the 5-nucleotide gap. d. Single-nucleotide-deletion fidelity of the pol λ triple mutant. The genetic assay used for measuring single-nucleotide deletions is described in the Methods section.
The binding pocket influences the fidelity of synthesis
The processivity data implies that the binding of an uncopied template base in the pocket contributes to the stability of the scrunched intermediate. Lower stability of a scrunched intermediate in the triple mutant predicts that it might affect the fidelity of DNA synthesis by pol λ, based on the following logic. Wild type pol λ is particularly prone to generate single base deletion intermediates during DNA synthesis23. This property is thought to reflect its normal biological function, i.e., the ability to participate in NHEJ by filling short gaps formed when two broken ends are aligned with as little as a single correct terminal base pair7,23,24. The mechanism for producing single base deletions is thought to involve dNTP-induced misalignment of the template-strand relative to the primer strand during catalytic cycling25. This generates an intermediate with an extrahelical template nucleotide in the duplex DNA upstream of the polymerase active site24,26. A wealth of evidence indicates that the ability of a polymerase to extend a misaligned intermediate to ultimately yield a single base deletion depends on the stability of the misaligned intermediate. Thus, as dNTP binding induces scrunching when filling gaps longer than a single nucleotide, the destabilization imparted by the triple mutant may disfavor extension of a misaligned intermediate more so than extension of an aligned intermediate, thereby reducing the rate at which pol λ generates single base deletions. To test this prediction, we compared the ability of wild type and triple mutant pol λ to generate single base deletions during synthesis to fill a 6-nucleotide gap containing a LacZ template TTTT run23 (Fig. 3d). As expected based on earlier studies23, wild type pol λ is inaccurate (lacZ mutant frequency 620 × 10−4). In comparison, the triple mutant is 4-fold more accurate (lacZ mutant frequency 160 × 10−4). This is consistent with the idea that the triple mutant destabilizes scrunched intermediates in a manner that disfavors extension of misaligned template-primers.
Structural effects of the triple substitution
To further examine the role of the binding pocket for the second nucleotide in the gap, we obtained a 3Å crystal structure of the triple mutant using the same substrate used with wild-type pol λ (Table 1). Four molecules of the triple mutant are present in the asymmetric unit. Two of these are in a conformation similar to that observed in the wild-type pol λ structure. Scrunching is observed, i.e., the 3'-OH and 5'-PO4 groups maintain a similar distance to what is observed in a single-nucleotide gap. However, while the backbone of the template strand adopts the same conformation as in the wild type structure, the absence of the binding pocket results in an increase in the conformational flexibility of the base of the second nucleotide in the gap. The other two molecules in the asymmetric unit are in a conformation reminiscent of that observed in the original two-nucleotide structure (1RZT). The incoming dNTP is present, but the base is not bound in its usual conformation (see Fig. 4a), i.e., the protein is in the inactive conformation. This indicates that correct geometry between the incoming nucleotide and the templating base is required to transition to an active, scrunched structure, and suggests that the scrunched intermediate is less stable in the triple mutant than in wild type pol λ, an interpretation consistent with the biochemical data.
Figure 4.
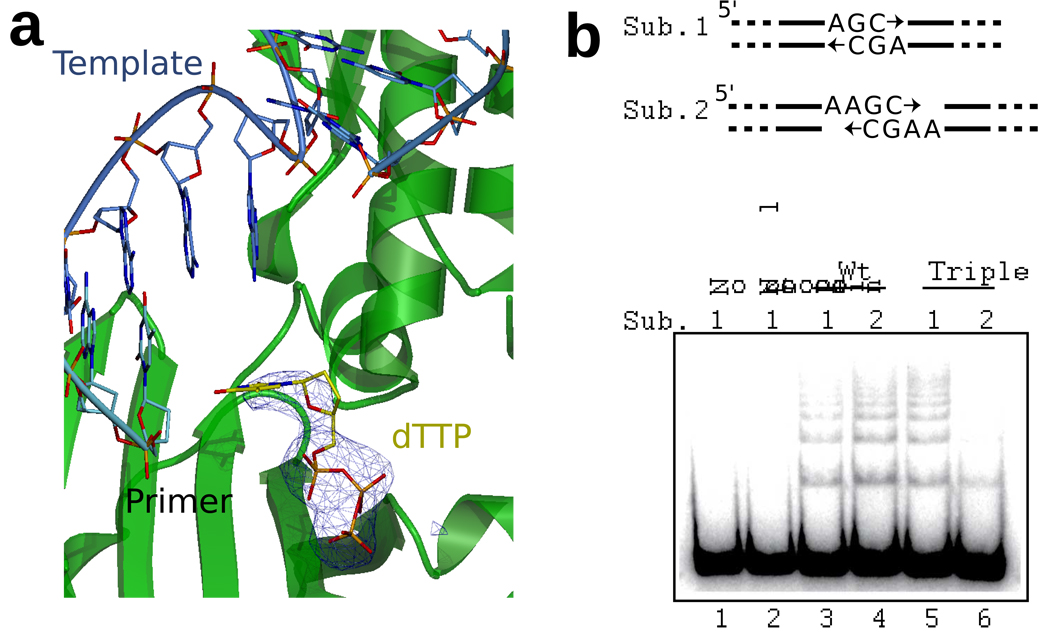
a. Scrunching in the triple alanine mutant. Aberrant nucleotide binding in the triple mutant structure. In two of the molecules in the asymmetric unit the nucleotide is only partly bound. As a consequence the polymerase has not adopted a pre-catalytic conformation and is only engaging the 5' end of the gap. A simulated annealing Fo-Fc omit density map is shown, contoured at 3σ. b. NHEJ reaction. 280 bp linear fragments were used as substrates that, after aligning partially complementary termini, require fill-in of one (Sub. 1) or two (Sub. 2) nucleotide gaps before ligation can occur. Ku, XRCC4-ligase IV, and polymerase were included as noted in reactions for 3 minutes (Substrate 1) or 15 minutes (Substrate 2).
“Scrunching” and NHEJ
The ability of the polymerase to “scrunch” the template strand and conduct processive gap-filling could be important for NHEJ reactions when the ends to be joined contain gaps greater than a single nucleotide. To test this possibility, we performed NHEJ reactions with Ku heterodimer, XRCC4, DNA ligase IV and either wild type pol λ or the triple mutant pol λ. We used two different linear 280 base pair substrates with overhanging sequences such that end alignment results in either a 1-nucleotide or a 2-nucleotide gap (Substrates 1 and 2, respectively, Fig. 4b). These gaps must be filled by pol λ before concatemer ligation products are generated (e.g., Fig. 4b, compare lanes 2 and 3). Using the substrate that requires filling a one-nucleotide gap, and where scrunching would not be necessary (Substrate 1), the triple mutant was slightly more efficient than wild type pol λ at promoting end-joining (Fig. 4b, compare lanes 3 and 5). Thus the triple mutant enzyme is fully capable of performing one-nucleotide gap filling during NHEJ. In contrast, using a substrate requiring filling a two-nucleotide gap (Substrate 2), where the scrunching observed in the crystal structure would be relevant, the efficiency of end joining using the triple mutant pol λ was 8-fold lower than for wild type pol λ (Fig. 4b, compare lanes 4 and 6). This indicates that the pocket that binds the uncopied template base is important for efficient NHEJ when end joining requires the filling of a 2-nucleotide gap.
Relevance of scrunching to longer gaps
The difference in termination by wild type and triple mutant pol λ seen at the +3 position as a 5-nucleotide gap is filled (Fig. 3b/c), and the fact that the triple mutant is more accurate than wild type pol λ during synthesis to fill a 6-nucleotide gap (Fig. 3d) both suggest that the binding pocket for the second template nucleotide in the gap may also be relevant to filling gaps of at least three nucleotides. For this reason, efforts were made to obtain crystal structures involving 3-, 4- and 5-nucleotide gaps. These attempts all failed. This prompted three modeling studies using the scrunched dA structure (Fig. 1) as an initial template. In one case, one dAMP was added 5' to the scrunched nucleotide. In another case, the dAMP was added 3' to the scrunched nucleotide. In the final case, two adenines were added 5' of the scrunched nucleotide. We then performed simulations (see Methods) on each of these models to investigate the position of the nucleotides in the gap throughout the simulation. In all cases, the simulations indicated that the nucleotide immediately 5' to the templating nucleotide is preferentially bound in the scrunching pocket, and that additional nucleotides 5' to this one could indeed be accommodated by the polymerase (Fig. 5). These models are consistent with the idea that pol ⌊ can accommodate additional uncopied template nucleotides while maintaining a conformation similar to that observed in the crystal structures. The models further suggest that, when given multiple choices, the scrunching pocket is more likely to accommodate the nucleotide immediately adjacent to the templating nucleotide.
Figure 5.
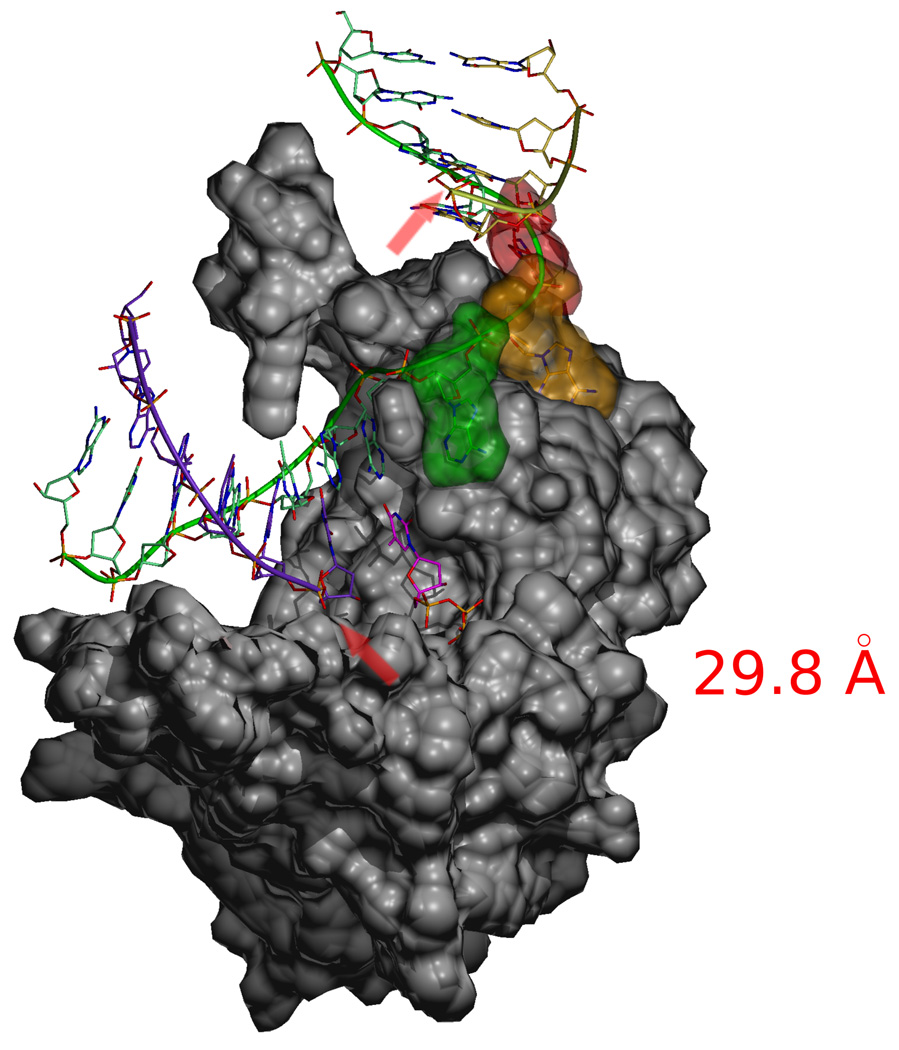
Final conformation of the template strand after molecular dynamics simulations with a 4-nucleotide gap substrate. The single-stranded nucleotides in the gap are shown with their van der Waals surfaces colored. The distance between the C3′ atom of the primer terminal nucleotide and the C5' atom of the 5'-nucleotide of the downstream primer (red arrows) is kept almost constant (29.8 Å versus 28.3 Å for a single nucleotide gap) despite the presence of two additional template nucleotides.
Discussion
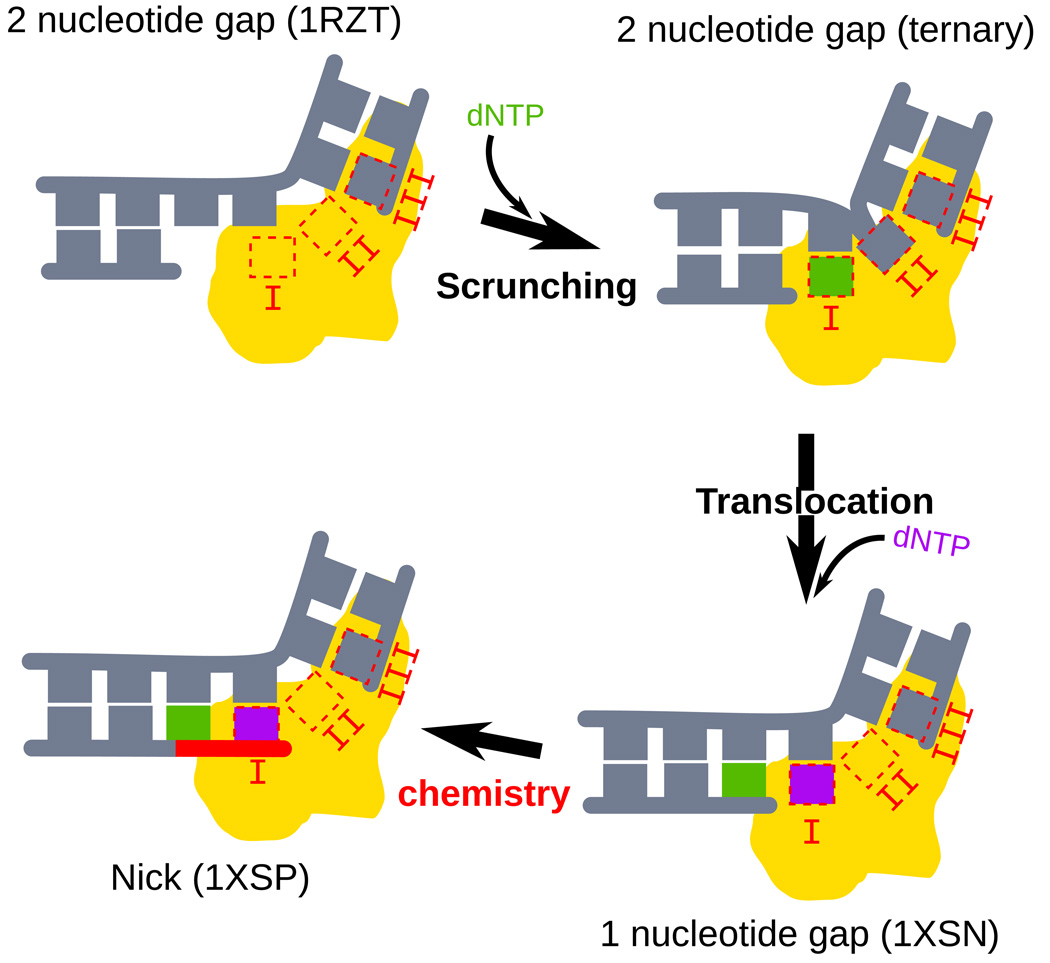
First proposed by Ikeda and Richardson22 as a mechanism by which T7 RNA polymerase initiates transcription, DNA scrunching was first captured structurally by Cheetham and Steitz20, and was more recently demonstrated for T727 and E. coli RNA polymerase21. Scrunching appears to be a general mechanism for initiation of transcription, presumably allowing the polymerase to proceed with RNA synthesis without needing to dissociate from the initiation site. We show here that scrunching of the DNA template also appears to be used by DNA polymerase λ during DNA synthesis associated with DNA repair that fills gaps longer than one nucleotide. This scrunching is accompanied by repositioning of the uncopied template base to an extrahelical position, where it binds within a pocket comprised of three conserved amino acids. This pocket appears to contribute to processive gap filling that may help to prevent premature release of repair intermediates that are potentially toxic and/or mutagenic. The identification of this binding pocket for the second template nucleotide in the gap brings to three the number of DNA substrate binding pockets observed in Pol λ, the other pockets being for the nascent base pair and the phosphate at the 5′ end of the gap (Fig. 6). From the currently available structures, a picture of gap filling emerges (Fig. 6 and Supplementary Video) wherein correct dNTP binding induces a number of conformational changes in the polymerase and the DNA, most especially in the template strand. These changes result in assembly the binding pockets for the nascent base pair and the second nucleotide in the gap. Apparently it is this remarkable degree of specialization that enables Pol λ to fulfill its biological roles in filling short gaps during DNA repair.
Figure 6.
Scrunching during gap-filling. A simplified model of the steps during gap-filling by pol λ is shown. Each of the steps corresponds to a crystallized intermediate. The DNA polymerase is represented as a yellow surface, and each of the three DNA binding sites (I, II and III) are represented. The PDB code corresponding to each intermediate is indicated above each panel11,19. An animated reconstruction of the reaction pathway can be found in the Supplementary Materials.
Methods
Protein expression and crystallization
We purified proteins as described19. Oligos T12A (5'-CGGCAAATACTG), T12C (5’-CGGCCAATACTG), P (5'-CAGTA) and D (5'-GCCG) were from Oligos Etc. ddTTP was from GE. We crystallized pol λ with T12A/T12C, P, D and ddTTP. Incorporation of ddTTP generates a dideoxyterminated two-nucleotide gap. We grew crystals at 4°C in a solution containing 10%(v/v) 2-propanol, 0.2 M sodium citrate and 100 mM sodium cacodylate, pH 5.5 (Pol λ + T12A), 2 M sodium formate (Pol λ + T12C) or 5% (v/v) 2-propanol, 25 mM ammonium acetate, 15 mM magnesium acetate and 100 mM cacodylic acid pH 6.5 (Pol λ L277A, R511A, H514A + T12A).
Data collection, processing and refinement
We collected data (T12A and T12C) at −178°C with a Saturn92 CCD detector and MicroMax-007HF (Rigaku) generator equipped with Varimax HF mirrors, and for the mutant at the SER-CAT 22-ID beamline at the APS, ANL. We processed all data using HKL200028.
Pol λ + T12A
We created a search model from PDB entry 2bcq24. We performed molecular replacement using MOLREP29, refined the solution with CNS30 and built the model using O31. Residues 289–294 and 313–328 are disordered, as can be judged from the high B-factors in those regions. The quality of the model was assessed using Molprobity32 (100% allowed, 97.48% favored).
Pol λ + T12C
We performed molecular replacement using the solved structure of Pol λ + T12A using MOLREP29 and refinement using O31 and CNS30. The T12C structure also showed disorder in residues 289–294 and 313–328. The quality of the model was assessed using Molprobity32 (99.70% allowed, 97.20% favored).
Pol λ L277A, R511A, H514A + T12A
We performed molecular replacement using the solved structure of Pol λ + T12A with MOLREP29. We used O31 and Coot33 for model building and CNS30 for refinement. Regions 246–251, 464–471 and 535–547 showed disorder. The quality of the model was assessed using Molprobity32 (99.00% allowed, 90.60% favored).
Steady-state analysis of nucleotide incorporation
We prepared the substrate by hybridizing 32P-5'-end-labeled P17 (GTACGACTGAGCAGTAT) with DP14 (5'P-GCCGGACGACGGTG) and T1g (CTCCGTCGTCCGGCAATACTGCTCAGTCGTAC). Reactions (10 µl) contained 50 mM Tris, pH 7.5, 1mM dithiothreitol, 4% glycerol, 0.1 mg·ml−1 BSA, 2.5 mM MgCl2, 200 nM DNA, and 1.5 nM full length wt or 3 nM full length triple mutant pol λ. Reactions were initiated by adding dTTP (0.1, 0.2, 0.5, 1, 2, 5, 10, 15 or 20 µM) and incubated at 37°C for 2.5 or 3 minutes (wt or triple mutant, respectively). Products were resolved by PAGE and quantified by autoradiography. The data were fit to the Michaelis-Menten equation using nonlinear regression.
Processivity
We constructed a Pol λ triple mutant with Quikchange (Stratagene). Measurements were carried out as previously described34. 32P 5'-labeled P (5'-GTACGACTGAGCAGTAT) was hybridized to T (5'-CTCCGTCGTCCGGCACGTAATACTGCTCAGTCGTAC), or to T and DT (5'-GCCGGACGACGGTG). Reactions (20 µl) contained 50 mM Tris-HCl, pH 7.5, 1 mM DTT, 4% (v/v) glycerol, 0.1 mg·ml−1 BSA, 10 mM MgCl2, 200nM of the DNA duplex and 2nM or 7 nM wt or triple mutant pol λ, respectively, were initiated by adding dNTPs (100 µM), incubated at 37°C and stopped at 1, 2, 3 and 4 minutes. We quantified product bands and calculated the probability of terminating processive synthesis. The DNA to enzyme ratios used prevent synthesis on previously used substrates: termination probabilities did not change with reaction time35.
Short-gap reversion assay
The substrate has been described36 and encodes a colorless M13 plaque phenotype. Frameshift mutations that restore the reading frame result in blue plaques. Reaction mixtures (20 µl) contained 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol, 2 µg of BSA, 4 % glycerol, 1.6 nM DNA, 50 µM dNTPs, 400 units of T4 DNA ligase and 100 nM wt or 50 nM triple mutant Pol λ. Following 1 h incubation at 37°C, reactions were terminated and products separated on an agarose gel, electroeluted, introduced into E. coli MC1061 and plated. We scored revertants and total plaques.
NHEJ assay
We incubated 25 nM Ku, 10 nM XRCC4-LigaseIV, and 10 nM full length pol λ polymerase with 5 nM DNA substrate (generated as in7) in a reaction buffer with 33 mM Tris-HCl (pH 7.5), 1mM DTT, 150 mM KCl, 200 ng BSA, 3% glycerol, 0.1 mM EDTA, 12.5% (w/v) polyethylene glycol (MW > 8000kDa), 1 µM dNTPs, 5 mM MgCl2 and 1 µg supercoiled plasmid DNA (Litmus38; New England Biolabs, Ipswich, MA). Reactions were stopped after 3 (Sub. 1) or 15 minutes (Sub. 2) at 37°C, deproteinized, analyzed by non-denaturing 5% PAGE, and quantified by densitometry (Imagequant v 5.1; GE biosciences).
Molecular dynamics
We created three models using SYBYL 8.0 (Tripos Inc.), modifying the crystal structure by adding extra nucleotides prior to the scrunched nucleotide; (i) two adenine nucleotides (4-gap) (ii) one adenine nucleotide (3-gap) (iii) one adenine nucleotide with sugar-phosphate backbones switched between the scrunched and added nucleotides (3-gap). The structures were energy minimized in vacuum using AMBER. We used the Amber.ff03 force field37 for amino acid residues and Parmbsc038 under the AMBER force field for nucleic acids. Prior to equilibration, we subjected the systems to several stages of minimization and relaxation under constant volume. 100ps belly dynamics runs on water molecules were used to relax their initial positions while keeping the protein and nucleotides frozen, followed by an energy minimization run, and a low temperature NPT (constant temperature/constant volume) run. After a complete energy minimization (10,000 conjugate gradient steps), and a step-wise heating procedure at constant volume (from 0 to 300K in 200ps), we carried out constant volume-constant temperature (NVT) equilibration runs for four nanoseconds at 300K for each system. We calculated final trajectories at 300K under NPT for about 11ns. All final MD runs were carried out with time steps of 1.0fs and the particle mesh Ewald method39 was used to treat long range electrostatics in all simulations. The PMEMD module of the Amber10 molecular dynamics package40 was used for all energy minimizations and MD trajectory calculations.
Supplementary Material
Acknowledgements
We thank Lee Pedersen, William Beard and Bennett Van Houten for critical reading of the manuscript. This work was partly supported by Project Z01 ES065070 to TAK from the Division of Intramural Research of the National Institutes of Health, National Institute of Environmental Health Sciences and by NIH grant CA 097096 to Dale Ramsden. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
Author Information Coordinates for the models and structure factors are available from the Protein Data Bank (www.pdb.org) under accession numbers 3HWT, 3HW8 and 3HX0.
References
- 1.Moon AF, et al. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair (Amst) 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase Beta. Chem Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 3.García-Díaz M, et al. DNA polymerase lambda, a novel DNA repair enzyme in human cells. J Biol Chem. 2002;277:13184–13191. doi: 10.1074/jbc.M111601200. [DOI] [PubMed] [Google Scholar]
- 4.García-Díaz M, Bebenek K, Kunkel TA, Blanco L. Identification of an intrinsic 5'-deoxyribose-5-phosphate lyase activity in human DNA polymerase lambda: a possible role in base excision repair. J Biol Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 5.Braithwaite EK, et al. DNA polymerase lambda protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J Biol Chem. 2005;280:31641–31647. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- 6.Tano K, et al. Interplay between DNA polymerases beta and lambda in repair of oxidation DNA damage in chicken DT40 cells. DNA Repair (Amst) 2007;6:869–875. doi: 10.1016/j.dnarep.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nick McElhinny SA, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Lee JW, et al. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem. 2004;279:805–811. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 9.Lieber MR, Lu H, Gu J, Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–133. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- 10.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nat Struct Mol Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 12.Singhal RK, Wilson SH. Short gap-filling synthesis by DNA polymerase beta is processive. J Biol Chem. 1993;268:15906–15911. [PubMed] [Google Scholar]
- 13.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 14.Moon AF, et al. Structural insight into the substrate specificity of DNA Polymerase mu. Nat Struct Mol Biol. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 15.van Gent DC, van der Burg M. Non-homologous end-joining, a sticky affair. Oncogene. 2007;26:7731–7740. doi: 10.1038/sj.onc.1210871. [DOI] [PubMed] [Google Scholar]
- 16.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis BJ, Havener JM, Ramsden DA. End-bridging is required for pol mu to efficiently promote repair of noncomplementary ends by nonhomologous end joining. Nucleic Acids Res. 2008;36:3085–3094. doi: 10.1093/nar/gkn164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brissett NC, et al. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Diaz M, et al. A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology. Mol Cell. 2004;13:561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 20.Cheetham GM, Steitz TA. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 21.Kapanidis AN, et al. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda RA, Richardson CC. Interactions of the RNA polymerase of bacteriophage T7 with its promoter during binding and initiation of transcription. Proc Natl Acad Sci U S A. 1986;83:3614–3618. doi: 10.1073/pnas.83.11.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda. Implications for function. J Biol Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 25.Bebenek K, et al. Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda. EMBO Rep. 2008;9:459–464. doi: 10.1038/embor.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Diaz M, Kunkel TA. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Tang GQ, Roy R, Ha T, Patel SS. Transcription initiation in a singlesubunit RNA polymerase proceeds through DNA scrunching and rotation of the N-terminal subdomains. Mol. Cell. 30:567–577. doi: 10.1016/j.molcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode, Methods in Enzymology, Volume 276: Macromolecular Crystallography, part A. In: Carter CW Jr, Sweet RM, editors. 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 29.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30:1022–1025. [Google Scholar]
- 30.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Bebenek K, Garcia-Diaz M, Patishall SR, Kunkel TA. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J Biol Chem. 2005;280:20051–20058. doi: 10.1074/jbc.M501981200. [DOI] [PubMed] [Google Scholar]
- 35.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 36.Osheroff WP, Jung HK, Beard WA, Wilson SH, Kunkel TA. The fidelity of DNA polymerase beta during distributive and processive DNA synthesis. J Biol Chem. 1999;274:3642–3650. doi: 10.1074/jbc.274.6.3642. [DOI] [PubMed] [Google Scholar]
- 37.Pérez A, et al. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan Y, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 39.Essmann U, et al. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 40.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke Luo RE, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. University of California, San Francisco: AMBER 10; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.