Abstract
Objective
To report a novel prion disease characterized by distinct histopathological and immunostaining features, and associated with an abnormal isoform of the prion protein (PrP) that, contrary to the common prion diseases, is predominantly sensitive to protease digestion.
Methods
Eleven subjects were investigated at the National Prion Disease Pathology Surveillance Center (NPDPSC) for clinical, histopathological, immunohistochemical, genotypical and PrP characteristics.
Results
Patients presented with behavioral and psychiatric manifestations on average at 62 years while mean disease duration was 20 months. The type of spongiform degeneration, the PrP immunostaining pattern and the presence of micro-plaques distinguished these cases from those with known prion diseases. Typical protease-resistant PrP was undetectable in the cerebral neocortex with standard diagnostic procedures. Following enrichment, abnormal PrP was detected at concentrations 16 times lower than common prion diseases; it included nearly four times less protease-resistant PrP, which formed a distinct electrophoretic profile. The subjects examined comprised about 3% of sporadic cases evaluated by the NPDPSC. Although several subjects had family history of dementia, no mutations were found in the PrP gene open reading frame (ORF).
Interpretation
The distinct histopathological, PrP immunohistochemical and physical-chemical features along with the homogeneous genotype indicate that this is a previously unidentified type of disease involving the prion protein, which we designated “protease-sensitive prionopathy or PSPr”. PSPr is not very rare among prion diseases, and might be even more prevalent than our data indicate since PSPr cases are likely to be also classified within the group of non-Alzheimer dementias.
Human prion diseases or transmissible spongiform encephalopathies may be sporadic, inherited or acquired by infection. 1 Creutzfeldt-Jakob disease (CJD) is the most common phenotype and occurs in all three forms. In the sporadic form, CJD is classified into five subtypes, which can be readily distinguished based on clinical features, type and distribution of brain lesions, and pattern of prion protein (PrP) immunostaining. 2, 3 Fatal insomnia (FI), a much rarer phenotype, includes sporadic and inherited forms and is characterized by loss of ability to sleep and preferential thalamic degeneration. 4 Gerstmann-Sträussler-Scheinker disease (GSS), the third phenotype, occurs exclusively as a heritable disease invariably associated with a mutation in the PrP gene open reading frame (ORF) and is characterized by the presence of prion amyloid plaques. 4
In spite of their heterogeneity, all sporadic human prion diseases described to date have been associated with abnormal PrP (commonly called PrPSc but henceforth referred to as PrPr ), which is resistant to treatment with proteases and is considered the diagnostic hallmark of these diseases. 1 PrPr is derived from normal or cellular PrP (PrPC) via a post-translational transition from α-helical-to β-sheet-rich conformations. PrPC and PrPr are quite different. While PrPC is soluble in non-denaturing detergents and is completely digested when exposed to appropriate concentrations of proteinase K (PK), PrPr is detergent insoluble and its C-terminal region resists PK-treatment. 5 Based on the size of their PK-resistant fragments, at least three major PrPr types are recognized, which co-distribute with specific disease phenotypes: i) PrPr type 1, which upon PK-treatment generates a ~21 kilo Daltons (kDa) fragment; ii) PrPr type 2, generating a ~19 kDa fragment; and iii) PrP7-8, a PrP internal fragment of 7–8 kDa. 4–6 Both PrPr types 1 and 2 have been observed associated with distinct subtypes of CJD. To date, PrP7-8 has been consistently observed only in GSS. Therefore, the conformational changes, which render PrPr pathogenic and in many, but not in all cases, infectious, may engender different species or strains of PrPr that can be recognized based on their distinct protease-resistant fragments and by their associated clinical-pathological phenotype. 5, 7–12
Studies mostly based on experimental models, have recently shown that PK-resistant PrP (PrPr) is associated with varying quantities of a PrP isoform that, as PrPr, is detergent-insoluble but sensitive to protease digestion (PrPs).11–15 The relationship of PrPs with PrPr and the role that PrPs plays in the pathogenesis of prion diseases remains uncertain. 16–18
Here we report eleven subjects with a human disease characterized by the presence of detergent insoluble PrP that is predominantly sensitive to protease digestion and forms unusual immunohistochemical patterns. Furthermore, the small amount of PrPr present generates a distinct profile on immunoblot. Several affected subjects have family history of dementia but lack mutations in the PrP gene ORF. We refer to this condition as protease-sensitive prionopathy (PSPr). PSPr broadens the spectrum of human prion diseases and raises several important issues related to the very nature of these diseases when they are associated with different PrP isoforms. Among prion diseases PSPr is not very rare. Since the presenting clinical signs often suggest the diagnosis of non-Alzheimer dementia, PSPr might be even more prevalent than our data indicate because many PSPr cases might currently be classified within this group of dementias. Parts of this study have been previously presented. 19
SUBJECTS AND METHODS
Subjects
The eleven (ten autopsy and one biopsy) subjects and the controls were referred to the NPDPSC between May 2002 and January 2006. Consent was obtained to use tissues for research, including genetic analyses.
General Tissue Processing
Fixed and frozen brain tissues were obtained from all subjects and processed as previously described. 20
Histopathology and Immunohistochemistry
Samples obtained from up to 18 brain regions were processed as previously described. 2, 3 Lesion profiles were constructed using semi-quantitative evaluation of spongiform degeneration and astrogliosis in twelve brain regions from six subjects and four or five regions from two subjects. SD and astrogliosis were scored (Fig 1 legend) and the scores from each of the brain regions were summed for each subject separately; values averaged, standard deviations determined, and plotted according to the brain region. 2 Vacuoles with > 4 µm diameter were measured individually on random photomicrographs of frontal neocortex (10/subject, 180 X) using Spotsoftware version 4.6 after calibration (Diagnostic Instruments, Inc). Sections from the frontal and occipital neocortices, hippocampus, basal ganglia, thalamus, cerebellar hemisphere and midbrain were processed for PrP immunohistochemistry with the monoclonal antibody (mAb) 3F4 or 1E4 (Cell Sciences, Inc., Canton, MA). 2, 20–23 Selected brain regions were also immunostained with the mAbs 4G8 to amyloid β (Aβ).24
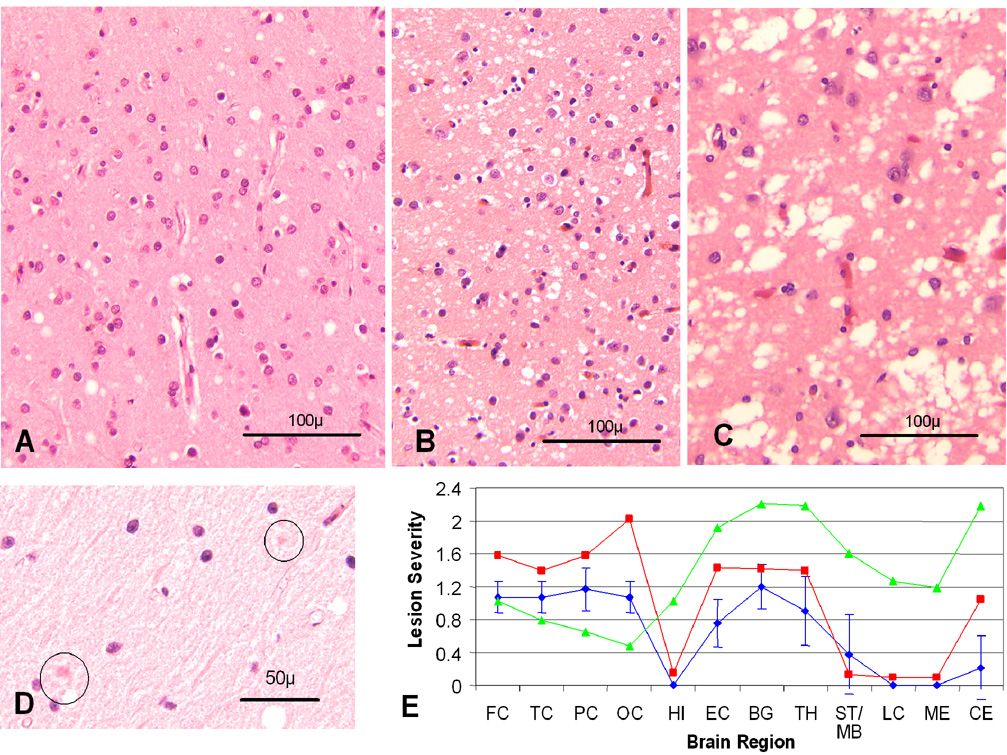
Fig 1.
Histopathology and lesion profile The spongiform degeneration of PSPr (A) is characterized by a mixture of small and intermediate size vacuoles while the vacuoles of two subtypes of sporadic (s) Creutzfeldt-Jakob disease (CJD), sCJDMM1 (B) and sCJDMM2 (C), are mostly small (sCJDMM1) or much larger and confluent (sCJDMM2); D: Eosinophilic micro-structures surrounded by a pale halo (circle) in the cerebellar molecular layer; (A–D: H.E). E: Lesion profiles of PSPr ( ), sCJDMM1 (
), sCJDMM1 ( ) and sCJDVV2 (
) and sCJDVV2 ( ); (FC, TC, PC and OC: frontal, temporal, parietal and occipital cortices; HI: CA1 of hippocampus; EC: entorhinal cortex; BG: basal ganglia; TH: thalamus medial-dorsal nucleus; MB/ST: mid brain in PSPr, substantia nigra in sCJDMM1 and sCJDVV2; LC: pons; ME: medulla; CE: cerebellar cortex). The vertical bars refer to standard deviations. In sCJDMM1 and sCJDVV2, for which data were adapted from Parchi et al., standard deviations were omitted for clarity. 2 Spongiform degeneration was scored on a 0 to 4 scale (0-not detectable, 1-mild, 2-moderate, 3-severe, and 4-confluent); astrogliosis on a 0 to 3 scale (0-not detectable, 1-mild, 2-moderate, and 3-severe).
); (FC, TC, PC and OC: frontal, temporal, parietal and occipital cortices; HI: CA1 of hippocampus; EC: entorhinal cortex; BG: basal ganglia; TH: thalamus medial-dorsal nucleus; MB/ST: mid brain in PSPr, substantia nigra in sCJDMM1 and sCJDVV2; LC: pons; ME: medulla; CE: cerebellar cortex). The vertical bars refer to standard deviations. In sCJDMM1 and sCJDVV2, for which data were adapted from Parchi et al., standard deviations were omitted for clarity. 2 Spongiform degeneration was scored on a 0 to 4 scale (0-not detectable, 1-mild, 2-moderate, 3-severe, and 4-confluent); astrogliosis on a 0 to 3 scale (0-not detectable, 1-mild, 2-moderate, and 3-severe).
Electron Microscopy
Formalin fixed postmortem brain tissue was processed for conventional electron microscopy (EM) and for PrP immunohistochemistry according to standard techniques using peroxidase-antiperoxidase (PAP) mAb 3F4 to PrP. 25
Molecular Genetics
The entire PrP ORF was amplified by PCR using genomic DNA extracted from unfixed brain tissue or blood and the primers PrPO-F [GTCATYATGGCGAACCTTGG (Y=C+T)] and PrPO-R [CTCATCCCACKATCAGGAAG (K=T+G)]; sequencing was done directly or after cloning into plasmid pSTBlue 1 (Novagen, Madison, WI) by automated sequencing. 22
Prion Protein Characterization
Conventional immunoblot
Five to 20 µl 10% W/V brain homogenates (BH) with or without proteinase K (PK) digestion (Sigma Chemical Co., St. Louis, MO), were loaded onto 15% Tris-HCl Criterion pre-cast gels (Bio-Rad Laboratories, Hercules, CA) for SDS-PAGE and immunoblotted with 3F4 and 1E4 to human PrP residues 109–112 and 97–108, respectively. 23 PrP was deglycosylated with PNGase F (New England Biolabs, Beverly, MA) following manufacturer’s instructions.
Enrichment of the abnormal PrP
Two procedures were utilized: 1) capture of the abnormal PrP with the gene 5 protein (g5p) as previously described; 13, 23 and 2) abnormal PrP precipitation with sodium phosphotungstate (NaPTA). 26
Sedimentation of PrP in sucrose gradients
Brain homogenates were incubated with 2% Sarkosyl for 30 min on ice, loaded atop a 10–60% step sucrose gradient and centrifuged 1 hour at 200,000 g in a SW55 rotor (Beckman Coulter, Fullerton, CA). 16, 23, 27
Statistics
Analyses were performed with the two-tail Student-t test.
RESULTS
Clinical Features
Mean age of onset and disease duration were 62 years (range 48 – 71) and 20 months (range 10 – 60), respectively (Table 1). Presentation and course were dominated by neurobehavioral and psychiatric signs, with progressive motor and cognitive decline. Seven patients were ataxic. Other consistent features included absence of periodic complexes on the electroencephalogram (EEG) and non diagnostic 14-3-3 protein test in the cerebrospinal fluid. Magnetic resonance imaging revealed diffuse atrophy without restricted diffusion signals (DWI) in all the 10 patients examined. No subject had known history of prion exposure; probable familial occurrence of dementia was reported in six out of ten investigated patients (Table 1).
Table 1.
Clinical findings
| Case No | Sex | Age (yr.) |
1Disease duration (mo.) |
2Symptoms at onset | Symptoms during illness evolution |
EEG | MRI atrophy/ diffusion |
Family history of dementia |
Other information |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 69 | 60 | Behavioral and mood swings, psychosis (patient diagnosed with bipolar illness) |
Dementia, aphasia, ataxia, and seizure |
Slowing right >left |
+/− | Mother died of dementia at age 70 |
(1) Right hemispheric hypoperfusion on SPECT study; (2) CSF 14-3-3 (not performed) |
| 2 | F | 71 | 33 | Depression and dementia |
Dementia, ataxia, and Parkinsonism |
Normal | +/− | Mother with dementia |
CSF 14-3-3 (not performed) |
| 3 | M | 70 | 12 | Dementia and apathy |
Aphasia, Parkinsonism, hyperreflexia and prominent frontal release signs |
Normal | +/− | Father with dementia at age 60 |
(1) Negative CSF 14-3-3; (2) ↑ CSF proteins 175 mg/dl without cells |
| 4 | M | 50 | 7 (died in a fall) |
Dementia and mood swings |
Psychosis, aphasia, patient fell and died of subdural hematoma |
Diffuse slowing |
NA | NA | Ambiguous CSF 14-3-3 |
| 5 | F | 67 | 11 | Dementia and aphasia |
Ataxia and depression | Not performed |
+/− | Dementia in a paternal aunt and sister died of dementia at age 69 |
CSF 14-3-3 (not performed) |
| 6 | M | 60 | 13 | Dementia | Ataxia, psychosis, and incontinence |
NA | +/− | No family history of dementia |
CSF 14-3-3 (not performed) |
| 7 | F | 48 | 17 | Dementia, emotional labiality, and outbursts |
Motor decline | Diffuse slowing |
+/− | Mother with early dementia at age 60 |
(1) Negative CSF 14-3-3; (2) Patient had VP shunt without response |
| 8 | F | 64 | 10 | Dementia, depression and psychosis |
Ataxia, Parkinsonism, and tremor |
Diffuse slowing |
+/− | Mother with dementia |
Negative CSF 14-3-3 |
| 9 | M | 63 | 23 (patient alive) |
Dementia, personality and behavioral changes |
Motor decline, Parkinsonism and psychosis |
Diffuse slowing |
+/− | Mother died at age 83 with mild dementia |
(1) Global hypoperfusion on SPECT study; (2) Negative CSF 14-3-3; (3) ↑ CSF protein 126 mg/dl without cells |
| 10 | F | 68 | 17 | Insomnia, tremor and slurred speech |
Dementia, ataxia, worsening depression with psychosis and agitation, hyperreflexia |
Diffuse slowing |
+/− | No family history of dementia |
History of bipolar illness with suicidal attempts |
| 11 | M | 52 | 13 | ↓ Verbal output, and progressive motor decline |
Dementia, ataxia, and Parkinsonism |
Normal | +/− | No family history of dementia |
NA |
EEG = electroencephalography; MRI = magnetic resonance imaging; plus and minus signs, respectively, indicate the presence and absence of atrophy or restricted diffusion signals on brain MRI; SPECT = single photon emission computerized tomography; NA = not available; CSF = cerebrospinal fluid; VP = ventriculo-peritoneal.
Average disease duration (20.4 ± 15.4) excludes patients #4, who died of subdural hematoma caused by a fall, and #9 still alive at last report;
The neurobehavioral and psychiatric manifestations included insomnia, apathy, personality changes, mood swings, emotional outburst, depression, and psychosis.
Neurohistopathology
Spongiform degeneration (SD) and astrogliosis of moderate severity were present in the cerebral cortex, basal ganglia, and thalamus of the PSPr cases without severe neuronal loss. SD comprised a mixture of fine vacuoles, comparable to those seen in sCJDMM1 (the most common sCJD subtype), and slightly larger vacuoles that resulted in a mean vacuolar diameter greater than that of sCJDMM1 (7.8 ± 2.7 µm vs. 5.8 ± 1.2 µm). But the “larger” vacuoles clearly were smaller than the “coarse” vacuoles characteristic of sCJDMM2 (Fig 1A – C). 2, 3 The hippocampal pyramidal cell layer appeared unaffected; the molecular layer of the dentate gyrus and the stratum lacunosum moleculare showed mild SD, which extended into the subiculum, the entorhinal and inferior temporal neocortices. No kuru plaques or multicore plaques were detected. In some subjects, structures suggestive of micro-plaques were observed in the molecular layer of the cerebellum (Fig 1D). Lesion profiling identified the cerebral neocortex, basal ganglia and thalamus as the regions most severely affected, while the brain stem and cerebellum were apparently spared (Fig 1E). Congo red staining of selected cerebral and cerebellar cortices was negative.
Immunohistochemistry
PrP immunostaining with mAbs 3F4 and 1E4 of the cerebral cortex, basal ganglia and thalamus from the PSPr cases was strong and in the hippocampal formation very selective with strong immunoreactivity in the molecular layer of the dentate and stratum lacunosum moleculare, without pyramidal cell layer staining (Fig 2A, B). The staining pattern in the cerebrum was characterized by round, loose clusters of coarse granules quite evenly distributed over a background of smaller granules (Fig 2C). The size of the cluster-forming granules often increased progressively toward the cluster’s center, which generally contained a larger granule or a tight aggregate of small granules (Fig 2D). Strongly immunostained globular structures were occasionally seen, rarely also in the white matter (Fig 2D, inset). Immunoreactivity in cerebellum and brain stem was limited to minute, rounded structures or aggregates of a few granules in the cerebellar molecular layer and midbrain colliculi, except for one subject that displayed a large number of these structures (Fig 2H). The immunostained clusters and globules could not be correlated with histologically detectable lesions except for the intense immunostaining of possible micro plaques in the cerebellum of some cases (Fig 1D and Fig 2H). The pattern of PrP immunostaining of cerebrum and cerebellum in the PSPr cases was readily distinguishable from those of sCJD subtypes, and non-prion disease controls (Fig 2E – J). Furthermore, on paraffin embedded tissues, PrP immunoreactivity was virtually removed with PK treatment (50 µg/ml, 37°C, 1 h) in the present cases whereas it was only reduced in sCJD (data not shown). Aβ immunostaining revealed mostly diffuse plaques apparently compatible with the subjects’ age.
Fig 2.
PrP immunohistochemistry A: Intense and widespread PrP immunostain of the cerebral cortex and (B) distinctive PrP immunostaining pattern in the hippocampal gyrus with staining of the molecular layers (arrows) but not of the pyramidal cell layer or of the end-plate (*). C and D: The cortical staining consists of coarse granules forming loose clusters with larger granules or a tighter aggregate of granules at the center; D inset: heavily stained globular structures are also present; (A – D: PSPr). E and F: Immunostaining patterns of the cerebral cortex in sCJDVV2 (E) and sCJDVV1 (F) showing laminar staining and occasional perineuronal staining in sCJDVV2 and very weak and fine widespread staining in sCJDVV1. G: No immunostaining is detectable in the cerebral cortex of a non-prion disease control. H – J: Cerebellar immunostaining patterns in PSPr (H), sCJDVV2 (I) and sCJDVV1 (J). There is intense and exclusive staining of large granules in the molecular layers in PSPr (H), presumably corresponding to the eosinophilic micro-structures surrounded by a pale halo shown in Fig 1D; staining of irregular deposit limited to the granule cell layer in sCJDVV2 (I); no detectable staining in sCJDVV1 (the staining of the granule cell nuclei is non-specific) (J). (A – I: mAb 3F4).
Electron Microscopy
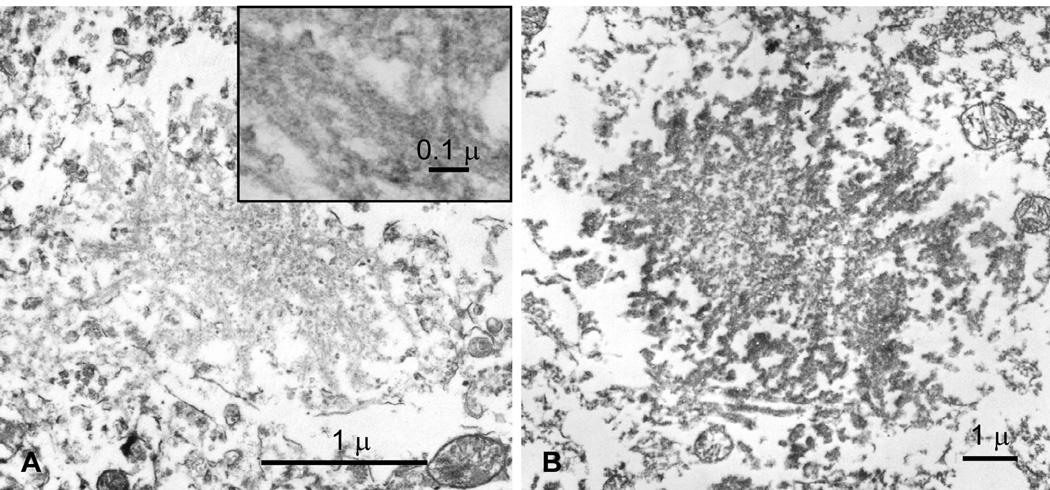
The ultrastructural examination of the cerebellar molecular layer from the case shown in Fig 1D revealed poorly defined rounded structures with barely detectable filament-like profiles which were embedded in an amorphous-granular matrix. These formations strongly reacted with antibodies to PrP and overall had the features of poorly formed or immature PrP micro plaques (Fig 3A and B).
Fig 3.
Electron microscopy (EM) of brain microstructures of PSPr A: EM of the eosinophilic microstructures observed at light microscopy (Fig 1D) reveals plaque-like formations with fuzzy filamentous appearance (inset). These structures are strongly reactive with antibodies to PrP (B) consistent with PrP micro plaques (Peroxidase-anti peroxidase with 3F4).
Genetic Findings
All PSPr subjects were homozygous for valine at codon 129 of the PrP gene, and none carried mutations in the PrP gene ORF; three subjects had silent polymorphisms (two at codon 117 and one at codon 122).
Prion Protein Characterization: Detergent insoluble, protease-resistant and protease-sensitive PrP
The total PrP immunoblot profile from all PSPr subjects was indistinguishable from that of non-prion disease controls (Fig 4A). The glycoform ratios of the three PrP bands from the two groups were similar. Measured by densitometry in arbitrary units, the diglycosylated or upper band was 10.44 ± 1.78 (n = 3) in PSPr vs. 7.83 ± 3.64 (n = 5) in non-prion disease controls (p = 0.30); the monoglycosylated or intermediate band was 4.40 ±1.88 (n = 3) in PSPr vs. 3.40 ± 2.74 (n = 5) in controls (p = 0.79). Under our conditions, the unglycosylated or lower band was not measurable in both PSPr and controls (Fig 4A). Furthermore, the mean amount of total PrP present in 6 subjects apparently did not significantly differ from that of the non-prion disease controls (n = 7) (1.69 ± 0.28 vs. 1.57 ± 0.39, p = 0.53) and from that of cases with prion disease (n = 3) (1.69 ± 0.28 vs. 2.03 ± 0.46, p = 0.20).
Fig 4.
Characterization of PrP in PSPr A: On conventional immunoblots, PK-resistant PrP is undetectable in non-prion disease controls (non-PrD) and the present subjects (PSPr), while it is prominent in sCJD. B: PK-resistant PrP from non-PrD and PSPr is not detectable even after treatment with very low PK concentrations but only in sCJD control when probed with the mAb 3F4. C: Sub-cortical regions of three PSPr cases treated with PK at 50 µg/ml prior to Western blot analysis with 3F4 showed various amounts of PK-resistant PrP in three PSPr cases. T1: PrPr type 1 control; T2: PrPr type 2 control; SN: substantia nigra; Pu: putamen; and Th: thalamus. Samples from temporal cortex (Tc) were used as controls. BH: brain homogenate. D: When the same samples used in B are probed with 1E4, moderately PK-resistant PrP fragments forming a ladder are observed. A, B and D tissues are from the frontal cortex.
In conventional diagnostic immunoblot procedures using mAb 3F4, classical PrPr (PrP27-30) was undetectable in the brain homogenates from the frontal cortex of all 11 subjects and from the occipital and cerebellar cortices of the 7 subjects in which these brain regions were tested (Fig 4A). Treatment with various doses of PK showed no consistent difference between these subjects and non-prion disease controls in these brain regions (Fig 4B). Barely detectable amounts of ~6 kDa PK-resistant PrP (PrP~6) were present in the temporal cortex of three of the eight tested subjects. Of the eight subjects where sub-cortical regions (substantia nigra, putamen and thalamus) were available, significant quantity of PK-resistant PrP27-30 was found in one case, and minimal amounts in two others (one showed small amounts of PrP~6 only), while no PrPr could be definitely detected in the other five subjects (Fig 4C). In contrast, probing with mAb 1E4 revealed a ladder of PK-resistant PrP fragments ranging from ~29 kDa to ~6 kDa in all PSPr cases examined (Fig 4D). The ladder-like electrophoretic mobility of the PrPr fragments did not match those associated with common subtypes of CJD, except for an approximately 20 kDa fragment, which, following deglycosylation, was tentatively identified as the unglycosylated form of PrPr (Fig 4C and data not shown).2 The ~6 kDa fragment was also unglycosylated and was reminiscent of the PrP~7 fragment of GSS.1 These fragments were most obvious at PK concentrations of 5–10 µg/ml and decreased at higher PK concentrations. The ladder-like electrophoretic profile of PrP treated with PK was highly reproducible and was observed in all eleven PSPr cases examined. In contrast, the PrPr fragments from sCJD were clearly detectable with both 3F4 and 1E4 mAb only after treatment with more than 10 µg/ml of PK and increased with higher PK concentrations (Fig 4C and data not shown). Therefore, a very small amount of PrPr detectable with mAb 3F4 is present mostly in the sub-cortical regions of these subjects. Moreover, most of the PrPr appears to have a different conformation from that of typical PrPr since upon PK digestion it generates a unique set of fragments that are detected by 1E4 but not by 3F4.
Total abnormal PrP and the PrPr conformers were further characterized in abnormal PrP-enriched preparations following the capture of the abnormal PrP with g5p, a single-stranded DNA binding protein with a high affinity for abnormal PrP regardless of its PK-resistance. 13, 23 The amount of PrP captured by g5p in the PSPr subjects was three times greater than the amount of PrP captured in non-prion disease controls (data not shown) but it was nearly 16 times less than the g5p-captured PrP in typical sCJD. As measured by densitometry in arbitrary units, the mean PrP captured by g5p in 8 of PSPr subjects was 3.44 ± 2.8% of the total PrP detected by direct gel loading compared to 53.55 ± 24.6% in sCJD (n = 3); (p = 0.00015) (Fig 5A). Furthermore, while nearly 90% of the g5p-captured PrP was resistant to PK digestion in sCJD, the PrPr accounted for only 24% of the total abnormal PrP captured in the PSPr subjects (87.59 ± 26.8% in 4 sCJD cases vs. 24.23 ± 14.9% in 9 PSPr cases; p = 0.0001) (Fig 5A). The PK-resistant PrP obtained after PrP enrichment from the subjects distributed in three major bands of ~26 kDa, ~20 kDa, and ~6 kDa, which were detected by both 3F4 and 1E4 and matched the major bands of the immunoblot ladder detected with 1E4 on direct loading (Fig 5A and data not shown). A similar PrP banding pattern was obtained following NaPTA precipitation, another method of abnormal PrP enrichment. 11, 26 It was detected by both 3F4 and 1E4, although the bands were much more prominent when probed with 1E4 (Fig 5B). The abnormal PrP enrichment experiments confirm that in PSPr subjects there is much less abnormal PrP than in sCJD, and that the proportion of abnormal PrP that is PK-resistant is much smaller.
Fig 5.
Capture by g5p (A) and NaPTA (B) of PrP from PSPr and sCJDMM1 (sCJD). Probing with 3F4 or 1E4 after stripping. The same ladder of PK-resistant PrP as in Fig 4D is detectable in PSPr preparations after heavy loading of the gel. S1: Supernatant of brain homogenate obtained after low speed centrifugation (1,000 g for 10 min).
PrP sedimentation in sucrose gradients
Following sucrose gradient sedimentation, 30% of the total PrP from the PSPr subjects was recovered in fractions 7–11 containing large aggregates, while these fractions accounted for only 5% of the total PrP in non-prion disease subjects (Fig 6A, B, and E). The same fractions contained about 24% and 58% of the total PrP in GSS with the A117V mutation and sCJDVV1, respectively (Fig 6C, D, and E). Also the percentages of PrP recovered in fractions 2, 3, differed significantly between PSPr and non-prion disease. PSPr differed from GSS in fractions 7 and 8 and from sCJD in fractions 1, 2, and 9 – 11 (Fig 6E). In addition to the quantitative differences, also the electrophoretic profiles of the high molecular weight aggregates from PSPr differed from those of non-PrD, GSS and sCJDVV1: the lower band was double in PSPr but single in other conditions (Fig 6A – D). Comparable data were obtained after gel filtration fractionation, which demonstrated that PrP aggregates exceeding 2,000 kDa were more abundant in PSPr than in non-prion disease controls but much fewer than in sCJD (data not shown).
Fig 6.
PrP profiles in sucrose gradient sedimentation A: Non-prion disease (Non-PrD); B: PSPr; C: GSS with the A117V mutation (GSSA117V); D: sCJDVV1; E: PrP distribution in the fractions plotted as percentages of the total PrP. While the amounts of PrP from PSPr are similar to those of non-PrD subjects in fractions 1 and 4 – 6, they differ significantly in fractions 2, 3 and 7 – 11 and also clearly differ from GSSA117V in fractions 7 and 8 and sCJDVV1 in fractions 1, 2 and 9 – 11. PSPr fractions 8–11 have also a distinctive low double band (B, arrowheads) not present in the fractions from non-PrD, GSS and sCJDVV1. Non-PrD (n = 6); PSPr (n = 6); GSS (n = 3) and sCJDVV1 (n = 7). The vertical bars refer to standard deviations. The asterisks denote PrP fractions from non-PrD, GSS and sCJDVV1 that by statistical analysis are significantly different from corresponding PSPr fractions. *: p < 0.05; **: p < 0.01 and ***: p < 0.001.
DISCUSSION
We report eleven patients affected by a disease that involves abnormal PrP and has homogeneous and distinctive features (Table 2). Based on several lines of evidence we argue that these features allow for the separation of this condition from all known forms of human prion disease. Firstly, the abnormal PrP associated with this disease is predominantly – and in several brain regions almost exclusively – sensitive to protease or PrPs, and the PK-resistant PrP isoform or PrPr has a very distinctive electrophoretic profile. The high sensitivity to PK and the distinctive electrophoretic profile of the abnormal PrP clearly distinguish the present cases from each of the five subtypes of sCJD and from sFI, the known human sporadic prion diseases. 1 For example, compared to sCJDMM1, the most common and typical sCJD, 2 the present cases have 16-times less total abnormal PrP, and the fraction of the total abnormal PrP that is PK-resistant is nearly 4-times less. Furthermore, the ladder-like electrophoretic profile of the PrPr associated with this condition has not been observed in either sCJD or sFI, all of which instead are characterized by the presence of the well known PrPr type 1 or type 2. 1 When present, the traditional PrPr, commonly called PrP27-30, was located in sub-cortical regions and was of type 1 another combination not observed in sporadic human prion diseases. 1 Secondly, the present cases are also homogeneous as for the PrP coding genotype since they are all homozygous for valine at codon 129 of the PrP gene, the site of a common methionine/valine polymorphism.28 Valine homozygosity in Caucasians is the rarest 129 genotype being found only in 12% of the population. 28 The sCJD subtypes associated with valine homozygosity, sCJDVV1 and sCJDVV2, have been well characterized and differ from the present cases phenotypically and for the characteristics of the abnormal PrP. 1 Thirdly, the pattern of PrP immunostaining and the presence of structures with the features of poorly formed plaques that we observed in the cerebellum are to our knowledge unprecedented. Lastly, the clinical presentation and initial course that prominently features relatively slow cognitive deterioration, occasional gait impairment and incontinence has evoked the diagnoses of normal pressure hydrocephalus, diffuse Lewy body disease or frontotemporal dementia while prion disease was only suspected at a later stage based on the relatively short duration.
Table 2.
Summary of PSPr common features
| Mean age at onset (range) |
Mean duration (range) |
Clinical Presentation |
Histopathology | PrP IHC | Abnormal PrP | Family History |
PrP Genetics |
|---|---|---|---|---|---|---|---|
| 62 (48–71)yrs | 20 (10–60)mo1 | Cognitive decline (8/11)2 and mood/ behavioral changes (7/11)2 |
Minimal spongiform degeneration with vacuoles larger than typical CJD; and minimal or no astrogliosis |
Intense staining with distinct target pattern in cerebral gray matter; and dot pattern in cerebellar molecular layer |
Minimal amount of PK-resistant PrP forming a ladder-like pattern on Western Blot |
Dementia (8/10)2. Dementia with onset < 61 (2/4)2 |
Valine homozygosity at codon 129. No mutation in the PrP gene coding region |
One patient alive after 23-month duration and one dead 7 months from onset for other causes excluded.
positive cases/total number of cases.
Although the present cases can be easily distinguished from sporadic prion diseases, some of their features such as overrepresentation of PrPs and the multiple PK-resistant PrP fragments have been reported in GSS. 4 However, all cases of GSS reported to date are associated with a mutation in the coding region of the PrP gene or immediately adjacent to it. 4 None of the present cases carried such mutation. Moreover, the ladder-like PK-resistant PrP fragments observed in our cases are preferentially detected with 1E4 but not with 3F4, which obviously separates the present cases from GSS carrying the multiple PK-resistant PrP fragments. In a recent study we observed that although 1E4 and 3F4 have adjacent epitopes along human PrP residues 97–112, their accessibility to these epitopes is different due to different neighboring N-terminal residues. 29 It is possible that the 1E4-selectively detected PK-resistant PrP fragments may have N-terminal starting sites that are different from those of the well-characterized PrPr type 1 and 2. The above evidence clearly indicates that the present condition differs from GSS although the possibility that it represents the long sought sporadic form of GSS remains to be excluded. Six of the ten subjects with obtainable pedigree had a family history of dementia that cannot be ignored, yet none carried a mutation in the PrP gene ORF. Therefore, at least in some cases a causative mutation might be located outside the ORF of the PrP gene, a condition never observed in human prion diseases. 1
All these considerations argue that the eleven patients were affected by a novel condition involving the PrP that cannot be classified within the spectrum of currently known human prion diseases. We suggest the designation of protease-sensitive prionopathy or PSPr to emphasize a major distinctive feature (Table 2).
Compared with other human prion diseases, PSPr is not exceedingly rare, as it accounts for about 3% of all sCJD and 16% of all valine homozygous CJD accessioned by the NPDPSC during the same time period as these eleven subjects, making PSPr about as common as some of the well known sporadic prion diseases (such as sCJDMM2, sFI and sCJDVV1). 2 Furthermore, since the clinical presentation and the duration of PSPr often do not point to the diagnosis of prion disease, some cases of PSPr might currently be classified within the group of non-Alzheimer’s dementias and not be investigated further. Should this be the case, PSPr might be more common than the present study suggests.
The small amount of PrPr associated with PSPr and the finding that about 76% of the detectable abnormal PrP is PK-sensitive not only hinders the diagnosis but also has implications concerning origin, pathogenicity, infectivity and classification of PSPr.
The discovery of PrPs has opened a new chapter in prion diseases.11–15 The demonstration that PrPs forms smaller aggregates than the PrPr counterpart 16 and that it is competent to convert PrPC to PrPr in vitro, as well as to seed the polymerization of recombinant PrP into amyloid 17, 18 suggests that PrPs shares defining features with PrPr. However, the pathogenetic mechanisms of PrPs in the absence of PrPr and, therefore, the very nature of the prion diseases associate with PrPs currently remain conjectural.
Prion diseases associated with PrPs, in the presence of minimal or no PrPr, have been modeled and studied in detail in a variety of transgenic (Tg) mouse lines carrying mouse homologues of human PrP gene mutants or over-expressing PrPC. 12, 30–33 Two Tg mouse models appear relevant to the present cases.
In the first model, Tg mice expressing high levels of mouse PrP carrying the P101L mutation, the mouse equivalent of the human P102L mutation associated with a GSS phenotype, 4, 34, 35 spontaneously developed a neurodegenerative process characterized by spongiform degeneration and prion plaque formation. Following inoculation, they transmitted a disease phenotypically similar to P101L mutated Tg mice but not to wild type mice. As in the present cases, the affected mice had PrPs but no, or minimal amounts of, PrPr indicating that PrPs can be associated with a prion disease that is partially transmissible and has a histopathological phenotype displaying general features of prion diseases. 12
In the second model, Tg mice carrying the P101L mutation were inoculated with brain homogenate from patients affected by a subtype of GSS P102L characterized by the exclusive presence of a ~8 kDa PK-resistant fragment reminiscent of the ~6 kDa fragment observed in small amount in our cases. The inoculated Tg mice remained largely asymptomatic but at histological examination they displayed PrP plaques and had minimal amounts of PrPr. 33 They failed to transmit the disease to wild-type mice but inoculation to P101L mutated mice resulted in the formation of PrP plaques in the absence of clinical disease.
These mouse models and now the present cases raise issues with the definition of prion diseases. Currently, it is unclear whether PSPr is transmissible because time-consuming transmissibility experiments to different lines of Tg mice and in vitro PrP replication are still ongoing. Should PSPr not be transmissible, the question is whether or not it is a prion disease. A similar question can be raised for GSS of which to date only one subtype has been shown to be consistently transmissible.4 The issue is further compounded by the recent evidence that amyloid β, the pathogenic peptide of Alzheimer’s disease (AD) has the propensity to replicate following inoculation into susceptible Tg mice in a conformation-dependent fashion reminiscent of prions.36 These findings seem to blur the once tight association of prion diseases and transmissibility. It may be more practical to apply the label of prion diseases to all conditions in which the prion protein is abnormal and appears to play a central role in the pathology, as in all prion diseases known to date and in PSPr. 37 In contrast, one might reserve the qualification of transmissible to those prion diseases that can be transmitted to recipients expressing relatively normal amounts of wild type PrP. 36
The finding that several PSPr patients had first degree relatives diagnosed with dementia necessitates a search for an underlying genetic cause. In AD, the discovery of mutations outside the gene of the amyloid precursor protein (the central protein in AD, as PrP is in prion diseases) has provided a wealth of information regarding pathogenetic mechanisms of AD. 38 Similarly, the discovery of a mutation outside the PrP gene ORF capable of generating a prion disease might greatly expand our understanding of pathogenetic mechanisms and the role of PrP in prion diseases.
Acknowledgments
John McGeehan, Ph.D. and Geoff Kneale, Ph.D. from University of Portsmouth, United Kingdom kindly provided g5p. John Hedreen, M.D., the Harvard Brain Tissue Resource Center (Mclean Hospital, Belmont, MA), Lawrence S. Honig, M.D., Ph.D., Christopher S. Calder, M.D., Ph.D., Lawrence P. Goldstick, M. D., and William Longstreth, M.D. helped in obtaining the cases. Mrs. Phyllis Scalzo and Diane Kofskey provided skillful histological and immunohistochemical preparations. Mr Bikram Chakraborty assisted in the preparation of the manuscript and illustrations. Supported by NIH grants AG14359 and NS049173, CDC Grant CCU 515004, Columbia University ADRC Grant AG08702, NIH, the Britton Fund and the CJD Foundation.
REFERENCES
- 1.Gambetti P, Kong Q, Zou WQ, et al. Sporadic and familial CJD: classification and characterisation. Br Med Bull. 2003;66:213–239. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- 2.Parchi P, Giese A, Capellari S, et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol. 1999;46:224–233. [PubMed] [Google Scholar]
- 3.Parchi P, Castellani R, Capellari S, et al. Molecular basis of phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 1996;39:767–778. doi: 10.1002/ana.410390613. [DOI] [PubMed] [Google Scholar]
- 4.Kong Q, Surewicz WK, Peterson RB, et al. Inherited prion diseases. In: Prusiner SB, editor. Prion Biology and Disease. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory press; 2004. pp. 673–775. [Google Scholar]
- 5.Parchi P, Zou WQ, Wang W, et al. Genetic influence on the structural variations of the abnormal prion protein. Proc Natl Acad Sci USA. 2000;97:10168–10172. doi: 10.1073/pnas.97.18.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parchi P, Chen SG, Brown P, et al. Different patterns of truncated prion protein fragments correlate with distinct phenotypes in P102L Gerstmann-Straussler-Scheinker disease. Proc Natl Acad Sci USA. 1998;95:8322–8327. doi: 10.1073/pnas.95.14.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pattison IH, Millson GC. Scrapie produced experimentally in goats with special reference to the clinical syndrome. J Comp Pathol. 1961;71:101–108. doi: 10.1016/s0368-1742(61)80013-1. [DOI] [PubMed] [Google Scholar]
- 8.Bruce ME, Dickinson AG. Biological evidence that scrapie agent has an independent genome. J Gen Virol. 1987;68:79–89. doi: 10.1099/0022-1317-68-1-79. [DOI] [PubMed] [Google Scholar]
- 9.Kimberlin RH, Cole S, Walker CA. Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J Gen Virol. 1987;68:1875–1881. doi: 10.1099/0022-1317-68-7-1875. [DOI] [PubMed] [Google Scholar]
- 10.Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safar J, Wille H, Itri V, et al. Eight prion strains have PrPSc molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 12.Tremblay P, Ball HL, Kaneko K, et al. Mutant PrPSc conformers induced by a synthetic peptide and several prion strains. J Virol. 2004;78:2088–2099. doi: 10.1128/JVI.78.4.2088-2099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou WQ, Zheng J, Gray DM, et al. Antibody to DNA specifically detects scrapie but not normal prion protein. Proc Natl Acad Sci USA. 2004;101:1380–1385. doi: 10.1073/pnas.0307825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Safar JG, Geschwind MD, Deering C, et al. Diagnosis of human prion disease. Proc Natl Acad Sci USA. 2005;102:3501–3506. doi: 10.1073/pnas.0409651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klingeborn M, Wik L, Simonsson M, et al. Characterization of proteinase K-resistant N- and C-terminally truncated PrP in Nor98 atypical scrapie. J Gen Virol. 2006;87:1751–1760. doi: 10.1099/vir.0.81618-0. [DOI] [PubMed] [Google Scholar]
- 16.Tzaban S, Friedlander G, Schonberger O, et al. Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry. 2002;41:12868–12875. doi: 10.1021/bi025958g. [DOI] [PubMed] [Google Scholar]
- 17.Pastrana MA, Sajnani G, Onisko B, et al. Isolation and characterization of a proteinase K-sensitive PrPSc fraction. Biochemistry. 2006;45:15710–15717. doi: 10.1021/bi0615442. [DOI] [PubMed] [Google Scholar]
- 18.Colby DW, Zhang Q, Wang S, et al. Prion detection by an amyloid seeding assay. Proc Natl Acad Sci USA. 2007;104:20914–20919. doi: 10.1073/pnas.0710152105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gambetti P. Prion strains in human prion diseases. NeuroPrion Network of Excellence, Prion. 2006 Abstracts http://www.neuroprion.com.
- 20.Pastore M, Chin SS, Bell KL, et al. Creutzfeldt-Jakob Disease (CJD) with a mutation at codon 148 of prion protein gene: relationship with sporadic CJD. Am J Pathol. 2005;167:1729–1738. doi: 10.1016/S0002-9440(10)61254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kascsak RJ, Rubenstein R, Merz PA, et al. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong Q, Huang S, Zou WQ, et al. Chronic wasting disease of elk: transmissibility to humans examined by transgenic mouse models. J Neurosci. 2005;25:7944–7949. doi: 10.1523/JNEUROSCI.2467-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan J, Xiao X, McGeehan J, et al. Insoluble aggregates and protease-resistant conformers of prion protein in uninfected human brains. J Biol Chem. 2006;281:34848–34858. doi: 10.1074/jbc.M602238200. [DOI] [PubMed] [Google Scholar]
- 24.Kim KS, Wisniewski HM, Wen GY. Comparison of four staining methods on the detection of Neuritic plaques. Acta Neuropathol (Berl) 1989;79:22–27. doi: 10.1007/BF00687398. [DOI] [PubMed] [Google Scholar]
- 25.Hunter E. Practical Electron Microscopy. New York: Cambridge University Press; 1993. [Google Scholar]
- 26.Wadsworth JD, Joiner S, Hill AF, et al. Tissue distribution of protease resistant prion protein in variant Creutzfeldt-Jakob disease using a highly sensitive immunoblotting assay. Lancet. 2001;358:171–180. doi: 10.1016/s0140-6736(01)05403-4. [DOI] [PubMed] [Google Scholar]
- 27.Pan T, Change B, Wong P, et al. An aggregation-specific enzyme-linked immunosorbent assay: detection of conformational differences between recombinant PrP protein dimers and PrPSc aggregates. J Virol. 2005;79:12355–12364. doi: 10.1128/JVI.79.19.12355-12364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collinge J, Palmer MS, Dryden AJ. Genetic predisposition to iatrogenic Creeutzfeldt-Jakob disease. Lancet. 1991;337:1441–1442. doi: 10.1016/0140-6736(91)93128-v. [DOI] [PubMed] [Google Scholar]
- 29.Yuan J, Dong Z, Guo JP, et al. Accessibility of a critical prion protein region involved in strain recognition and its implications for the early detection of prions. Cell Mol Life Sci. 2008;65:631–643. doi: 10.1007/s00018-007-7478-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegde RS, Mastrianni JA, Scott MR, et al. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 31.Hegde RS, Tremblay P, Groth D, et al. Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature. 1999;402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]
- 32.Westaway D, DeArmond SJ, Cayetano-Canlas J, et al. Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell. 1994;76:117–129. doi: 10.1016/0092-8674(94)90177-5. [DOI] [PubMed] [Google Scholar]
- 33.Piccardo P, Manson JC, King D, et al. Accumulation of prion protein in the brain that is not associated with transmissible disease. Proc Natl Acad Sci USA. 2007;104:4712–4717. doi: 10.1073/pnas.0609241104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsiao K, Baker HF, Crow TJ, et al. Linkage of a prion protein missense variant to Gertmann-Straussler syndrome. Nature. 1989;338:342–345. doi: 10.1038/338342a0. [DOI] [PubMed] [Google Scholar]
- 35.Hsiao KK, Scott M, Foster D, et al. Spontaneous neurodegeneration in transgenic mice with mutant prion protein. Science. 1990;250:1509–1510. doi: 10.1126/science.1980379. [DOI] [PubMed] [Google Scholar]
- 36.Meyer-Luehmann M, Coomaraswamy J, Bolmont T, et al. Exogenous induction of cerebral beta-amyloidogenisis is govered by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 37.Zou WQ. Transmissible spongiform encephalopathy and beyond. Science. 2007 www.sciencemag.org/cgi/eletters/308/5727/1420.
- 38.Selkoe DJ, Podlisny MB. Deciphering the genetic basis of Alzheimer's disease. Annu Rev Genomics Hum Genet. 2002;3:67–99. doi: 10.1146/annurev.genom.3.022502.103022. [DOI] [PubMed] [Google Scholar]