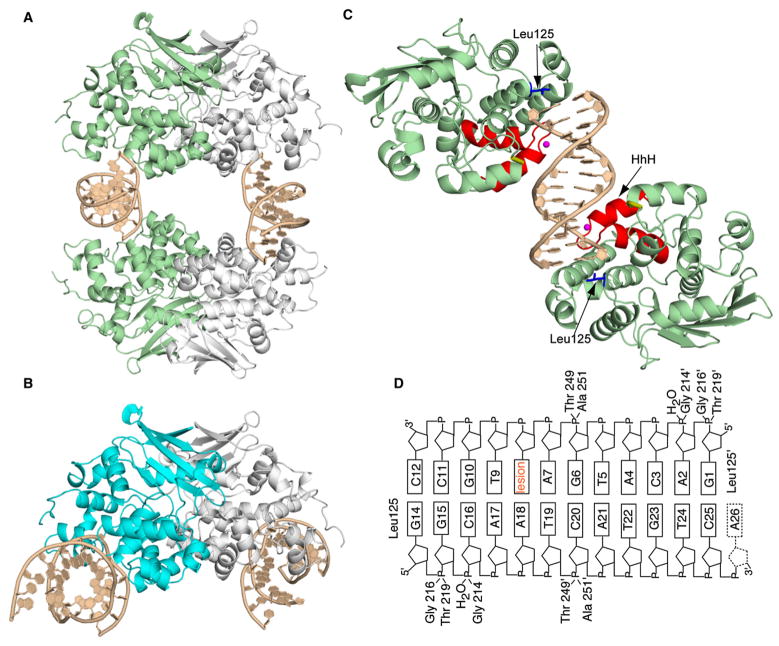
Figure 1. Structure of the AlkA:DNA Host/Guest Complex.
(A) Ribbon representation of the asymmetric unit in the AlkA:DNA host/guest complex (HGC) structure, with four AlkA monomers bookending two DNA duplexes. The two AlkA monomers in green are those that interact with the DNA duplex comprising chains G and H; this half of the structure is slightly more ordered than the other half and hence was used for the analyses throughout this work.
(B) Ribbon representation of the AlkA azaribose lesion-recognition complex (Hollis et al., 2000). One of the AlkA monomers is colored cyan. Note the difference in the DNA-binding mode between (A) and (B).
(C) Ribbon representation of the two AlkA monomers interacting with DNA chains G and H, (same as in [A]). Shown in red is the signature helix-hairpin-helix DNA-binding motif common to all members of the superfamily to which AlkA belongs. The loop colored in gold contains residues 249 and 251, which also participate in interactions with the DNA. The water molecule mediating protein:DNA interactions is colored in magenta, whereas the side chain of Leu125 is shown in blue.
(D) Schematic representation of the AlkA:DNA interactions. The prime sign (′) represents residues interacting from the second AlkA protomer.