Abstract
In colorectal cancer, BRAF and KRAS oncogenes are mutated in about 15% and 35% respectively at approximately the same stage of the adenoma-carcinoma sequence. Since these two mutations rarely coexist, further analysis to dissect their function of transformation in colon cancer is required. Caco-2 human colon adenocarcinoma cells were stably transfected with BRAFV600E (Caco-BR cells) or KRASG12V (Caco-K cells) oncogenes. BRAFV600E is more efficient in transforming Caco-2 cells and altering their morphology. The dominant nature of BRAFV600E is evident by its ability to render Caco-2 cells tumorigenic in vivo all be it through selective extracellular signal-related kinase (ERK) 2 phosphorylation and high levels of cyclin D1. As a consequence, the cell cycle distribution of parental cells is altered and microsatellite instability is introduced. Attenuated ERK activation observed correlated with KSR downregulation by BRAFV600E without further implications to signaling. Highly activated ERK in case of KRASG12V (Caco-K cells) leads to mild transformation causing Caco-K cells to express premature senescence-related markers and acquire growth factor-dependent viability. Interestingly, BRAFWT gets equally activated by upstream KRAS mutations present in colon adenocarcinoma cells such as DLD-1 and SW620. Taken together, these results suggest that the two oncogenes have different transforming capability in colon cancer, although they both use the mitogen-activated protein (MAP) kinase pathway to carry out their effect. In general, BRAFV600E presents greater potential in mediating tumorigenic effect as compared to KRASG12V both in vivo and in vitro. These findings may have implications in personalised diagnosis and targeted therapeutics.
Introduction
Colorectal cancer (CRC) can be either hereditary or sporadic characterized by deregulated signal pathways and cellular programs through gain and/or loss of function mutations that accumulate in a stepwise manner. Among the most hurtful of all genetic abnormalities that appear in CRC development are KRAS and its downstream effector BRAF because they result to abnormal extracellular signal-related kinase (ERK) signaling. Approximately 13% and 35% of CRC contain activating point mutations in the BRAF and KRAS genes, respectively [1–4]. Mutant RAS proteins end up losing their initially high GTPase enzymatic activity [5], with mutations in codon 12 by far the most frequent compared with codon 13 [1,6]. Recent evidence suggests that different mutations in KRAS have different biological consequences in vivo. In addition, although G12D seems to be more frequent compared with G12V in colon cancer, KRASG12V has been associated with more aggressive colorectal carcinomas and greater mortality than other codon 12 or 13 mutations [7–9]. Downstream the RAS-RAF-mitogen-activated protein kinase (MAPK) pathway is BRAF, which is activated through binding to RAS-GTP.
Activated RAF kinases phosphorylate MAPK/ERK kinase (MEK), which in turn phosphorylate and activates ERK. BRAF has been identified as an oncogene in human cancer especially in malignant melanoma and colon carcinoma. Analysis of mutations in human colorectal tumors showed that more than 80% of the BRAF mutations were BRAFV600E (constitutive activation of BRAF kinase) and that this mutation greatly increased ERK and nuclear factor κB signaling when transformed in NIH3T3 cells [10]. It is believed that activated BRAF and KRAS have a causal role in the development of cancer and cell transformation [5,11,12]. Various RAS inhibitors have been examined for their ability to block signaling in these constitutive active mutants, and isoprenylation inhibitors have been proven to be ineffective in clinical trials [13]. Therefore, greater focus has been placed on the downstream RAF molecules.
BRAF and KRAS have the potential to regulate similar as well as different pathways. Similarly, although phosphatidylinositol 3-kinase (PI3K) is activated by Ras in fibroblasts, Ras is not an activator of the PI3K/Akt pathway in T lymphocytes [14].
Mutational activation of oncogenes such as KRAS and BRAF results to constitutive ERK signaling, which can lead to cell cycle changes that have a profound influence on the G1/S transition by regulating the expression of a number of cell cycle proteins such as cyclins D and E and inhibitors such as p21Cip1 and p27kip [15,16]. Induction of senescence or tumorigenesis depends on the expression of cell cycle regulatory proteins (p21Cip1, p16INK4a, p14ARF, p15INK4b, p19ARF, and p53). Similarly, cyclin D1 is equally important for the development and progression of several cancers, including colon cancer, and it has been found that it is significantly upregulated and related to a more aggressive tumor phenotype and poor prognosis [17,18]. Cyclin E, often found overexpressed in cancer, can efficiently induce the phosphorylation of Rb, thus causing its functional inactivation, which contributes to tumorigenesis. Similarly, high levels of the p21Cip1 inhibitor have been associated with RAS signaling and with the clinical progression of malignant melanoma [19]. All these incidents have in common a mutated BRAF or KRAS gene and result to high cell proliferation through increased ERK signaling. Nevertheless, hyperactivation of the ERK pathway can also lead to the opposite situation, cell cycle arrest, and p53-dependent or -independent senescence [20–22]. More specifically, sustained BRAFV600E expression in human melanocytes induces cell cycle arrest, along with induction of both p16INK4a and senescence-associated β-galactosidase (SA-β-Gal) activity [23]. By contrast, the expression of oncogenic RAS in primary human or rodent cells results in premature senescence and allows proliferation to continue unabated in the presence of oncogenic stimuli [24].
Oncogenes have differential effects in colon cancer progression [25]. Because BRAF and KRAS are oncogenes in the same pathway and yet are mutually exclusive in colon cancer [26], further analysis to dissect their function of transformation in the colonic epithelium may reveal marked functional differences about their oncogenic potential and tissue-specific pathway activation. Caco-2 cells are derived from a typical colon adenocarcinoma and are often used in the study of enterocyte differentiation because on reaching confluence, the cells differentiate into a mature enterocyte phenotype, characterized by the formation of microvilli and brush borders [27]. Caco-2 cells harbor a truncating mutation in the APC gene [28], a Smad4 C-terminal missense mutations [29], and are further mutated at the p53 locus, generating a stop codon at amino acid 204, resulting in no detectable p53 expression [30]. Thus, these cells represent an ideal model system in which to study the transforming effects of BRAF and KRAS activating mutations in a transformation-permissive genetic background. Overexpression of these oncogenes in Caco-2 will also allow to study how the genetic background of a cell affects the outcome of an activating mutation and all that in comparison with commercially available human colon cancer cell lines carrying a more compromised mutated background, such as HT29V600E, Colo205V600E, DLD-1G13D, and SW620G12V. Briefly, we established stable Caco-2 cell lines constitutively expressing BRAFV600E (Caco-BR13, -BR23) or KRASG12V (Caco-K6, -K15) and a control cell line stably transfected with the empty expression cassette (Caco-Neo). Clone selection was based on the protein expression levels for the two oncogenes, and their characterization was performed both in vivo and in vitro.
Materials and Methods
Stable Transfection and Characterization of Caco-2 Cells with BRAFV600E and KRASG12V
The plasmids pH8-BRAFV600E, kindly provided by Dr. Tsuneo Ikenoue (Department of Gastroenterology, Tokyo, Japan), and pcDNA3-KRASG12V [31] were transfected into Caco-2 cells using calcium phosphate. As a control, Caco-2 cells were also transfected with an empty pcDNA3 expression vector, and the arising clones were established by clonal selection with 0.5 mg/ml geneticin (Sigma-Aldrich, Poole, UK). Successful clones were further selected on the basis of protein expression for each oncogene. New BRAFV600E and KRASG12V clones were named Caco-BR (or briefly BR) and Caco-K (or briefly K), respectively.
The growth rate of newly established clones was determined in replicate wells of a six-well dish seeded at 1 x 105 cells per well. Cells were harvested by trypsinization and counted using a Coulter counter (Model Z2; Coulter, Miami, FL) on days 1 to 5. For the anchorage-independent growth on soft agar, a layer of 0.5% agar in growth medium was first set in each well of a six-well dish. Growth medium containing 0.3% agar was mixed with 1 x 103 cells per well and overlaid onto the first layer of agar. Cells were allowed to form colonies in 5% CO2 for 2 weeks at 37°C, fixed with methanol for 10 minutes at room temperature, and stained with 0.01% crystal violet. The colony formation was examined with a x30 objective of an inverted light microscope (Axiovert 25; Zeiss, Jena, Germany), and the total number of colonies in the top layer of agar was determined. To investigate the tumorigenicity of the clones, approximately 1 x 106 cells (Caco-2, Caco-NEO9, Caco-BR13, Caco-BR23, Caco-K6, Caco-K15, HT29, and DLD-1) in a total volume of 100 µl of PBS were subcutaneously injected into each flank of a severe combined immunodeficient (SCID) mouse (Charles River Laboratories, Inc., L'Arbresle, Cedex, France) and monitored for tumor formation for a period of 5 months. All animal experiments were approved by the Animal Ethics Committee of the Institute of Biological Research and Biotechnology, National Hellenic Research Foundation, and procedures were according to the guidelines approved by theUnited Kingdom Coordinating Committee of Cancer Research [32]. Tumors were isolated from each condition after 2 weeks of their appearance, and their weight was determined.
Protein Immunoblot Analysis
Whole cell lysates were prepared with lysis buffer containing protease and phosphatase inhibitors and were subjected to Western blot analysis. Methodology and antibody information is listed in Supplementary Materials and Methods.
Adherent Cell Proliferation Assay
Cells were plated into a 96-well plate at a density of 2 x 103 per well and left to grow for 24 hours. Cells were treated with two different concentrations of UO126 (30 and 60 µM; ALX-270-237-M001; Alexis Biochemicals, Lausen, Switzerland) in triplicates. In each instance, cells were left to grow for 72 hours before being treated with 100 µl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (M2003; Sigma, St. Louis, MO) for 4 hours. MTT crystals were solubilized using isopropanol. The resulting absorbance was read in a Tecan Safire2 Microplate Reader (Tecan Austria GmbH, Grodig, Austria) at 560 nm. Absorbance readings were subtracted from the value of blank wells; the reduction in cell growth was calculated as a percentage of control absorbance in the absence of any inhibitor.
Senescence-Associated β-Galactosidase
Cells were washed twice with PBS, fixed with 2% formaldehyde and 0.2% glutaraldehyde prepared in PBS for 5 minutes, and washed twice with PBS. Cells were stained with X-Gal solution pH 6 (1 mg/ml X-Gal, 5 mM K3Fe[CN]6, 5 mM K4Fe[CN]6, 2 mM MgCl2, 150 mM NaCl, 40 mM citric acid prepared in sodium phosphate buffer) prepared in PBS. Staining was performed at 37°C for approximately 16 hours without CO2 [33]. Images were obtained using a Nikon Eclipse T-200 (Tokyo, Japan) inverted phase-contrast microscope equipped with an Olympus digital camera (Olympus, SP-51OU2, Hamburg, Germany). The objective lens used was x30.
Immunofluorescence in Cultured Cells
Cells were grown on cover slips and fixed using 2% paraformaldehyde in PBS for 10 minutes at room temperature. Permeabilized cells were blocked with 4% FBS at room temperature for 1 hour and stained with anti-PML antibody (clone PGM3, SC-966; Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C. Secondary antibody Alexa Fluor 488 goat anti-mouse (1:600, A11001; Molecular Probes, Eugene, OR) was applied to the cells for 1 hour at room temperature. The nuclei were stained with Hoechst No. 33342 (B2261; Sigma) for 10 minutes and coverslips were mounted on glass slides in Gelvatol/DABCO aqueous medium (Sigma-Aldrich). Samples were visualized with a Leica TCS SPE confocal laser scanning microscope, and image acquisition was via the LAS AF software, both from Leica Lasertechnik, Heidelberg, Germany. The objective lens used was x63.
Flow Cytometric Analysis of Cell Cycle
For flow cytometry, 5 x 106 cells washed with PBS and fixed overnight with ice-cold 70% ethanol at -20°C. After fixation, cells were stained with 1 mg/ml propidium iodide in PBS solution containing 20 µg/ml RNaseA and incubated at 37°C for 20 minutes. Samples were analyzed by flow cytometry (Calibur, Becton Dickinson, UK). To further study cell cycle checkpoints, cells were treated with growth medium containing 0.1 µM nocodazole (M1404; Sigma), which is known to block cell cycle progression in G2-M through disruption of mitotic spindles, for 2, 8 and 16 hours.
Gene Expression Reverse Transcription-Polymerase Chain Reaction Analysis
Total RNA (3 µg) was reverse-transcribed into complementary DNA using the SuperScript II Reverse Transcriptase (Invitrogen, Karlsruhe, Germany) according to the manufacturer. Additional details are provided in Supplementary Materials and Methods.
Microsatellite Instability Analysis
Both Caco-BR cell lines, the parental cell line Caco-2 and microsatellite instability (MSI)-positive colon cancer cell lines DLD-1 and HCT116 were used to independently study MSI status by allelic profiling of mononucleotide repeat sequences (NR-21, NR-22, NR-24, BAT-25, and BAT-26) within the 3′ and 5′ untranslated regions of five different genes as previously described [34]. Briefly, one primer in each pair was labeled with one of the fluorescent markers 6-carboxyfluorescein (FAM), 6-carboxytetramethylrhodamine (TAMRA), or dichloro-2′,7′-dimethoxy-6-carboxyfluorescein (JOE). Each mononucleotide repeat was amplified individually. Polymerase chain reaction (PCR) products were purified with Gene Clean Turbo PCR Kit (Bio101; QBiogene, Illkirch, Strasbourg, France) according to the manufacturer's instructions and were subsequently quantified on a 2% gel against molecular weight markers. Purified PCR products were directly gel-sequenced using an ABI PRISM 3100 genetic analyzer (MWG-Biotech; Ebersberg, Germany).
Results
Characterization of Caco-2 Clones Expressing Mutant BRAFV600E (Caco-BR) and KRASG12V Oncogenes (Caco-K)
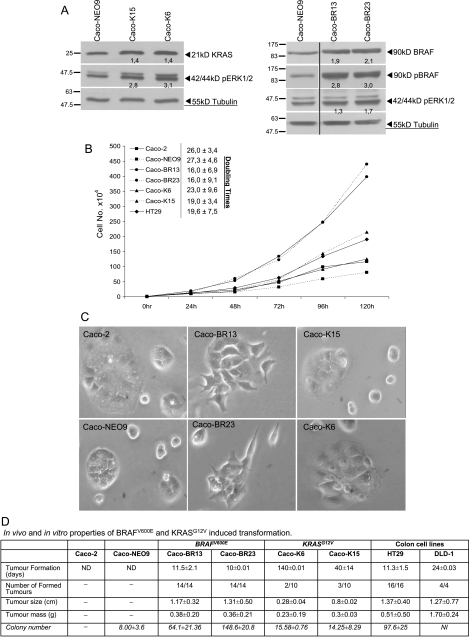
Plasmid (pH8-BRAFV600E and pcDNA3-KRASG12V) concentrations were selected to provide an efficient protein expression of BRAFV600E and KRASG12V. Initially, 10 different clones were selected for each oncogene and empty vector and checked for presence of the plasmid (data not shown). Although we isolated clones expressing varying amounts of mutant protein (data not shown), we avoided further analysis of clones that expressed very high or very low levels (lower compared to controls) of transgene. For further analysis Caco-K6 and -K15 [31], clones were selected in case of KRASG12V transformation, whereas Caco-BR13 and -BR23 were the selected clones that expressed relatively low levels of highly phosphorylated BRAFV600E (Figure 1A). In both cases of transformation, the level of oncoprotein was of similar expression compared with the control. To examine how the two mutations related to the downstream activities of signaling components known to be regulated by the RAS-RAF-MAPK pathway, we examined the phosphorylation status of ERK1/2 where both oncogenes managed to induce significant activation of this kinase (Figure 1A).
Figure 1.
BRAFV600E significantly alters morphology and growth characteristics of Caco-2 cells. (A) Western blot analysis from selected Caco-2 clones stably transfected with KRASG12V (Caco-K6 and Caco-K15) and BRAFV600E (Caco-BR13 and Caco-BR23) oncogenes. Left panel shows the expression levels of total KRAS and phosphorylation status of ERK1/2. Right panel shows the expression levels of total BRAF, active pBRAF, and phosphorylation status of ERK1/2. The vertical dividing black line indicates multiple fields taken from the same image. (B) Growth rate of Caco-2, Caco-NEO9, Caco-BR13, Caco-BR23, Caco-K6, and Caco-K15 cells. Results presented are average expression levels from at least three independent experiments. (C) Photographs illustrate the altered spindle-like morphology induced by BRAFV600E. Representative images. Original magnification, x30. (D) BRAFV600E efficiently transforms Caco-2 cells both in vivo and in vitro. Tumor formation efficiency of different Caco-BR and Caco-K cells and control colon cancer cells was assessed in SCID mice during a period of 5 months after cell injection. Tumor number represents successful tumor formed per group of cell line used. Tumor size (cm) and weight (g) describe the isolated tumors 2 weeks after their appearance per group of cell line used. For the SCID mice experiment, two or three 5-week-old female athymic mice were used per cell line. The values are mean ± SD of three individual experiments performed in duplicates and triplicates. Accordingly, their ability to form colonies on 0.3% soft agar for 2 weeks was assessed and is represented in the table under the colony number. For soft agar assays, data were accumulated from two independent experiments, each clone in triplicate. ND indicates not determined; NI, not included in the experiment.
BRAFV600E Alters Cell Growth and Cellular Morphology and Mediates Cell Transformation More Efficiently than KRASG12V
We noted that Caco-BR13 and -BR23 cell lines grew with different kinetics to Caco-K6 and -K15 and control cell lines (Caco-2 and NEO9) exhibiting a high proliferation rate and significantly quicker doubling times (Figure 1B). Caco-2 and NEO9 cell lines grew with similar kinetics. Photographs of Caco-2, NEO9, Caco-BR, and Caco-K cells illustrate the altered morphology specifically induced by the BRAFV600E transformation (Figure 1C). Caco-BR cells seem to have adopted a spindle-shaped morphology and stopped forming large islets compared with Caco-2. However, even the Caco-2 clones expressing the highest levels of KRASG12V did not adopt this altered morphology, as previously shown [31]. In comparison, Caco-K cells formed tight contacts evidenced by their preference to grow in colonies with distinct boundaries between each cell (Figure 1C). In summary, our results demonstrate that the establishment of an altered more transformed morphology was restricted to expression of the mutant BRAF.
The tumor-inducing potential of each oncogene in vivo was examined in SCID mice, in which Caco-BR13 and -BR23 allowed the formation of significantly larger tumors than Caco-K6 and -K15 in significantly less time, whereas tumor formation of Caco-K cells varied from 1 to 5 months in some animals (Figure 1D). Moreover, both oncogenes (BRAFV600E and KRASG12V) significantly increased the ability of Caco-2 cells to grow in soft agar (Figure 1D). Similarly, Caco-BR cells consistently formed colonies more efficiently than Caco-K cells and generated the same number of colonies on average with HT-29 cancer cell line (Figure 1D). BRAFV600E confers higher transforming activity to Caco-2 cells compared with KRASG12V both in vivo and in vitro.
BRAFV600E and KRASG12V Downstream Signaling
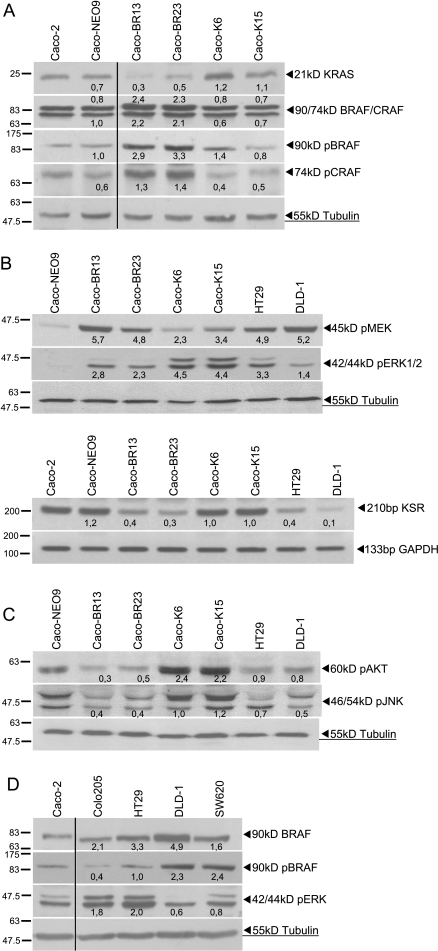
Analysis of the RAS-RAF-MEK-ERK pathway showed more efficient downstream ERK phosphorylation in the presence of KRASG12V rather than BRAFV600E. Nevertheless significant was the activation of ERK2 (lower pERK1/2 band) in Caco-BR cells. In contrast, upstream ERK1/2 activator MEK was more activated in the Caco-BR cells, suggesting that other factors may be implicated in BRAFV600E signaling, considering that the family member Raf-1 (CRAF) was found to be equally expressed and activated as BRAF in Caco-BR cells (Figure 2, A and B). To elucidate differential ERK activity, molecules that regulate its activation were analyzed. It was found that in Caco-BR cells (and not in Caco-K cells), kinase suppressor of RAS (KSR), a positive regulator of ERK activation, was significantly downregulated (Figure 2B). However, no significant change was observed in the RAF-1 kinase inhibitory protein (RKIP) gene suggesting no involvement (Figure W1). Therefore, limited KSR availability in the BRAFV600E system may be responsible for the moderate ERK activation regardless of increased BRAF-MEK activity.
Figure 2.
The MAPK pathway is activated in the Caco-BR and Caco-K cells. (A) Expression analysis of KRAS/RAF and biochemical detection of activated RAF proteins. (B) Signaling downstream of KRASG12V and BRAFV600E and mRNA levels of KSR-positive regulator of pERK. (C) RAS effector pathways (PI3K and JNK) differentially regulated by KRASG12V and BRAFV600E, respectively. (D) Biochemical detection of activated BRAF and its downstream effector ERK in control cell lines. Results presented are average expression levels among three independent experiments. Verticals dividing black lines indicate multiple fields taken from the same image.
Analysis of other RAS effector pathways showed that the survival pathway PI3K (pAKT) was significantly activated in the Caco-K cells, whereas the Jun N-terminal kinase ( JNK) pathway was found to be downregulated in the Caco-BR cells (Figure 2C). Finally, the Ral pathway (RalA-GTP) remained unaffected in either case of oncogenic transformation (Figure W2).
Comparison of the RAS-RAF-MAPK signaling pathway in HT29 and Colo205 cells (V600E) versus DLD-1 and SW620 cells (G13D and G12V, respectively) revealed that, although pBRAF was higher in KRAS mutant cells, the pathway was more active in the BRAF mutant cell lines (Figure 2D).
Pharmacologic Inhibition of MAPK Signaling Pathway
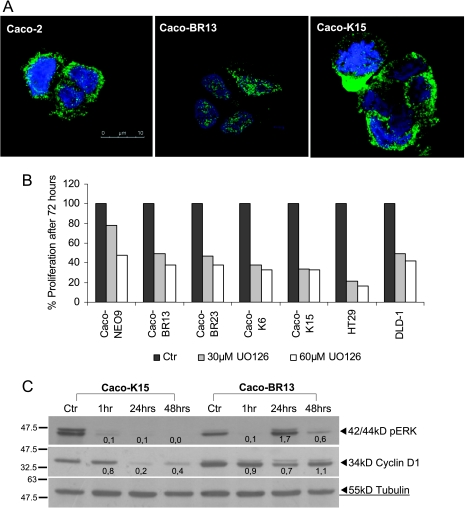
To further explore ERK signaling, we analyzed its localization by immunostaining and discovered that, in Caco-BR cells, pERK was mainly localized in the nucleus compared with Caco-K and control cells, where it was found in the cytoplasm (Figure 3A). Pharmacological inhibition of the biochemical activities caused by mutant BRAF and KRAS was achieved using the specific MEK inhibitor UO126 in the presence of which cell growth of both Caco-BR and Caco-K cells was significantly reduced by approximately 50% to 60%, respectively, as shown by the MTT assay (Figure 3B). Under adherent culture conditions, the MEK inhibitor UO126 blocked growth through down-regulation of pERK within 1 hour and determined that cyclin D1 is the transcriptional target of KRASG12V. However, UO126 seemed insufficient in blocking cyclin D1 in Caco-BR cells in which pERK seemed to have a dynamic response to the treatment and recovers rapidly (Figure 3C).
Figure 3.
ERK localization and the pharmacological inhibition of MAPK pathway. (A) Nuclear localization of pERK in Caco-BR cells. Representative confocal immunofluorescence images, double labeled with Hoechst (blue) and pERK (green). Original magnification, x63. (B) Pharmacological inhibition of cell proliferation using the MEK inhibitor UO126 at 30 and 60 µM for 72 hours and analyzed by MTT assay in Caco-BR and Caco-K cells. The inhibitor was replaced every 24 hours, and data in triplicates were normalized to the negative controls. (C) Determination of the inhibitory effect of 60 µM UO126 on pERK and cyclin D1 for various time points at the protein levels. Results were consistent among three independent experiments.
Differential Expression of Senescence Markers by Oncogenic KRASG12V
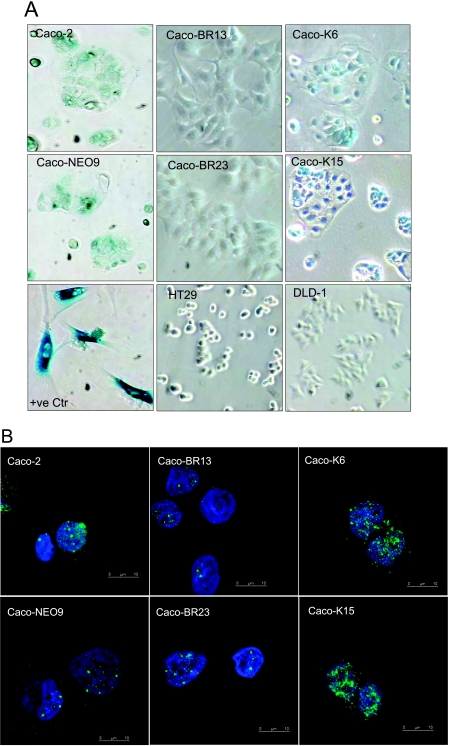
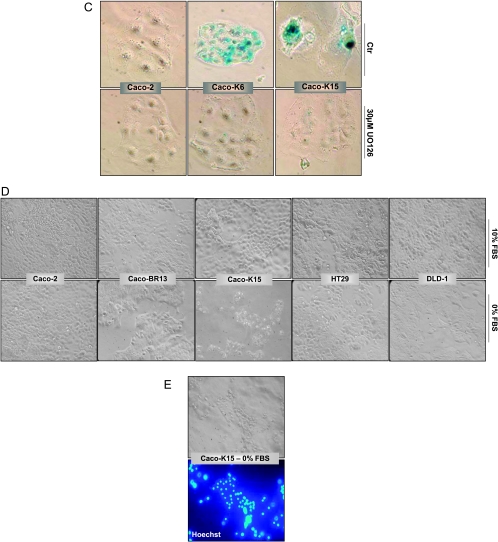
Overexpression of KRASG12V but not BRAFV600E in Caco-2 cells led to the acquisition of premature senescence-related markers as determined by SA-β-Gal staining (Figure 4A) and increased promyelocytic leukemia (PML) gene product localization, which has been previously associated with cellular senescence in RAS-arrested cells, and can provide further evidence of senescence activity [35] (Figure 4B). Regardless of senescence appearance, the cells did not arrest but continued proliferating and even increased their growth rate compared with Caco-2 cells (Figure 1B). Pigmentation that appears in the Caco-2 and NEO9 control cells after SA-β-Gal staining is considered as background staining because it is not comparable to the positive control staining (human embryonic lung fibroblasts HFL1 at PD55; Figure 4A). To determine whether oncogene-induced senescence in Caco-2 cells is mediated by the MAPK signaling pathway, pharmacological inhibition of ERK1/2 using UO126 was performed. Blocking of the MAPK pathway for 48 hours in Caco-K cells resulted in regression of the senescence-activated β-gal activity (Figure 4C).
Figure 4.
Induction of KRASG12V induces senescence-related markers in Caco-2 colon epithelial cells as determined by (A) SA-β-Gal (representative images; original magnification, x30) and (B) PML bodies staining (representative confocal immunofluorescence images, double labeled with Hoechst (blue) and PML (green); original magnification, x63). (C) Pharmacologic inhibition of ERK1/2 using 30 µM UO126 for 48 hours resulted in regression of the senescence-activated β-gal activity in Caco-K cells. The inhibitor was replaced every 24 hours. Representative images. Original magnification, x30. (D) Cell growth on growth factor depletion. Significant cell death was observed microscopically in Caco-K cells after 5 days of serum starvation, whereas Caco-BR and control cells (Caco-2, HT29, DLD-1) continued proliferating normally. All cells were cultured in triplicates with 0% and 10% FBS in parallel for comparison, and medium was replaced every day. (E) Hoechst staining showed apoptotic characteristics in the Caco-K cells after serum starvation. Representative images. Original magnification, x30.
In a parallel experiment, cell growth on growth factor depletion was analyzed for 3 to 5 days after which significant cell death was observed microscopically in Caco-K cells when Caco-BR cells and control cells continued proliferating normally (Figure 4D). This is of interest, considering that Caco-2 cells after KRASG12V transformation acquired senescence markers and at the same time growth factor-dependent viability. Hoechst staining of Caco-K cells grown in the absence of serum was used to show death by apoptosis (Figure 4E).
These experiments show a clear difference in cell senescence marker staining between Caco-K and Caco-BR cells, which probably dramatically affects differential viability of these cells under growth factor deprivation.
Rb Inactivation and p19ARF Collaborate with BRAFV600E in Oncogenic Transformation
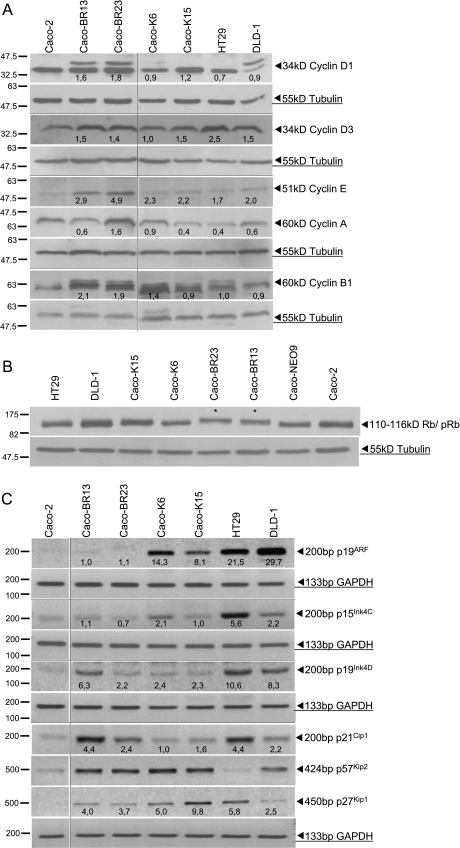
D-type cyclins (D1 and D3; G1-phase cyclins), which prepare chromosomes for replication and initiate Rb phosphorylation, were significantly overexpressed in the Caco-BR transformed cells (Figure 5A). Nevertheless, both cyclins A and E (S-phase cyclins) were also significantly overexpressed in Caco-BR cells. These proteins initiate S phase as soon as M phase is over, supporting DNA replication along with E2Fs, and inactivate the Rb protein by completing its phosphorylation. These results come to support the finding that the Rb is hyperphosphorylated and therefore inactivated in Caco-BR and not in Caco-K cells (Figure 5B). Nonphosphorylated Rb protein in the Caco-K cells is consistent with the appearance of senescent characteristics in those cells as described earlier (Figure 5, A and B).
Figure 5.
Differentially expressed cell cycle regulators in Caco-BR and Caco-K cells. (A) Protein analysis of cyclins A, B1, D1, D3, and E1 as well as (B) phosphorylation status of the Rb protein in KRASG12V versus BRAFV600E clones and control cancer cells (HT29 and DLD-1). (C) Transcription analysis by RT-PCR of cell cycle inhibitors. Results were consistent among three independent experiments. Horizontal dividing black lines indicate group of images acquired with exact same conditions on a separate experiment. Vertical dividing black lines indicate multiple fields taken from the same image.
Activation of the RAS-RAF pathway is known to mediate some of its effects through the activation of p16Ink4A and p19ARF [36]. Because p16Ink4A is hypermethylated in Caco-2 cells [37], analysis was performed for the alternative reading frame of the same locus, the p19ARF transcript along with other cell cycle regulators at transcriptional level (Figure 5C). Two members of the Ink family (p19ARF and p15Ink4C) were upregulated only in Caco-K cells, whereas the p27Kip1 member of the Cip/Kip family was upregulated in both cases of oncogenic transformation. Interestingly, p21Cip1 and p19Ink4D were upregulated in Caco-BR cells. The cell cycle inhibitor p57Kip2 was the only gene not altered between the two oncogenes but upregulated compared with Caco-2 parental cell line (Figure 5C). Control cell lines HT29 (bearing endogenous BRAFV600E) and DLD-1 (bearing endogenous KRASG13D) were also analyzed. The analysis regarding cell cycle inhibitors was limited to reverse transcription-PCR (RT-PCR) because of the poor quality results yielded by the available antibodies.
BRAFV600E Induces MSI in Caco-2 Cells by Altering DNA Content and Cell Cycle Distribution
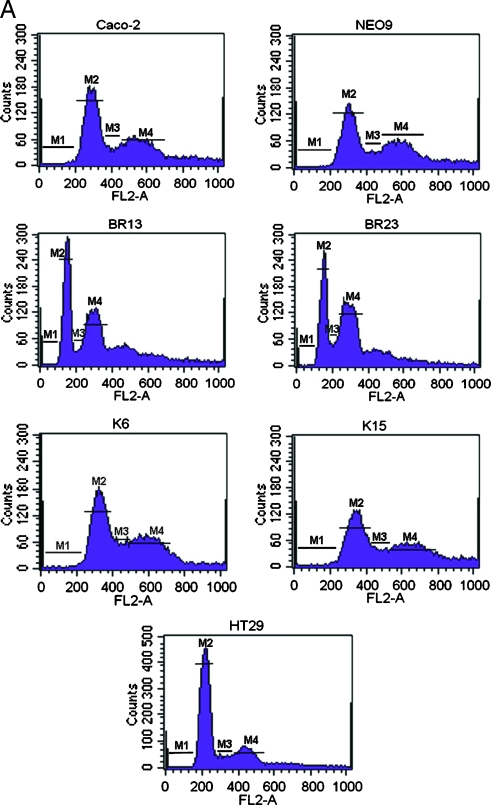
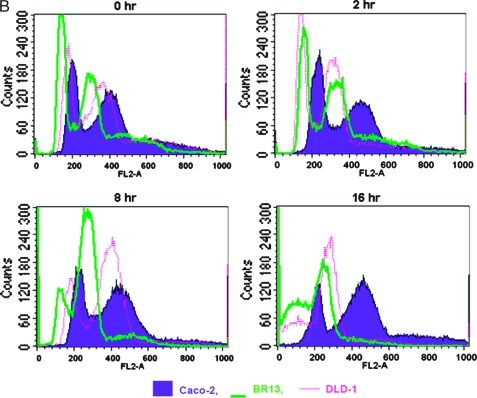
The ability of transfected oncogenes to modulate the cell cycle distribution of Caco-2 cells was measured by FACS analysis. BRAFV600E oncogene caused major cell cycle alterations when compared with parental Caco-2 cells, whereas no alterations were incurred in the presence of activated KRAS. Of interest is the fact that, after BRAFV600E transformation, a new population of cells was generated that seems to contain a hypodiploid number of chromosomes and therefore a lower amount of DNA content. This is depicted by a shift to the left, on the FL2 axis, of the corresponding G1 phase in both Caco-BR clones (hypodiploid peak; Figure 6A). To exclude the possibility of this being an apoptotic cell population, cells were treated with nocodazole for various time points after which there was a time-dependent reduction in the hypodiploid peak with a gradual shift of the cell populations toward the right on the FL2 axis (Figure 6B). Cell population movement of the hypodiploid peak suggests ongoing cell cycle and is confirmed by statistical analysis of the percent of gated cells at each time point (data not shown). It is worth mentioning that during the 2-hour treatment with nocodazole, Caco-BR dividing cells colocalized with the G1-phase peak of Caco-2-untreated cells. This observation could further support the lower DNA content in these cells. Therefore, the novel finding presented here suggests that BRAFV600E oncogene may induce MSI characteristics to Caco-2 cells (Figure 6A).
Figure 6.
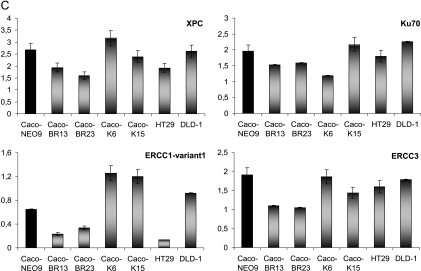
BRAFV600E induce MSI in Caco-2-transformed cells. (A) FACS analysis of cell populations after propidium iodide staining reveals altered cell cycle distribution in BR- cells. M1 indicates sub-G1 phase; M2, G1 phase; M3, S phase; M4, G2/M phase. (B) Cells challenged with 0.1 µM nocodazole to establish a time dependent reduction in the hypodiploid peak with a gradual shift of the cell populations toward the right on the FL2 axis. Cell cycle progression toward the G2/M phase was monitored at the indicated time points and progressive appearance of dead cells at 8 and 16 hours and excluded the presence of apoptosis, which only appears after the 16-hour treatment point. Results were consistent among three independent experiments. (C) Relative RNA levels of DNA repair genes in Caco-BR and Caco-K cells and control HT29 and DLD-1 human colon cancer cell lines were evaluated by RT-PCR analysis. The analysis was performed in triplicates and the mean ± SD is shown. DNA repair genes downregulated as a result of BRAFV600E-induced transformation in Caco-2 cells. Columns indicate relative RNA level normalized to glyceraldehyde 3-phosphate dehydrogenase. (D) Electropherogram of the mononucleotide repeat markers NR-21, NR-22, NR-24, BAT-25, and BAT-26 of BRAFV600E-transformed cells, their parental cell line Caco-2, and MSI-positive cell lines DLD-1 and HCT116. The appearance of numerous mononucleotide repeats of lower molecular weight in BR- cells in more than two MSI markers indicates MSI-H. Dashed line indicates the reference point relative to Caco-2 cells.
Regarding the hypothesis of BRAFV600E-induced MSI, specific DNA repair (MSH2 and Ku70) as well as nucleotide excision repair (NER) genes (ERCC1-variant 1, ERCC3, and XPC), found to be significantly downregulated in a microarray analysis (Joyce et al., unpublished observations) in Caco-BR cells, were validated by RT-PCR analysis to further support this hypothesis (Figure 6C).
The distinct role of BRAFV600E with respect to mediating MSI was further supported by the allelic profiling of NR-21, NR-22, and NR-24 in which Caco-BR cells presented lower molecular weight fragments, visible as peaks that had shifted to left and varied in 4 bp or greater (Table W2) compared with the parental cell line Caco-2. When comparing Caco-BR cells to MSI-positive cell lines DLD-1 and HCT116 [6,38]; the eletrophoretic pattern was only comparable with that observed in NR-22. In case of NR-21 and NR-42 loci, DLD-1 cells were represented by two distinct cell populations, one has been represented by normal-sized alleles and the other by peaks shifted to left, whereas the HCT116 were only represented by the lower molecular weight fragments. Both situations refer to MSI (Figure 6D). In contrast, the shift observed in BAT-25 and BAT-26 loci was bellow the cutoff level (≥4 bp) for Caco-BR23 but not Caco-BR13 and was considered to represent a polymorphism or somatic alteration. As previously, HCT116 were once again represented by a new cell population of lower molecular weight in BAT-26 locus, whereas a shift to the left (≥4 bp) was observed in BAT-25. Similarly, DLD-1 cells showed evidence of MSI in both BAT-25 and BAT-26 loci that were represented by shifts to the left (≥4 bp; Figure 6D).
Discussion
Differential Human Colon Cell Transformation by Oncogenic BRAF and KRAS
Several studies have investigated the role of BRAFV600E and KRASG12V acting as oncogenes, but no study so far has compared these two activating mutations for their relevant transforming capabilities in colon cancer cells. Although it has been reported that BRAFV600E is oncogenic when transfected into melanocytes [12], others have shown that BRAFV600E transforms NIH3T3 immortal fibroblasts less efficiently than HRASG12V [11] and that its expression confers rat thyroid cells with little growth advantage because of concomitant activation of DNA synthesis and apoptosis [39]. Results obtained in the present study point toward the presence of a potent machinery carrying out aggressive transformation in the Caco-2 cells transfected with the BRAFV600E oncogene.
BRAFV600E and KRASG12V Signal Through ERK1/2 to Transform Caco-2 Colon Cells
Although KRASG12V could activate ERK more efficiently, it was BRAFV600E that mediated complete transformation of colon epithelial cells. However, although MEK activation in Caco-BR cells was more potent, it did not lead to significantly increased ERK signaling as previously described for BRAFV600E [40]. In a similar study where BRAFV600E was compared to HRASG12V, pERK levels were comparable to basal in the case of BRAFV600E [41]. Furthermore, it was observed that, in Caco-BR cells, there was a marked difference between the two ERK isoforms, namely, ERK1 and ERK2, with the latter being far more activated. The important role of ERK2 in tumor growth has been recently revealed through RNAi-mediated knockdown, where ERK2 but not ERK1 inhibited growth of liver cancer cells in vitro and in vivo and silencing of ERK2 in NIH3T3 cells slowed down cell proliferation [42,43]. Taking into account that Caco-BR cells have enhanced cell growth, this selective ERK2 activation may play a crucial role in the regulation of cell proliferation in the BRAFV600E-transformed cells regardless of moderate ERK activity. In addition, localization of pERK in the nucleus explains why moderate ERK activity is sufficient for the cell transformation of Caco-BR cells because activated ERK is translocated to the nucleus where it can phosphorylate and activate several molecules, such as cyclin D1, which is significantly activated and hyperphosphorylated in these cells. Increased kinase activation was also recorded for CRAF, coexpression of which can induce a B/C RAF complex in a RAS-dependent manner, inducing distinct biochemical properties [44]. A different explanation for the moderate but efficient ERK activation may lay with the fact that excessive amount of ERK signaling can lead to cell cycle arrest or senescence, and cells have been shown to switch from BRAF to CRAF signaling to avoid this [45]. Pharmacologic inhibition of the MAPK signaling pathway with respect to the hyperproliferative phenotype of Caco BR cells, and to a lesser extent, of Caco K cells was indicative that MAPK pathway is mediating cell transformation. ERK inhibition was more potent in case of KRASG12V cell transformation and had a profound effect on the transcriptional target cyclin D1.
Other RAS effector pathways such as the JNK and the PI3K were found differentially regulated between the two oncogenes, and no modulation was detected for Ral. These observations indicate that the expression of these oncoproteins induces abundant as well as variable downstream signaling in the colonic epithelium.
Also of interest is the significant activation of BRAF kinase in both DLD-1 and SW620 colon cancer cells, as a result of the upstream KRAS mutation present in these colon adenocarcinomas [46], which, however, does not induce pERK. However, the presence of BRAFV600E in HT29 and Colo205 cells induces low pBRAF and high pERK. It is worth noting that the genetic background of these cell lines, apart from a KRAS or BRAF mutation, includes several other (p53, APC, PI3K, SMAD4). Here, we show the differential properties of two frequently mutated oncogenes in a permissive genetic background (in Caco-2 cell line) and how much these properties vary in the context of other mutations (e.g., HT29 and DLD-1 cell lines). This finding may have in vivo implications for personalized therapeutics in the future, considering how these mutations may influence pathway cross talk.
Attenuation of ERK Activation Due to KSR Down-regulation by BRAFV600E
Nevertheless, the RAS-RAF-MAPK signaling pathway is not a simple one because many molecules have been identified to regulate its function. For example, signal flow from RAF to MEK is believed to be regulated by scaffold proteins such as KSR and protein interaction disrupters such as RKIP [47]. KSR is a putative scaffolding protein for the RAF-MEK-ERK module that positively regulates ERK activation by binding to MEK and ERK constitutively [48]. In the present study, KSR was found significantly downregulated in the Caco-BR cells, and this could account for the moderate levels of ERK activation detected. Studies that associate KSR with cancer progression led to assume that pharmacological inactivation of KSR may serve as a treatment for RAS-driven malignancies, such as pancreatic cancer [49].
Presence of Rb and p19ARF Overexpression Is Not Sufficient for RASG12V-Induced Transformation
We report here that colon epithelial cells transformed by BRAFV600E presented no sign of senescence, whereas premature senescence was detected in the Caco-K cells, which also had ongoing cell cycle. To identify the exact mechanism by which KRASG12V switches from promoting proliferation to inducing senescence mechanism, the MAPK pathway was pharmacologically blocked, and replicative senescence was successfully prevented. Therefore, hyperactivation of the ERK pathway in Caco-K cells seems to lead to the opposite situation, which are the induction of a senescence program, growth factor-dependent cell viability, and finally not efficient cell transformation, which are not observed in Caco-BR cells.
Because in the parental Caco-2 cells the p53 is mutated and the p16Ink4 tumor suppressor is hypermethylated, the remaining possible mediator of senescence in the KRASG12V is the Rb pathway. Studies have shown that overexpression of RASG12V or Raf1 by retroviral delivery in normal human fibroblasts disrupted the Rb pathway [21,50]. In the present study, although the Rb was found to be selectively hyperphosphorylated and therefore deactivated in the Caco-BR cells, it was still functional in the Caco-K cells in which the p19ARF was also significantly overexpressed. Together, these data show that both the Rb and the p19ARF are working to induce senescence-related markers in Caco-K cells, which, however, escape from growth arrest. Cell survival could also be facilitated by the selective activation of AKT observed in the Caco-K cells. RAS-induced senescence has not been fully elucidated and is considered to be the coordinated output of cell type-specific signaling networks [51]. This is further supported by the fact that additional growth factor extrinsically regulated signaling pathways are important for the viability of Caco-2 cells, whereas Caco-BR cell viability is not dependent on extrinsic growth factors. Increased proliferation of Caco-BR cells could be well explained by the increased expression of most cyclins including cyclin D1, D3, A, and E. Significantly increased expression of D cyclins could also account for the increased p21Cip1 because it has been shown that cyclin D1 specifically inhibits its degradation [19]. Both oncogenes (BRAFV600E and KRASG12V) increase cyclin D1, which, when combined with Cdk4 and Cdk6, phosphorylates and inactivates Rb; however, this is not the case for the Caco-K cells. Accumulated p27Kip1 manages to take advantage of the hypophosphorylated Rb in the Caco-K cells and silences cyclin D/Cdk2 complex that may be present in the early G1 phase. Working along this hypothesis, KRASG12V signaling is efficient in inducing cyclin B1, the most powerful mediator of cell cycle. Cyclin B1 is the cyclin localized in the cytoplasm, and its activation may be a direct signaling from increased cytoplasmic ERK in Caco-K cells.
BRAFV600E-Mediated MSI in Caco-2 Cells
Cell cycle distribution of Caco-2 cells was notably affected after BRAFV600E transformation because it was represented by a general shift to the left of the FL2 axis. This was indicative of a cell population with a lower DNA content due to a hypodiploid number of chromosomes. Nocodazole treatment of cells revealed that these cells were normally proliferating because they progressed through the M and G2 phases. However, their dividing capacity was diminished, characteristic of cells containing a hypodiploid number of chromosomes. In a recent study, BRAFV600E overexpression in a rat thyroid cell (PCCL3) among others managed to induce chromosomal instability (CIN) [39]. Caco-2 cells that are characterized by CIN acquire MSI characteristics in the presence of BRAFV600E, a hypothesis also supported by a microarray analysis in the Caco-BR cells (Joyce et al., unpublished observations). The microarray study revealed down-regulation of essential DNA repair (MSH2 and Ku70) and NER genes (ERCC1-variant 1, ERCC3, and XPC). Validation of array results confirmed down-regulation of ERCC1, ERCC3, and XPC in Caco-BR cells, all members of the NER subfamily. Deregulation in one of these NER members may result to failure of DNA repair initiation and cause defects in the mismatch repair mechanisms and its gene members [52]. Notably, DLD-1, already described to have MSI, also presents a significant down-regulation of ERCC1-variant 1 gene, and their FACS profile is comparable to that of Caco-BR cells'. BRAF mutations have long been connected with MSI sporadic colorectal tumors because mismatch repair-deficient tumors have a very high incidence of BRAF mutations, and its overexpression in a rat cell line of thyroid origin (PCCL3) induced CIN [39,53–55]. The role of BRAFV600E in MSI is further elucidated through the allelic profiling of certain mononucleotide repeats widely used to determine the microsatellite status. In particular, more than two of five analyzed loci (NR-21, NR-22, NR-24, BAT-25, and BAT-26) show instability in the Caco-BR cells, and therefore, we classify them as MSI-high (MSI-H) [56]. Because MSI cell lines display higher sensitivity to chemotherapeutics (CPT-11) than MSS cells, this could be later exploited to tailor chemotherapy based on MSI status [57,58].
We propose that the effect of BRAFV600E, elicited on mitotic impairment, triggers chromosome breakage in the presence of a defective DNA damage response mechanism. As a result, cells continue cycling and acquire, within a single cell cycle, both chromosome rearrangements and abnormal chromosome numbers that remarkably mimic the complex genetic hallmark of tumorigenesis [59].
Conclusions
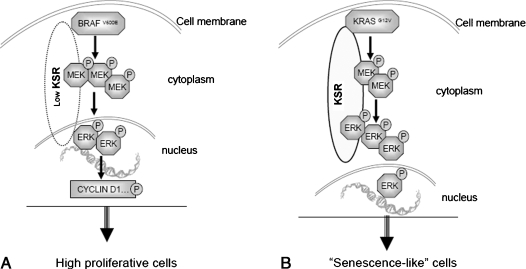
Our study demonstrates that BRAFV600E is amore potent oncogene than KRASG12V in transforming colon epithelial cells, whereas KRASG12V managed to confer premature senescence to the same cell system. It was established that BRAFV600E transformation was facilitated by the moderate activation of pERK, as a consequence of low KSR levels, which, however, translocated to the nucleus driving the increased proliferation rate in theses cells through the hyperphosphorylation of cyclin D1 (Figure 7A). Conversely, the same MAPK pathway, only being more activated, was the mediator of KRASG12V oncogene-induced senescence markers and growth factor-dependent viability (Figure 7B). Within the different biochemical properties of these two oncogenes lays also the induction of MSI by BRAFV600E rendering Caco-2 MSI-H.
Figure 7.
Differential effects of KRASG12V and BRAFV600E on cell cycle. BRAFV600E drives cells to high proliferation rate through nuclear pERK and the consequent phosphorylation of cyclin D1 (A), whereas KRASG12V generates senescence-related characteristics mediated by high cytoplasmic levels of pERK (B).
Supplementary Materials and Methods
Protein Immunoblot Analysis and Glutathione S-Transferase Pull-down Assay
Whole cell lysates were prepared with lysis buffer containing 50 mM Tris-HCl (pH 7.4), 250 mM sucrose, 1 mM EDTA, 10 mM NaF, 1 mM EGTA, 1% (v/v) Triton X-100, 1 mM sodium orthovanadate, 10 µg/ml leupeptin, and 0.2 mM PMSF. Cells were lysed by swelling in hypotonic buffer (10 mM Hepes pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA). For Western blot analysis, protein concentrations were determined by the Bradford assay (Bio-Rad Laboratories, Hercules, CA) using bovine serum albumin as a standard. Extracts were resolved on SDS-PAGE (10% or 12% wt/vol acrylamide), transferred to nitrocellulose membrane (Whatman; Scheiche & Schuell, Dassel, Germany). Antibodies used were as follows: K-Ras (sc-30), B-Raf (sc-5284), cyclin D1 (sc-718), pTyr204 ERK (sc-7383), and α-tubulin (sc-8035) that were purchased from Santa Cruz; pSer445 B-Raf (2696), pSer473 Akt (9271), pSer217/221 MEK1/2 (9121), and pThr183/185 SAPK/JNK (9251) that were purchased from Cell Signaling (Danvers, MA); cyclin E (3512-1), cyclin A (3507-1), cyclin B1 (3508-1), and cyclin D3 (3571-1) that were purchased from Clontech (Heidelberg, Germany); C-Raf (610151), Rb (554136), and Ral A (610221) that were purchased from BD (Franklin Lakes, NJ); and pSer338-Raf-1 (05-538; Upstate, Lake Placid, NY).
For the glutathione S-transferase (GST) pull-down assay, 200 µg of the identical protein extract used for Western analysis was incubated with GST-Ral binding protein to glutathione agarose beads for 1 hour by rotating at 4°C. Beads were washed four times in cell lysis buffer before being loaded on 12% wt/vol SDS-PAGE. The construct was kindly provided by Johannes L. Bos (Centre for Biomedical Genetics, Utrecht, the Netherlands). Fold expression of all proteins analyzed was determined after the band intensity was established using Molecular Dynamics Image Quant Software (Amersham Biosciences, Uppsala, Sweden).
Gene Expression RT-PCR Analysis
For RT-PCR amplification, 2.5 µl of complementary DNA template, from all clones and control cells lines, was analyzed against different primers (20 pmol each) (Table W1) for a total of 25 µl of reaction. Each sample was amplified using the MJ thermocycler PTC-200 (Bio-Rad, Hercules, CA). Intensity values were measured using Molecular Dynamics Image Quant Software. All PCR products were normalized to GAPDH expression.
Footnotes
This work is supported by the European Union grants LSHC-CT-2006-037278 “ONCODEATH,” MTKD-CT-2004-509836 “MACROGENEX,” MRTN-CT-2004-504228 “TAF-Chromatin,” and GSRT grant 03 ED562 to A.P.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 2.Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, Kambara T, MacPhee DG, Young J, Leggett BA, et al. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol. 2004;22:4584–4594. doi: 10.1200/JCO.2004.02.154. [DOI] [PubMed] [Google Scholar]
- 3.Fransen K, Klintenas M, Osterstrom A, Dimberg J, Monstein HJ, Soderkvist P. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis. 2004;25:527–533. doi: 10.1093/carcin/bgh049. [DOI] [PubMed] [Google Scholar]
- 4.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 5.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 6.COSMIC Database. [May 11, 2009]. Available at: http://www.sanger.ac.uk/
- 7.Al-Mulla F, Milner-White EJ, Going JJ, Birnie GD. Structural differences between valine-12 and aspartate-12 Ras proteins may modify carcinoma aggression. J Pathol. 1999;187:433–438. doi: 10.1002/(SICI)1096-9896(199903)187:4<433::AID-PATH273>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 8.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 9.Winder T, Mundlein A, Rhomberg S, Dirschmid K, Hartmann BL, Knauer M, Drexel H, Wenzl E, De Vries A, Lang A. Different types of K-Ras mutations are conversely associated with overall survival in patients with colorectal cancer. Oncol Rep. 2009;21:1283–1287. doi: 10.3892/or_00000352. [DOI] [PubMed] [Google Scholar]
- 10.Ikenoue T, Hikiba Y, Kanai F, Tanaka Y, Imamura J, Imamura T, Ohta M, Ijichi H, Tateishi K, Kawakami T, et al. Functional analysis of mutations within the kinase activation segment of B-Raf in human colorectal tumors. Cancer Res. 2003;63:8132–8137. [PubMed] [Google Scholar]
- 11.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 12.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 13.Rao S, Cunningham D, de Gramont A, Scheithauer W, Smakal M, Humblet Y, Kourteva G, Iveson T, Andre T, Dostalova J, et al. Phase III double-blind placebo-controlled study of farnesyl transferase inhibitor R115777 in patients with refractory advanced colorectal cancer. J Clin Oncol. 2004;22:3950–3957. doi: 10.1200/JCO.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 14.Genot E, Reif K, Beach S, Kramer I, Cantrell D. p21ras initiates Rac-1 but not phosphatidyl inositol 3 kinase/PKB, mediated signaling pathways in T lymphocytes. Oncogene. 1998;17:1731–1738. doi: 10.1038/sj.onc.1202101. [DOI] [PubMed] [Google Scholar]
- 15.Woods D, Parry D, Cherwinski H, Bosch E, Lees E, McMahon M. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasid U, Dritschilo A. RAF antisense oligonucleotide as a tumor radiosensitizer. Oncogene. 2003;22:5876–5884. doi: 10.1038/sj.onc.1206700. [DOI] [PubMed] [Google Scholar]
- 17.Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mermelshtein A, Gerson A, Walfisch S, Delgado B, Shechter-Maor G, Delgado J, Fich A, Gheber L. Expression of D-type cyclins in colon cancer and in cell lines from colon carcinomas. Br J Cancer. 2005;93:338–345. doi: 10.1038/sj.bjc.6602709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coleman ML, Marshall CJ, Olson MF. Ras promotes p21(Waf1/Cip1) protein stability via a cyclin D1-imposed block in proteasome-mediated degradation. EMBO J. 2003;22:2036–2046. doi: 10.1093/emboj/cdg189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravi RK, Weber E, McMahon M, Williams JR, Baylin S, Mal A, Harter ML, Dillehay LE, Claudio PP, Giordano A, et al. Activated Raf-1 causes growth arrest in human small cell lung cancer cells. J Clin Invest. 1998;101:153–159. doi: 10.1172/JCI831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janderova-Rossmeislova L, Novakova Z, Vlasakova J, Philimonenko V, Hozak P, Hodny Z. PML protein association with specific nucleolar structures differs in normal, tumor and senescent human cells. J Struct Biol. 2007;159:56–70. doi: 10.1016/j.jsb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 24.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 25.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6:313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Zweibaum A, Triadou N, Kedinger M, Augeron C, Robine-Leon S, Pinto M, Rousset M, Haffen K. Sucrase-isomaltase: a marker of foetal and malignant epithelial cells of the human colon. Int J Cancer. 1983;32:407–412. doi: 10.1002/ijc.2910320403. [DOI] [PubMed] [Google Scholar]
- 28.Ilyas M, Tomlinson IP, Rowan A, Pignatelli M, Bodmer WF. β-Catenin mutations in cell lines established from human colorectal cancers. Proc Natl Acad Sci USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Bosscher K, Hill CS, Nicolas FJ. Molecular and functional consequences of Smad4 C-terminal missense mutations in colorectal tumour cells. Biochem J. 2004;379:209–216. doi: 10.1042/BJ20031886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Djelloul S, Forgue-Lafitte ME, Hermelin B, Mareel M, Bruyneel E, Baldi A, Giordano A, Chastre E, Gespach C. Enterocyte differentiation is compatible with SV40 large T expression and loss of p53 function in human colonic Caco-2 cells. Status of the pRb1 and pRb2 tumor suppressor gene products. FEBS Lett. 1997;406:234–242. doi: 10.1016/s0014-5793(97)00208-1. [DOI] [PubMed] [Google Scholar]
- 31.Roberts ML, Drosopoulos KG, Vasileiou I, Stricker M, Taoufik E, Maercker C, Guialis A, Alexis MN, Pintzas A. Microarray analysis of the differential transformation mediated by Kirsten and Harvey Ras oncogenes in a human colorectal adenocarcinoma cell line. Int J Cancer. 2006;118:616–627. doi: 10.1002/ijc.21386. [DOI] [PubMed] [Google Scholar]
- 32.Workman P, Twentyman P, Balkwill F, Balmain A, Chaplin D, Double J, Embleton J, Newell D, Raymond R, Stables J, et al. United Kingdom Co-ordinating Committee on Cancer Research (UKCCR) guidelines for the welfare of animals in experimental neoplasia (second edition) Br J Cancer. 1998;77:1–10. doi: 10.1038/bjc.1998.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, Seruca R, Iacopetta B, Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804–1811. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 35.Ferbeyre G. Barriers to Ras transformation. Nat Cell Biol. 2007;9:483–485. doi: 10.1038/ncb0507-483. [DOI] [PubMed] [Google Scholar]
- 36.Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 37.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 38.Russo MT, Blasi MF, Chiera F, Fortini P, Degan P, Macpherson P, Furuichi M, Nakabeppu Y, Karran P, Aquilina G, et al. The oxidized deoxynucleoside triphosphate pool is a significant contributor to genetic instability in mismatch repair-deficient cells. Mol Cell Biol. 2004;24:465–474. doi: 10.1128/MCB.24.1.465-474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitsutake N, Knauf JA, Mitsutake S, Mesa C, Jr, Zhang L, Fagin JA. Conditional BRAFV600E expression induces DNA synthesis, apoptosis, dedifferentiation, and chromosomal instability in thyroid PCCL3 cells. Cancer Res. 2005;65:2465–2473. doi: 10.1158/0008-5472.CAN-04-3314. [DOI] [PubMed] [Google Scholar]
- 40.Dhillon AS, Kolch W. Oncogenic B-Raf mutations: crystal clear at last. Cancer Cell. 2004;5:303–304. doi: 10.1016/s1535-6108(04)00087-x. [DOI] [PubMed] [Google Scholar]
- 41.Benjamin CL, Ananthaswamy HN. Oncogenic potential of BRAF versus RAS. Cancer Lett. 2008;261:137–146. doi: 10.1016/j.canlet.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 42.Bessard A, Fremin C, Ezan F, Fautrel A, Gailhouste L, Baffet G. RNAi-mediated ERK2 knockdown inhibits growth of tumor cells in vitro and in vivo. Oncogene. 2008;27:5315–5325. doi: 10.1038/onc.2008.163. [DOI] [PubMed] [Google Scholar]
- 43.Lefloch R, Pouyssegur J, Lenormand P. Single and combined silencing of ERK1 and ERK2 reveals their positive contribution to growth signaling depending on their expression levels. Mol Cell Biol. 2008;28:511–527. doi: 10.1128/MCB.00800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garnett MJ, Rana S, Paterson H, Barford D, Marais R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell. 2005;20:963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 45.Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, Bastian BC, Springer C, Marais R. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66:9483–9491. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 46.Banerji U, Affolter A, Judson I, Marais R, Workman P. BRAF and NRAS mutations in melanoma: potential relationships to clinical response to HSP90 inhibitors. Mol Cancer Ther. 2008;7:737–739. doi: 10.1158/1535-7163.MCT-08-0145. [DOI] [PubMed] [Google Scholar]
- 47.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. 2000;351(Pt 2):289–305. [PMC free article] [PubMed] [Google Scholar]
- 48.Razidlo GL, Kortum RL, Haferbier JL, Lewis RE. Phosphorylation regulates KSR1 stability, ERK activation, and cell proliferation. J Biol Chem. 2004;279:47808–47814. doi: 10.1074/jbc.M406395200. [DOI] [PubMed] [Google Scholar]
- 49.Xing HR, Cordon-Cardo C, Deng X, Tong W, Campodonico L, Fuks Z, Kolesnick R. Pharmacologic inactivation of kinase suppressor of ras-1 abrogates Ras-mediated pancreatic cancer. Nat Med. 2003;9:1266–1268. doi: 10.1038/nm927. [DOI] [PubMed] [Google Scholar]
- 50.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang G, Dombkowski A, Chuang L, Xu XX. The involvement of XPC protein in the cisplatin DNA damaging treatment-mediated cellular response. Cell Res. 2004;14:303–314. doi: 10.1038/sj.cr.7290375. [DOI] [PubMed] [Google Scholar]
- 53.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. T umorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 54.Oliveira C, Velho S, Domingo E, Preto A, Hofstra RM, Hamelin R, Yamamoto H, Seruca R, Schwartz S., Jr Concomitant RASSF1A hypermethylation and KRAS/BRAF mutations occur preferentially in MSI sporadic colorectal cancer. Oncogene. 2005;24:7630–7634. doi: 10.1038/sj.onc.1208906. [DOI] [PubMed] [Google Scholar]
- 55.Vilar E, Scaltriti M, Balmana J, Saura C, Guzman M, Arribas J, Baselga J, Tabernero J. Microsatellite instability due to hMLH1 deficiency is associated with increased cytotoxicity to irinotecan in human colorectal cancer cell lines. Br J Cancer. 2008;99:1607–1612. doi: 10.1038/sj.bjc.6604691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 57.Bertagnolli MM, Niedzwiecki D, Compton CC, Hahn HP, Hall M, Damas B, Jewell SD, Mayer RJ, Goldberg RM, Saltz LB, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J Clin Oncol. 2009;27:1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Des GG, Uzzan B, Nicolas P, Schischmanoff O, Morere JF. Microsatellite instability: a predictive marker in metastatic colorectal cancer? Target Oncol. 2009;4:57–62. doi: 10.1007/s11523-008-0103-8. [DOI] [PubMed] [Google Scholar]
- 59.Dunican DS, McWilliam P, Tighe O, Parle-McDermott A, Croke DT. Gene expression differences between the microsatellite instability (MIN) and chromosomal instability (CIN) phenotypes in colorectal cancer revealed by high-density cDNA array hybridization. Oncogene. 2002;21:3253–3257. doi: 10.1038/sj.onc.1205431. [DOI] [PubMed] [Google Scholar]
- 60.Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20(15):2471–2472. doi: 10.1093/bioinformatics/bth254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.