Abstract
Infection with the Epstein-Barr virus (EBV) is a strong predisposing factor in the development of nasopharyngeal carcinoma (NPC). Many viral gene products including EBNA1, LMP1, and LMP2 have been implicated in NPC tumorigenesis, although the de novo control of these viral oncoproteins remains largely unclear. The recent discovery of EBV-encoded viral microRNA (miRNA) in lymphoid malignancies has prompted us to examine the NPC-associated EBV miRNA. Using large-scale cloning analysis on EBV-positive NPC cells, two novel EBV miRNA, now named miR-BART21 and miR-BART22, were identified. These two EBV-encoded miRNA are abundantly expressed in most NPC samples. We found two nucleotide variations in the primary transcript of miR-BART22, which we experimentally confirmed to augment its biogenesis in vitro and thus may underline the high and consistent expression of miR-BART22 in NPC tumors. More importantly, we determined that the EBV latent membrane protein 2A (LMP2A) is the putative target of miR-BART22. LMP2A is a potent immunogenic viral antigen that is recognized by the cytotoxic T cells; down-modulation of LMP2A expression by miR-BART22 may permit escape of EBV-infected cells from host immune surveillance. Taken together, we demonstrated that two newly identified EBV-encoded miRNA are highly expressed in NPC. Specific sequence variations on the prevalent EBV strain in our locality might contribute to the higher miR-BART22 expression level in our NPC samples. Our findings emphasize the role of miR-BART22 in modulating LMP2A expression, which may facilitate NPC carcinogenesis by evading the host immune response.
Introduction
MicroRNA (miRNA) are a group of small, noncoding RNA with a size of approximately 18 to 24 nucleotides. They are produced by endogenous enzymatic (Drosha/DGCR8 and Dicer) digestion of RNA transcripts containing hairpin structure. Mature miRNA function as negative gene regulators through complementary sequence pairing to the 3′ untranslated region (3′UTR) of the target gene by inducing either messenger RNA (mRNA) degradation or translational repression [1]. As gene regulators, mammalian miRNA play key roles not only in various biological processes including development and differentiation but also in cancer development.
Infection with the Epstein-Barr virus (EBV) is common among most adults worldwide. Whereas most people will recover from the symptoms of acute infection, latent infection with EBV is known to be associated with a number of lymphoid and epithelial malignancies, such as Hodgkin disease (HD), Burkitt lymphoma, nasopharyngeal carcinoma (NPC), and a subset of gastric carcinoma. The epithelial cancer NPC is uniquely prevalent in Southern China. Clonal EBV genomes can be detected in both high-grade dysplastic lesions and invasive carcinoma, implying an important etiologic role for EBV in NPC carcinogenesis [2]. EBV resides in NPC as latency II infection, where only few viral latent proteins including EBV nuclear antigen 1 (EBNA1) and latent membrane proteins 1 and 2A (LMP1 and LMP2A) are expressed. These viral proteins have thus been proposed in the NPC development by their disruption of multiple signaling pathways and cellular mechanisms [3]. Two types of noncoding RNA, EBV-encoded RNA (EBERs) and BamH1-A rightward transcripts (BARTs), are expressed in EBV-positive NPC cells. EBERs are small, nonpolyadenylated RNA that are consistently expressed in all forms of viral latency. BARTs are multispliced transcripts that were originally discovered in NPC and later reported in other EBV-infected individuals [4–6]. Because expression of BARTs is selectively high in EBV-positive NPC and gastric cancer cells, it has been long speculated that BARTs have important functional roles in these EBV-associated epithelial malignancies [7]. Recent studies on viral miRNA have identified 23 EBV miRNA (ebv-miRNA) from latently infected B cells [7–9]. Among them, 20 miRNA (miR-BART1 to miR-BART20) are located in two clusters within the BARTs region, which is generally highly expressed in most EBV infected epithelial cells [7]. Because only a few viral latent proteins are expressed in NPC, it is plausible that EBV augments cancer development through its viral-encoded miRNA. Whereas in most instances the function of these EBV-miRNA remains unknown, recent discoveries suggest much importance for miR-BARTs in modulating both viral and cellular gene expressions [10,11]. In line with these reports, our group has recently demonstrated that miR-BART1, 16, and 17 can regulate the LMP1 viral oncogene expression in NPC by translational repression [12]. Furthermore, miR-BART5 can alter the expression of the p53-upregulated modulator of apoptosis (PUMA), which promotes NPC cell survival [13]. On the basis of perfect complementary to the 3′UTR of BALF5 mRNA, miR-BART2 could downregulate this viral lytic protein production by mRNA degradation [14].
Whereas miR-BARTs hold much importance in complex virus-host interactions and contribute to human carcinogenesis, currently identified ebv-miRNA were cloned from EBV-positive lymphoma cell lines and confirmed for expression in these cell lines after computational prediction from the EBV sequence [7–9]. Thus, EBV-encoded miRNA involved in NPC may have escaped detection in previous discovery processes. In this study, we have systemically examined the NPC-associated EBV genome for viral-encoded miRNA expression. By constructing a small complementary DNA (cDNA) library from a native EBV-positive NPC cell line (C666-1) and a xenograft (X2117), we identified within the BARTs region two novel ebv-miRNA, which now named miR-BART21 and miR-BART22. We found that these two miRNA are highly expressed in NPC cell lines, xenografts, and primary tumor biopsies. By comparing sequences between different EBV strains, we identified two nucleotide variations upstream of mature miR-BART22, which have likely favored its biogenesis by Drosha/DGCR8 processing. This finding could in part explain the high expression of miR-BART22 in endemic NPC samples. By bioinformatics analysis and functional screening, we found that LMP2A, an important viral latent gene, is one of the downstream targets of miR-BART22. Among all EBV latent genes expressed in NPC cells, the LMP2A-encoded protein exhibits relatively strong immunogenicity toward cytotoxic T cells (CTLs). In this context, LMP2A expression in NPC is tightly regulated to promote tumor cell survival by escaping the host immune response. The findings in this report highlight that specific ebv-miRNA are present in NPC and that miR-BART22 may well have contributed to NPC development through its modulation of LMP2A expression.
Materials and Methods
Plasmid Constructs
The renilla luciferase reporter control plasmid, pRL-CMV, and the enhanced green fluorescence protein (EGFP) expression plasmid, pEGFP-C1, were purchased from Promega Corp (Madison, WI) and Clontech (Mountain View, CA), respectively. The LMP2A expression plasmid contained the complete LMP2A ORF + 3′UTR and the miRNA expression plasmids contained miRNA flanking sequence (∼300 nt). They were generated by polymerase chain reaction (PCR) and cloned into the pcDNA3.1 (Invitrogen, Carlsbad, CA). Construction of the firefly luciferase reporter plasmids by using pMIR-REPORT (Ambion, Inc, Austin, TX) was described previously [15]. The sequences of the primers used are listed in SI Methods.
Cell Lines, Xenografts, and Tumor Samples
All cell lines were cultured in RPMI plus 10% FBS unless otherwise specified. 293FT and HEK293 cells were cultured in minimum essential medium plus 10% FBS. NP69 and NP460 were grown in keratinocyte serum-free medium and Defined keratinocyte serum-free medium (Invitrogen), respectively. All nasopharyngeal (NP), NPC, infectious mononucleosis (IM), and HD biopsies were obtained from the Prince of Wales Hospital, of the Chinese University of Hong Kong. The study was approved by the local ethics committee.
Transfection of HEK293 cells was performed using GeneJuice transfection reagent (Novagen, Madison, WI) following the manufacturer's protocol. LMP2A expressing HEK293 stable clones were isolated as single colonies and selected in medium containing 400 µg/ml of G418 (Invitrogen) for 12 weeks before analysis.
Transfection of siRNA and miRNA Mimic
SiRNA duplexes and miRNA mimics were purchased from Qiagen (Hilden, Germany). The target sequence of siLMP2A with dTdT overhangs modification was AACUCCCAAUAUCCAUCUGCU [16]. The sequences of the miRNA mimics used are listed in Figure 4B. Allstars negative control (Qiagen) was used in both siRNA and miRNA transfection experiments. In the experiments, 20 nM of siRNA and miRNA mimics were used to transfect cells in six-well plate format. Protein and RNA were extracted 24 hours after transfection. All transfections were carried out by Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
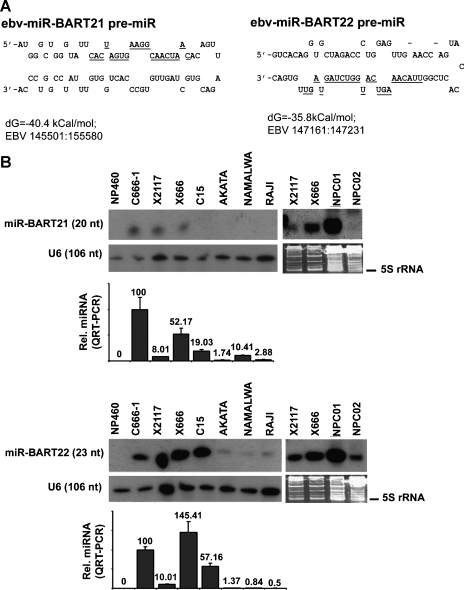
Figure 1.
Hairpin structures and expression of novel ebv-miRNA. (A) Stem-loop structures of miR-BART21 and miR-BART22 are illustrated. The cloned mature miRNA sequences are indicated by underlining. The folding energy (dG) and EBV genomic sequences are listed below the figures. (B) Representative Northern blot for cell lines, NPC xenografts and NPC biopsies showing the expression of miR-BART21 and miR-BART22 are displayed. The normal epithelial cell line NP460 was included as a negative control. U6 RNA or SYBR Gold staining PAGE was used to control for RNA loading. The miRNA expression in the same sample was assayed by QRT-PCR and was compared with C666-1 (set at 100). The analysis of each sample was performed in triplicate with the mean ± SD shown.
RNA Extraction and Small cDNA Library Construction
Total RNA was extracted by TRIzol reagent (Invitrogen) without any ethanol washing step. The enriched small RNA fraction was collected with miVana miRNA Isolation Kit according to the manufacturer's instructions (Ambion). Twenty micrograms of enriched RNA was used for library construction by using the miRCat cloning kit (Integrated DNA Technologies, Coralville, IA). Sequence analysis was performed using an ABI PRISM 3130xl DNA Sequencer.
miRNA Identification and RNA Structure Prediction
Isolated RNA fragments longer than 18 nt were annotated to the genome. Known miRNA were identified by blasting the sequences to “miRBase” [9]. The remaining sequences were individually blasted to the National Center for Biotechnology Information databases. Putative precursor sequences of the cloned EBV fragments were extracted from the EBV genome (AJ507799) and further examined using the MFOLD program to predict their pre-miRNA structures [17]. The most stable predicted structure was trimmed to a size of less than 80 nucleotides.
Northern Blot Analysis
Ten micrograms total of RNA was electrophoresed by 12% PAGE and transferred onto Nytran SPC membrane (Whatman, Inc, Piscataway, NJ) for blot analysis. Oligonucleotides complementary to the mature miRNA were end-labeled with (γ-32P) as a probe. Hybridization was performed in ULTRAhyb-Oligo hybridization buffer (Ambion). The sequences of the probes are listed in SI Methods. The density of the Northern blot signal was measured by Image J software (download from http://rsb.info.nih.gov/ij/).
Quantitative Reverse Transcription-Polymerase Chain Reaction
For normal reverse transcription-polymerase chain reaction (RT-PCR), 1 µg of RNA was reverse-transcribed with Superscript III using random primers (Invitrogen), and quantitative RT-PCR (QRT-PCR) products were amplified by using the SYBR Green PCR Master Mix Kit (Applied Biosystems, Foster City, CA). QRT-PCR for LMP2A was performed as described in Bell et al. [18] except that total cDNA was used for PCR. Expression levels of viral and cellular genes were normalized to actin unless otherwise specified. For miRNA quantification, miScript Reverse Transcription kit and miScript SYBR Green PCR Kit (Qiagen) were used according to the manufacturer's instructions. The in-house designed miRNA specific primer sequences are listed in SI Methods. Expression levels of miRNA were normalized to RNU6B snRNA. Analysis of each sample was performed in triplicate.
In Vitro Assay for Drosha Complex Digestion
The digestion substrate was prepared by in vitro transcription from a T7-added miR-BART22 PCR product (∼300 nt) using MAXIscript kit (Ambion). The primer sequences are listed in SI Methods. For in vitro digestion, Drosha and Flag-DGCR8 expression vectors [19] were cotransfected into 293FT cells at a 2:1 ratio. The Drosha/DGCR8 enzymatic complex was purified using the Flag-Tagged Protein immunoprecipitation kit (Sigma, St Louis, MO) 48 hours after transfection. Digestion was performed by mixing 100 ng of RNA with the precipitated complex at 37°C for 1.5 hours. Digested products were visualized on 8% PAGE with SYBR Gold staining (Invitrogen) and Northern blot analysis.
Target Prediction
To obtain more putative targets, we loosened the stringency in MiRanda prediction by adjusting the energy threshold to -15 kcal/mol and the cutoff score to 90. We used default settings for the RNAhybrid program prediction. The LMP2A reference sequence for target prediction was extracted from the National Center for Biotechnology Information (AB290724).
Luciferase Reporter Assay
293FT cells (1 x 105) grown in 24-well plates were cotransfected with miRNA and reporter constructs for analysis. Cells were harvested after 2 days for luciferase activity analysis using Dual Luciferase Reporter Kit (Promega) as described previously [15]. See SI Methods for the details of transfection.
Immunohistochemical Staining of LMP2A
Immunohistochemical (IHC) staining was performed by modified methods as previously described [20]. In brief, the paraffin sections were dewaxed in xylene and rehydrated in ethanol and peroxidase-blocked with 3% H2O2 in room temperature for 20 minutes. Antigen retrieval was performed by using pressure cooker with 10 nM citrate buffer (pH 6.0) for 4 minutes. Primary antibody against LMP2A (1:50 dilution, 15F9; AbD SeroTec, Raleigh, NC) was applied and incubated for 2 hours. The section was incubated with biotinylated secondary antibody (1:100 dilution; DAKO, Glostrup, Denmark) for 1 hour. The signal was visualized by using DAB Detection Kit (Sigma). The presence of the EBV in the paraffin sections was confirmed by EBER in situ hybridization, which carried out with an EBV probe ISH kit (Novocastra, Newcastle, UK).
Western Blot Analysis
Western blot analysis was performed as previously described [15]. The primary antibodies used were LMP2A (1:2000 dilution, 15F9; AbD SeroTec), AKT (1:1000 dilution; Cell Signaling Technology, Danvers, MA), phospho-AKT (ser473; 1:1000; Cell Signaling). EGFP (1:20,000 dilution; Clontech) and β-actin (1:30,000 dilution; Sigma). All horseradish peroxidase-conjugated secondary antibodies used were purchased from DAKO (1:20,000 dilution). The density of Western blot signals was measured by Image J software.
Results
Identification and Expression of Novel EBV-Encoded miRNA in NPC
Two independent cDNA libraries were constructed from C666-1 and X2117 cells. A total of 2928 clones with sequences longer than 18 nucleotides were obtained, including 1813 clones from C666-1 and 1115 clones from X2117 (Table W1). The known miRNA and human sequences were annotated by blasting individual extracted sequences against miRBASE [21] and GenBank databases. Sequences that matched the EBV genome were identified from BLAST search against the full-length EBV genome (accession number: AJ507799).
In the C666-1 library, 615 and 277 clones were matched to known EBV and human miRNA, respectively, based on miRBase (Version 11.0) search. In addition to the known miRNA, 811 clones were human sequences that mostly corresponded to rRNA and tRNA fragments. Of interest, 32 sequences that aligned to the EBV genome showed no match to any known miRNA previously reported in lymphoid cells. According to their sequence similarity, these novel EBV transcripts could be classified into three groups (Table 1). Groups 1 and 2 represented EBV fragments at positions 145515-145534 and 147203-147215, respectively. Group 3 contained multiple short EBV fragments with homology to EBERs. Their sequences and cloning frequency are listed in Table 1. Distribution of small RNA in the X2117 library was similar to C666-1, excepted that a higher percentage of human cellular miRNA was found in X2117 (Table W1). On the whole, the most abundant EBV miRNA cloned were miR-BART9 (n = 467) and miR-BART10 (n = 251). However, miR-BART15 and miR-BART20 were not detected in our study. In fact, our previous study suggested that miR-BART15 was undetectable by Northern blot in all native infected, reinfected epithelial cell lines as well as in HeLa cells overexpressing EBV RNA transcripts from the C666-1 strain [12]. Nevertheless, Cosmopoulos et al. [22] have recently reported the detection of BART15 and BART20 expression in C666-1 cell using real time RT-PCR method.
Table 1.
Sequence and Genomic Location (AJ507799) of EBV Encode RNA Not Matching to Known miRNA.
| RNA Fragment | Sequence (5′–3′) | Length | C666-1 | X2117 | Position | |
| Group 1 | miR-BART21 | CACUAGUGAAGGCAACUAAC | 20 | 1 | 0 | 145515:145534 |
| Group 2 | miR-BART22 | UUACAAAGUCAUGGUCUAGUA(GU)* | 21–23 | 14 | 1 | 147203:147215 |
| Group 3 | EBER1–5′ | (A)AGGACCUACGCUGCCCU(A) | 17–19 | 8 | 0 | 6629:6646 |
| EBER1 | GACUCUGCUUUCUGCCGU | 18 | 1 | 0 | 6742:6759 | |
| EBER1 | GGGAGGUUCUCUCGGGGCCACGCC | 24 | 1 | 0 | 6659:6676 | |
| EBER1 3′ | (UACC)AGCUGGUGGUCCGCAUGUUU | 20–24 | 5 | 1 | 6772:6794 | |
| EBER2 | UAGUGGUUUCGGACACAC | 18 | 1 | 0 | 6972:6989 | |
| EBER2 | GAGAAGGGUAUUCGGCUUGU | 20 | 2 | 0 | 7098:7117 | |
| EBER2 | AGGACAGCCGUUGCCCUAGUGGUUU | 25 | 0 | 1 | 6956:6980 |
Sequence variations of the cloned fragments are indicated by parentheses.
Of 15 sequences, 2 had two nucleotides shorter.
To establish whether some of the isolated RNA fragments could represent potential novel ebv-miRNA, putative precursor sequences with sizes of around 70 nucleotides contained the cloned fragments from the EBV genome were subjected to structure predictions using the MFOLD program [17]. A precursor miRNA (pre-miRNA) hairpin structure was produced from each of the putative precursor sequences from Groups 1 and 2 EBV fragments, which now named miR-BART21 and miR-BART22, respectively (Figure 1A).
The expressions of miR-BART21 and miR-BART22 in EBV-infected NPC tumors were validated by Northern blot. The cell line C666-1, three xenografts (X666, X2117, C15) and two primary tumor biopsies were examined. The EBV-negative immortalized normal nasopharyngeal epithelial cell, NP460, was included as negative control. EBV-positive lymphoid cell lines Akata, Namalwa, and Raji were also examined for comparison. Expression of miR-BART21 was detected in C666-1, X666, X2117, and one of the two primary NPC biopsies (Figure 1B, upper panel). In contrast, miR-BART22 was expressed in all EBV-positive cells and particularly highly in NPC samples (Figure 1B, lower panel). These results suggested that the two novel EBV miRNA were preferentially expressed inNPC cells. Because only a small amount of RNA could be isolated from the primary NPC biopsies, we designed a sensitive QRT-PCR assay for the detection of these novel EBV miRNA in tissue specimens. The relative expression levels of miR-BART21 and miR-BART22 in cell lines and xenografts as measured by QRT-PCR were similar to those determined by Northern blot analysis (Figure 1B). By using QRT-PCR, we confirmed high and consistent expression of these two EBV miRNA in primary NPC tumors (Table 2).
Table 2.
Expression of Novel ebv-miRNA and LMP2A in NPC Samples.
| Sample | BART21 | BART22 | LMP2A mRNA* | LMP2A IHC† |
| C666-1 | 100 | 100 | 100 | - |
| NPC1 | 9.17 | 36.39 | <1 | - |
| NPC2 | 52.57 | 123.18 | 226 | - |
| NPC3 | 31.85 | 41.07 | 242 | - |
| NPC4 | 26.97 | 18.85 | 258 | - |
| NPC5 | 47.44 | 76.54 | 257 | - |
| NPC6 | 3.15 | 2.06 | <1 | + |
| NPC7 | 29.36 | 39.17 | 233 | - |
| NPC8 | 25.81 | 28.05 | 335 | + |
| NPC9 | 8.71 | 30.68 | 264 | - |
| NPC10 | 61.72 | 64.04 | NA | - |
| NPC11 | 12 | 55.2 | 183 | + |
| NP1 | 0 | 0 | 0 | - |
| NP2 | 0 | 0 | 0 | - |
| NP3 | 0 | 0 | 0 | - |
Expression level of C666-1 was set at 100.
NA indicates not analyzed.
Expression level was normalized to EBNA1.
LMP2A-negative (-) or-positive (+) were indicated.
Nucleotide Polymorphism Is Important for Drosha Processing of miR-BART22
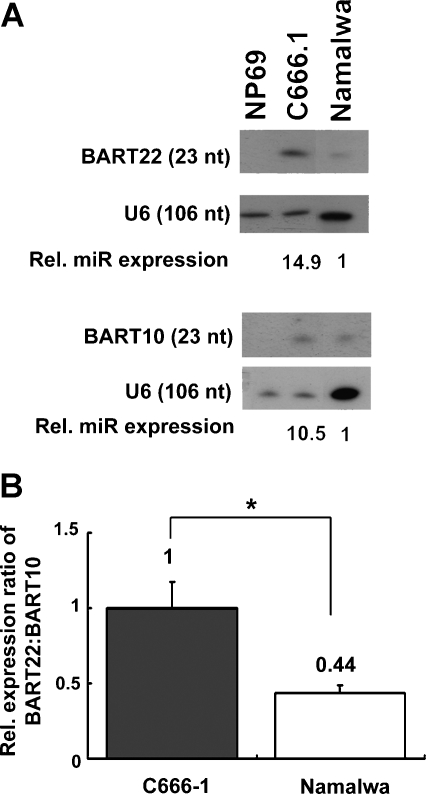
Because pre-miR-BART10 (AJ507799; 147304-147380) and pre-miR-BART22 (147161-147231) are in close proximity in the EBV genome, miR-BART10 and miR-BART22 are thought to share the same RNA transcript in miRNA biogenesis. However, the expression ratio of miR-BART22 to miR-BART10 was significantly higher in C666-1 than Namalwa cells (P < .05) in both Northern blot analysis and QRT-PCR analysis (Figure 2). This observation suggests that the biogenesis of miR-BART10 and miR-BART22 differs between these two cell types. To evaluate if nucleotide variation(s) plays a role in the biogenesis of these novel miRNA, the flanking sequences of miR-BART21 and miR-BART22 from C666-1 (EU828629) and Raji (AJ507799) were compared. We deployed the Raji-EBV sequence for analysis because Raji is commonly accepted as reference EBV sequence. On the basis of matching analysis, we found two nucleotide variations in miR-BART21 and four nucleotide changes in miR-BART22 (Figure 3A).
Figure 2.
miR-BART10 and miR-BART22 expression in C666-1 and Namalwa cells. (A) Northern blot analysis demonstrates that the expression of miR-BART22 is significantly higher than miR-BART10 in C666-1 (P < .05) but not in Namalwa cells. Representative results from three independent experiments are shown. The relative expression level for each miRNA is calculated with reference to Namalwa (set as 1) after normalization with U6 level. (B) Expression ratio of miR-BART22 to miR-BART10 is significantly higher in C666-1 than in Namalwa cells as measured by QRT-PCR (P < .05). The expression of miR-BART22 is compared with miR-BART10 after normalization with EBNA1. The results shown are the mean ± SD from three independent experiments.
Figure 3.
The nucleotide polymorphisms in pri-miR-BART-21 and 22 from C666-1-EBV. (A) The nucleotide variations in pri-miR-BART21 (AJ507799, 145435:145614; left) and pri-miR-BART22 (147126:147305; right) are shown in shaded color. The mature miRNA sequences are shown in gray boxes. (B) The predicted secondary structures of C666-1-EBV miR-BART22 (EU828629, 8087:8267; left), Raji-EBV miR-BART22 (AJ507799, 147137:147456; middle), and the two nucleotide variations of Raji-EBV, Raji-EBV-2nt mut (147144 A > T and 147146 C > A; right) are shown. The folding energy (dG) with units (kcal/mol) is indicated. (C) In vitro Drosha/DGCR8 processing of the pri-miR-BART22 transcript. C666-1-EBV and Namalwa-EBV pri-miR-BART22 transcripts (pri-RNA) (AJ507799, 147137:147456) were in vitro-transcribed and incubated with bead-bound Flag-tagged Drosha/DGCR8 complex (+) or beads containing empty vector transfected cell extract (-). The digested products were separated on 8% PAGE for analysis (left). The presence of pre-miR-BART22 with a size around 70 nt was confirmed by Northern blot with a miR-BART22 complementary oligonucleotide probe (right). The representative result from at least three individual blots is shown in panel C.
By MFOLD analysis, the predicted secondary structures of the miR-BART21 primary transcript (∼300 nucleotides; pri-miR-BART21) from Raji-EBV and C666-EBV strains were highly similar. In contrast, the secondary structure predicted for miR-BART22 was dissimilar between the Raji-EBV and C666-EBV sequences. Folding predictions for pri-miR-BART22 (147137:147456) from Raji-EBV suggested the presence of a small side-branched stem-loop adjacent to the mature miR-BART22 sequence (Figure 3B, left panel). However, this stem-loop was not predicted for the C666-EBV derived miR-BART22 sequence (Figure 3B, middle panel). It is plausible that the nucleotide variations found in the C666-EBV associated miR-BART22 underlie the distinct structural difference predicted between the EBV strains. Although all four nucleotide variations in miR-BART22 are positioned distal from the hairpin structure, changes of two nucleotides (147144 A > T and 147146 C > A) could readily affect the stem-loop formation (Figure 3B, right panel). Remarkably, these two specific nucleotide variations were identified in all 17 primary tumors tested from Hong Kong NPC patients (Figure W1). According to the “junction anchoring” model proposed for pri-miRNA processing [23], formation of the small-side stem-loop could impair miRNA maturation by concealing the DGCR8 recognition site during pri-miRNA processing. To elucidate whether this nucleotide polymorphism of pri-miR-BART22 affects miRNA maturation, we examined the digestion efficiency of Drosha/DGCR8 enzymatic complex by incubating immunoprecipitated flag-tagged Drosha/DGCR8 with in vitro-transcribed EBV RNA substrates (147137-147456) from C666-1 and Namalwa EBV genome, which share the same polymorphism as Raji (Figure W1). PAGE analysis showed digested products of ∼70 bp, which could indicate the presence of either miR-BART10 or miR-BART22 pre-miRNA (Figure 3, B and C). To further confirm that the digested product contained pre-miR-BART22, Northern blot analysis with a complementary miR-BART22 oligonucleotide was carried out on the membrane-transferred PAGE gel. We found that in vitro digestion of pre-miR-BART22 from C666-1 was much facilitated in Drosha/DGCR8 processing compared with Namalwa (Figure 3C). Our findings lead us to hypothesize that nucleotide polymorphisms within the pri-miR-BART22 transcript can augment its maturation in NPC and explain at least in part the varying transcript levels in different EBV strains.
LMP2A Is a Potential Target of miR-BART22
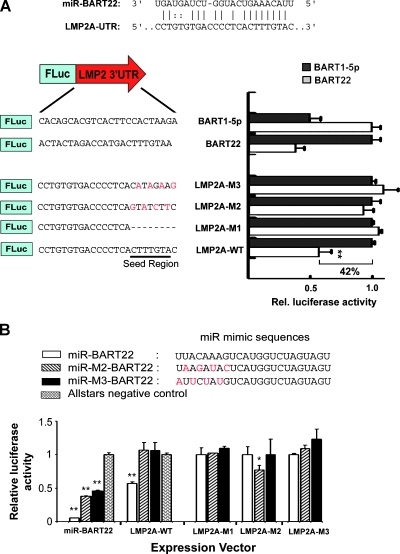
We attempted prediction of potential viral and cellular targets of miR-BART21 and miR-BART22 by MiRanda [24,25] and RNA-hybrid [26] programs using standard parameters. A number of potential cellular mRNA targets were suggested (Table W2), although many failed to be validated by luciferase reporter assay (data not shown). However, we found a putative miR-BART22 binding site in the LMP2A-3′ UTR (AJ507799; 5546:5568). The predicted putative LMP2A-3′UTR target site was highly conserved between different EBV strains (Figure W2). A total of 16 nucleotides including the seed sequences (2–8 nt) of miR-BART22 are perfectly complementary to the target site (Figure 4A). To study the predicted suppressive effect of miR-BART22 on LMP2A, a series of dual luciferase reporter assays were performed in 293FT cells. The reporter constructs contained synthetic oligonucleotides of the predicted or mutated binding sequence cloned into the 3′ UTR of pMIR-REPORT plasmid. Using a miRNA expression vector in reporter assays, we found that miR-BART22 exerted strong inhibitory effect on the LMP2A 3′UTR (42%, P < .001). Nevertheless, repression was eliminated when the complementarities of the seed region were either deleted or mutated, LMP2A-M1-M3 in Figure 4A. This observation suggested that the repressive property of BART22 on the LMP2A 3′UTR was both functional and specific.
Figure 4.
Viral LMP2A is a potential target of miR-BART22. (A) Base complementarity suggests that the putative miR-BART22 target site is in the 3′UTR of LMP2A (AJ507799, 5545:5568; upper). The wild-type (LMP2A-WT), truncated (LMP2A-M1) and mutated (LMP2A-M2 and -M3) LMP2A sequences are shown. The seed-binding region is underlined and the base substitutions a remarked in red (lower). (B) The sequences of miRNA mimics are displayed. The base substitutions made to restore seed mutant complementarity are labeled in red (miR-M2-BART22 and miR-M3-BART22). The relative firefly luciferase activity was normalized to the renilla luciferase control, and results were taken from at least three independent experiments. Data shown are the mean ± SD. Statistical analysis by Student's t test was used. **P < .001.
To further confirm the importance of seed sequence complementarity in the BART22-LMP2A interaction, we performed additional luciferase assays using LMP2A-M1-M3 reporter plasmids with miR-BART22 and two miRNA mimics, miR-M2-BART22 and miR-M3-BART22, designed to compensate for the mutated seed regions of the LMP2A-M2 and M3 reporters (Figure 4B). The miRNA mimics exerted differing levels of suppression with the BART22 3′UTR, indicating that the miRNA mimics were functionally active. However, in cotransfection with the LMP2A-WT3′UTR reporter, only miR-BART22 was able to successfully reduce translation. Although seed binding may be critical for initiating repression, mutant miR-M3-BART22 with restored complementarity to LMP2A-M3 failed to exert an inhibitory effect on its corresponding mutated reporter. Whereas miR-M2-BART22 significantly inhibited translation of the LMP2A-M2 mutated reporter, it expressed only slightly below the control level and clearly did not exhibit a profound repression ability (Figure 4B). Together, our data suggest that the seed interaction between miR-BART22 and LMP2A-3′UTR is unique and that replacing seed pairing with other complementary sequences yields negligible or only partial suppressive effects. We further investigated whether LMP2A-3′UTR might contain binding sites for other miR-BARTs. By loosening the MiRanda program parameters for prediction, target sites for miR-BART1, 12, 14, and 16 were suggested but none could be successfully validated by luciferase reporter assay (Figure W3). This further suggests the uniqueness of the interaction between miR-BART22 and the LMP2A-3′UTR and thus of the controlled expression of LMP2A.
Differential Expression of LMP2A in NPC
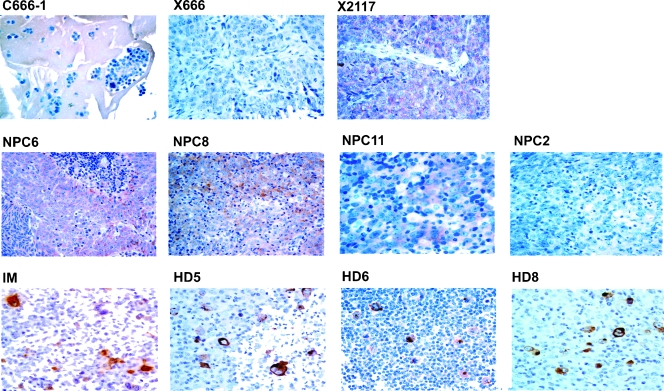
To investigate whether LMP2A is commonly expressed in NPC, we performed both RT-PCR and Western Blot on a panel of NPC samples from Southern China. Although the LMP2A RNA transcript could be detected in C666-1 cells and in NPC xenografts X666 and X2117, none of them showed detectable LMP2A protein levels in Western blots (Figure W4). Nevertheless, we were able to detect weak focally expressed LMP2A in X2117 by IHC. As a control, strong LMP2A staining was demonstrated in an EBV-positive IM and six of the eight HD biopsies. This finding suggested that IHC analysis has a higher sensitivity for detection of LMP2A expression. By IHC, we were able to detect weak LMP2A expression in 6 (23%) of 26 primary NPC tumors (Figure 5). The expression levels of miR-BART22 and LMP2A mRNA have also been determined in ten of these tumors (Table 2). Interestingly, the LMP2A mRNA expression level did not directly correlate with the protein expression, and relatively low expression levels of miR-BART22 miRNA were observed in all three LMP2A-positive primary tumors, as well as in X2117 (NPC6, 8, and 11 in Table 2 and Figure 5). This finding supports the possible regulatory role of miR-BART22 in LMP2A expression.
Figure 5.
LMP2A protein expression in NPC, IM, and HD specimens. Upper panel: EBV-positive NPC cell line and xenografts. LMP2A expression was not detected in NPC cell line C666-1 and xenograft X666. Only weak and focally LMP2A expression was detected in xenograft X2117. Middle panel: Primary NPC biopsies. Weak expression of LMP2A was noted in three NPC samples (NPC6, 8, 11). A negative example (NPC2) was included. Lower panel: IM and HD. Strong LMP2A expression was detected in IM. Three positive examples of HD were included.
Suppression of LMP2A Protein Expression by miR-BART22
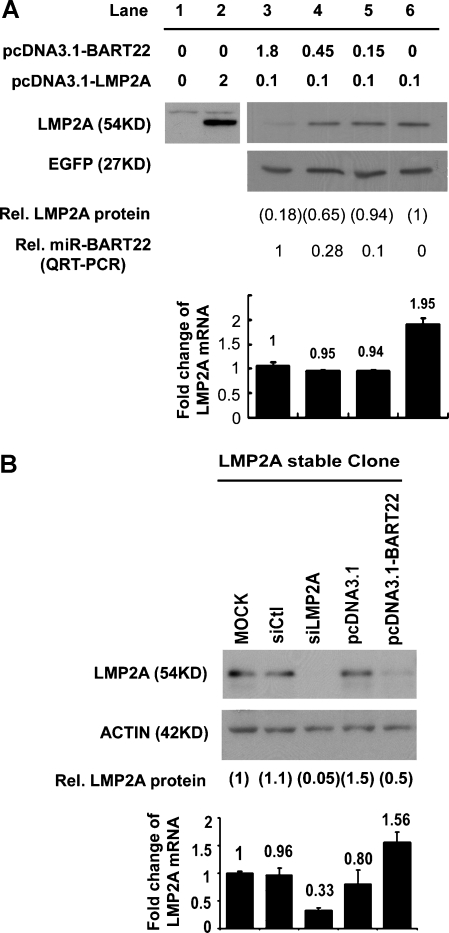
To establish the strong interaction between miR-BART22 and the LMP2A-3′UTR, two supportive experiments were designed. First, the dose effect of miR-BART22 on LMP2A expression was studied by cotransfection of different amounts of miR-BART22 with the LMP2A expression vector that included the full length of LMP2A 3′UTR. MiR-BART22 suppressed the LMP2A protein level in a dose-dependent manner without an apparent effect on LMP2A mRNA levels (Figure 6A). MiR-BART22 expression had also no obvious effect on the EGFP control protein. Second, transfection of miR-BART22 into HEK293 cells that had been stably transfected with pcDNA3.1-LMP2A (Figure 6B) readily suppressed levels of the LMP2A protein. The transfection again had no significant effect on LMP2A mRNA expression. These results strongly suggest that LMP2A is a direct target of miR-BART22, which specifically represses LMP2A expression at the posttranscriptional level.
Figure 6.
Suppression of LMP2A protein expression by miR-BART22. (A) Western blot of LMP2A and EGFP in 293FT cells transiently cotransfected with 0.1 µg per well pEGFP-C1 plasmid in six-well plate format, the amount of expression plasmids as indicated (lanes 3–6), and 2 µg of total DNA made up with pcDNA3.1 as carrier. LMP2A expression level was normalized to EGFP. miR-BART22 and LMP2A RNA expression levels in the same sample were analyzed by QRTPCR and are shown relative to lane 3. (B) The representative Western blot results indicate that miR-BART22 represses LMP2A expression in HEK293-LMP2A stable clones. The stable clones transfected with the siRNA control (siCtl) and LMP2A specified siRNA (siLMP2A) were included as controls. The LMP2A expression level was normalized to actin and the level relative to mock transfection (set at 1) was calculated. LMP2A mRNA expression in the same sample was assayed by QRT-PCR.
Discussion
Besides the production of viral latent proteins, it is believed that EBV contributes to NPC development through expressing abundant noncoding viral RNA, including EBERs and miRNA. In this study, we report the identification of two new ebv-miRNA, miR-BART21 and miR-BART22, from screening 2928 clones by traditional small RNA library cloning method in the NPC cell line and xenograft. These two newly identified miRNA are consistently expressed in NPC cells and primary tissues. In line with our finding, Zhu et al. [27] have recently reported the identification of several novel EBV miRNA by massive sequencing of 47,000 clones that were derived from two NPC biopsies. We note that two identified EBV miRNA from their study, ebv-miR-BART21-3p and ebv-miR-BART22, are identical in sequence to our newly discovered miR-BART21 and miR-BART22, respectively. Our study clearly establishes the relevance of miR-BART21 and miR-BART22 in NPC and demonstrates the feasibility of direct cloning of small cDNA libraries as an efficient approach for novel discovery of viral miRNA. Nevertheless, the study by Meister's group failed to address the prevalence and functional implications of miR-BART21 and miR-BART22 in NPC. In this study, we demonstrate the presence of a strain specific EBV polymorphism that predisposes to the enhanced expression of miR-BART22.
Of the two new ebv-miRNA, miR-BART22 was found to be highly expressed in almost all NPC samples studied. We found polymorphisms in EBV strains from Southern China, especially at nt147144 A > T and nt147146 C > A, which favor the biogenesis of miR-BART22 and thus its expression level (Figure 3). Although the LMP2A transcript was previously reported to be detected in most NPC samples [20,28,29], our IHC data indicate only 23% of primary NPC tumors from our locality have detectable LMP2A protein expression. Transcriptional regulation of LMP2A in EBV-infected cells by epigenetic and viral latent protein mechanisms has been reported previously [30–32]. In this study, we showed that LMP2A can also be modulated at the translational level by miR-BART22. Moreover, whereas seed complementarity of the LMP2A-3′UTR with other known ebv-miRNA has been suggested, we have not been able to confirm target association in a reporter assay (Figure W3), implicating a unique role of miR-BART22 in the translational regulation of LMP2A. We were able to demonstrate LMP2A protein expression in NPC biopsies, which tend to have relatively low expression of miR-BART22 (Table 2). However, not all biopsies with low miR-BART22 expression have detectable LMP2A, indicating that LMP2A expression might also be regulated by other pathways. Such multiple regulatory mechanisms have also been implicated in LMP1 modulation [12].
Previous reports suggested that expression of LMP2A can induce AKT activity in C666-1 cells [33] and activate the Notch signaling pathway in epithelial cells [34]. We showed that transient transfection of miR-BART22 in LMP2A-expressing HEK293 stable clones could suppress Notch-1 mRNA expression and mildly suppress AKT activation (Figure W5). Although suppression of both Notch and AKT pathways may not be in favor of cancer cell growth, forced expression of miR-BART22 in normal epithelial cells did not induce any change in cell proliferation (unpublished data). These observations are consistent with the recent finding that overexpression of LMP2A in C666-1 cells did not affect cell growth and migration, although it can activate the PI3K/Akt pathway [33]. Thus, we believed that down-regulation of LMP2A by miR-BART22 may contribute to cancer development by other means, such as permitting the escape of infected cells from the host immune surveillance.
The potential benefits of suppressing LMP2A expression during NPC development can be many. Previous reports showed that NPC cells are capable of processing and presenting endogenously synthesized proteins to CTLs. LMP2A, in particular, has a stronger immunogenicity than two other NPC expressed viral genes, namely, EBNA1 and LMP1 [29,35–37]. In this regard, limiting LMP2A protein expression would have potential advantages for NPC cells to escape host immune surveillance, and thus, LMP2A expression in NPC is predictably low. In fact, immunomodulatory effects of other ebv-miRNA have recently been demonstrated. For example, down-regulation of LMP1 expression by miR-BARTs may favor immune escape by decreasing the antigen-processing function of NPC cells [12]. In primary effusion lymphoma, miR-BHRF1-3 can target CXCL-11/I-TAC, an IFN-inducible T-cell attracting chemokine [38]. Furthermore, miR-BART2-5p can repress the expression of the cellular stress-induced immune molecule, MICB [39]. Apart from immunogenicity, LMP2A, as opposed to LMP1, could suppress nuclear factor-κB levels resulting in an antiproliferative effect [40]. Moreover, LMP2A-expressing epithelial cells could also inhibit telomerase reverse transcription activity, an enzyme important for cell immortalization and transformation [41]. Because LMP2A has diverse functional roles in epithelial cells, its expression level is therefore required to be tightly regulated during the development of NPC.
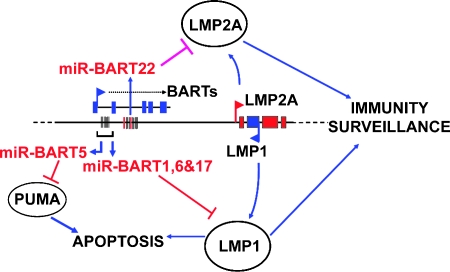
Our group has previously reported that miR-BARTs regulate LMP1 expression [12] and that miR-BART5 affects the expression of the cellular target gene PUMA [13]. In this study, we further identified miR-BART22 as a modulator of an important oncogenic and immunogenic viral gene, LMP2A (Figure 7). In concordance with our previous reports, we have thus provided evidence for the vital roles of EBV-encoded miRNA in regulating oncogenic and immunogenic viral latent protein expression, which holds importance in the progression and survival of EBV-infected NPC.
Figure 7.
Function of ebv-BART-miRNA on host cell survival. miR-BART1, 16, and 17 can regulate the viral LMP1 protein expression [12] and miR-BART5 can alter the expression of the p53-upregulated modulator of apoptosis (PUMA) [13]. And in this article, miR-BART22 is demonstrated as a modulator of the viral gene, LMP2A.
Supporting Information (SI)
SI Methods
Plasmid constructs
The miRNA expression plasmids were made by inserting the PCR product that contains miRNA flanking sequence (∼300 nt) into pcDNA3.1 expression vector through Hind III and either XbaI or XhoI sites. All the PCR products were generated using C666-1 DNA as template, and the primer sequence used for PCR amplification are listed below:
BART1-F: 5′-GGTAAGCTTATGCTGCTGGTGT-3′;
BART1-R: 5′-GGCTCTAGATGGTCATGTTTCCCT-3′;
BART14-F: 5′-GGTAAGCTTGGACGGCTGAC-3′;
BART14-R: 5′-GGCTCTAGAAAAGGCCTGCTGT-3′;
BART16-F: 5′-GGTAAGCTTCTGATGCTCTGTGG-3′;
BART16-R: 5′-GGCTCTAGATGGATTGGACCAAC-3′;
BART21-F: 5′-AGCCAAGCTTGCTGGGCAGAGAATGTTTGT- 3′;
BART21-R: 5′-ACCGCTCGAGTAAGGGGAGGGGAAAGCTAAA- 3′;
BART22-F: 5′-AGCCAAGCTTACTTCATGGGTCCCGTAGTG- 3′;
BART22-R: 5′-ACCGCTCGAGCCACACTGCTAAGGCAGTCA- 3′.
The pcDNA3.1-LMP2A expression plasmid was constructed by inserting 2 kb of RT-PCR product that included a complete LMP2A ORF and 3′UTR into pcDNA3.1 expression vector through EcoRI and XbaI sites. The RT-PCR product was amplified from RNA in B95-8 cells by using the LMP2A-F primer: 5′-ACG GAA TTC TGC TGC AGC TAT G-3′ (EBV coordination: 166092-166105) and LMP2A-R primer: 5′-CGT TCT AGA GCA CAT TGG GTT TAT TGG-3′ (EBVcoordination: 5840-5856). The presence of LMP2A coding sequence and the complete 3′UTR was confirmed by sequencing.
Construction of firefly luciferase reporter vector was described previously [1]. In brief, two micrograms of sense (S) and antisense (AS) oligonucleotides that encoded a single-miRNA tested target were annealed in 30 mM HEPES buffer (pH 7.4) containing 100 nM potassium acetate and 2 mM magnesium acetate and cloned down-stream of the cytomegalovirus (CMV) promoter-driven firefly luciferase cassette in a pMIR-REPORT vector (Ambion, Inc) through SpeI and HindIII sites. The sequences of oligonucleotides used for cloning are listed as below:
T1-S: 5′-CTAGTGATCGCCTGCCACTTCCACAGCAA-3′
T1-AS: 5′-AGCTTTGCTGTGGAAGTGGCAGGCGATCA-3′
T2-S: 5′-CTAGTAACCCCACGGAGCAGGGCAACATTGCAGGGA- 3′
T2-AS: 5′-AGCTTCCCTGCAATGTTGCCCTGCTCCGTGGGGTTA-3′
T3-S: 5′-CTAGTAGACTATGCATACACTGAATTTAGA-3′
T3-AS: 5′-AGCTTCTAAATTCAGTGTATGCATAGTCTA-3′
T4-S: 5′-CTAGTAGACCTGTGTGCTGTATTTAA-3′
T4-AS: 5′-AGCTTTAAATACAGCACACAGGTCTA-3′
BART12-S: 5′-CTAGTAACCACACCAAACACCACAGGAA-3′
BART12-AS: 5′-AGCTTTCCTGTGGTGTTTGGTGTGGTTA-3′
BART14-S: 5′-CTAGTATCCCTACTACTGCAGCATTTAA-3′
BART14-AS: 5′-AGCTTTAAATGCTGCAGTAGTAGGGATA-3′
BART16-S: 5′-CTAGTAGAGCACACACCCACTCTATCTAAA-3′
BART16-AS: 5′-AGCTTTTAGATAGAGTGGGTGTGTGCTCTA-3′
BART21-S: 5′-CTAGTGTTAGTTGCCTTCACTAGTGA-3′
BART21-AS: 5′-AGCTTCACTAGTGAAGGCAACTAACA-3′
BART22-S: 5′-CTAGTACTACTAGACCATGACTTTGTAAA-3′
BART22-AS: 5′-AGCTTTTACAAAGTCATGGTCTAGTAGTA-3′
BART1-5p-S:5′-CTAGTCACAGCACGTCACTTCCACTAAGAA-3′
BART1-5p-AS: 5′-AGCTTTCTTAGTGGAAGTGACGTGCTGTGA-3′
LMP2A-WT-S: 5′-CTAGTCCTGTGTGACCCCTCACTTTGTACA-3′
LMP2A-WT-AS:5′-AGCTTGTACAAAGTGAGGGGTCACACAGGA-3′
LMP2A-M1-S: 5′-CTAGTCCTGTGTGACCCCTCAA-3′
LMP2A-M1-AS: 5′-AGCTTTGAGGGGTCACACAGGA-3′
LMP2A-M2-S: 5′-CTAGTCCTGTGTGACCCCTCAGTATCTTCA-3′
LMP2A-M2-AS: 5′-AGCTTGAAGATACTGAGGGGTCACACAGGA-3′
LMP2A-M3-S: 5′-CTAGTCCTGTGTGACCCCTCACATAGAAGA-3′
LMP2A-M3-AS: 5′-AGCTTCTTCTATGTGAGGGGTCACACAGGA-3′
Transfection for luciferase analysis
Cells grown in 24-well plates were cotransfected with miRNA and reporter construct for analysis. All miRNA expression plasmid transfection experiments were carried out by GeneJuice transfection reagent (Novagen). Transfection complex containing 50 ng of reporter plasmid, 2 ng of pRL-CMV-control reporter plasmid, and 200 ng of indicated miRNA expression plasmid was prepared for each transfection. For miRNA mimic transfection, a mixture containing 50 nM of miRNA mimic, 800 ng reporter vector, and 80 ng control reporter vector was prepared with Lipofectamine 2000 (Invitrogen). The luciferase activities were assayed by Dual Luciferase Reporter Kit (Promega). Three separate experiments were performed, and Student's t test was used for statistical analysis. P < .05 was considered as statistically significant.
Others oligonucleotide sequences used in this study Northern blot:
BART10 NB: 5′-TGTACAGAACCAAAGAGGTGGC-3′
BART21 NB: 5′-GTTAGTTGCCTTCACTAGTG-3′
BART22 NB: 5′-ACTACTAGACCATGACTTTGTAA-3′
U6NB: 5′-GCAGGGGCCATGCTAATCTTCTCTGTATCG-3′
In vitro transcription PCR:
T7-C666-BART22-F: 5′-TAATACGACTCACTATAGGGCTAATATCA-3′
T7-Nam-BART22-F: 5′-TAATACGACTCACTATAGGGCTAATAACC-3′
T7-BART22-R: 5′-CCCAAGGCAGGTAAACATTG-3′
RT-PCR for LMP2A and actin:
LMP2A-Exon 1-F: 5′-CGGGATCACTCATCTGAACACATA-3′
LMP2A-Exon 3-R: 5′-CATGTTAGGCAAATTGCAAA-3′
Actin-207F: 5′-TAAGGAGAAGCTGTGCTACGTC-3′
Actin-207R: 5′-GGAGTTGAAGGTAGTTTCGTGG-3′
QRT-PCR (miRNA):
BART10: 5′-TACATAACCATGGAGTTGGCTGT-3′
BART21: 5′-CACTAGTGAAGGCAACTAAC-3′
BART22: 5′-TTACAAAGTCATGGTCTAGTAGT-3′
snU6: 5′-ACGCAAATTCGTGAAGCGTT-3′
QRT-PCR (SYBR Green):
Actin 69F: 5′-CTGGCACCCAGCACAATG-3′
Actin 69R: 5′-GCCGATCCACACGGAGTACT-3′
EBNA1-1162F: 5′-TCATCATCATCCGGGTCTCC-3′
EBNA1-1229R: 5′-CCTACAGGGTGGAAAAATGGC-3′
EGFP 82F: 5′-CGACAACCACTACCTGAGCA-3′
EGFP 82R: 5′-GAACTCCAGCAGGACCATGT-3′
Notch 1-F: 5′-CCGCAGTTGTGCTCCTGAA-3′
Notch 1-R: 5′-ACCTTGGCGGTGTCGTAGCT-3′
QRT-PCR for LMP2A [2]:
LMP2A-Exon 1-F: 5′-CGGGATCACTCATCTGAACACATA-3′
LMP2A-Exon 2-R: 5′-GGCGGTCACAACGGTACTAACT-3′
TaqMan Probe: (FAM)-CAGTATGCCTGCCTGTAATTGTTGCG-(TAMRA)
Acknowledgments
The authors thank Joan A. Steitz (Yale University), Kasandra Riley (Yale University), Steve P. Lee (University of Birmingham), and Alan B. Rickinson (University of Birmingham) for comments and suggestions for the manuscript; and Nathalie Wong (CUHK) for critical reading of the manuscript.
Footnotes
This research was supported by Hong Kong Government Research Fund for the Control of Infectious Diseases (Project Code 06060372 and 07060242) and in part by UGC Collaborated Research Fund (CUHK04/CRF/08).
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 to W5 and are available online at www.neoplasia.com.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5:423–428. doi: 10.1016/s1535-6108(04)00119-9. [DOI] [PubMed] [Google Scholar]
- 3.Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol. 2002;12:431–441. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- 4.Karran L, Gao Y, Smith PR, Griffin BE. Expression of a family of complementary-strand transcripts in Epstein-Barr virus-infected cells. Proc Natl Acad Sci USA. 1992;89:8058–8062. doi: 10.1073/pnas.89.17.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hitt MM, Allday MJ, Hara T, Karran L, Jones MD, Busson P, Tursz T, Ernberg I, Griffin BE. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H, Lee JM, Wang Y, Huang DP, Ambinder RF, Hayward SD. The Epstein-Barr virus latency BamHI-Q promoter is positively regulated by STATs and Zta interference with JAK/STAT activation leads to loss of BamHI-Q promoter activity. Proc Natl Acad Sci USA. 1999;96:9339–9344. doi: 10.1073/pnas.96.16.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 9.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen BR. Viral and cellular messenger RNA targets of viral microRNAs. Nature. 2009;457:421–425. doi: 10.1038/nature07757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2009;37:1035–1048. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci USA. 2007;104:16164–16169. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Guasparri I, Bubman D, Cesarman E. EBV LMP2A affects LMP1-mediated NF-kappaB signaling and survival of lymphoma cells by regulating TRAF2 expression. Blood. 2008;111:3813–3820. doi: 10.1182/blood-2007-03-080309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuker M. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell AI, Groves K, Kelly GL, Croom-Carter D, Hui E, Chan AT, Rickinson AB. Analysis of Epstein-Barr virus latent gene expression in endemic Burkitt's lymphoma and nasopharyngeal carcinoma tumour cells by using quantitative real-time PCR assays. J Gen Virol. 2006;87:2885–2890. doi: 10.1099/vir.0.81906-0. [DOI] [PubMed] [Google Scholar]
- 19.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Heussinger N, Buttner M, Ott G, Brachtel E, Pilch BZ, Kremmer E, Niedobitek G. Expression of the Epstein-Barr virus (EBV)-encoded latent membrane protein 2A (LMP2A) in EBV-associated nasopharyngeal carcinoma. J Pathol. 2004;203:696–699. doi: 10.1002/path.1569. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cosmopoulos K, Pegtel M, Hawkins J, Moffett H, Novina C, Middeldorp J, Thorley-Lawson DA. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J Virol. 2009;83:2357–2367. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 24.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. Micro-RNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grasser F, Meister G. Identification of novel Epstein-Barr virus miRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83:3333–3341. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busson P, McCoy R, Sadler R, Gilligan K, Tursz T, Raab-Traub N. Consistent transcription of the Epstein-Barr virus LMP2 gene in nasopharyngeal carcinoma. J Virol. 1992;66:3257–3262. doi: 10.1128/jvi.66.5.3257-3262.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks L, Yao QY, Rickinson AB, Young LS. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol. 1992;66:2689–2697. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerle B, Koroknai A, Fejer G, Bakos A, Banati F, Szenthe K, olf H, Niller HH, Minarovits J, Salamon D. Acetylated histone H3 and H4 mark the upregulated LMP2A promoter of Epstein-Barr virus in lymphoid cells. J Virol. 2007;81:13242–13247. doi: 10.1128/JVI.01396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson LJ, Longnecker R. An auto-regulatory loop for EBV LMP2A involves activation of Notch. Virology. 2008;371:257–266. doi: 10.1016/j.virol.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez AF, Rosales C, Lopez-Nieva P, Grana O, Ballestar E, Ropero S, Espada J, Melo SA, Lujambio A, Fraga MF, et al. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 2009;19:438–451. doi: 10.1101/gr.083550.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shair KH, Schnegg CI, Raab-Traub N. EBV latent membrane protein 1 effects on plakoglobin, cell growth, and migration. Cancer Res. 2008;68:6997–7005. doi: 10.1158/0008-5472.CAN-08-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson LJ, Longnecker R. Epstein-Barr virus latent membrane protein 2A exploits Notch1 to alter B-cell identity in vivo. Blood. 2009;113:108–116. doi: 10.1182/blood-2008-06-160937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khanna R, Busson P, Burrows SR, Raffoux C, Moss DJ, Nicholls JM, Cooper L. Molecular characterization of antigen-processing function in nasopharyngeal carcinoma (NPC): evidence for efficient presentation of Epstein-Barr virus cytotoxic T-cell epitopes by NPC cells. Cancer Res. 1998;58:310–314. [PubMed] [Google Scholar]
- 36.Lee SP, Chan AT, Cheung ST, Thomas WA, CroomCarter D, Dawson CW, Tsai CH, Leung SF, Johnson PJ, Huang DP. CTL control of EBV in nasopharyngeal carcinoma (NPC): EBV-specific CTL responses in the blood and tumors of NPC patients and the antigen-processing function of the tumor cells. J Immunol. 2000;165:573–582. doi: 10.4049/jimmunol.165.1.573. [DOI] [PubMed] [Google Scholar]
- 37.Leen A, Meij P, Redchenko I, Middeldorp J, Bloemena E, Rickinson A, Blake N. Differential immunogenicity of Epstein-Barr virus latent-cycle proteins for human CD4(+) T-helper 1 responses. J Virol. 2001;75:8649–8659. doi: 10.1128/JVI.75.18.8649-8659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–385. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Stewart S, Dawson CW, Takada K, Curnow J, Moody CA, Sixbey JW, Young LS. Epstein-Barr virus-encoded LMP2A regulates viral and cellular gene expression by modulation of the NF-kappaB transcription factor pathway. Proc Natl Acad Sci USA. 2004;101:15730–15735. doi: 10.1073/pnas.0402135101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen F, Liu C, Lindvall C, Xu D, Ernberg I. Epstein-Barr virus latent membrane 2A (LMP2A) down-regulates telomerase reverse transcriptase (hTERT) in epithelial cell lines. Int J Cancer. 2005;113:284–289. doi: 10.1002/ijc.20594. [DOI] [PubMed] [Google Scholar]
References
- 1.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135(1):257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Bell AI, Groves K, Kelly GL, Croom-Carter D, Hui E, Chan AT, Rickinson AB. Analysis of Epstein-Barr virus latent gene expression in endemic Burkitt's lymphoma and nasopharyngeal carcinoma tumour cells by using quantitative realtime PCR assays. J Gen Virol. 2006;87(Pt 10):2885–2890. doi: 10.1099/vir.0.81906-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.