Abstract
PURPOSE: We aimed to identify putative predictive protein biomarkers of radioresistance. EXPERIMENTAL DESIGN: Three breast cancer cell lines (MCF7, MDA-MB-231, and T47D) were used as in vitro models to study radioresistance. Inherent radiosensitivities were examined using a clonogenic survival assay. It was revealed that each cell line differed in their response to radiotherapy. These parental breast cancer cell lines were used to establish novel derivatives (MCF7RR, MDA-MB-231RR, and T47DRR) displaying significant resistance to ionizing radiation. Derivative cells were compared with parental cells to identify putative biomarkers associated with the radioresistant phenotype. To identify these biomarkers, complementary proteomic screening approaches were exploited encompassing two-dimensional gel electrophoresis in combination with mass spectrometry, liquid chromatography coupled with tandem mass spectrometry and quantitative proteomics using iTRAQ technology. RESULTS: A large number of potential biomarkers were identified, and several of these were confirmed using Western blot analysis. In particular, a decrease in the expression of the 26S proteasome was found in all radioresistant derivatives when compared with the respective parent cells. Decreased expression of this target was also found to be associated with radioresistant laryngeal tumors (P = .05) in a small pilot immunohistochemical study. CONCLUSIONS: These findings suggest that the 26S proteasome may provide a general predictive biomarker for radiotherapy outcome.
Introduction
Radiotherapy is a common treatment strategy used for the local control of many malignant disorders, including breast cancer, with 40% to 60% of all cancer patients receiving radiation treatment. In addition, radiotherapy is used with potentially curative intent for some early-stage cancers (e.g., laryngeal squamous cell carcinoma). Radiotherapy involves the fractionated delivery of high-energy x-rays, which exert a cytotoxic effect by producing free radicals within target tissues. Such radicals are the source of reactive oxygen species and reactive nitrogen species, which create DNA damage by forming both single-stranded DNA breaks (SSBs) and double-stranded DNA breaks (DSBs). Cellular enzymes are able to perform the efficient repair of SSBs; however, DSBs are more difficult to repair or are irreparable and thus trigger apoptosis. X-rays can also produce damage to individual DNA bases, which increase the complexity of both SSBs and DSBs and their subsequent repair.
Unfortunately, not all patients derive a therapeutic benefit from radiotherapy because cancer cells may be refractory to treatment. This may manifest not only as distant metastatic spread but also as disease recurrence at the primary site. Moreover, radiotherapy is associated with many unpleasant side effects. Some of these adverse effects are serious and are potentially life threatening. In the curative setting, for example in early laryngeal cancer, radiotherapy is administered in multiple fractions during a period of several weeks, and therefore, failure to respond to a course of treatment may also delay the implementation of a more appropriate treatment strategy [1]. During this delay, tumor progression may occur, which may impact significantly on patient survival. The identification of novel biomarkers that correlate with radiotherapy response would not only increase our understanding on the mechanisms of resistance but also allow therapy to be tailored on an individual patient basis. Ultimately, those patients unlikely to respond to radiation treatment would be spared the serious life-threatening side effects for no therapeutic gain. Furthermore, clinicians would be able to select the most appropriate strategy from the onset of treatment. Such biomarkers may also provide information on new drug targets for future therapeutic intervention such as new sensitizing agents to increase the efficacy of radiotherapy.
Many studies have been conducted to further elucidate the mechanisms of radioresistance and identify such predictive biomarkers in human cancers. For example, it has been found that the phosphoinositide-3 kinase pathway is associated with both in vivo and in vitro radioresistance in several human cancers including larynx, uterine cervix, head and neck, bladder, colon, breast, and fibrosarcoma [2–5]. In addition, the association of BCL-2 with radioresistance is well reported [1,6]. However, the mechanisms underlying radioresistance remain largely obscure, and it is advantageous to use global approaches to study such complex phenomena. These techniques allow the analysis of many targets simultaneously to obtain a comprehensive view of complex biologic systems. This allows the expression of individual and/or combinations of previously unknown targets to be associated with particular disease phenotypes.
In this study, we aimed to establish novel cell line derivatives displaying significant radioresistance and to use global proteomic methodologies to identify proteins that may be associated with this phenotype.
Materials and Methods
Cell Lines
Breast cancer cell lines MCF7, MDA-MB-231, and T47D, representing different breast cancer subtypes, were maintained in RPMI 1640 media supplemented with 10% vol/vol fetal calf serum, 2 mM glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. MCF7 and T47D cells represent luminal breast tumors, demonstrating estrogen receptor (ER)-positive and progesterone receptor (PR)-positive phenotypes. In contrast, MDA-MB-231 cells are ER-negative, PR-negative, and HER2-negative and represent “triple-negative,” basal-type breast tumors.
Establishment of Novel Radioresistant Cell Line Derivatives
Novel derivatives displaying significant resistance to radiotherapy (hereafter named MCF7RR, MDA-MB-231RR, and T47DRR) were established from their parental breast cancer cell lines (MCF7, MDA-MB-231, and T47D, respectively) as described below. Cells were seeded at a concentration of 1 x 106 cells in 5 ml of supplemented RPMI 1640 media in a screw-capped vial with a diameter of 18 mm. Irradiation was performed at the Princess Royal Hospital, Hull, UK, by a radiation physicist (A.W.B.) using the 6 MV beam on a linear accelerator. A simple cradle was manufactured, which allowed the screw-capped vial to be suspended inside a water-filled vessel (phantom). The phantom used was a glass jar shaped with flat surfaces such that the water irradiated by the x-ray beam was a simple cuboid volume. The cradle was designed such that the screw-capped vial containing the cells was mounted precisely at the center of the phantom. Given the depth of the water between the phantom surface and the center of the screw-capped vial (5.5 cm), the size of the irradiation beam used (8 cm x 8 cm), the distance from the radiation “source” to the center of the screw-capped vial (100 cm), and the energy of the x-ray beam (6 MV), it was possible to calculate the irradiation time required to give a specific dose to that point. It was assumed that the dose given to any cells at the center of the screw-capped vial represented that imparted to cells throughout its volume. Using this experimental setup, it was possible to deliver consistent doses to the cell samples throughout the course of the study. On the basis of fractionated treatment protocols exploited within the clinic to ensure that a degree of clinical relevance was applied to this in vitro study, each original cell line population received a total of 40 Gy. This was administered in 20 sequential fractions of 2 Gy/wk, allowing the irradiated cell populations a period to recover between exposures. The novel radioresistant derivatives (MCF7RR, MDA-MB-231RR, and T47DRR) were established from the surviving populations of their parental breast cancer cells (MCF7, MDA-MB-231, and T47D, respectively).
Assessment of In Vitro Response to Radiotherapy
A modified colony counting assay was used to measure the in vitro response to ionizing radiation. Parental and derivative cells were irradiated with single fractions of 0, 2, 4, 6, 8, and 10 Gy as previously described. After irradiation, the cells from each vial were seeded at a concentration of 1000 cells per well in triplicate wells in six-well tissue culture plates. The cells were cultured for 12 to 14 days on which, all media were aspirated, cell colonies were fixed with 3:1 methanol/acetic acid, stained with 0.05% Gentian violet, and counted. A colony was defined as a group of 50 cells or more, and plating efficiency was calculated as described elsewhere [7].
Two-dimensional Gel Electrophoresis and Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry
Two-dimensional Gel Electrophoresis (2DE) and peptide mass fingerprinting by matrix-assisted laser desorption/ionization (MALDI) time of flight (TOF) MS were performed as described previously [8]. Briefly, proteins were extracted in 1 ml of isoelectric focusing (IEF) buffer (7 M urea, 2 M thiourea, 4% CHAPS, 50 mM dithiothreitol [DTT], 1% protease inhibitor mix [80-6501-23; GE Healthcare, Bucks, UK], 0.2% 100x Bio-Lyte 3/10 Ampholyte [163-2094; Bio-Rad, Hemel Hempstead, UK], 0.002% bromophenol blue) and quantified using the 2D Quant Kit (80-6483-56; GE Healthcare). Samples were prepared using the 2D Clean-Up Kit (163-2130; Bio-Rad) in accordance with the manufacturer's protocol. Separation in the first dimension was carried out using 11 cm pH 4 to 7 and pH 7 to 10 ReadyPrep immobilized pH gradient (IPG) strips (163-2015 and 163-2019; Bio-Rad). IPG strips were passively rehydrated in the presence of 200 µg of total protein for 16 hours. IEF was performed using the Protean IEF Cell (Bio-Rad) at 250 V for 20 minutes, ramped up to 8000 V for 150 minutes, and then maintained at 8000 V for an additional 20,000 V-h. After equilibration, strips were transferred to the top of 11-cm Criterion gels (345-0105; Bio-Rad) with an 8% to 16% gradient and embedded in 1% hot agarose. Electrophoresis was performed using a Criterion cell (Bio-Rad) at 200 V for 65 minutes after which proteins were visualized with Bio-Safe Coomassie Blue (161-0787; Bio-Rad). Three independent gels were prepared for each cell line to account for experimental variations. Gels were digitized with a GS-800 laser densitometer (Bio-Rad), and image analysis was performed using PDQuest (version 7.2.0; Bio-Rad). Normalization was performed using the total quantity in valid spots method, and the Students t-test (P < .05) was used to identify differentially expressed proteins (at least two-fold change in expression).
Differentially expressed proteins were excised and destained using a series of ammonium bicarbonate/acetonitrile (ACN) washing steps. Proteins were reduced with DTT, alkylated with iodoacetamide (IAA), and dried in a vacuum centrifuge. Gel pieces were rehydrated in 25 µl of 40 mM ammonium bicarbonate/10% ACN containing 25 µg of porcine methylated Trypsin Gold (V5280; Promega, Southampton, UK) and incubated at 37°C overnight. Peptides were subsequently extracted using ACN and 5% formic acid. Peptide samples were spotted onto an anchorchip MALDI target (Bruker Daltonics, Coventry, UK) in a saturated matrix solution (2,5 dihydroxybenzoic acid in 50% ACN with 1% formic acid), and mass spectra were recorded in the reflectron mode on a Bruker Daltonics Reflex IV MALDI-TOF MS equipped with a 337-nm nitrogen laser. Ions were accelerated through a potential of 20 kV into the TOF mass analyzer, and ions within the mass-to-charge (m/z) range of 900 to 2500 were detected. Data were acquired using the Flex Control (version 2.4; Bruker Daltonics) program and processed with Flex Analysis (version 2.4; Bruker Daltonics) and Biotools (version 3.0; Bruker Daltonics). Peaks were deisotoped, and monoisotopic peptide masses were assigned. Contaminating ions from the autolysis of trypsin and keratin were removed. The resultant peptide mass fingerprints were searched against the NCBInr database (containing 4,649,506 sequences and 1,604,431,067 residues) using the MASCOT search engine (www.matrixscience.com) for protein identification. The search was restricted to human taxonomy (191,507 sequences). Fixed carbamidomethyl modifications and variable methionine oxidation modifications were considered. A maximum of one missed tryptic cleavage was considered, and the mass tolerance for the monoisotopic peptide masses was set at ±0.1%. Mowse scores were used to measure the level of significance of each match, whereby scores greater than 64 were considered a significant match (P < .05). In addition, a minimum of 25% sequence coverage was required for each significant match, and the estimated molecular weight and pI information (from gel positioning) were compared with each match to further ensure correct identifications were assigned. Further analysis by tandem mass spectrometry (MS/MS) was performed using the ABI 4800 Plus MALDI-TOF/TOF analyzer (Applied Biosystems, Warrington, UK) as described later.
In-solution Digestion and Liquid Chromatographic Separation
Two hundred micrograms of total protein was precipitated using the 2D Clean-Up Kit (163-2130; Bio-Rad) and resuspended in 100 mM ammonium bicarbonate to a final concentration of 2 mg/ml. Proteins were reduced with DTT and alkylated with IAA. Porcine methylated Trypsin Gold (V5280; Promega) was added at a ratio of 20:1 (wt/wt, protein/enzyme) and incubated overnight at 37°C. The reaction was ceased by the addition of formic acid to a final concentration of 0.1%, and samples were dried in a vacuum centrifuge. Digested peptide mixtures were separated using an Ultimate 3000 Nano liquid chromatography (LC) system (Dionex-LC Packings, Sunnyvale, CA) equipped with a Probot Micro-fraction Collector (Dionex-LC Packings). Peptide samples were resuspended with LC Solvent A (98% water, 2% ACN, 0.05% trifluoroacetic acid [TFA]), and 10 µg of each peptide mixture was injected onto a C18 LC column (C18 PepMap 100, 75-µm ID; 160321; Dionex-LC Packings) for reverse phase separation. The flow rate of the system was 0.25 µl/min, and the gradient used was as follows: 0 to 5 minutes 2% LC Solvent B (10% water, 90% ACN, 0.05% TFA), 5 to 50 minutes 2% to 30% LC Solvent B, 50 to 64 minutes 30% to 60% LC Solvent B, 64 to 65 minutes 60% to 95% LC Solvent B, 65 to 75 minutes 95% LC Solvent B, 75 to 76 minutes 95% to 5% LC Solvent B, and 76 to 90 minutes 5% LC Solvent B. The LC microfractions were mixed with MALDI matrix (2 mg/ml α-cyano-4-hydroxycinnamic acid in 70% ACN and 0.1% TFA containing 10 fmol/µl Glu-Fib as an internal standard) before spotting onto an Opti-TOF LC MALDI target plate (Applied Biosystems) with a speed of one spot per 20 seconds. MS/MS was performed using the ABI 4800 Plus MALDI-TOF/TOF analyzer (Applied Biosystems) as described later.
Quantitative Proteomics Using iTRAQ Technology
Two hundred micrograms of total protein was precipitated using the 2D Clean-Up Kit (163-2130; Bio-Rad) and resuspended in 500 mM triethylammonium bicarbonate buffer with 0.1% sodium dodecyl sulfate (SDS) to a final concentration of 2 mg/ml. Fifty micrograms of each sample was independently reduced using DTT, alkylated using IAA, and digested with trypsin. Digested proteins were labeled with iTRAQ reagents following the manufacturer's instructions (4390812; Applied Biosystems). Briefly, protein samples from parental cells were labeled with iTRAQ Reagent 114, and protein samples from the radioresistant derivatives were labeled with iTRAQ Reagent 115. Labeling was allowed to proceed at room temperature for 1 hour. Labeled samples from each cell line pair were combined, and 10 µg of each combined sample was subjected to strong cation exchange chromatography. Five separate fractions from each cell line pair were collected as previously described and analyzed independently by MS/MS performed using the ABI 4800 Plus MALDI-TOF/TOF analyzer (Applied Biosystems) as described later.
MS/MS
MS/MS was performed using the ABI 4800 Plus MALDI-TOF/TOF analyzer (Applied Biosystems). Tryptic digests from 2DE analyses were reconstituted with 50% ACN with 1% formic acid, prepared with MALDI matrix (5 mg/ml α-cyano-4-hydroxycinnamic acid in 50% ACN with 0.1% TFA), and spotted onto an Opti-TOF 384-well MALDI plate (Applied Biosystems).
Samples on the Opti-TOF 384-well MALDI plate and the Opti-TOF LC MALDI plate were analyzed using an ABI 4800 Plus MALDI-TOF/TOF analyzer (Applied Biosystems) equipped with a Nd:YAG laser. For MS analyses, 1000 shots were accumulated per spectrum, and ions within the m/z range of 1000 to 4000 were detected. For MS/MS analyses, 4000 shots were accumulated per spectrum using stop conditions. A maximum of 10 precursors per spot were selected for MS/MS analyses and 12 precursors per spot were selected for iTRAQ analyses; exclusion lists were created to ensure trypsin autolysis peaks, and matrix clusters and Glu-Fib peaks (for LC-based analyses) were not selected for sequencing. The GPS Explorer Software (Applied Biosystems) was used to create and search combined MS and MS/MS ion files (for 2DE-based analyses) or MS/MS ion files (for LC-based analyses) against the NCBInr database (containing 2,367,365 sequences and 802,797,248 residues) using the MASCOT search engine for peptide and protein identification. The search was restricted to human taxonomy (129,848 sequences). Fixed carbamidomethyl modifications and variable methionine oxidation modifications were considered. The mass tolerance for monoisotopic peptide masses was set at ±0.1% (for 2DE-based analyses) and ±50 ppm (for LC-based analyses), and the mass tolerance for peptide fragments was set at ±0.3 Da. A maximum of one missed tryptic cleavage was also considered. For 2DE-based analyses, mowse scores of greater than 64 were significant (P < .05). A minimum of 25% sequence coverage was required for each significant match, and the estimated molecular weight and pI information (from gel positioning) were compared with each match to further ensure correct protein identification. For LC-based analyses, a minimum total ion confidence score (CI) of greater than 98% was required for protein identification.
Expression Microarray Analysis
Expression microarray (MA) slides were constructed using the Array-Ready Human Cancer Subset v3.0 (806103; Operon, Ebersberg, Germany). The subset consisted of 2876 oligonucleotides (70-mer) with 12 control housekeeping genes designed from the UniGene Database Version Hs 147 and the Human Reference Sequence (RefSeq) Database (www.ncbi.nlm.nih.gov). A QArrayMini microarray printer equipped with 16 split-tungsten MA pins of 100 µm in diameter (Genetix, New Milton, UK) was used to print oligonucleotides in a triplicate random pattern onto epoxy-coated Nexterion slide E glass microscope slides (1066643; Schott AG, Mainz, Germany). Slides were printed at 8°C in 50% humidity and were checked for quality before use. The full list of genes in the Human Cancer Subset v3.0 can be viewed online (http://omad.operon.com/download/index.php). Total RNA was extracted using the Qiagen RNeasy Mini Animal Cell RNA Extraction Kit (74104; Qiagen, Crawley, UK) according to the manufacturer's protocol. RNA was quantified and assessed using the RNA 6000 Nano Kit (5067-1511; Agilent Technologies, Stockport, UK) and Bioanalyser (Agilent Technologies), and an RNA integrity number of 8.5 or greater was used to select those samples of sufficient quality for MA analysis. Expression MA analysis was performed as described elsewhere [9]. Briefly, indirect complementary DNA (cDNA) labeling was performed using the FairPlay II Microarray Labeling Kit (252006; Stratagene, La Jolla, CA) using Cy3 and Cy5 monofunctional reactive dyes (PA23001 and PA25001; Amersham Biosciences, Bucks, UK). Labeled cDNA samples were pooled, combined with 2 µl of 1 µg/ml human Cot-1 DNA (15279011; Invitrogen, Paisley, UK), 2 µl of 1 µg/µl Oligo d(A) (POLYA.GF; Invitrogen), 2 µl of 1 µl/µl yeast tRNA (15401011; Invitrogen), and 390 µl of 10 mM Tris base (pH 8.5) and concentrated to 5 µl using a Microcon YM-100 Centrifugal Filter Device (42413; Millipore, Watford, UK) according to the manufacturer's protocol. Slides were washed once in Triton X-100 (0.1% vol/vol) for 5 minutes, twice in two separate washes of 1 mM HCl for 2 minutes, once in 100 mM KCl solution for 10 minutes, and finally in distilled water for 1 minute. Slides were blocked by incubating in 1x Nexterion Blocking Solution (1066071; Schott) for 15 minutes at 50°C, followed by rinsing in distilled water for 1 minute. Slides were dried by centrifugation at 2200g for 2 minutes. Subsequently, a 22 x 40-mm glass Lifterslip (22X40I-2-4710; Erie Scientific, Portsmouth, NH) was applied to the slide over the print area. The labeled cDNA pool (5 µl) was mixed with 1 µl of 7.5% wt/vol (0.2% final concentration) BSA (15260037; Invitrogen) and 19 µl of Nexterion Hybridisation Buffer E (1066075; Schott) to make a total hybridization volume of 25 µl. The hybridization mix was denatured at 95°C for 3 minutes in a thermal cycler before application to the MA slide. Slides were sealed into humidified array hybridization chambers (4120-000002; MWG-Biotech, Ebersberg, Germany) and incubated at 65°C for 16 to 18 hours in a dark hybridization oven. The coverslip was subsequently removed by gentle agitation in 0.2-µm filtered 4x SSC. Slides were washed for 5 minutes in filtered 2x SSC/0.1% wt/vol SDS. Slides were dipped in filtered 2x SSC to remove residual SDS and then washed for 5 minutes in filtered 2x SSC. Finally, slides were dipped in filtered 0.2x SSC and washed for 5 minutes in filtered 0.2x SSC. Slides were dried by centrifugation at 2200g for 2 minutes. Images were acquired using the GenePix 4100A Personal Microarray Scanner in combination with Genepix Pro (version 4.1) image collection software (Molecular Devices Corporation, Sunnyvale, CA). The MA slides were scanned at a resolution of 5 µm per pixel with the photomultiplier tube gains manually set for each dye to ensure a normal ratio across the whole array of 1 (±0.1). After application of the .gal file, the results files were imported into Acuity v4.0 (Molecular Devices Corporation) and normalized using the nonlinear Lowess normalization method. A data set was created by filtering each array to include only those features with the parameter “Flags ≥ 0.” This ensured that only those features that were “found” and not “bad” or “not found” were included in the analyses. Once filtered, the data were analyzed according to the following parameters: low interarray variance (P ≤ .05) and absolute log (635/532) value ≥1.0/-1.0 in three of four independent experiments. A log ratio value of ≥1.0/-1.0 was chosen for the cutoff level because this represented an actual fold change of two-fold or greater, which is a standard for MA analysis [10].
Real-time Quantitative Polymerase Chain Reaction
Real-time quantitative polymerase chain reaction (RT-qPCR) was performed as described elsewhere [9]. Briefly, 2 µg of total RNA were treated with 2 U of amplification grade DNase 1 (AMP-D1; Sigma-Aldrich, Poole, UK) at room temperature for 15 minutes and then reverse-transcribed into cDNA using the iScript cDNA Synthesis Kit (170-8890; Bio-Rad). RT-qPCR was carried out with 50 ng of equivalent RNA template using the AB Power SYBR Green Supermix (4367659; Applied Biosystems) in a 20-µl reaction volume on a MyiQ Single-Color Real-time PCR Detection System (Bio-Rad). Primer details are shown in Table 1 and synthesized by Applied Biosystems. Data were collected using the MyiQ Optical System Software (Bio- Rad), and data analysis was carried out according to the method of Pfaffl [11].
Table 1.
Primers Used Successfully for RT-qPCR.
| Gene | Sense Primer | Antisense Primer | Amplicon Length (bp) |
| ER | 5′-GGAGGGCAGGGGTGAA-3′ | 5′-GGCCAGGCTGTTCTTCTTAG-3′ | 100 |
| GAPDH | 5′-CATCTTCCAGGAGCGAGATCC-3′ | 5′-GTGGTGAAGACGCCAGTGG-3′ | 87 |
| GSTM3 | 5′-GCGATACTGGCATTTCCCTA-3′ | 5′-TCTCCAACTGTGCAATCTCG-3′ | 104 |
| PGR | 5′-CGCGCTCTACCCTGCACTC-3′ | 5′-TGAATCCGGCCTCAGGTAGTT-3′ | 121 |
| PSME1 | 5′-GTGGATGTGTTTCGTGAAGACC-3′ | 5′-CTTGGGGAAATAGCTCCCGAG-3′ | 66 |
| PSME2 | 5′-GCAAACAGGTGGAGGTCTTCAGG-3′ | 5′-CATTACATGAGTCTCCTTGGAGG-3′ | 515 |
Sequences were selected using RTPrimerDB (http://medgen.ugent.be/rtprimerdb/) or PrimerBank (http://pga.mgh.harvard.edu/primerbank/). The sequences for PSME2 were previously used by Maksymowych et al. [33]. GAPDH was used as the reference gene.
Western Blot Analysis
Proteins were extracted in 1 ml of Laemmli buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 5% β-mercaptoethanol, 1% protease inhibitor mix, 0.00125% bromophenol blue) and quantified using the RC-DC Protein Assay Kit (500-0122; Bio-Rad). Equal amounts of protein (15–25 µg per lane) were loaded onto a 12% homogenous NuPage gel (EC6005BOX; Invitrogen). Electrophoresis was performed at 200 V for 60 minutes, and proteins were transferred to a 0.45-µm nitrocellulose membrane (162-0115; Bio-Rad) at 400 mA for 60 minutes at 4°C. Membranes were incubated in blocking buffer (5% nonfat dry milk, 0.05% Tween 20 in Tris-buffered saline [TBS]) for 60 minutes. Aldolase A (1:200; SC-30082; Santa Cruz Biotechnology, Santa Cruz, CA), aspartyl-tRNA synthetase (DARS, 1:1000; H00001615-M01; Abnova Corporation, Taipei City, Taiwan), glucose-regulated protein/GRP78 (1:400; ab21685; Abcam, Cambridge, UK), GSTM3 (1:2000; kindly provided by Professor John Hayes, Biomedical Research Centre, University of Dundee, UK), HSP90 Beta (1:100; 37-9400; Zymed, Paisley, UK), keratin 17 (1:50; ab2502; Abcam), moesin (1:200; SC-13122; Santa Cruz Biotechnology), p53 (1:200; 554293; BD Pharmingen, Oxford, UK), L-plastin (1:100; MS-1326; Neomarkers, Loughborough, UK), 26S proteasome (1:100; ab21165; Abcam), proteasome alpha 7 subunit/PSMA7 (1:200;H00005688-M01; Abnova Corporation), QARS (1:500; H00005859-M01; Abnova Corporation), triosephosphate isomerase 1 (1:1000; H00007167-M02A; Abnova Corporation), TRAP1/HSP75 (1:100; MS-1309;Neomarkers), E1 ubiquitin activating enzyme (1:5000; ab24623; Abcam), or the glyceraldehyde 3 phosphate dehydrogenase (GAPDH) loading control (1:10,000; ab9485; Abcam) were added for 2 hours at room temperature. The appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (antimouse or antirabbit; SC-2031 or SC-2030; Santa Cruz Biotechnology) was then incubated with the membrane for 1 hour. Proteins were detected using the SuperSignal West Pico Chemiluminescence Substrate (PN34078; Pierce, Loughborough, UK) according to the manufacturer's protocol. Films were scanned using a GS-800 laser densitometer, and protein expression was measured using Quantity One software (version 4.6.1; Bio-Rad).
Immunohistochemistry
Local research ethics committee approval was obtained for the collection of clinical data and archival pretreatment biopsy material. Patients whose conditions were diagnosed with early-stage (T1-T2 N0) laryngeal cancer and treated with single-modality radiotherapy with curative intent (55–60 Gy in 20–25 fractions) were identified from databases held in ENT departments in England [1]. Patients were identified as having radiosensitive or radioresistant tumors according to their response to radiotherapy. The radiosensitive and radioresistant groups were matched with regards to T stage, laryngeal subsite, and smoking history to reduce confounding variables. Tumors were staged according to the TNM classification and were all clinically nodal negative (N0) and metastatic negative (M0) at the time of treatment.
The radiosensitive group of tumors consisted of pretreatment biopsies from 22 patients. The criteria for a radiosensitive tumor were as follows: 1) the radiotherapy had to have been given as a single-modality treatment with curative intent for a biopsy-proven squamous cell carcinoma of the larynx; 2) after treatment, patients had a minimum follow-up of 3 years with no evidence of a recurrent laryngeal tumor.
The radioresistant group of tumors consisted of pretreatment biopsies from 22 patients. The criteria for a radioresistant tumor were as follows: 1) the radiotherapy had to have been given as a single-modality treatment with curative intent for a biopsy-proven squamous cell carcinoma of the larynx; 2) biopsy-proven recurrent squamous cell carcinoma, the recurrence occurring at the original anatomical site within 12 months of finishing a course of radiotherapy.
Immunohistochemical assays were carried out as described previously [12]. Briefly, 4 µm sections were cut onto Superfrost Plus microscope slides (Menzel-Glaser, Braunschweig, Germany) and incubated at 37°C overnight. Sections were dewaxed in Histoclear (National Diagnostics, Hull, UK) and rehydrated in alcohol before blocking endogenous peroxidase by incubating in 400 ml of methanol containing 8 ml of hydrogen peroxide (30% vol/vol). Antigenic sites were retrieved by boiling slides in 1500 ml of distilled water with 15 ml of Antigen Unmasking Solution (H-3300; Vector Laboratories, Peterborough, UK) in a pressure cooker for 3 minutes at 15 psi. Non-specific protein was blocked with 1x casein (SP-5020; Vector Laboratories) diluted in TBS for 10 minutes, and endogenous avidin and biotin were blocked by 15-minute incubation periods with biotin and avidin, respectively (Avidin Biotin Blocking Kit; SP-2001; Vector Laboratories). The 26S proteasome primary antibody (ab21165; Abcam), as used for Western blot analysis, was prepared to a dilution of 1:50 with 0.2x casein in TBS and applied at room temperature for 2 hours. A negative control was also included, in which the primary antibody was omitted. Antibody detection was carried out using an HRP-conjugated Streptavidin-Biotin Complex Duet Kit (K0492; DakoCytomation, Ely, UK) according to the manufacturer's protocol. Immunoreactivity was visualized using 0.05% 3′,3-diaminobenzidine as the chromogen, and 0.01% hydrogen peroxide was used as the HRP substrate. Sections were counterstained with Harris hematoxylin, dehydrated in alcohol, and mounted with Histomount (National Diagnostics). Slides were scored by two independent investigators, and any discrepancies in the results were resolved by consensus following reassessment by both investigators. Sections were scored as negative if expression of the 26S proteasome was lost in more than 50% of the tumor cells. This is because loss of expression was considered as detrimental. Association between protein expression and response to radiotherapy was determined using the χ2 test using SPSS (version 14, SPSS Inc, Chicago, IL).
Results
Inherent Sensitivity of Parental Cell Lines to Radiotherapy
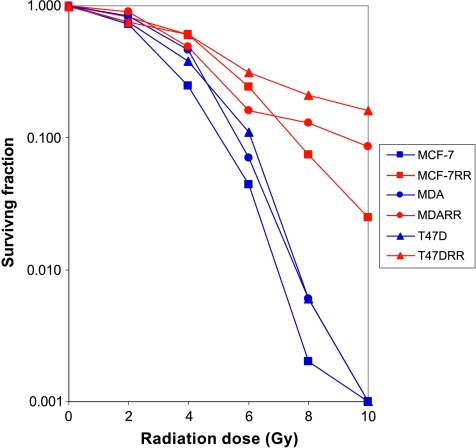
Figure 1 demonstrates the relative sensitivity of each of the parent human breast cancer cell lines MCF7, MDA-MB-231, and T47D, to ionizing radiation given as single fractions over the dose range of 0 to 10 Gy. The three cell lines had plating efficiencies of 30.9 ± 4.16%, 11 ± 0.47%, and 13.5 ± 0.40%, respectively. There was no significant difference in cell survival between the three parent cell lines based on the survival fraction at 2 Gy. When the survival fraction at 4 Gy was calculated, there was a significant difference between survival of MCF7 versus MDA-MB-231 and MCF7 versus T47D (P < .05 by ANOVA). MDA-MB-231 demonstrated the most inherent radioresistance of the three parent cell lines.
Figure 1.
A log-linear plot demonstrating the relative sensitivity of the three parental breast cancer cell lines (MCF7, MDA-MB-231, and T47D) and their novel radioresistant derivatives (MCF7RR, MDA-MB-231RR, and T47DRR, respectively) to ionizing radiation over the range of 0 to 10 Gy as determined by a modified colony counting assay. Each point represents the mean of six replicates over two independent experiments. Compared with the respective parental cells, MCF7RR and T47DRR demonstrated a significant increase in survival at 4, 6, 8, and 10 Gy (P ≤ .01; ANOVA). MDA-MB-231RR cells demonstrated a significant increase in survival at 6, 8 and 10 Gy compared with parental MDA-MB-231 cells (P ≤ .01; ANOVA). Compared with the respective parental cells, the overall maximum resistance demonstrated by MCF7RR, MDA-MB-231RR, and T47DRR was 37-fold, 22-fold, and 34-fold, respectively, all of which where observed at 8 Gy (P ≤ .01; ANOVA).
Establishment of Radioresistant Derivatives
The three parental breast cancer cell lines (MCF7, MDA-MB-231, and T47D) were subjected to fractionated radiotherapy to a total dose of 40 Gy, and the novel radioresistant derivatives (hereafter named MCF7RR, MDA-MB-231RR, and T47DRR, respectively) were assayed using the modified colony counting assay (Figure 1). The MCF7RR and T47DRR cell lines were significantly more resistant to radiotherapy than their parent cell lines (MCF7 and T47D, respectively) at 4, 6, 8, and 10 Gy (P ≤ .01; ANOVA) with maximal 37-fold and 34-fold increases, respectively, in resistance observed at 8 Gy. The MDA-MB-231RR cell line was significantly more resistant than its MDA-MB-231 parent at 6, 8, and 10 Gy (P ≤ .01; ANOVA) with a maximal 22-fold increase in resistance observed at 8 Gy. Western blot analysis revealed that parent and derivative cells demonstrated similar expression levels of p53. Similarly, RT-qPCR analysis revealed that both MCF7 and T47D cells retained their ER- and PR-positive status following treatment. RT-qPCR analysis was also able to detect only trace amounts of ER and PR messenger RNA (mRNA) in MDA-MB-231 cells both before and after treatment, indicating that treatment with radiotherapy had not altered the phenotypic characterization of these cell lines.
2DE/MS Analysis
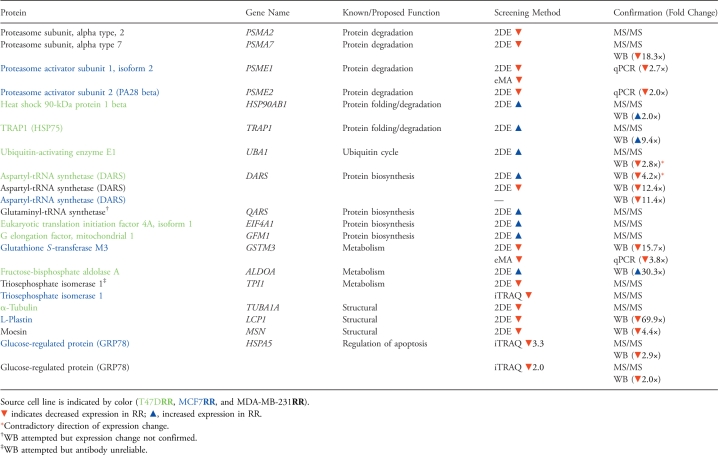
The protein profiles of three parental breast cancer cell lines (MCF7, MDA-MB-231, and T47D) were compared with the protein profiles of their novel radioresistant derivatives (MCF7RR, MDA-MB-231RR, and T47DRR, respectively) using 2DE. Proteins were fractionated using medium-pH range IPG strips (pH 4–7 and pH 7–10) to maximize resolution. A representative gel image is shown in Figure 2. In total, a significant change in the expression of 65 proteins (down-regulation of 25; up-regulation of 40) was detected in one or more of the radioresistant derivatives. The expression of 18 proteins (16 proteins in the pH 4–7 range and 2 proteins in the pH 7–10 range; 12 downregulated proteins and 6 upregulated proteins) was significantly altered in the MCF7RR cell line compared with the MCF7 parental cell line. The expression of 18 proteins (14 proteins in the pH 4–7 range and 4 proteins in the pH 7–10 range; 9 downregulated proteins and 9 upregulated proteins) was significantly altered in the MDA-MB-231RR cell line compared with the MDA-MB-231 parental cell line. The expression of 29 proteins (23 proteins in the pH 4–7 range and 6 proteins in the pH 7–10 range; 4 downregulated proteins and 25 upregulated proteins) was significantly altered in the T47DRR cell line compared with the T47D parental cell line. Figure 3 shows several examples of differentially expressed proteins from the analysis of each cell line pair. A total of 65 protein spots were excised and subjected to tryptic digestion. The resulting peptides were extracted and analyzed by MALDI-TOF and/or MALDI-TOF/TOF MS. Assignments were made for 50 of these 65 protein spots (Table 2). After classification according to their function, many of these proteins played a role in metabolism, protein biosynthesis, or the ubiquitin-proteasome protein degradation pathway (Table 2).
Figure 2.
Representative 2D gaussian patterns for the MCF7 (upper panel) and MCF7RR (lower panel) cell lines in the pH range of 4 to 7.
Figure 3.
Representative 2DE gel analysis showing several examples of proteins identified by mass spectrometry (Table 2) that were differentially expressed (at least two-fold) between the radioresistant derivatives (MCF7RR, MDA-MB-231RR, and T47DRR) and their parental cell lines (MCF7, MDA-MB-231, and T47D, respectively). The histograms show relative spot quantities; the first three bars represent the quantity of that spot in each of the three parental biologic replicates and the last three bars represent the quantity of that spot in each of the three radioresistant biologic replicates. Spots were significantly differentially expressed as determined by the Student's t test (P < .05) and were all tested by Western blot analysis (Figure 4). GST-M3 indicates glutathione-S-transferase mu-3; PSMA7, proteosome alpha 7 subunit; TRAP1, TNF receptor-associated protein-1/HSP75.
Table 2.
Proteomics Data, Ranked by Mowse Score, for All Assigned Proteins Associated with In Vitro Radioresistance in MCF7RR, T47DRR, and MDA-MB-231RR Cells Identified Using 2DE in Combination with MALDI-TOF or MALDI-TOF/TOF MS.
| Protein | NCBI Accession | Gene Name | Known/Proposed Function | RR Cell Line | pH Gradient | Expression Level | Mowse Score |
| Heat shock 90-kDa protein 1 beta*,† | 34304590 | HSP90AB1 | Protein folding/degradation | T47D | 4–7 | ▴ | 787 |
| Eukaryotic translation initiation factor 4A, isoform 1*,‡ | 54696624 | EIF4A1 | Protein biosynthesis | T47D | 4–7 | ▴ | 725 |
| Ubiquitin activating enzyme E1*,†,‡ | 35830 | UBA1 | Ubiquitin cycle | T47D | 4–7 | ▴ | 492 |
| α-Tubulin* | 37492 | TUBA1A | Structural | T47D | 4–7 | ▾ | 473 |
| Glutaminyl-tRNA synthetase*,† | 4826960 | QARS | Protein biosynthesis | MDA | 7–10 | ▴ | 309 |
| TRAP1*,† | 3273383 | TRAP1/HSP75 | Protein folding/degradation | T47D | 4–7 | ▴ | 302 |
| G elongation factor, mitochondrial 1* | 18390331 | GFM1 | Protein biosynthesis | T47D | 4–7 | ▴ | 177 |
| Triosephosphate isomerase 1*,† | 17389815 | TPI1 | Metabolism | MDA | 7–10 | ▾ | 177 |
| Lymphocyte cytosolic protein 1 (l-plastin)† | 14043359 | LCP1 | Structural | MCF7 | 4–7 | ▾ | 155 |
| Mitochondrial malate dehydrogenase precursor‡ | 21735621 | MDH2 | Metabolism | T47D | 7–10 | ▴ | 149 |
| Hypothetical (PGM2) | 12052930 | PGM2 | Metabolism | MDA | 4–7 | ▾ | 149 |
| HSPC263 | 6841176 | OTUB1 | Ubiquitin cycle | T47D | 4–7 | ▴ | 148 |
| Vinculin isoform VCL | 4507877 | VCL | Cell adhesion | T47D | 4–7 | ▴ | 144 |
| Actinin, alpha 4 | 12025678 | ACTN4 | Structural | MCF7 | 4–7 | ▴ | 133 |
| Eukaryotic translation elongation factor 2 | 4503483 | EEF2 | Protein biosynthesis | T47D | 4–7 | ▴ | 133 |
| Solute carrier family 25, member 24 | 46249805 | SLC25A24 | Transport | MDA | 4–7 | ▴ | 131 |
| Fructose-bisphosphate aldolase A† | 4557305 | ALDOA | Metabolism | T47D | 7–10 | ▴ | 125 |
| Proteasome subunit, alpha type, 7*,† | 30583169 | PSMA7 | Protein degradation | MDA | 7–10 | ▾ | 124 |
| Voltage-dependent anion channel 1 | 4507879 | VDAC1 | Transport | T47D | 7–10 | ▴ | 121 |
| Aspartyl-tRNA synthetase† | 179102 | DARS | Protein biosynthesis | T47D | 4–7 | ▴ | 118 |
| Aspartyl-tRNA synthetase† | 62896511 | MDA | ▾ | 95 | |||
| Glutathione S-transferase M3†,§ | 14250650 | GSTM3 | Metabolism | MCF7 | 4–7 | ▾ | 118 |
| Keratin 17† | 48735384 | KRT17 | Structural | MCF7 | 4–7 | ▾ | 117 |
| Pyrophosphatase 1 | 85397510 | PPA1 | Metabolism | MCF7 | 4–7 | ▾ | 115 |
| Moesin† | 4505257 | MSN | Structural | MDA | 4–7 | ▾ | 108 |
| Leucine-rich PPR motif-containing protein | 31621305 | LRPPRC | Transport | MCF7 | 4–7 | ▴ | 108 |
| N-Acetyltransferase ARD1 | 10835057 | ARD1A | Protein modification | MCF7 | 4–7 | ▾ | 105 |
| Proteasome activator subunit 2 (PA28 beta)§ | 13325110 | PSME2 | Protein degradation | MCF7 | 4–7 | ▾ | 105 |
| Glyoxalase domain containing 4 | 16198390 | GLOD4 | Unknown | MCF7 | 4–7 | ▾ | 101 |
| EPB41 | 24657823 | EPB41 | Structural | MDA | 4–7 | ▾ | 101 |
| Proteasome subunit, alpha type, 2* | 51094758 | PSMA2 | Protein degradation | MDA | 7–10 | ▾ | 101 |
| Spermidine synthase | 63253298 | SRM | Spermidine biosynthesis | MDA | 4–7 | ▾ | 99 |
| Heterogeneous nuclear ribonucleoprotein A2/B1 isoform A2 | 4504447 | HNRNPA2B1 | mRNA processing | T47D | 7–10 | ▾ | 96 |
| ER lipid raft associated 2 isoform 1 | 6005721 | ERLIN2 | Unknown | T47D | 4–7 | ▴ | 95 |
| STARD10 | 31455229 | STARD10 | Unknown | MCF7 | 4–7 | ▾ | 95 |
| S-Adenosylhomocysteine hydrolase | 9951915 | AHCY | Metabolism | T47D | 4–7 | ▴ | 94 |
| Isocitrate dehydrogenase 3 (NAD+), alpha precursor | 5031777 | IDH3A | Metabolism | MDA | 4–7 | ▴ | 91 |
| Ribosomal protein, large, P0 | 12654583 | RPLP0 | Protein biosynthesis | MCF7 | 4–7 | ▾ | 90 |
| Enoyl coenzyme A hydratase, short chain, 1, mitochondrial | 14286220 | ECHS1 | Metabolism | MDA | 4–7 | ▴ | 88 |
| Eukaryotic translation initiation factor 3, subunit 2 beta | 54696064 | EIF3I | Protein biosynthesis | MCF7 | 4–7 | ▾ | 82 |
| Proteasome activator subunit 1 isoform 2§ | 30581141 | PSME1 | Protein degradation | MCF7 | 4–7 | ▾ | 81 |
| Growth inhibiting gene 5 protein | 41350397 | CACYBP | Ubiquitin cycle | MCF7 | 7–10 | ▾ | 81 |
| Mitochondrial aldehyde dehydrogenase 2 precursor | 25777732 | ALDH2 | Metabolism | MDA | 4–7 | ▴ | 80 |
| Pro-alpha 1 (II) collagen | 30097 | COL2A1 | Structural | MDA | 4–7 | ▾ | 75 |
| CNDP dipeptidase 2 (metallopeptidase M20 family), isoform CRA_e | 119586960 | CNDP2 | Peptidolysis | MDA | 4–7 | ▴ | 72 |
| Hypothetical protein LOC80007 | 13376421 | C10orf88 | ATPase activity | MDA | 4–7 | ▴ | 70 |
| Lysyl-tRNA synthetase | 11095909 | KARS | Protein biosynthesis | MCF7 | 4–7 | ▴ | 69 |
Expression levels represent the up- (▴) or down- (▾) regulation of proteins by at least two-fold in the radioresistant derivatives relative to parental cells. Gene ontology information was derived from the GOA database (http://www.ebi.ac.uk/GOA/).
A summary of all confirmed data is shown in Table 5.
Proteins for which MS/MS data were obtained.
Targets selected for Western blot analysis (Figure 4).
Proteins identified in two separate but closely mapped spots on the same gel.
Targets selected for analysis by RT-qPCR.
LC-MS/MS Analysis
LC-MS/MS was used as a complementary approach to 2DE-MS for the analysis of all proteins present in T47D and T47DRR cells. A total of 586 and 652 proteins were identified in T47D and T47DRR cells, respectively, using a minimum CI score of greater than 98%. Those proteins identified in both cell lines and any redundant entries were removed to reveal those proteins, which were unique to each cell sample. In total, 242 unique proteins were identified in T47D cells (that could not be identified in T47DRR cells) with MASCOT ion scores ranking from 556 to 37. Forty of these proteins were “unknown,” “unnamed,” “hypothetical,” or “predicted.” Likewise, a total of 310 unique proteins were identified in T47DRR cells (that could not be identified in T47D cells) with MASCOT ion scores ranking from 757 to 37. Seventy-five of these proteins were “unknown,” “unnamed,” “hypothetical,” or “predicted.” The proteins from each cell line were categorized into functional groups, and several changes were observed between cell lines. Of note, several proteins involved in the ubiquitin-proteasome degradation pathway were missing in T47DRR cells compared with parental cells (Table 3). This is in accordance with our 2DE-MS results, which also identified the downregulation of several proteasome-associated proteins (Table 2).
Table 3.
Proteins Associated with the Ubiquitin-Proteasome Pathway of Protein Degradation Which Were Identified in T47D Parent Cells But Not in T47DRR Cells Using LC-MALDI-TOF/TOF MS.
| Protein | NCBI Accession | Gene | Known/Proposed Function | Total Ion Score | Total Ion CI (%) |
| Proteasome alpha 1 subunit, isoform 2 | 16307002 | PSMA1 | Protein degradation | 80 | 100 |
| 26S proteasome subunit p40.5 | 3618343 | PSMD13 | Protein degradation | 65 | 99.997 |
| Proteasome (prosome, macropain) subunit, beta type, 2 | 30583113 | PSMB2 | Protein degradation | 41 | 99.322 |
| Ubiquitin-associated protein 2-like | 13111995 | UBAP2L | Ubiquitin cycle | 100 | 100 |
| Hyd protein (ubiquitin protein ligase E3 component n-recognin 5 | 9545980 | UBR5 | Ubiquitin cycle | 44 | 99.626 |
Quantitative Proteomics
A complementary quantitative approach for the identification of biomarkers associated with radioresistance in this panel of novel breast cancer cell lines was performed using iTRAQ technology. A minimum CI of greater than 98% was used for the identification of proteins, and at least two peptide sequences were matched to most identified proteins. A total of 1370 proteins were identified in the analysis of the MCF7/MCF7RR cell line pair with MASCOT ion scores ranking from 2717 to 39. Likewise, 1025 proteins were identified in the MDA-MB-231/MDA-MB-231RR cell line pair with MASCOT ion scores ranking from 2336 to 39. Finally, 1252 proteins were identified in the T47D/T47DRR cell line pair with MASCOT ion scores ranking from 3320 to 39. Using a standard two-fold change or greater in expression, these iTRAQ analyses revealed significant changes in the expression of 40 characterized proteins in one or more of the radioresistant derivatives (Table 4). In the MCF7RR cell line, relative to the MCF7 parent, 23 proteins demonstrated a decrease in expression (ranging from 2.0- to 6.2-fold change) and 5 proteins demonstrated an increase in expression (ranging from 2.0- to 3.0-fold change). In the MDA-MB-231RR cell line, relative to the MDA-MB-231 parent, seven proteins demonstrated a decrease in expression (ranging from 2.0- to 3.5-fold change). In the T47DRR cell line, relative to the T47D parent, four proteins demonstrated a decrease in expression (ranging from 2.0- to 2.5-fold change) and one protein demonstrated an increase in expression (2.4-fold change). A decrease in the expression of glucose-regulated protein (GRP78) and filamin A alpha was found in both MCF7RR and MDA-MB-231RR cells in relation to their parental cells (MCF7 and MDA-MB-231, respectively).
Table 4.
Proteomics Data, Ranked by Fold Change, for All Differentially Expressed (at Least Two-Fold) Characterized Proteins Associated with Radioresistance in One or More of the Radioresistant Derivatives Identified Using iTRAQ Technology.
| Protein | NCBI Accession | Gene Name | Function | Cell Line | Total Ion Score | Total Ion Score CI (%) | Average iTRAQ Ratio (115:114) | Fold Change |
| Keratin 8 | 49256423 | KRT8 | Structural | MCF7 | 2717 | 100.0 | 0.161 | ▾6.2 |
| Keratin 19 | 6729681 | KRT19 | Structural | MCF7 | 2387 | 100.0 | 0.278 | ▾3.6 |
| Vimentin | 7576229 | VIM | Structural | MDA | 2336 | 100.0 | 0.288 | ▾3.5 |
| Glucose-regulated protein (GRP78)* | 6900104 | HSPA5 | Regulation of apoptosis | MCF7 | 2241 | 100.0 | 0.304 | ▾3.3 |
| MDA | 1882 | 100.0 | 0.491 | ▾2.0 | ||||
| Heat shock 70-kDa protein 1A | 55962551 | HSPA1A | Protein folding | MCF7 | 2183 | 100.0 | 0.315 | ▾3.2 |
| Cytokeratin 18 (424 AA) | 30311 | KRT18 | Structural | MCF7 | 2183 | 100.0 | 0.327 | ▾3.1 |
| ACSF3 protein | 47938904 | ACSF3 | Unknown | MCF7 | 39 | 98.02 | 3.0 | ▴3.0 |
| Heat shock protein 90-alpha | 72219 | HSP90AA1 | Protein folding | MCF7 | 2161 | 100.0 | 0.349 | ▾2.9 |
| Plectin 1 isoform 3 | 41322908 | PLEC1 | Structural | MDA | 2118 | 100.0 | 0.352 | ▾2.8 |
| Filamin A, alpha (actin binding protein 280) | 57284201 | FLNA | Structural | MCF7 | 2088 | 100.0 | 0.353 | ▾2.8 |
| Filamin A, alpha (actin binding protein 280) | 57284203 | MDA | 2094 | 100.0 | 0.371 | ▾2.7 | ||
| Nucleolin | 55956788 | NCL | Angiogenesis | MCF7 | 1895 | 100.0 | 0.371 | ▾2.7 |
| Heat shock 70-kDa protein 9 precursor | 24234688 | HSPA9 | Protein folding | MCF7 | 1824 | 100.0 | 0.402 | ▾2.5 |
| Structural maintenance of chromosomes 3 | 28958118 | SMC3 | Cell Division | T47D | 60 | 99.98 | 0.403 | ▾2.5 |
| Desmoyokin | 627367 | AHNAK | Nervous system development | MDA | 2014 | 100.0 | 0.417 | ▾2.4 |
| OS9 | 17986213 | OS9 | Unknown | T47D | 97 | 100.0 | 0.418 | ▾2.4 |
| Fatty acid synthase | 41872631 | FASN | Metabolism | MCF7 | 1774 | 100.0 | 0.420 | ▾2.4 |
| PRKC, apoptosis, WT1, regulator | 55769533 | PAWR | Regulation of transcription | T47D | 51 | 99.88 | 2.4 | ▴2.4 |
| Lamin A/C transcript variant 1 | 57014043 | LMNA | Structural | MCF7 | 1652 | 100.0 | 0.429 | ▾2.3 |
| Prolyl 4-hydroxylase, beta polypeptide | 48735337 | P4HB | Protein folding | MCF7 | 1650 | 100.0 | 0.432 | ▾2.3 |
| MYH9 | 47678583 | MYH9 | Structural | MDA | 1955 | 100.0 | 0.442 | ▾2.3 |
| Mitochondrial ATP synthase beta subunit precursor | 32189394 | ATP5B | Transport | MCF7 | 1626 | 100.0 | 0.444 | ▾2.3 |
| Chaperonin 60, Hsp60 | 6996447 | HSPD1 | Protein folding | MCF7 | 1567 | 100.0 | 0.444 | ▾2.3 |
| Surfeit 1 | 55958186 | SURF1 | Unknown | T47D | 56 | 99.96 | 0.445 | ▾2.2 |
| Heat shock protein 90-kDa beta (Grp94), member 1 | 44890631 | HSP90B1 | Protein folding | MCF7 | 1556 | 100.0 | 0.449 | ▾2.2 |
| Cytokeratin | 1419564 | KRT8 | Structural | MCF7 | 1443 | 100.0 | 0.450 | ▾2.2 |
| ACTB protein | 15277503 | ACTB | Structural | MCF7 | 1440 | 100.0 | 0.458 | ▾2.2 |
| Heterogeneous nuclear ribonucleoprotein A2/B1 isoform B1 | 14043072 | HNRNPA2B1 | mRNA processing | MCF7 | 1350 | 100.0 | 0.462 | ▾2.2 |
| Pyruvate kinase, muscle | 31416989 | PKM2 | Metabolism | MCF7 | 1322 | 100.0 | 0.467 | ▾2.1 |
| ER-60 protein | 2245365 | PDIA3 | Transport | MCF7 | 1315 | 100.0 | 0.469 | ▾2.1 |
| Aldolase A | 49456715 | ALDOA | Metabolism | MCF7 | 1242 | 100.0 | 0.479 | ▾2.1 |
| Protein phosphatase 1, regulatory (inhibitor) subunit 8 | 56204167 | PPP1R8 | Metabolism | MCF7 | 39 | 98.03 | 2.1 | ▴2.1 |
| MDH2 | 49168580 | MDH2 | Metabolism | MCF7 | 1183 | 100.0 | 0.490 | ▾2.0 |
| Triosephosphate isomerase 1 | 15929332 | TPI1 | Metabolism | MCF7 | 1180 | 100.0 | 0.490 | ▾2.0 |
| Josephin MJD1 | 2262199 | ATXN3 | DNA repair | T47D | 83 | 99.99 | 0.493 | ▾2.0 |
| Actinin, alpha 4 | 12025678 | ACTN4 | Structural | MDA | 1805 | 100.0 | 0.497 | ▾2.0 |
| TPA: ubiquitin-specific protease 17-like protein | 33186790 | LOC401447 | Ubiquitin cycle | MCF7 | 39 | 98.16 | 2.0 | ▴2.0 |
| Ribonucleotide reductase M1 | 13676332 | RRM1 | DNA replication | MCF7 | 39 | 98.13 | 2.0 | ▴2.0 |
| alpha1,6 Fucosyltransferase | 7288173 | FUT8 | Metabolism | MCF7 | 39 | 98.10 | 2.0 | ▴2.0 |
Gene ontology information was derived from the GOA database (http://www.ebi.ac.uk/GOA/).
Expression MA Analysis
A targeted expression MA screening approach was used for the complementary analysis of the MCF7 and MCF7RR cell lines. This analysis independently confirmed the decrease in expression of GSTM3 (by 3.6-fold) and PSME1 (by 2.8-fold) in the MCF7RR cells, which had been identified by the 2DE-MS analysis of these cells. RT-qPCR was used to confirm the expression MA data and revealed a 3.8-fold decrease in the expression of GSTM3 and a 2.7-fold decrease in the expression of PSME1 in MCF7RR cells relative to parental cells (Table 5). In addition, PSME2, a related target also identified by 2DE-MS, was selected for analysis by RT-qPCR analysis and exhibited a 2.0-fold decrease in the expression in MCF7RR cells relative to parental cells (Table 5).
Table 5.
Summary of Proteins Identified by 2DE-MS or iTRAQ, Grouped by Function, Where Additional Confirmation Was Provided by MS/MS from a Second Instrument, Western Blot (WB) Analysis or Expression MA (eMA) with RT-qPCR (qPCR).
Western Blot Analysis
To validate the proteomics data, several targets were selected for confirmation by Western blot analysis. Each band was normalized to GAPDH, and protein expression was quantitated using densitometric analysis. Because α-tubulin was identified as a differentially expressed protein in the 2DE-MS study, GAPDH was selected as the loading control after equal expression of this protein was observed between cell line pairs after careful protein quantification and equal lane loading. Fold changes were calculated using the normalized quantities from three biologic replicates for each cell line.
Several targets identified by 2DE-MS from each cell line pair were selected for Western blot analysis (Table 2), and confirmation of significant (at least two-fold) differential protein expression was established for a number of targets (Figure 4; Table 5).
Figure 4.
Several targets identified by 2DE-MS from each pair of cell lines were selected for confirmation using Western blot analysis. Each band was quantitated by densitometry using Quantity One software (Bio-Rad) and normalized to a GAPDH loading control. A change in expression of at least two-fold was taken to be significant. Analysis revealed the down-regulation of DARS in all radioresistant derivatives in comparison with the respective parent cells (MCF7RR, 11.4-fold decrease in expression; MDA-MB-231RR, 12.4-fold decrease; T47DRR, 4.2-fold decrease). The down-regulation of l-plastin (69.9-fold decrease) and GST-M3 (15.7-fold decrease) in MCF7RR cells relative to parental MCF7 cells was confirmed. The down-regulation of keratin 17 reached only a 1.7-fold decrease in MCF7RR cells relative to parental MCF7 cells. The down-regulation of PSMA7 (18.3-fold decrease) and Moesin (4.4-fold decrease) in MDA-MB-231RR cells relative to parental MDA-MB-231 cells was confirmed. The up-regulation of HSP90 beta (2.0-fold increas), TRAP1 (9.4-fold increase), and aldolase A (30.3-fold increase) was also confirmed. See Figure 3 for abbreviations. See Table 5 for summary.
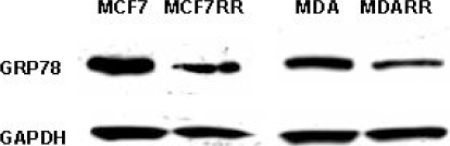
From the iTRAQ analysis, glucose-regulated protein (GRP78) was selected for Western blot analysis because this protein was identified from two radioresistant cell lines. A significant decrease in expression of this protein was observed in MCF7RR cells (2.9-fold decrease) and MDA-MB-231RR cells (2.0-fold decrease), thereby confirming this iTRAQ result (Figure 5; Table 5).
Figure 5.
After iTRAQ analysis, the down-regulation of glucose-regulated protein (GRP78) in MCF7RR cells (2.9-fold decrease) and MDA-MB-231RR cells (2.0-fold decrease) relative to their parents (MCF7 and MDA-MB-231, respectively) was confirmed by Western blot analysis. Each band was quantitated by densitometry using Quantity One software (Bio-Rad) and normalized to a GAPDH loading control.
The 26S Proteasome
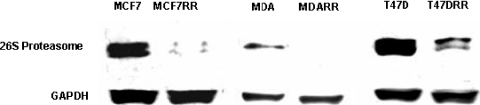
Data obtained for all cell lines demonstrated a trend for decreased expression of protein components from the 26S proteasome in radioresistant cells. Results from the 2DE analyses indicated that a decrease in the expression of proteasome subunit alpha type 2 (PSMA2) and proteasome subunit alpha type 7 (PSMA7) was associated with radioresistance (Table 5). Furthermore, the LC-MALDI MS/MS analysis of the T47D and T47DRR cell lines revealed that proteasome alpha 1 subunit isoform 2 (PSMA1), proteasome subunit beta type 2 (PSMB2), and 26S proteasome subunit p40.5 (PSMD13) were present in parental cells but were not detected in T47DRR cells, suggesting possible down-regulation of these proteins in radioresistant cells. To analyze this trend further, the expression of the 26S proteasome in all cell lines was examined by Western blot analysis. The antibody used recognized the 20S subcomplex within the 26S hetero-oligomeric protein complex and also recognized the free cytosolic form of the 20S subcomplex. A decrease in the expression of the 26S proteasome was observed in all radioresistant derivatives in comparison with their parents (Figure 6). Analysis revealed a decrease in expression by 7.7-fold, 45.1-fold, and 6.3-fold in MCF7RR, MDA-MB-231RR, and T47DRR cells, respectively.
Figure 6.
Western blot analysis of the 26S proteasome in the three radioresistant derivatives in comparison with parental cell lines. Each band was quantitated by densitometry using Quantity One software (Bio-Rad) and normalized to a GAPDH loading control. The 26S proteasome antibody (ab21165; Abcam), which recognizes the 20S component of the 26S proteasome, produced the expected band size of approximately 32 kDa. A significant decrease in the expression of the 26S proteasome was observed in all radioresistant derivatives in comparison with parental cell lines: MCF7RR, 7.7-fold decrease; MDA-MB-231RR, 45.1-fold decrease; T47DRR, 6.3-fold decrease.
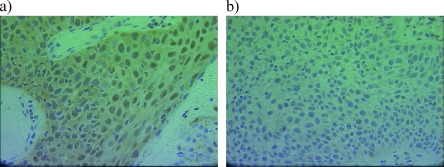
Immunohistochemistry was subsequently performed using this same antibody to determine whether expression of the 26S proteasome also correlated with response to radiotherapy in vivo. Pretreatment biopsies were obtained for a series of patients diagnosed with early stage laryngeal cancer. These patients were treated with single-modality radiotherapy with curative intent, with documented radiosensitivity or radioresistance, and therefore represented a suitable series with which to determine whether expression of the 26S proteasome correlated with radioresistance. Such a series is difficult to obtain for breast cancer patients, where radiotherapy is used in the adjuvant setting, because of a potentially large number of confounding variables. In total, 44 laryngeal squamous cell carcinomas were selected for a pilot analysis, classified into radiosensitive (n = 22) and radioresistant (n = 22) groups matched as closely as possible to reduce the number of confounding variables. Staining with the 26S proteasome, when evident, was present in both the cytoplasm and the nuclei of tumor cells. In the radioresistant group, 18 (81%) of 22 tumors were negative for cytoplasmic expression of the 26S proteasome. In contrast, only 12 (54%) of 22 radiosensitive tumors were negative. Loss of expression in the cytoplasm was associated with radioresistant tumors (P = .05) in this pilot series (Figure 7).
Figure 7.
Immunohistochemistry to detect the 26S proteasome (Ab21165; Abcam) was performed using a pilot series of 44 biopsies taken from early stage laryngeal tumors before treatment with primary radiotherapy. Loss of expression in the cytoplasm was significantly associated with radioresistant tumors (P = .05). (a) Representative image of a tumor with positive cytoplasmic staining. (b) Tumor lacking expression of the 26S proteasome.
Discussion
Radioresistance is a major problem in the effective management of human cancers. We examined the inherent radiosensitivities of three well-described breast cancer cell lines using a clonogenic survival assay and revealed that each cell line differed in their response to radiotherapy. The ER-positive, PR-positive (luminal subtype)MCF7, and T47D cells were most sensitive to radiotherapy. Interestingly, the MDA-MB-231 cells were the least sensitive. These cells have an ER-negative, PR-negative, HER2-negative phenotype, which is representative of breast cancers of the triple-negative molecular subtype. Triple-negative tumors are more aggressive and have higher local recurrence rates after adjuvant radiotherapy than that seen in luminal subtypes [13,14] and therefore may benefit from a therapeutic strategy that involves radiosensitization.
Novel derivative cell sublines (MCF7RR, MDA-MB-231RR, and T47DRR) were developed, which displayed significant resistance to radiotherapy. These derivative cells were compared with parent cells to identify putative biomarkers associated with the radioresistant phenotype. This area of breast cancer research has recently been highlighted as a significant knowledge “gap” by the Breast Cancer Campaign [15]. In 2005, Wang et al. [16] established a radioresistant derivative from MCF7 parent cells and used a global 2DE-MS proteomic approach to reveal an up-regulation of peroxiredoxin II in radioresistant cells. The silencing of peroxiredoxin II using small interfering RNA only partially restored sensitivity to radiotherapy, suggesting that other factors are associated with response [16]. Our study is the first of its kind using a range of complementary proteomic methods to analyze radioresistance in breast cancer cells. We identified very few overlapping targets from data produced by 2DE-MS, iTRAQ, and LC-MALDI MS/MS approaches, highlighting the importance of using a comprehensive, complementary screening approach in biomarker discovery.
A summary of the putative protein biomarkers associated with radioresistance in this study, where some confirmatory evidence has been obtained, is given in Table 5, and these proteins are discussed later. A number of other interesting observations remain to be confirmed including the independent identification of the down-regulation of filamin A alpha in two radioresistant cell lines by iTRAQ analysis and the identification of a number of proteins from the heat shock protein family, which exhibited down-regulation in MCF7-RR cells by iTRAQ analysis (Table 4).
Proteins Identified from More than One RR Cell Line
DARS was identified by 2DE-MS in two of three radioresistant cell lines (upregulated in T47DRR cells but downregulated in MDA-MB-231RR cells). Western blot analysis revealed a significant decrease in expression in all three radioresistant cell lines (Table 5). The reason for the discrepancies between these techniques is unclear but highlights the importance of independent data confirmation. DARS is involved in protein biosynthesis and has no previous link to radioresistance.
Glucose-regulated protein (GRP78) was identified by iTRAQ in two of three radioresistant cell lines (downregulated in MCF7RR and MDA-MB-231RR cells). Western blot analysis confirmed a significant decrease in the expression of both of these radioresistant cell lines (Table 5). Glucose-regulated protein is a molecular chaperone, and overexpression has been associated with cell survival and chemoresistance in a number of cancer types [17–19]. No association with radioresistance has been previously identified.
Proteins Identified by Both 2DE-MS and iTRAQ
The down-regulation of triosephosphate isomerase 1 (TPI1) was identified in MCF7RR cells by iTRAQ and in MDA-MB-231RR cells by 2DE-MS (Table 5). A reliable antibody against triosephosphate isomerase 1 was not found, and therefore, Western blot analysis confirmation has not been possible.
Proteins Identified by Both iTRAQ and LC-MALDI
The expression of the structural maintenance of chromosomes 3 (SMC3) protein was found to be downregulated by a factor of 2.5-fold in T47DRR cells compared with parental cells. This result is supported by the LC-MALDI MS/MS results, which identified the structural maintenance of chromosomes 3 protein in T47D cells but not in T47DRR cells, which may suggest that this protein has been downregulated. The structural maintenance of chromosomes 3 protein is involved in a protein complex that detects and repairs DNA DSBs.
The 26S Proteasome
Our findings suggested that the down-regulation of the 26S proteasome may be associated with radioresistant phenotype. The downregulation of PSMA2) and PSMA7 was identified by 2DE-MS/MS analysis in the MDA-MB-231RR cells (Table 5). LC-MALDI-MS/MS analysis of the T47DRR cells also provided some contributory evidence, suggesting down-regulation of PSMA1 and PSMB2, as well as down-regulation of the PA700/19S regulatory subunit (PSMD13; Table 3). In a pilot immunohistochemical study of early laryngeal carcinomas, from patients treated preoperatively with radiotherapy, the use of the 26S proteasome antibody demonstrated down-regulation of this protein complex. This was significantly associated with the radioresistant tumor subset in this pilot series (P = .05). This putative biomarker may represent a general predictive test for radiotherapy outcome and therefore warrants further investigation in a larger series of tumors.
The primary catalytic site of the 26S proteasome is a 20S core; a barrel-shaped complex composed of rings of alpha and beta subunits [20]. In isolation, the 20S Proteasome is inactive because substrates are unable to access the active sites. Thus, the 26S proteasome requires activation by the PA700/19S, which binds to both ends of the 20S core to form the 26S proteasome. The 26S proteasome recognizes and degrades proteins that have been modified by polyubiquitination.
The mechanisms of how the down-regulation of the 26S proteasome may contribute to radioresistance remain unclear because this complex is involved in the degradation of proteins from a wide variety of functional pathways. Compromising the function of the 26S proteasome may result in the increased stability of those proteins required for the development of resistance to radiotherapy. Key cell cycle regulators (e.g., cyclins and cyclin-dependent kinase inhibitors) are major substrates for proteasomal degradation. Thus, it is possible that the inefficient degradation of these proteins will prevent cell cycle arrest in response to radiotherapy-induced DNA damage. The turnover of antiapoptotic proteins such as Mcl-1 and Bfl-1/A1 is also under the influence of the 26S proteasome [21,22]. Thus, defective proteasomal degradation may result in the inappropriate stabilization of those proteins harboring cell survival properties.
It has also been shown that the ubiquitin-proteasome system is functionally linked with autophagy, a second major intracellular protein degradation system [23]. Autophagy is usually responsible for the degradation of long-lived proteins and other cellular components, but evidence suggests that this system also plays a compensatory role and is activated by proteasome inhibition [24]. Furthermore, numerous studies have shown that the induction of autophagy is protective against cell death [23–27] and inhibition of autophagy concurrent with irradiation has been shown to enhance cytotoxicity of radiotherapy in resistant cancer cells [27]. This provides a possible mechanism by which down-regulation of the 26S proteasome may result in radioresistance.
It is also known that interactions occur between the 26S proteasome and major DNA repair pathways, including the base excision repair and DNA DSB repair pathways [28–30]. The 26S proteasome may degrade proteins involved in the DSB repair pathway [28], and therefore, the abnormal down-regulation of the proteasome in radioresistant cells may facilitate an increased DNA rate of repair of the damage induced by irradiation.
There is previous evidence to suggest that the inactivation or down-regulation of the 26S proteasome may be associated with resistance to anticancer therapy [31], and this phenotype has recently been identified as a marker of cancer stem cells [32]. Functional studies using RNAi and/or pharmacological inhibitors are now required to determine whether the down-regulation of the 26S proteasome is a cause or consequence of radioresistance in our in vitro cell culture models because this would be counterintuitive to the clinical use of proteasome inhibitors (e.g., bortezomib) for the radiosensitization of tumor cells to the cytotoxic effects of radiotherapy.
Acknowledgments
The authors thank John Hayes of the Biomedical Research Centre, University of Dundee, UK, for providing the GSTM3 antibody. J. K. A. Jameel assisted with the initial development of the radioresistant cell line model. P. Nix and N.D. Stafford provided clinical information for the archival series of laryngeal tumors.
Abbreviations
- ACN
acetonitrile
- CI
total ion confidence score
- 2DE
two-dimensional gel electrophoresis
- DSBs
double-stranded DNA breaks
- DTT
dithiothreitol
- ER
estrogen receptor
- HRP
horseradish peroxidase
- IAA
iodoacetamide
- IEF
isoelectric focusing
- IPG
immobilized pH gradient
- LC
liquid chromatography
- MA
microarray
- MALDI
matrix-assisted laser desorption/ionization
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- m/z
mass to charge
- PR
progesterone receptor
- RT-qPCR
real-time quantitative polymerase chain reaction
- SDS
sodium dodecyl sulfate
- SSBs
single-stranded DNA breaks
- TFA
trifluoroacetic acid
- TOF
time of flight
Footnotes
Parts of this work were funded by small grants to Mr. O. Qutob and L. Griffiths from the Peel Medical Research Trust.
References
- 1.Nix P, Cawkwell L, Patmore H, Greenman J, Stafford N. Bcl-2 expression predicts radiotherapy failure in laryngeal cancer. Br J Cancer. 2005;92:2185–2189. doi: 10.1038/sj.bjc.6602647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta AK, McKenna WG, Weber CN, Feldman MD, Goldsmith JD, Mick R, Machtay M, Rosenthal DI, Bakanauskas VJ, Cerniglia GJ, et al. Local recurrence in head and neck cancer: relationship to radiation resistance and signal transduction. Clin Cancer Res. 2002;8:885–892. [PubMed] [Google Scholar]
- 3.Ma BB, Bristow RG, Kim J, Siu LL. Combined-modality treatment of solid tumors using radiotherapy and molecular targeted agents. J Clin Oncol. 2003;21:2760–2776. doi: 10.1200/JCO.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 4.McKenna WG, Muschel RJ, Gupta AK, Hahn SM, Bernhard EJ. The RAS signal transduction pathway and its role in radiation sensitivity. Oncogene. 2003;22:5866–5875. doi: 10.1038/sj.onc.1206699. [DOI] [PubMed] [Google Scholar]
- 5.Bussink J, van der Kogel AJ, Kaanders JH. Activation of the PI3-K/AKT pathway and implications for radioresistance mechanisms in head and neck cancer. Lancet Oncol. 2008;9:288–296. doi: 10.1016/S1470-2045(08)70073-1. [DOI] [PubMed] [Google Scholar]
- 6.An J, Chervin AS, Nie A, Ducoff HS, Huang Z. Overcoming the radioresistance of prostate cancer cells with a novel Bcl-2 inhibitor. Oncogene. 2007;26:652–661. doi: 10.1038/sj.onc.1209830. [DOI] [PubMed] [Google Scholar]
- 7.Blumenthal RD. Chemosensitivity. Totowa, NJ: Humana Press; 2005. [Google Scholar]
- 8.Smith L, Welham KJ, Watson MB, Drew PJ, Lind MJ, Cawkwell L. The proteomic analysis of cisplatin resistance in breast cancer cells. Oncol Res. 2007;16:497–506. doi: 10.3727/096504007783438358. [DOI] [PubMed] [Google Scholar]
- 9.Watson MB, Lind MJ, Smith L, Drew PJ, Cawkwell L. Expression microarray analysis reveals genes associated with in vitro resistance to cisplatin in a cell line model. Acta Oncol. 2007;46:651–658. doi: 10.1080/02841860601156157. [DOI] [PubMed] [Google Scholar]
- 10.Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cawkwell L, Gray S, Murgatroyd H, Sutherland F, Haine L, Longfellow M, O'Loughlin S, Cross D, Kronborg O, Fenger C, et al. Choice of management strategy for colorectal cancer based on a diagnostic immunohistochemical test for defective mismatch repair. Gut. 1999;45:409–415. doi: 10.1136/gut.45.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyndi M, Sørensen FB, Knudsen H, Overgaard M, Nielsen HM, Overgaard J Danish Breast Cancer Cooperative Group, author. Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: the Danish Breast Cancer Cooperative Group. J Clin Oncol. 2008;26:1419–1426. doi: 10.1200/JCO.2007.14.5565. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen PL, Taghian AG, Katz MS, Niemierko A, Abi Raad RF, Boon WL, Bellon JR, Wong JS, Smith BL, Harris JR. Breast cancer subtype approximated by estrogen receptor, progesterone receptor, and HER-2 is associated with local and distant recurrence after breast-conserving therapy. J Clin Oncol. 2008;26:2373–2378. doi: 10.1200/JCO.2007.14.4287. [DOI] [PubMed] [Google Scholar]
- 15.Thompson A, Brennan K, Cox A, Gee J, Harcourt D, Harris A, Harvie M, Holen I, Howell A, Nicholson R, et al. Evaluation of the current knowledge limitations in breast cancer research: a gap analysis. Br Cancer Res. 2008;10:R26. doi: 10.1186/bcr1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang T, Tamae D, LeBon T, Shively JE, Yen Y, Li JJ. The role of peroxiredoxin II in radiation-resistant MCF-7 breast cancer cells. Cancer Res. 2005;65:10338–10346. doi: 10.1158/0008-5472.CAN-04-4614. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Wang S, Wangtao, Wang Y, Wang J, Jiang L, Li S, Hu X, Wang Q. Upregulation of GRP78 and GRP94 and its function in chemotherapy resistance to VP-16 in human lung cancer cell line SK-MES-1. Cancer Invest. 2009;27:453–458. doi: 10.1080/07357900802527239. [DOI] [PubMed] [Google Scholar]
- 18.Pyrko P, Schönthal AH, Hofman FM, Chen TC, Lee AS. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res. 2007;67:9809–9816. doi: 10.1158/0008-5472.CAN-07-0625. [DOI] [PubMed] [Google Scholar]
- 19.Lee E, Nichols P, Spicer D, Groshen S, Yu MC, Lee AS. GRP78 as a novel predictor of responsiveness to chemotherapy in breast cancer. Cancer Res. 2006;66:7849–7853. doi: 10.1158/0008-5472.CAN-06-1660. [DOI] [PubMed] [Google Scholar]
- 20.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 21.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Kucharczak JF, Simmons MJ, Duckett CS, Gelinas C. Constitutive proteasome-mediated turnover of Bfl-1/A1 and its processing in response to TNF receptor activation in FL512 pro-B cells convert it into a prodeath factor. Cell Death Differ. 2005;12:1225–1239. doi: 10.1038/sj.cdd.4401684. [DOI] [PubMed] [Google Scholar]
- 23.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 24.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. inking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 28.Gudmundsdottir K, Lord CJ, Ashworth A. The proteasome is involved in determining differential utilization of double-strand break repair pathways. Oncogene. 2007;26:7601–7606. doi: 10.1038/sj.onc.1210579. [DOI] [PubMed] [Google Scholar]
- 29.Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 30.Motegi A, Murakawa Y, Takeda S. The vital link between the ubiquitin-proteasome pathway and DNA repair: impact on cancer therapy. Cancer Lett. 2009;283:1–9. doi: 10.1016/j.canlet.2008.12.030. [DOI] [PubMed] [Google Scholar]
- 31.Smith L, Lind MJ, Drew PJ, Cawkwell L. The putative roles of the ubiquitin/proteasome pathway in resistance to anticancer therapy. Eur J Cancer. 2007;43:2330–2338. doi: 10.1016/j.ejca.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 32.Vlashi E, Kim K, Lagadec C, Donna LD, McDonald JT, Eghbali M, Sayre JW, Stefani E, McBride W, Pajonk F. In vivo imaging, tracking, and targeting of cancer stem cells. J Natl Cancer Inst. 2009;101:350–359. doi: 10.1093/jnci/djn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maksymowych WP, Ikawa T, Yamaguchi A, Ikeda M, McDonald D, Laouar L, Lahesmaa R, Tamura N, Khuong A, Yu DT, et al. Invasion by Salmonella typhimurium induces increased expression of the LMP, MECL, and PA28 proteasome genes and changes in the peptide repertoire of HLA-B27. Infect Immun. 1998;66:4624–4632. doi: 10.1128/iai.66.10.4624-4632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]