Abstract
We have generated and made publicly available two very large networks of molecular interactions: 49,493 mouse-specific and 52,518 human-specific interactions. These networks were generated through automated analysis of 368,331 full-text research articles and 8,039,972 article abstracts from the PubMed database, using the GeneWays system. Our networks cover a wide spectrum of molecular interactions, such as bind, phosphorylate, glycosylate, and activate; 207 of these interaction types occur more than 1,000 times in our unfiltered, multi-species data set. Because mouse and human genes are linked through an orthological relationship, human and mouse networks are amenable to straightforward, joint computational analysis. Using our newly generated networks and known associations between mouse genes and cerebellar malformation phenotypes, we predicted a number of new associations between genes and five cerebellar phenotypes (small cerebellum, absent cerebellum, cerebellar degeneration, abnormal foliation, and abnormal vermis). Using a battery of statistical tests, we showed that genes that are associated with cerebellar phenotypes tend to form compact network clusters. Further, we observed that cerebellar malformation phenotypes tend to be associated with highly connected genes. This tendency was stronger for developmental phenotypes and weaker for cerebellar degeneration.
Author Summary
We described and made publicly available the largest existing set of text-mined statements; we also presented its application to an important biological problem. We have extracted and purified two large molecular networks, one for humans and one for mouse. We characterized the data sets, described the methods we used to generate them, and presented a novel biological application of the networks to study the etiology of five cerebellum phenotypes. We demonstrated quantitatively that the development-related malformations differ in their system-level properties from degeneration-related genes. We showed that there is a high degree of overlap among the genes implicated in the developmental malformations, that these genes have a strong tendency to be highly connected within the molecular network, and that they also tend to be clustered together, forming a compact molecular network neighborhood. In contrast, the genes involved in malformations due to degeneration do not have a high degree of connectivity, are not strongly clustered in the network, and do not overlap significantly with the development related genes. In addition, taking into account the above-mentioned system-level properties and the gene-specific network interactions, we made highly confident predictions about novel genes that are likely also involved in the etiology of the analyzed phenotypes.
Introduction
A quarter of century ago a (former) Hewlett-Packard executive famously complained: “If only HP knew what HP knows” [1]. This inability to access invaluable “collective wisdom” is by no means specific to a single community. It is felt acutely in every present-day endeavor involving multi-human exploration of complex phenomena. The problem is especially dramatic in the case of the explosively expanding molecular biology literature. There are thousands of existing biological periodicals and millions of potentially useful publications. New journals are emerging on a weekly basis and new articles accumulate as if deposited by an avalanche.
Understandably, no omniscient repository exists that lists all known (published) molecular events (such as protein–protein interactions) detected in human or murine cells. Although current text-mining tools are imperfect in their extraction accuracy and recall, they do help us to process huge amounts of unstructured text in nearly real time (which humans cannot do), moving us a bit closer to total awareness about the current state of knowledge [2].
Here we describe and make available two large new data sets derived through mining one-third of a million full-text research articles and a complete and up-to-date PubMed collection of journal abstracts. These data sets comprise mouse- and human-specific molecular interactions between genes and/or their products. We present here only the subset of text-mined interaction assertions that involve gene or protein names that we can link to unique identifiers in the standard sequence databases. This choice is determined by the goal of making our data immediately useful for applications that would have difficulty handling ambiguity in gene identity. The complete data are available through the Columbia University (http://wiki.c2b2.columbia.edu/workbench) and the University of Chicago (http://anya.igsb.anl.gov/genewaysApp).
We use our newly generated data to analyze genetic variation related to abnormal cerebellum phenotypes in mouse and human. Our analysis results in a compact set of statistically significant predictions that can be tested experimentally.
Results/Discussion
Gene-centric networks
Text mining with the GeneWays system [3],[4] allows us to capture multiple classes of relationships among biological entities, such as “A phosphorylates B,” “C activates D,” and “E is a part of F.” Table S1 displays the full list of relations that we can extract currently. The system also can recognize multiple classes of biological entities (terms) mentioned in the text: genes, proteins, mRNAs, small molecules, processes (such as cell death and proliferation), tissues, cell types, and phenotypes (such as diabetes and hypertension). While one can immediately think of a wide spectrum of applications where the full diversity of entities must be used, most of the current experimental methods are either gene-centric or genetic loci-centric (e.g., gene expression arrays, ChIP-on-chip, yeast two-hybrid, and genetic linkage or association data).
For this reason, the molecular networks we present here are gene-centric. This means that a given node in the network represents the union of the gene and its products (mRNA(s) and protein(s), if any); we exclude all other types of nodes (such as small molecules and phenotypes). Our practice of collapsing multiple nodes to a single node (gene plus mRNA plus protein) does not lead to a loss of information, because most of the physical interactions are defined for specific types of molecules. For example, in our restricted network relationship, “phosphorylate” can link only a pair of proteins, one acting as a protein kinase and another as the kinase's substrate, but not a gene and an mRNA. Furthermore, each original sentence used to extract the relation is preserved in the data set, along with the extracted fact and the reference to the appropriate paper, so that additional disambiguation can be conducted later, if required. We refer to each pair of extracted relationships and the original snippet of text as an action mention, as opposed to action, which is a relation disconnected from the source text and potentially mapped to multiple distinct action mentions.
A single pair of nodes in our text-mined network can be connected with multiple edges. These edges (interactions) can be undirected (we treat “A binds B” and “B binds A” as identical) or directed (“C activates D” is not the same as “D activates C”). We also subdivide edge types into two groups: logical and physical. Logical interactions include a family of regulatory relations that can be either direct (physical contact between two molecules) or indirect (mediated by one or more other molecules), such as activate, inhibit, and regulate (see Table S1). Physical interactions are by definition direct, such as methylate, bind, glycosylate, and cleave (see Table S1). The distinction between physical and logical interactions is important in understanding the data sets that we describe here. GeneWays ontology [5] includes a number of relationships between molecules that are neither physical nor logical interactions (for example, A is an ortholog of B, or C is part of D). We call this class of relations other.
Reducing the noise level
In typical free text, gene names are dissociated from any references to gene-annotation databases. Furthermore, the “raw” text-mined molecular-interaction data are vast (GeneWays 7.0 comprises more than 8 million action mentions) but rather noisy: the error rate is close to 35% [6]. To get to smaller, cleaner, species-specific networks, we performed the following steps.
First, out of the complete network we retained only those gene names that can be linked to either human or mouse sequence database entries (normalization step) (see “Mapping names to genes” in the Text S1). Second, we filtered out relationships that are not molecular interactions and collapsed multiple edges between two nodes into a single edge. Third, we weeded out “raw” text-mined statements that did not meet our precision threshold (precision is defined as the proportion of correctly extracted statements among all those automatically extracted by a system). The third step was conducted automatically, using our automated curator engine [6], which has near-human curation precision (see the “Quality-of-extraction assessment” section in the Text S1.).
The first step resulted in the H70 and M70 networks (human- and mouse-specific GeneWays 7.0), in which nodes can be connected by multiple directed or undirected edges. The second step led to generation of the H70-PL and M70-PL networks (PL stands for physical and logical), where direction of edges was abandoned. The third step, assigned a precision threshold of 0.9 (90% of action mentions are correct), produced even smaller data sets, H70-PL0.9 and M70-PL0.9. Table 1 provides an overview of these networks at different levels of granularity. All intermediate data sets in this pipeline of data filtering are available for third-party computational analyses (see Datasets S2 to S5)
Table 1. Molecular networks and their properties.
| Network tested in our analysis | How each network was generated | Ordered relations between genes | Node type | Node count | Interaction count | Action mention count (instances of relations mentioned in text) |
| GW70 | Text-mining | Y | Name | 1,759,377 | 5,934,024 | 8,424,449 |
| H70 | Filtering of GW70: all human-specific relations | Y | Gene | 9,501 | 223,425 | 431,326 |
| H70-PL | Filtering of GW70: all physical and logical human-specific interactions | N | Gene | 8,186 | 63,449 L: 42,791 P: 8,934 L,P: 11,724 | 306,531 |
| H70-PL0.9 | Filtering of GW70: physical and logical human-specific interactions, 90% precision | N | Gene | 7,793 | 52,518 L: 35,811 P: 7,385 L,P: 9,322 | 261,733 |
| H70-P0.9 | Filtering of GW70: physical human-specific interactions, 90% precision | N | Gene | 5,453 | 16,707 | 61,826 |
| M70 | Filtering of GW70: all mouse-specific relations | Y | Gene | 8,049 | 250,774 | 492,122 |
| M70-PL | Filtering of GW70: all physical and logical mouse-specific interactions | N | Gene | 7,975 | 70,445 L: 47,723 P: 9,586 L,P: 13,136 | 357,958 |
| M70-PL0.9 | Filtering of GW70: physical and logical mouse-specific interactions, 90% precision | N | Gene | 7,600 | 57,786 L: 39,534 P: 7,860 L,P: 10,392 | 305,446 |
| M70-P0.9 | Filtering of GW70: physical mouse-specific interactions, 90% precision | N | Gene | 5,356 | 18,252 | 69,360 |
| HRPD | Manual curation of literature [7] | N | Gene | 9,460 | 37,081 | ∼45,000 |
In addition, we produced networks with non-redundant edges and solely physical interactions, H70-P0.9 and M70-P0.9. As in the previous data sets, to filter these networks we used a precision threshold of 0.9.
Evaluating the precision of the data
To evaluate the quality of the H70-PL0.9 network, we chose two random sets of logical and physical action mentions, a hundred mentions each, and asked an expert to evaluate their correctness. The expert commented on two steps of the process: whether the action mention is correctly extracted by the GeneWays system and, if the answer was “yes,” whether the corresponding gene names were correctly mapped to sequence identifiers. This allowed us to measure the absolute precision of the H70-PL0.9 network, the precision of term mapping, and the overall precision over the information extraction and term mapping stages.
The physical action mentions set indicated a precision of 0.8, with a confidence interval (CI) of [0.71, 0.87]. (We use CI at the 95% level of significance consistently throughout this paper.) The logical action mentions set showed a higher precision of 0.91, CI: [0.84, 0.95]. Because in our data set the number of logical interactions exceeds the number of physical interactions by more than two-to-one (2.49∶1), the overall precision of the HL70-PL0.9 data set is close to the target value of 0.9 (0.88). Term-to-sequence mapping precision was 0.89 (CI: [0.84, 0.93]) and 0.87 (CI: [0.81, 0.91]) for physical and logical action mentions, respectively (see Table 2).
Table 2. Evaluation of the precision of the H70-PL0.9 dataset.
| Evaluation Set | Information extraction precision | Gene name mapping precision | Overall precision |
| Physical 100 action mentions from H70-PL0.9 | 0.80 (0.71, 0.87) 80/100 correctly extracted | 0.89 (0.84, 0.93) 143/160 terms correctly mapped | 0.66 (0.56, 0.74) 66/100 correctly extracted and mapped |
| Logical 100 action mentions from H70-PL0.9 | 0.91 (0.84, 0.95) 91/100 correctly extracted | 0.87 (0.81, 0.91) 158/182 terms correctly mapped | 0.69 (0.59, 0.77) 69/100 correctly extracted and mapped |
100 physical and 100 logical action mentions were evaluated. The two steps of processing—GeneWays system extraction and the mapping of gene names—were evaluated separately in addition to the evaluation of the overall process.
Despite the favorable precision of the GeneWays extraction and the per-term mapping, the precision over both steps is less impressive: 0.66 (CI: [0.56, 0.74]) and 0.69 (CI: [0.59, 0.77]) for the physical and logical datasets, respectively. The reason for the lower overall result is the multiplicative calculus of the probability of not making an error: The overall precision of a term-mapped logical action is a product of the information extraction precision and the precision of two independent term mappings: 0.91×0.87×0.87 = 0.69.
Thus far we have evaluated the quality of extraction and mapping of action mentions. Recall that the same relation (action) between a pair of genes can be independently extracted from multiple sentences, generating distinct action mentions. Intuitively, the precision of an action (because an action is correctly extracted if at least one of its associated action mentions is correctly extracted) should be at least as high (or higher) than precision of the corresponding action mentions. To evaluate this precision, we sampled a hundred random actions from the H70-PL0.9 dataset, asked an expert to evaluate them at the levels of extraction and term mapping, and obtained an estimate of action-level two-stage precision of 0.74, CI: [0.65, 0.82]. This estimate is higher than the estimate of two-stage action mention precision (0.66 or 0.69). We believe that the action-level precision is more relevant to real-life applications in which scientists tend to care primarily about the precision of actions (statements distilled from multiple sources) rather than about their individual instances linked to text.
Note that the precision discussed in this section reflects only properties of our information extraction system and not the verity of published data.
Comparison with HPRD
Several publicly accessible databases generated by manual analysis of research literature are available, including the Human Protein Reference Database (HPRD) [7],[8], Reactome [9], the Biomolecular Interaction Network Database [10], and the Database of Interacting Proteins [11]. These four data sets, along with a few others, were carefully compared in a recent study [12]. HPRD is by far the largest of the four.
As another quality control measure for our study, we compared our data with HPRD 7. The HPRD 7 network [7],[8] comprises 9,460 nodes (unique gene identifiers) and 37,081 edges, compared to 7,793 nodes and 52,518 edges in the H70-PL0.9 network. The H70-P0.9 network comprises 5,453 nodes and 16,707 edges; the node-wise and the edge-wise overlaps of H70-P0.9 with the HPRD networks are 4,543 and 4,877, respectively.
The HPRD and H70-PL0.9 networks share 5,945 unique gene-specific nodes. Out of the possible maximum of 17,668,540 interaction pairs between these nodes, the HPRD network has 23,662 and the H70-PL0.9 covers 43,496. We would expect a random overlap of about 58 interactions, while in reality we observe 7,577. The expected and the observed values are so far apart that the p-value (obtained with a hyper-geometric overlap test) is effectively zero—that is, the apparent overlap between the two sets of data is extremely non-random.
Gold standard evaluation
Because human-curated databases may still harbor errors [13], we also compared our literature-mined dataset to a small set of high quality interactions produced by careful manual verification of a set of interactions shared by several human-curated databases [13].
In a recent study, Cusick et al. sought to evaluate the ultimate (truth) quality of the molecular interaction datasets generated via manual curation of the literature [13]. The authors selected two sets of curated interactions: one consisted of interactions that were curated in multiple databases and that were supported by multiple manuscripts and the other consisted of interactions supported by a single publication. They then carefully recurated the selected interactions and were able to estimate the corresponding error rates. As a byproduct of the evaluation, the authors produced two relatively small datasets, LC-multiple and LC-single, with 110 and 92 interacting pairs respectively, of exceptionally high-quality curated interactions. The LC-multiple set contained the interactions that were supported by multiple manuscripts even after the recuration and the LC-single set contained the interactions with one supporting manuscript that was confirmed during the recuration. The LC-multiple set subsequently was used as a gold standard for the evaluation of high-throughput yeast two-hybrid assays in a second manuscript by the same group, in which an additional random set of 188 supposedly non-interacting pairs (the Negative set) was selected [14].
We used the LC-multiple, LC-single, and Negative sets as comparison standards for our own literature-mined networks (see Table 3). It is reassuring that our H70-PL network covers nearly 70% (75 out of 110 pairs) and that our most filtered human network, H70-P0.9, covers more than 55% of the well-supported interactions in the LC-multiple set. The more obscure interactions from the LC-single set are not covered as well (i.e., H70-P0.9 contains about 20% of the LC-single set). However, given that we have processed only a small portion of all of the scientific literature with a system that highly favors precision over recall, being able to recover 20% of the interactions supported by a single article is surprisingly high. Finally, our networks do not contain any of the interactions listed in the Negative set. For comparison, the last two lines in Table 3 give the results for the two high-throughput assays MAPPIT and Y2H-CCSB evaluated in Figure 2 of [14].
Table 3. Overlap with the comparison standards.
| Test Set | |||
| Network | LC-multiple (110 pairs) | LC-single (92 pairs) | Negative (188 pairs) |
| H70-PL | 75 | 28 | 0 |
| H70-PL0.9 | 73 | 27 | 0 |
| H70-P0.9 | 61 | 19 | 0 |
| MAPPIT | 19 | N/A | 2 |
| Y2H-CCSB | 15 | N/A | 0 |
Precision versus recall
The performance of text-mining methods is commonly evaluated using two metrics: precision and recall. For information-extraction systems, precision is defined as the proportion of correctly extracted statements among all those automatically extracted by a system. The recall is the ratio between the number of statements correctly extracted by the system and the total number of statements that can be extracted from the original text by a hypothetically perfect system. In a less than perfect system, recall and precision are antagonistic: one is increased at the expense of the other.
In this study we favored precision at the expense of recall: We explicitly used a statement precision threshold as a filtering criterion. We also excluded actions with ambiguous gene names and disqualified some 105 potentially useful instances of text-mined intramolecular relations that fit neither physical nor logical categories (such as contain and is a homolog of), thus worsening recall and improving precision. In addition, we used only those actions that involve either genes or their products (and no other entity classes).
While our human-specific network, which unifies physical and logical interactions (H70-PL0.9), is larger than HPRD 7, the relationship is reversed for the physical-interaction (H70-P0.9) data set and HPRD 7. This is because we filtered out from our data numerous physical action mentions that did not pass our precision threshold. (Note that HPRD 7 incorporates high-throughput interaction data that is probably distinct from the small-scale experimental data published in research papers, in terms of error patterns.) Nevertheless, the HPRD 7 data sets and our data sets are very different. The joint interaction coverage of HPRD 7, H70-P0.9, and M70-P0.9 ortholog data sets is more than twice as large as the coverage of HPRD 7 alone (Figure 1); this is enough to merit the use of a union of these networks in biological applications.
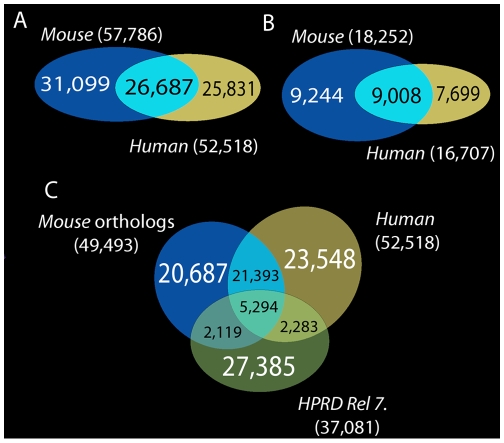
Figure 1. Network interaction-overlaps.
A. Overlap between the human and mouse PL (physical and logical) networks. B. Overlap between the human and mouse P (physical) networks. Interactions in A and B are compared through gene orthology. C. Composition of the union network (GeneWays human (H70-PL0.9), HPRD network, and GeneWays mouse (M70-PL0.9) orthology).
Because we are making the “raw” (unfiltered) statements publicly available, anyone interested in using our data can apply his/her own custom-made filtering process to achieve the desired balance between recall and precision in the output.
Human and mouse networks
Two genes residing in genomes of distinct species can either share a common origin (homology) or be unrelated. Homologs come in at least in two flavors [15]: orthologs and paralogs. Two genes in, say, human and mouse, are orthologs if they were separated by a speciation event. If, in addition to speciation, an intragenomic gene duplication occurred, separating two genes from a common ancestral gene, they are paralogs. For example, human and mouse embryonic β-globins are orthologs, but mouse α-globin is a paralog of human β-globin.
Physical interactions between molecules are not immutable over long evolutionary intervals [16]. Nevertheless, an interaction between two proteins discovered in one species has a reasonable chance of existing between orthologs of these proteins in another species if the two species are closely related. Therefore, if we know of interacting molecules in one species and can identify orthologous molecules in another species, we can formulate hypotheses about the existence of orthologous interactions in the latter species. All such computationally formed hypotheses are subject to experimental validation.
Mouse and human genomes are separated by more than 100 million years of independent evolution [17], but mouse genetics and molecular biology are commonly used to understand human phenotypes in health and disease. Therefore, we decided to compile a molecular-interaction network summarizing the wealth of knowledge for humans and mice. We used orthology-mapping of human and mouse genes to connect the two networks. (Reactome's developers [9] used a similar strategy with their manual compilation of data.) Such a network could potentially have a multitude of practical applications.
We assembled our network by combining mouse- and human-specific networks extracted from the biomedical literature using text-mining tools. We used human-to-mouse gene orthology mapping provided by the Mouse Genome Database [18],[19]. Some of the mouse interactions could not be mapped to corresponding human interactions because at least one of the involved genes lacked known human orthologs. We transferred by mouse-to-human orthology-mapping 49,493 and 16,317 interactions for physical-logical and physical networks, respectively. These orthology-mapped interactions are subsets of the 57,786 and 18,252 interactions in the physical-logical and physical networks, respectively. Although a large number of interactions occur both in humans and mice individually (see Figure 1), the double-confirmed overlap constitutes only about a third of all interactions in the union network (see Figure 1 A and B). Figure 1 C shows a three-way Venn diagram for our text-mined interactomes (human and mouse) and the HPRD dataset. Clearly, all three networks contain a substantial number of unique interactions that merit their joint consideration in biological applications (see Dataset S6).
To illustrate an application of our data to the analysis of mammalian phenotypes, we performed mapping of mouse cerebellar phenotypes (related to ataxia) to the three-data set network.
Mapping ataxia phenotypes to the mouse-human network
The word ataxia (αταξια—“lack of order”), in its English usage, refers to a lack of muscular coordination in an animal body. Humans with ataxia often have difficulty walking, talking, maintaining posture and balance, controlling eye movements, holding and manipulating objects, gesturing, and even swallowing food. In a mammalian brain, the cerebellum is predominantly responsible for spatial and temporal coordination of complex neuromuscular processes. Cerebellar function is also essential for cognition sensory discrimination [20]. Most cases of ataxia are associated with either environmental or genetic damage to this brain region. The typical environmental triggers of ataxia include head trauma, viral infections, and exposure to recreational or medicinal poisons, such as alcohol, lithium carbonate, tranquilizers, antipsychotics, and the anticonvulsant carbamazepine. Genetic factors include a diverse spectrum of genomic aberrations that cause abnormal development and/or premature degeneration of the cerebellum. Ataxia can be severely debilitating and, unfortunately, the phenotype is reversible in only a minority of cases (such as those caused by short-term alcohol intake).
Mouse and human geneticists who study brain phenotypes typically group developmental malformations by the anatomical structures that are affected. As brain topology in three-dimensional space does not lend itself readily to verbal description, we provide three projections of a typical mouse brain in Figure 2 (see also the interactive model in Figure S1). Moving front-to-back in the external view of the mouse brain, there are two olfactory bulbs followed by hemispheres of cerebral cortex that are immediately adjacent to the cerebellum and brainstem (see Figure 2 A–C). We focus here on the cerebellum (literally, “little brain”) that contains involuted cortex with narrow leaf-like gyri (“folia,” see Figures 3 A and C). Like the brain itself, the cerebellum has two hemispheres with a worm-like medial structure, the vermis, between them (Figure 3 A and B).
Figure 2. Morphology of mouse brain: olfactory bulbs, cerebral cortex, midbrain, cerebellum, and spinal cord are labeled.
A. Top view: a mouse brain is shown next to an outline of a mouse head. B. Side view of a mouse brain superimposed with a mouse head. C. Perspective view of a mouse brain and head.
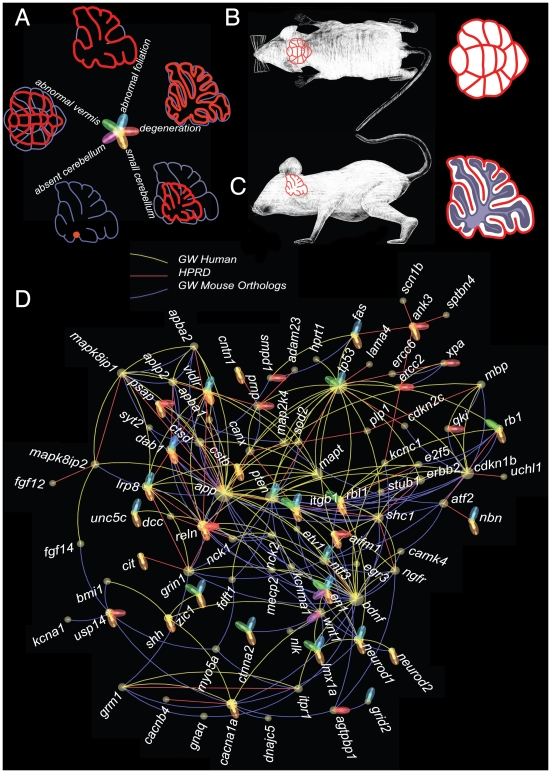
Figure 3. Molecular-interaction network integrating genes related to the ataxia phenotype.
The network was identified by selecting the human orthologs of the mouse genes associated with ataxia in the MGI database. Interactions from the three different sources are indicated with different edge colors. The flower-like design of nodes indicates the specific subset of cerebellar phenotypes associated with each gene. A. Schematic representation of the abnormal cerebellum phenotypes and assignment of petal phenotypes. The thin gray line represents what a normal cerebellum should look like and the red line shows the observed cerebellum. B and C. Schematic representation of a normal cerebellum in relation to whole mouse body in top view B and in side view C. D. The largest connected component of the ataxia sub-network. The abnormal cerebellum phenotype assignments are shown with the flower petals, the size of a node represents the number of interactions it is involved in, and the color of the edges represents the source network as defined in the legend.
In both humans and mice, a collection of genetic aberrations exist that are known to predispose the bearer to specific cerebellar abnormalities. For computational implementation it is convenient to represent phenotypic variations of cerebellar structure with hierarchically ordered categories in a mammalian phenotype (MP) ontology [21]. We focused on five broad anatomical/cerebellar causes of ataxia which can be observed as structural abnormalities in brain imaging studies (such as MRI scans) or histological analysis. These phenotypes are represented with MP concepts: degeneration (MP:0000876), abnormal foliation (MP:0000857 and MP:0000853), abnormal vermis (MP:0000864), small cerebellum/cerebellar hypoplasia (MP:0000852 and MP:0000851), and absent cerebellum (MP:0000850).
Cerebellar degeneration is abnormal death of cerebellar neurons—the cerebellar folia become narrower over time and are separated by irregular and wider spaces compared with those in a healthy brain (see Figure 3 A). As with other major insults to the cerebellum, degeneration reveals itself in abnormalities in body balance, jerky movements of limbs, unsteady (wide-legged) gait, and irregularities of speech (slurred or slow) and eye movement (nystagmus, or rapid involuntary eye movements). Most defined degenerative ataxias affect the fully mature cerebellum, but a small subset of degenerative ataxias have a developmental onset [22].
Abnormal foliation typically involves the absence of some of the cerebellar folia and irregular shape of those that are present. In normal individuals, cerebellar foliation is stereotypical, with the basic folial pattern conserved between mice and humans. Disruption of foliation disrupts the topographical map of incoming and outgoing neuronal connections [23].
An abnormal vermis is typically reduced (compressed and distorted) compared with a normal one, or it can even be completely missing (see Figure 3 A). Clinical outcome is variable [24]. Dandy-Walker malformation is one of the well-known birth defects in humans and mice that are defined by an abnormal vermis. In addition to an aberrant vermis, Dandy-Walker malformation frequently involves enlargement of the fourth brain ventricle and an increase in fluid-filled space around the brain [25]. It is not uncommon in clinical reports to find an abnormal vermis coupled with other cerebellar malformations [26],[27].
Small cerebellum, or cerebellar hypoplasia, refers to phenotypes in which the cerebellum, while present, does not develop to normal size (see Figure 3 A). In humans, cerebellar hypoplasia can lead to delayed or undeveloped speech, difficulties with walking and maintaining balance, mental retardation, floppy muscle tone, nystagmus, and seizures. In its worst forms, cerebellar hypoplasia can be completely debilitating and even lethal [28].
Absent cerebellum is infrequent in adult humans and mice, possibly because in most cases it causes early death. Rarely, individuals are only mildly affected. For example, a documented brain autopsy of a 38-year-old individual who accidentally electrocuted himself revealed a virtually absent cerebellum [29]. The individual was apparently functional and capable of conducting all common human activities, including gesturing, talking, performing complex manual work, and participating in social interactions. Some have proposed that an absent cerebellum is therefore less disabling than a present, but abnormal cerebellum [30].
Fortunately, the Mouse Genome Database (MGD, [18]) uses the MP ontology to link genetic variation in mouse genes to phenotypic aberrations that are causally related to known genomic changes. We were able to use the MGD to associate 244 human genes (with the help of the human and mouse orthology) with the five ataxia phenotypes described above and with ataxia (MP:0001393) (see Table S7). By integrating mouse (M70-PL0.9), human (H70-PL0.9), and HPRD (Release 7) networks through human–mouse gene orthology mapping, we obtained a larger network of interacting human genes with annotation of ataxia phenotypes generated in mouse studies. The largest connected component of the ataxia graph includes 88 human genes. These 88 genes are connected with 145, 147, and 72 interactions derived from human GeneWays, mouse GeneWays, and HPRD, respectively (see Figure 3 D).
Our analysis of ataxia-related phenotypes in the context of a molecular network generated rather curious and statistically significant results, as described in the following section.
Observations and predictions derived from analysis of ataxia phenotypes in the context of a molecular-interaction network
Same-gene co-association of phenotypes
First, we looked at how often genetic aberrations causing distinct cerebellar phenotypes occur within the same gene (see Tables S2 and S3). Within the five phenotypes considered here, four appear to be associated with errors in development of the organ (abnormalities of foliation and vermis, and small or absent cerebellum), while the fifth phenotype is related to degeneration of the already-developed organ.
Genetic variation causing cerebellar degeneration does not seem to occur at more than random frequency in the same gene as variation causing the “developmental” phenotypes. Put differently, cerebellar degeneration appears to be statistically independent of the other cerebellar malformation phenotypes in terms of genetic variation, given our current sample size. In contrast, the four developmental phenotypes are highly significantly correlated pair-wise in terms of their gene associations (see Tables S2 and S3). This indicates that the developmental phenotypes are genetically entangled, in that they are associated with highly overlapping sets of genes. It is likely that pleiotropic genetic variation exists that can cause more than one of the four phenotypes simultaneously. Although cell death is a normal part of development, the lack of overlap between these two phenotypic classes is not surprising given our current understanding of these disorders. Many genes causing cerebellar degeneration when mutated are involved in neuronal metabolism and are widely expressed throughout the brain. The high metabolic rate of the large cerebellar Purkinje neurons may predispose these specific neurons to lethal metabolic stress when they are not functioning normally [31]. In contrast, developmental cerebellar genes are enriched for cell signaling and signal transduction molecules.
Network clustering of phenotype-specific genes
We next asked if phenotype-specific genetic variation in our data set maps to non-randomly clustered groups of genes within our text-mined molecular network. To test the significance of gene clustering, we used three different approaches: a parametric test of clustering (see Text S1) and two non-parametric approaches. Both non-parametric tests are based on estimation of a background distribution of a test statistic (under a no-clustering scenario) by stochastic permutation of the original network. We used the total number of interactions among phenotype-specific genes as test statistics for both non-parametric tests.
The simpler non-parametric test (Ini) involved stochastic sampling of 1,000 gene sets of the same cardinality as the original set of phenotype-associated genes. The slightly more sophisticated non-parametric test (Rwr) generated 1,000 randomly re-wired molecular networks with both node identities (phenotype mapping) and node connectivity values preserved exactly as in the original network. We computed empirical p-values for both tests by comparing the real value of our statistic against the appropriate background distribution.
The three tests are based on different sets of assumptions about the background model. For example, non-parametric tests attempt to emulate the empirical connectivity distributions of genes, whereas the parametric test does not.
We applied the three tests to the five phenotypes chosen for this study and a set of molecular networks: the union of human–mouse GeneWays and HPRD (the union network), HPRD alone, and subset of the union network including only physical interactions (we assumed that all HPRD interactions are physical) (see Table S5). All tests indicated significant clustering of phenotype-specific genes within our networks; the re-wiring test generated the most conservative (high) p-values.
The HPRD network and the physical network allowed detection of highly significant gene clustering in all groups of phenotypes except for degeneration and absent cerebellum. The whole network showed highly significant gene clustering for all studied groups of phenotypes; significance was most impressive for the largest (union) network (see Table S5). It is reassuring that genes for the same phenotype are brought close together in our largest network and this can be used as an argument for the necessity of the extended network. This result might be due in part to a publication bias: Genes that were discovered earlier tend to accumulate a larger number of published logical interactions. However, when we repeated the analysis of the union network with logical interactions removed, the phenotype-specific clustering was still present and significant, albeit at a lower significance level (see Table S5). We observed higher clustering for genes specific to developmental phenotypes. Finally, the most conservative rewiring test failed to detect significant clustering of genes associated with abnormal vermis and absent cerebellum in physical-interaction and HPRD networks.
Cerebellar malformations tend to be caused by highly connected genes
While examining the lists of genes associated with cerebellar malformations, we suspected that the highly connected genes appear in these lists with greater than random frequency. A parametric test designed to test this hypothesis confirmed that this trend is highly significant for most phenotypes, with the exception of cerebellar degeneration and absent cerebellum, analyzed in the context of physical interactions (see Table S4 and Text S1 for a description of the test).
We came up with four possible explanations for this finding. First, the genes involved in embryogenesis often have high connectivity. Second, genetic variation harbored by highly connected genes has elevated the odds of affecting the morphology of the cerebellum. Third, our knowledge about molecular interactions is skewed by the history of the field—that is, genes that were studied earlier are more likely to accumulate a large number of known links (especially logical ones) compared to their peers that were examined much later [32]. By the same logic, well-studied genes are more likely to be tested for causal association with a phenotype than their more obscure counterparts. Another explanation is that more than one of the above factors contributed to the observed abundance of highly connected genes in our lists of genes relevant to cerebellar development.
While all of these hypotheses are testable, we have to wait for adequate data to be available to distinguish among them. We are well aware that the historical knowledge bias is real, especially for logical interactions of well studied and conserved genetic pathways. To test if our degree preference results are due solely to the literature bias, we ran the same test using the protein–protein network produced by fusing two large-scale yeast two-hybrid assays (Y2H with 2936 nodes and 5722 edges) [33],[34]. The strong significance for high-degree preference was diminished in the Y2H network, suggesting that the literature bias could be a factor, but did not disappear completely. (Empirical p-values calculated for the “all” and “abnormal vermis” gene sets within Y2H network were smaller that 0.05, leaving room for the first and second explanation.) Therefore, at the moment, we favor the fourth (composite cause) explanation.
Predicting new phenotype-specific genes
We used a molecular triangulation technique [35] to identify additional candidate genes relevant to each phenotype by studying clusters of phenotype-specific genes. The molecular triangulation technique is designed to analyze the output of a genetic linkage analysis: it detects non-random gene clustering within a molecular-interaction network. Triangulation uses a null model in which the linkage signals are uninformative (unlinked to the phenotype) and any observed gene clustering within a molecular network is accidental. The hypothesis competing with the null assumes that the linkage signal is associated with a group of genes forming a functional cluster within the molecular network. In this application we used as input to molecular triangulation analysis phenotype-specific causal genes (rather than genetic linkage signals). The triangulation analysis generated a surprisingly large number of statistically significant predictions (results are not shown).
We expect that a large number of genes—possibly thousands—contribute to shaping the architecture of an organ as complex as the human or mouse cerebellum. This is clearly supported by the large number of genes expressed in this region of the developing brain [36]. It is not productive, however, for an experimentalist to start with too many predictions. In addition, as long as we have firm evidence that highly connected nodes are over-represented among our phenotype-specific genes, some of the significant predictions may be artifacts of the excessive connectivity. For this reason we modified the triangulation technique (see Text S1) to take into account the apparent connectivities of genes within the molecular network in general and of potentially biased seed genes in particular. While retaining the major assumptions of the original molecular triangulation, this modified test involved node degree-preserving random rewiring of the network, not unlike our test applied to phenotype-specific gene clustering. The degree-preservation addition made the test much more conservative (see Tables 4 and S6 and Figure 4); instead of hundreds or thousands of predictions, only a few survived the significance threshold of a 0.5 level of false discovery rate (FDR) (see Tables 4 and S6 and Figure 4). (FDR is a computational technique used to adjust significance results of statistical testing for multiple tests [37],[38]) The 0.5 FDR threshold means that half our predictions are expected to be false positive. We also conducted a test with stricter FDR levels of 0.1 and 0.01 and indicated the genes that are significant at these levels with bold and underlined bolds gene symbols, respectively, in Tables 4 and S6. Further, the numerous genes that are significant at FDR level of 0.0001 based on our ini-trn analysis (see the Text S1) are listed in Table S9.
Table 4. A subset of the phenotype-specific gene predictions at 0.5 FDR level.
| Gene | Gene | p-value | ||||
| Phenotype | Id | symbol | Initial neighbors | whole | HPRD | physical |
| degeneration | 412 | sts | 2×10−5 | 0.396 | ||
| abnormal foliation | 7471 | wnt1 | ccnd1, ccnd2, ctnna2, en1, gas1, lmx1a, pten | 8×10−7 | 0.999 | 0.372 |
| abnormal vermis | 22943 | dkk1 | en1, fgfr1, msx2, tp53 | 5×10−6 | 0.962 | 0.983 |
| 5076 | pax2 | en2, fgf8, pax5, rb1, tp53 | 8×10−6 | 0.041 | 0.012 | |
| 2253 | fgf8* | en1, en2, fgf17, fgfr1, pax5, zic1 | 1×10−5 | 0.287 | 0.355 | |
| 4487 | msx1 | fgf8, msx2, tp53, zic1 | 1×10−5 | 0.003 | 0.020 | |
| 5077 | pax3 | fgf8, gli2, msx2, tp53, zic1 | 4×10−5 | 0.590 | 0.676 | |
| 8817 | fgf18 | en2, fgf8, fgfr1 | 6×10−5 | 0.115 | 0.194 | |
| 27330 | rps6ka6 | fgf8 | 7×10−5 | 0.332 | 0.294 | |
| 10253 | spry2 | fgf8, fgfr1 | 8×10−5 | 0.521 | 0.037 | |
| 81848 | spry4 | fgf8, fgfr1 | 9×10−5 | 0.391 | 0.026 | |
| 7471 | wnt1 | ctnna2, en1, fgf8, lmx1a | 9×10−5 | 0.999 | 0.113 | |
| 655 | bmp7 | en1, fgf8, fgfr1, msx2, zic1 | 1×10−6 | 0.956 | 0.542 | |
| 268 | amh | rbl1 | 0.0001 | 0.389 | 0.053 | |
| 1745 | dlx1 | fgf8 | 0.0002 | 0.143 | 0.035 | |
| 3223 | hoxc6 | fgf8, fgfr1 | 0.0002 | |||
| 5178 | peg3 | tp53 | 0.0003 | 0.582 | 0.380 | |
| 7476 | wnt7a | en1, fgf8, lmx1a | 0.0003 | 0.953 | 0.849 | |
| 2637 | gbx2 | fgf8, gli2 | 0.0004 | |||
| 2737 | gli3 | fgf8, fgfr1, gli2, zic1 | 0.0004 | 0.026 | 0.002 | |
| 4613 | mycn | pax5, rb1, tp53 | 0.0005 | 0.578 | 0.032 | |
| 429 | ascl1 | fgf8 | 0.0005 | 0.197 | 0.151 | |
| 5727 | ptch1 | fgfr1, gli2 | 0.0005 | 0.600 | 0.533 | |
| 17 | aavs1 | 0.0006 | 1×10−4 | |||
| 3222 | hoxc5 | fgf8 | 0.0006 | |||
| 2535 | fzd2 | 0.0006 | 0.486 | 0.133 | ||
| 8646 | chrd | en2, fgf8, fgfr1 | 0.0008 | 0.908 | 0.390 | |
| 54756 | il17rd | fgf8, fgfr1 | 0.0009 | 0.044 | 0.046 | |
| 5081 | pax7 | fgf8, gli2 | 0.0010 | 0.593 | 0.340 | |
| 985 | cdc2l2 | 0.0011 | 0.040 | |||
| 6677 | spam1 | tp53 | 0.0012 | |||
| small cerebellum | 1020 | cdk5 | ccnd1, ccnd2, cdk5r1, cdk5r2, dab1, dcx, erbb3, pura, rb1, reln | 7×10−6 | 1×10−5 | 9×10−7 |
| 7471 | wnt1 | ccnd1, ccnd2, ctnna2, en1, gas1, lmx1a, lmx1b, shh | 2×10−5 | 0.999 | 0.794 | |
The bold genes are significant at 0.1 FDR level and the bold and underlined genes are significant at 0.01 FDR level. For a complete list of our predictions see Table S6. The *Fgf8 is among our initial genes for the abnormal vermis phenotype.
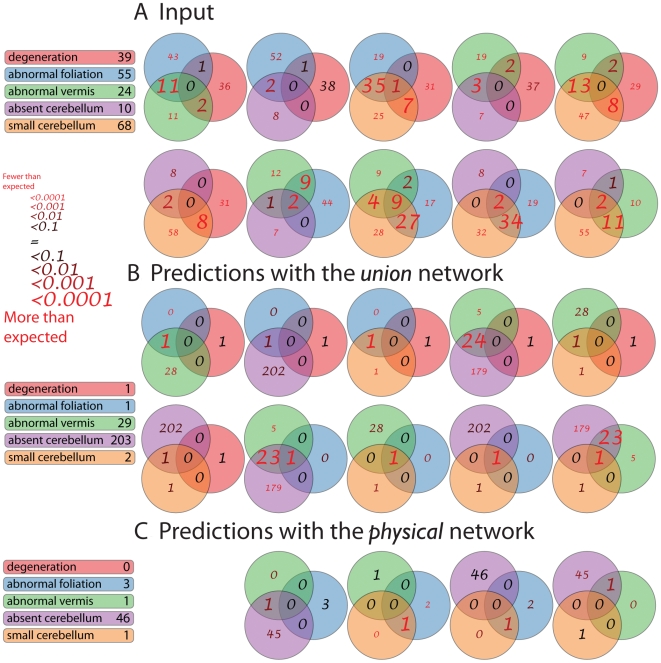
Figure 4. Overlap of genes associated with several cerebellar phenotypes.
A. Venn diagrams for phenotype-specific gene sets retrieved from the Mouse Genome Database. B. Similar Venn diagram for newly predicted candidate genes for the same phenotypes generated through analysis of the union network (both logical and physical interactions). C. Analysis of the union network with only physical interactions retained.
Our leave-one-out cross-validation experiments (see Text S1) demonstrated that the union network clearly outperformed the smaller networks in our comparison in predicting “missing” phenotype-associated gene sets.
As Figure 4 shows, absent and small cerebellum phenotypes were especially rich in gene candidate predictions, whereas cerebellar degeneration was the poorest. Following is an overview of a few selected genes that appeared in the prediction gene lists for both abnormal vermis and absent cerebellum phenotypes.
One of our top predictions, the Ascl1 gene, is involved in neuronal commitment and differentiation [39]. Another gene, Bmp7, encodes a member of the bone morphogenic protein (BMP) family; genes of this family are implicated in a wide spectrum of developmental processes in vertebrates, including bone development [40],[41]. Both genes are expressed in the developing cerebellum (informatics.jax.org), but no cerebellar phenotypes have been described in available mouse mutants. Homozygous Ascl1 mouse mutants die neonatally before extensive cerebellar development. Nevertheless, it has recently been reported that complete loss of Ascl1 alters the development of cerebellar interneurons, oligodendrocytes and astrocytes during late embryogenesis [42] validating our prediction. There is extensive published evidence that Bmp7 can influence multiple aspects of cerebellar development both in vitro and in vivo [43],[44], but again the cerebellum has not been the focus of published mouse mutant phenotypic analysis, hence Bmp7 is not yet associated with any MP category.
An exciting quadruplet of predictions (Fgf8, Fgf18, Spry2, and Spry4) is tightly linked to tissue differentiation pathways. Spry2 and Spry4 are inhibitors of fibroblast growth factor [45],[46] and of receptor-transduced mitogen-activated protein kinase (MAPK) signaling pathways [47]. Fibroblast growth factors (FGFs) and fibroblast growth factor receptors (FGFRs) participate in regulatory processes of tissue pattern formation and cell differentiation. They are implicated in the developmental regulation of all the major systems of the mammalian body, including the limbs, bones, and brain. There are multiple neurological phenotypes, including mood disorders and asocial behavior [48]–[50], linked to genetic variation harbored by genes representing these families. Fgfs have long been implicated in cerebellar development [23]. More recently, Basson et al. [51] have demonstrated that regulation of Fgf signaling by ectopic misexpression of Spry2 has profound effects on cerebellar vermis development. The endogenous roles of these proteins in cerebellar development have not yet been the focus of any phenotypic investigation in the mouse. Our analysis here suggests a second look.
We also found a triplet of PAX genes: Pax2, Pax3, and Pax7. PAX (paired-box) family genes form a small yet critical family of developmental genes encoding transcription factors that regulate cell proliferation, cell-lineage specification, migration, and survival [52],[53]. There is experimental evidence linking PAX genes to the development and function of the cerebellum [54]. Although no Pax3 cerebellar phenotype has yet been described in mice or humans, a Dandy-Walker malformation locus (abnormal cerebellar vermis) has recently been mapped to the upstream region of the Pax3 gene [55], suggesting that Pax3 misregulation may be involved in human cerebellar development. The superimposition of genetic locus information on the cerebellar malformation gene network thus is likely to generate lists of candidate genes for mapped phenotypes.
Yet another pair of predictions, the Hoxc5 and Hoxc6 genes, are neighbors on chromosome 12 in the human genome and belong to the HOX (homeo-box-containing) family. HOX genes are major players in vertebrate embryonic pattern formation, particularly (but not exclusively) in the central nervous system [56]. Again, no cerebellar phenotypes have been reported for these genes. Notably, however, these genes are never expressed in the developing cerebellum but do have well known roles in more posterior regions of the developing central nervous system. Thus, these particular Hox gene predictions highlight the importance of future work to integrate additional known biological information such as expression patterns (for example, GeneSat) into the gene network to generate more biologically relevant predictions.
Additional observations and predictions
See Text S1 and Dataset S1 for our Gene-set enrichment and Cross-Validation experiments.
Conclusion
We have provided two very large molecular-interaction sets for human and mouse genes (Datasets S2, S3, S4, S5). The sets were integrated through gene orthology and are immediately applicable to a spectrum of experimental data analysis tasks (Dataset S6). Our analysis of mouse mutant cerebellar phenotypes, with the aid of our text-mined networks, lead to a number of intuitively reasonable and biologically testable predictions.
Our present study shares its spirit, goals, and some methods with other efforts in the field. For example, one of the most recent studies succeeded in integrating a diverse array of approaches to design a tool generating new disease-related hypotheses [57]. This group was able to combine information extraction [58], biomedical ontology mining, statistical analysis of sequences of natural language tokens, probabilistic analysis of error patterns across data types, computational reasoning, understanding of large-scale experimental datasets, and exploratory visualization in one application.
Because we are releasing our complete set of annotated data to public domain, these data might be instrumental for direct comparison for analogous text-mining results produced elsewhere [59]–[67].
Automated reasoning over text-mined, experimental, and machine-deduced data (Reading, Reasoning, and Reporting, as [57] put it), is likely to become a dominant mode of science in the near future, as size of experimental datasets and complexity of natural system under scrutiny grows.
Methods
GeneWays system and GeneWays 7.0 database
GeneWays is an information extraction (text-mining) system: It ingests computer-encoded full-text research articles or journal abstracts and distills from them a collection of biological relations. The architecture and implementation of the system are described in great detail elsewhere [3], [4], [6], [68]–[71], so here we provide just a brief outline of the system.
In a simplified view, the processing pipeline inside the GeneWays system is a sequence of just two steps. The first step deals with recognition of words or phrases representing important biological entities (such as p53, Alzheimer's disease, or endoplasmic reticulum; computer scientists call this step named entity recognition, NER). The second step deals with detecting relations among the entities (such as p53 activates JAK) and is called information extraction (IE).
Our NER module (MarkIt, [72]) works by following a hierarchy of rules defined by human experts. Our IE module (GENIES, [3],[68]) also is based on a collection of expert-encoded rules, but the underlying mathematical model is a bit more sophisticated (a deterministic context-free grammar). We use MarkIt to identify a spectrum of biological entities, such as disease, process, gene, protein, RNA, small molecule, tissue, and species. We apply GENIES to each individual sentence, trying to reconstruct the most probable steps that led to generation of the sentence. This reconstruction process is called parsing; besides satisfying our academic curiosity, parsing is useful for capturing complex relationships between entities. The results of parsing depend strongly on the formal grammar implemented in the parser.
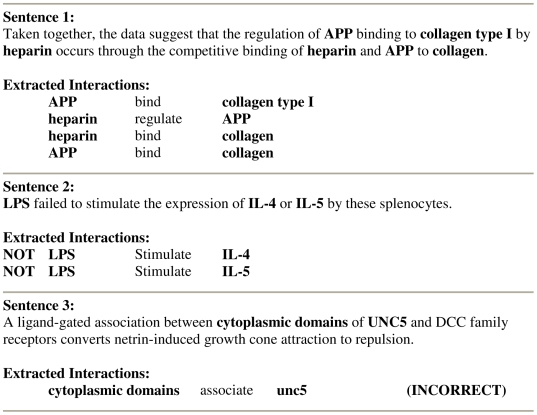
Most of the relations that we can extract from biomedical texts are directional (A activates B is not the same as B activates A) and binary (only two entities are involved, which we call upstream and downstream, according to their position within the relation). A sentence can contain any number between none and dozens of relations. We can think of a typical binary relation as a quadruplet of values: [negation, upstream entity, action, downstream entity] (see Figure 5). Negation allows us to capture negative statements (“In our experiment, AKT failed to phosphorylate BAD”→[not, AKT, phosphorylate, BAD]) as well as positive statements. Relation type (action) indicates semantic connection between the two entities (bind, activate, methylate, transport, is part of, is homolog of, etc.).
Figure 5. Examples of text-mined statements.
Sentence 1: four correctly extracted interactions from one sentence. Sentence 2: two correctly extracted negative interactions. Sentence 3: an incorrectly extracted interaction.
To facilitate automatic reasoning over semantic groups of relations, we arrange them into an acyclic directed graph, where graph arcs represent the relation “is a.” The GeneWays system currently recognizes 391 different action types, 207 of which are frequent (see Table S1).
Textual and text-mined data
To generate the molecular networks described in this study, we analyzed 368,331 full-text articles and 8,039,972 article abstracts from PubMed (see Table S8). The system identified 5,890,150 relations in the full text articles and 2,534,299 relations in the abstracts: in total, 5,934,024 unique relationships. The action types with the largest number of relationships are induce (695,615), bind (532,385), inhibit (386,523), associate (370,133), contain (366,654), and activate (332,336); the numbers in parentheses indicate the abundance of relations of each type in the GeneWays 7.0 database.
Supporting Information
Supplementary experiments; supplementary methods.
(0.34 MB DOC)
Interactive mouse brain head model.
(1.17 MB PDF)
Complete list of Geneways action types.
(0.07 MB XLS)
Pairwise phenotype overlap. The significance is measured based on a whole genome comprised of 25,000 genes.
(0.03 MB DOC)
Pairwise phenotype overlap. The significance is measured based on a ataxia sub-network comprising 172 genes.
(0.03 MB DOC)
Results of the tests for high connectivity of the phenotype genes.
(0.01 MB DOC)
Results of the tests for clustering of the phenotype genes.
(0.02 MB PDF)
Complete gene predictions.
(0.06 MB XLS)
Initial genes.
(0.04 MB XLS)
Text corpus description.
(0.03 MB XLS)
Complete gene predictions (ini-trn, FDR 0.0001).
(1.72 MB XLS)
All enrichment results.
(0.20 MB ZIP)
Human network files.
(5.50 MB DOC)
Human action mentions.
(8.87 MB ZIP)
Mouse network files.
(6.41 MB ZIP)
Mouse action mentions.
(10.07 MB ZIP)
Combined human network.
(6.42 MB ZIP)
Acknowledgments
The authors are grateful to Anna Divoli, Rita Rzhetsky, and Lixia Yao for comments on an earlier version of the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the National Institutes of Health (GM61372 and U54 CA121852-01A1), the National Science Foundation (0438291 and 0121687), and the Cure Autism Now Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Davenport TH, Prusak L. Working knowledge: how organizations manage what they know. Boston, Mass: Harvard Business School Press; 1998. p. xv, 199. [Google Scholar]
- 2.Rzhetsky A, Seringhaus M, Gerstein M. Seeking a new biology through text mining. Cell. 2008;134:9–13. doi: 10.1016/j.cell.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman C, Kra P, Yu H, Krauthammer M, Rzhetsky A. GENIES: a natural-language processing system for the extraction of molecular pathways from journal articles. Bioinformatics. 2001;17(Suppl 1):S74–S82. doi: 10.1093/bioinformatics/17.suppl_1.s74. [DOI] [PubMed] [Google Scholar]
- 4.Rzhetsky A, Iossifov I, Koike T, Krauthammer M, Kra P, et al. GeneWays: a system for extracting, analyzing, visualizing, and integrating molecular pathway data. J Biomed Inform. 2004;37:43–53. doi: 10.1016/j.jbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Rzhetsky A, Koike T, Kalachikov S, Gomez SM, Krauthammer M, et al. A knowledge model for analysis and simulation of regulatory networks. Bioinformatics. 2000;16:1120–1128. doi: 10.1093/bioinformatics/16.12.1120. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Esteban R, Iossifov I, Rzhetsky A. Imitating manual curation of text-mined facts in biomedicine. PLoS Comput Biol. 2006;2:e118. doi: 10.1371/journal.pcbi.0020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, et al. Human protein reference database–2006 update. Nucleic Acids Res. 2006;34:D411–414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vastrik I, D'Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathivanan S, Periaswamy B, Gandhi TK, Kandasamy K, Suresh S, et al. An evaluation of human protein-protein interaction data in the public domain. BMC Bioinformatics. 2006;7(Suppl 5):S19. doi: 10.1186/1471-2105-7-S5-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cusick ME, Yu H, Smolyar A, Venkatesan K, Carvunis AR, et al. Literature-curated protein interaction datasets. Nat Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 16.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 17.Nei M, Glazko GV. The Wilhelmine E. Key 2001 Invitational Lecture. Estimation of divergence times for a few mammalian and several primate species. J Hered. 2002;93:157–164. doi: 10.1093/jhered/93.3.157. [DOI] [PubMed] [Google Scholar]
- 18.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bult CJ, Blake JA, Richardson JE, Kadin JA, Eppig JT, et al. The Mouse Genome Database (MGD): integrating biology with the genome. Nucleic Acids Res. 2004;32 Database issue:D476–481. doi: 10.1093/nar/gkh125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmahmann JD. Disorders of the cerebellum: ataxia, dysmetria of thought, and the cerebellar cognitive affective syndrome. J Neuropsychiatry Clin Neurosci. 2004;16:367–378. doi: 10.1176/jnp.16.3.367. [DOI] [PubMed] [Google Scholar]
- 21.Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Millen KJ, Gleeson JG. Cerebellar development and disease. Curr Opin Neurobiol. 2008;18:12–19. doi: 10.1016/j.conb.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sillitoe RV, Joyner AL. Morphology, molecular codes, and circuitry produce the three-dimensional complexity of the cerebellum. Annu Rev Cell Dev Biol. 2007;23:549–577. doi: 10.1146/annurev.cellbio.23.090506.123237. [DOI] [PubMed] [Google Scholar]
- 24.Parisi MA, Dobyns WB. Human malformations of the midbrain and hindbrain: review and proposed classification scheme. Mol Genet Metab. 2003;80:36–53. doi: 10.1016/j.ymgme.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Ueno H, Yamaguchi H, Katakami H, Matsukura S. A case of Kallmann syndrome associated with Dandy-Walker malformation. Exp Clin Endocrinol Diabetes. 2004;112:62–67. doi: 10.1055/s-2004-815728. [DOI] [PubMed] [Google Scholar]
- 26.Russo R, Fallet-Bianco C. Isolated posterior cerebellar vermal defect: a morphological study of midsagittal cerebellar vermis in 4 fetuses–early stage of Dandy-Walker continuum or new vermal dysgenesis? J Child Neurol. 2007;22:492–500; discussion 501. doi: 10.1177/0883073807299862. [DOI] [PubMed] [Google Scholar]
- 27.Sztriha L, Johansen JG, Al-Gazali LI. Extreme microcephaly with agyria-pachygyria, partial agenesis of the corpus callosum, and pontocerebellar dysplasia. J Child Neurol. 2005;20:170–172. doi: 10.1177/08830738050200021301. [DOI] [PubMed] [Google Scholar]
- 28.Deda G, Caksen H, Icagasioglu D. A fatal case of cerebellar hypoplasia associated with anterior horn cell disease. Genet Couns. 2003;14:253–256. [PubMed] [Google Scholar]
- 29.Leestma JE, Torres JV. Unappreciated agenesis of cerebellum in an adult: case report of a 38-year-old man. Am J Forensic Med Pathol. 2000;21:155–161. doi: 10.1097/00000433-200006000-00013. [DOI] [PubMed] [Google Scholar]
- 30.LeDoux MS, Lorden JF, Ervin JM. Cerebellectomy eliminates the motor syndrome of the genetically dystonic rat. Exp Neurol. 1993;120:302–310. doi: 10.1006/exnr.1993.1064. [DOI] [PubMed] [Google Scholar]
- 31.Soong BW, Paulson HL. Spinocerebellar ataxias: an update. Curr Opin Neurol. 2007;20:438–446. doi: 10.1097/WCO.0b013e3281fbd3dd. [DOI] [PubMed] [Google Scholar]
- 32.Cokol M, Iossifov I, Weinreb C, Rzhetsky A. Emergent behavior of growing knowledge about molecular interactions. Nat Biotechnol. 2005;23:1243–1247. doi: 10.1038/nbt1005-1243. [DOI] [PubMed] [Google Scholar]
- 33.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 34.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 35.Krauthammer M, Kaufmann CA, Gilliam TC, Rzhetsky A. Molecular triangulation: Bridging linkage and molecular-network information for identifying candidate genes in Alzheimer's disease. Proc Natl Acad Sci U S A. 2004;101:15148–15153. doi: 10.1073/pnas.0404315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heintz N. Gene expression nervous system atlas (GENSAT). Nat Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 38.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
- 39.Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, et al. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9:770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- 40.Lee H, Stultz BG, Hursh DA. The Zic family member, odd-paired, regulates the Drosophila BMP, decapentaplegic, during adult head development. Development. 2007;134:1301–1310. doi: 10.1242/dev.02807. [DOI] [PubMed] [Google Scholar]
- 41.Waite KA, Eng C. From developmental disorder to heritable cancer: it's all in the BMP/TGF-beta family. Nat Rev Genet. 2003;4:763–773. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]
- 42.Grimaldi P, Parras C, Guillemot F, Rossi F, Wassef M. Origins and control of the differentiation of inhibitory interneurons and glia in the cerebellum. Dev Biol. 2009;328:422–433. doi: 10.1016/j.ydbio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Alder J, Cho NK, Hatten ME. Embryonic precursor cells from the rhombic lip are specified to a cerebellar granule neuron identity. Neuron. 1996;17:389–399. doi: 10.1016/s0896-6273(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 44.Chizhikov VV, Lindgren AG, Currle DS, Rose MF, Monuki ES, et al. The roof plate regulates cerebellar cell-type specification and proliferation. Development. 2006;133:2793–2804. doi: 10.1242/dev.02441. [DOI] [PubMed] [Google Scholar]
- 45.Glienke J, Fenten G, Seemann M, Sturz A, Thierauch KH. Human SPRY2 inhibits FGF2 signalling by a secreted factor. Mech Dev. 2000;96:91–99. doi: 10.1016/s0925-4773(00)00378-6. [DOI] [PubMed] [Google Scholar]
- 46.Katoh Y, Katoh M. FGF signaling inhibitor, SPRY4, is evolutionarily conserved target of WNT signaling pathway in progenitor cells. Int J Mol Med. 2006;17:529–532. [PubMed] [Google Scholar]
- 47.Tsavachidou D, Coleman ML, Athanasiadis G, Li S, Licht JD, et al. SPRY2 is an inhibitor of the ras/extracellular signal-regulated kinase pathway in melanocytes and melanoma cells with wild-type BRAF but not with the V599E mutant. Cancer Res. 2004;64:5556–5559. doi: 10.1158/0008-5472.CAN-04-1669. [DOI] [PubMed] [Google Scholar]
- 48.Akil H, Evans SJ, Turner CA, Perez J, Myers RM, et al. The fibroblast growth factor family and mood disorders. Novartis Found Symp. 2008;289:94–96; discussion 97–100, 193–105. doi: 10.1002/9780470751251.ch8. [DOI] [PubMed] [Google Scholar]
- 49.Scearce-Levie K, Roberson ED, Gerstein H, Cholfin JA, Mandiyan VS, et al. Abnormal social behaviors in mice lacking Fgf17. Genes Brain Behav. 2008;7:344–354. doi: 10.1111/j.1601-183X.2007.00357.x. [DOI] [PubMed] [Google Scholar]
- 50.Wilkie AO, Patey SJ, Kan SH, van den Ouweland AM, Hamel BC. FGFs, their receptors, and human limb malformations: clinical and molecular correlations. Am J Med Genet. 2002;112:266–278. doi: 10.1002/ajmg.10775. [DOI] [PubMed] [Google Scholar]
- 51.Basson MA, Echevarria D, Ahn CP, Sudarov A, Joyner AL, et al. Specific regions within the embryonic midbrain and cerebellum require different levels of FGF signaling during development. Development. 2008;135:889–898. doi: 10.1242/dev.011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lang D, Powell SK, Plummer RS, Young KP, Ruggeri BA. PAX genes: roles in development, pathophysiology, and cancer. Biochem Pharmacol. 2007;73:1–14. doi: 10.1016/j.bcp.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 53.Robson EJ, He SJ, Eccles MR. A PANorama of PAX genes in cancer and development. Nat Rev Cancer. 2006;6:52–62. doi: 10.1038/nrc1778. [DOI] [PubMed] [Google Scholar]
- 54.Kioussi C, Gruss P. Differential induction of Pax genes by NGF and BDNF in cerebellar primary cultures. J Cell Biol. 1994;125:417–425. doi: 10.1083/jcb.125.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jalali A, Aldinger KA, Chary A, McLone DG, Bowman RM, et al. Linkage to chromosome 2q36.1 in autosomal dominant Dandy-Walker malformation with occipital cephalocele and evidence for genetic heterogeneity. Hum Genet. 2008;123:237–245. doi: 10.1007/s00439-008-0467-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papageorgiou S. HOX gene expression. Austin, Texas New York: Landes Bioscience/Eurekah.com; Springer Science+Business Media; 2007. 149 [Google Scholar]
- 57.Leach SM, Tipney H, Feng W, Baumgartner WA, Kasliwal P, et al. Biomedical discovery acceleration, with applications to craniofacial development. PLoS Comput Biol. 2009;5:e1000215. doi: 10.1371/journal.pcbi.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunter L, Lu Z, Firby J, Baumgartner WA, Jr, Johnson HL, et al. OpenDMAP: an open source, ontology-driven concept analysis engine, with applications to capturing knowledge regarding protein transport, protein interactions and cell-type-specific gene expression. BMC Bioinformatics. 2008;9:78. doi: 10.1186/1471-2105-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li X, Cai H, Xu J, Ying S, Zhang Y. A mouse protein interactome through combined literature mining with multiple sources of interaction evidence. Amino Acids. 2009 doi: 10.1007/s00726-009-0335-7. [DOI] [PubMed] [Google Scholar]
- 60.Kabiljo R, Clegg AB, Shepherd AJ. A realistic assessment of methods for extracting gene/protein interactions from free text. BMC Bioinformatics. 2009;10:233. doi: 10.1186/1471-2105-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bandy J, Milward D, McQuay S. Mining Protein-Protein Interactions from Published Literature Using Linguamatics I2E. Methods Mol Biol. 2009;563:3–13. doi: 10.1007/978-1-60761-175-2_1. [DOI] [PubMed] [Google Scholar]
- 62.Krallinger M, Rodriguez-Penagos C, Tendulkar A, Valencia A. PLAN2L: a web tool for integrated text mining and literature-based bioentity relation extraction. Nucleic Acids Res. 2009;37:W160–165. doi: 10.1093/nar/gkp484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiao D, Wild DJ. Extraction of CYP chemical interactions from biomedical literature using natural language processing methods. J Chem Inf Model. 2009;49:263–269. doi: 10.1021/ci800332w. [DOI] [PubMed] [Google Scholar]
- 64.Chowdhary R, Zhang J, Liu JS. Bayesian inference of protein-protein interactions from biological literature. Bioinformatics. 2009;25:1536–1542. doi: 10.1093/bioinformatics/btp245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics. 2009;10(Suppl 2):S6. doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krallinger M, Leitner F, Rodriguez-Penagos C, Valencia A. Overview of the protein-protein interaction annotation extraction task of BioCreative II. Genome Biol. 2008;9(Suppl 2):S4. doi: 10.1186/gb-2008-9-s2-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang M, Ding S, Wang H, Zhu X. Mining physical protein-protein interactions from the literature. Genome Biol. 2008;9(Suppl 2):S12. doi: 10.1186/gb-2008-9-s2-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. J Biomed Inform. 2002;35:222–235. doi: 10.1016/s1532-0464(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 69.Krauthammer M, Kra P, Iossifov I, Gomez SM, Hripcsak G, et al. Of truth and pathways: chasing bits of information through myriads of articles. Bioinformatics. 2002;18(Suppl 1):S249–S257. doi: 10.1093/bioinformatics/18.suppl_1.s249. [DOI] [PubMed] [Google Scholar]
- 70.Rzhetsky A, Zheng T, Weinreb C. Self-correcting maps of molecular pathways. PLoS ONE. 2006;1:e61. doi: 10.1371/journal.pone.0000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rzhetsky A, Iossifov I, Loh JM, White KP. Microparadigms: chains of collective reasoning in publications about molecular interactions. Proc Natl Acad Sci U S A. 2006;103:4940–4945. doi: 10.1073/pnas.0600591103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krauthammer M, Nenadic G. Term identification in the biomedical literature. J Biomed Inform. 2004;37:512–526. doi: 10.1016/j.jbi.2004.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary experiments; supplementary methods.
(0.34 MB DOC)
Interactive mouse brain head model.
(1.17 MB PDF)
Complete list of Geneways action types.
(0.07 MB XLS)
Pairwise phenotype overlap. The significance is measured based on a whole genome comprised of 25,000 genes.
(0.03 MB DOC)
Pairwise phenotype overlap. The significance is measured based on a ataxia sub-network comprising 172 genes.
(0.03 MB DOC)
Results of the tests for high connectivity of the phenotype genes.
(0.01 MB DOC)
Results of the tests for clustering of the phenotype genes.
(0.02 MB PDF)
Complete gene predictions.
(0.06 MB XLS)
Initial genes.
(0.04 MB XLS)
Text corpus description.
(0.03 MB XLS)
Complete gene predictions (ini-trn, FDR 0.0001).
(1.72 MB XLS)
All enrichment results.
(0.20 MB ZIP)
Human network files.
(5.50 MB DOC)
Human action mentions.
(8.87 MB ZIP)
Mouse network files.
(6.41 MB ZIP)
Mouse action mentions.
(10.07 MB ZIP)
Combined human network.
(6.42 MB ZIP)