Abstract
Transcriptional activators and coactivators overcome repression by chromatin, but regulation of chromatin disassembly and coactivator binding to promoters is poorly understood. Activation of the yeast HO gene follows the sequential binding of both sequence specific DNA-binding proteins and coactivators during the cell cycle. Here we show that the nucleosome disassembly occurs in waves both along the length of the promoter and during the cell cycle. Different chromatin modifiers are required for chromatin disassembly at different regions of the promoter, with Swi/Snf, the FACT chromatin reorganizer, and the Asf1 histone chaperone each required for nucleosome eviction at distinct promoter regions. FACT and Asf1 both bind to upstream elements of the HO promoter well before the gene is transcribed. The Swi/Snf, SAGA, and Mediator coactivators bind first to the far upstream promoter region and subsequently to a promoter proximal region, and FACT and Asf1 are both required for this coactivator re-recruitment.
INTRODUCTION
Chromatin structure can repress transcription, and chromatin remodeling, histone modification, and nucleosome displacement all play important roles in gene activation (Li et al., 2007). Chromatin disassembly often occurs at promoters when a gene is activated, allowing transcription factors to access to DNA (Williams and Tyler, 2007; Workman, 2006). Transcriptional coactivators or chromatin factors can be required for nucleosome displacement at specific promoters (Adkins et al., 2004; Biddick et al., 2008; Schwabish and Struhl, 2007; Verdone et al., 2002). Swi/Snf, SAGA, and Mediator are transcriptional coactivator complexes that promote transcription activation by changing chromatin. The Swi/Snf complex uses ATP hydrolysis to remodel nucleosomes, thus increasing DNA accessibility and facilitating transcription. The SAGA complex contains the Gcn5 acetyl transferase that acetylates N-terminal histone lysine residues and thus promotes transcription, and it also has subunits that interact with the TATA-binding factor and with activators. It is believed that the Mediator complex recruits basal factors to the promoter and stimulates preinitiation complex formation; Mediator can also be recruited to upstream promoter elements in the absence of basal factors (Bhoite et al., 2001; Cosma et al., 2001; Govind et al., 2005).
The HO gene has a large promoter by yeast standards, with mapped promoter elements extending nearly 2 kb (Fig 1A). The upstream URS1 region of the promoter (−1900 to −1000) contains two binding sites for the Swi5 DNA-binding protein, while the URS2 region at −900 to −200 contains eight sites for SBF. Previous work has shown that Swi5 recruits the Swi/Snf and Mediator coactivators to URS1, and that SAGA is subsequently recruited to the URS2 region of the promoter (Bhoite et al., 2002; Cosma et al., 1999). SBF, composed of Swi4 and Swi6, binds to sites within URS2 and is thought to be the ultimate activator of HO (Cosma et al., 2001). Some previous studies were conducted in ash1 mutant strains (Cosma et al., 2001; Cosma et al., 1999); HO is normally expressed only in mother cells, and the ash1 mutation allows HO expression in daughters as well (Bobola et al., 1996; Sil and Herskowitz, 1996). The ash1 mutation was thought to only increase the HO signal, but Ash1 is now known to be associated with a histone deacetylase complex (Carrozza et al., 2005). The ash1 mutation changes HO regulation in mother cells, allowing Swi/Snf to bind to HO URS1 in the absence of the normally required Gcn5 acetyltransferase (Mitra et al., 2006). Here we use chromatin immunoprecipitation (ChIP) to examine factor binding under native conditions in ASH1 strains. We show that three coactivators bind first to URS1 and later at URS2, and that the subsequent binding at URS2 is dependent on SBF, the FACT chromatin reorganizing factor, and the Asf1 histone chaperone.
Fig 1. Nucleosome eviction occurs in waves along the HO Promoter.
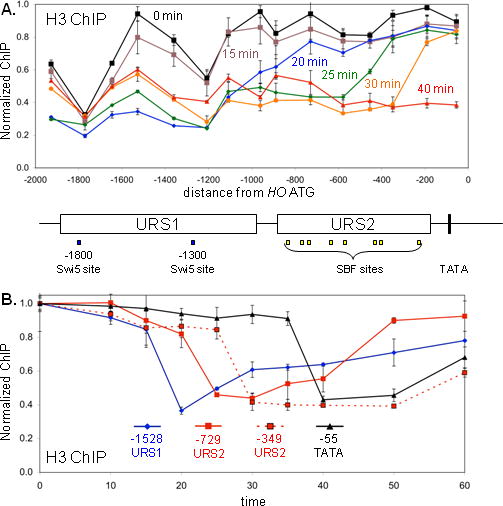
A. DY6669 cells (GALp::CDC20) with a GALp::CDC20 allele were synchronized in mitosis by removing galactose, followed by release by addition of galactose (t = 0). The CDC20 arrest is at the G2/M transition, and HO expression at 40 min following release corresponds to late G1 phase. Nucleosome occupancy was measured by H3 ChIP using samples taken at various times after the release. This data along with additional time points are shown in Suppl Fig S1. The DNA in the experiment was sheared to an average of 350 bp, as opposed to approximately 550 bp in the other ChIP experiments. URS1, URS2, the Swi5 and SBF binding sites are shown for the HO promoter, where the ATG represents +1 and the transcription start site is at −20. Error bars in ChIP assays reflect the standard deviation of three replicate PCRs.
B. The data from panel A, along with additional time points, is plotted as a function of time for PCR intervals centered at −1528, −729, −349, and −55.
We also show that FACT and Asf1 are required for other chromatin transitions that are important for normal HO expression. The yeast FACT complex consists of two subunits, Spt16 and Pob3, and an associated HMG protein Nhp6 (Formosa, 2008). SPT16 and POB3 are essential for viability, and alleles with visible phenotypes have been isolated. FACT changes the accessibility of DNA within nucleosomes without hydrolyzing ATP and without repositioning the histone octamer core relative to the DNA (Formosa et al., 2001; Rhoades et al., 2004); mechanistically this is quite different from the Swi/Snf family of chromatin remodeling factors which require ATP. Mutations affecting the FACT chromatin reorganizing complex were already known to reduce HO expression (Formosa et al., 2001).
Genetic and biochemical evidence indicate FACT has roles in both DNA replication and transcription (Formosa, 2008). Binding of FACT to replication factors, genetic interactions with replication mutants, and sensitivity of FACT mutants to replication inhibitors, all support a replication function (Gambus et al., 2006; VanDemark et al., 2006; Wittmeyer et al., 1999). Evidence for a role for FACT in promoting transcriptional elongation includes stimulation of transcription through a chromatin barrier in vitro (Orphanides et al., 1998), physical association of yFACT with elongation factors (Krogan et al., 2002; Simic et al., 2003), and binding of yFACT to transcribed regions of genes in vivo (Mason and Struhl, 2003; Saunders et al., 2003). Finally, a role for FACT in establishing the transcription preinitiation complex was suggested by genetic interactions between basal transcription factors and FACT, by FACT stimulation of TBP and TFIIA binding to a nucleosome in vitro, and by reduced TBP binding to promoters in vivo in a FACT mutant (Biswas et al., 2006a; Biswas et al., 2007; Biswas et al., 2006b; Biswas et al., 2005).
Asf1 is a histone chaperone, a protein that associates with histones and stimulates histone transfer reactions without being incorporated into nucleosomes (De Koning et al., 2007). Asf1 binds to a histone H3–H4 dimer (Agez et al., 2007; Antczak et al., 2006; English et al., 2006; Natsume et al., 2007), and is believed to promote nucleosome assembly during DNA synthesis (Mousson et al., 2007). Consistent with a replication role, asf1 mutants are sensitive to the replication inhibitor hydroxyurea (Sutton et al., 2001). We find that FACT asf1 double mutants show a strong additive sensitivity to hydroxyurea, suggesting that Asf1 and FACT both promote DNA replication. asf1 mutants are defective for changes in chromatin structure at the PHO5 promoter that accompany transcriptional activation (Adkins et al., 2004), and for exchange of histone H3 at promoters (Kim et al., 2007; Rufiange et al., 2007). Asf1 is required for H3 eviction during transcriptional elongation, and Asf1 binds to promoters and ORFs of active genes (Schwabish and Struhl, 2006). An asf1 mutation also reduces histone H3(K56) acetylation by the Rtt109 acetyltransferase (Collins et al., 2007; Driscoll et al., 2007; Han et al., 2007; Schneider et al., 2006; Tsubota et al., 2007).
Here we describe changes in chromatin structure that occur along the length of the HO promoter as the cell cycle progresses. The Swi/Snf, SAGA, and Mediator coactivators are recruited first to the URS1 region and nucleosomes are evicted here. Subsequently, nucleosomes are evicted from the downstream URS2 region, followed by coactivators binding to this region. Mutations affecting SBF, FACT or Asf1 eliminate both chromatin changes and coactivator binding to URS2, but events at URS1 are unaffected. Thus FACT and Asf1, which both bind to the URS2 region of the promoter, are required for these chromatin transitions that occur before recruitment of Pol II.
RESULTS
Changes in HO promoter chromatin structure during the cell cycle
Chromatin disassembly at promoters often accompanies transcription activation (Williams and Tyler, 2007), so we looked for changes in histone occupancy during HO activation during the cell cycle (Fig 1A). A histone H3 ChIP assay was performed using cells synchronized by GALp::CDC20 arrest and release. Using this synchrony protocol, HO is expressed maximally at 40 min following release (Fig 2D). We used PCR primers tiled across the HO promoter to interrogate the ChIP samples for each time point. At the G2 arrest (0 min) two Swi5 binding sites are in nucleosome free regions, and the nucleosomes in between these sites are evicted at 20 min and start to return at 30 min. There are nucleosomes covering the URS2 region at 0 min. Nucleosomes are evicted from the left end of URS2 between 20 and 25 min, and from the right end of URS2 at 30 min. This wave of nucleosome disassembly across URS2 can be clearly seen in Fig 1B, where histone loss at the PCR interval centered at −729 precedes loss at the −349 interval. Additionally, nucleosome reassembly at the left end of URS2 occurs sooner that at the right end. Finally, histone eviction at the TATA region only occurs at 40 min, a time when the gene is transcribed (see Fig 2D). This time course experiment clearly shows a wave of nucleosome disassembly that moves across the HO promoter during the cell cycle.
Fig 2. Histone acetylation, Swi5, and pol II ChIPs at the HO promoter.
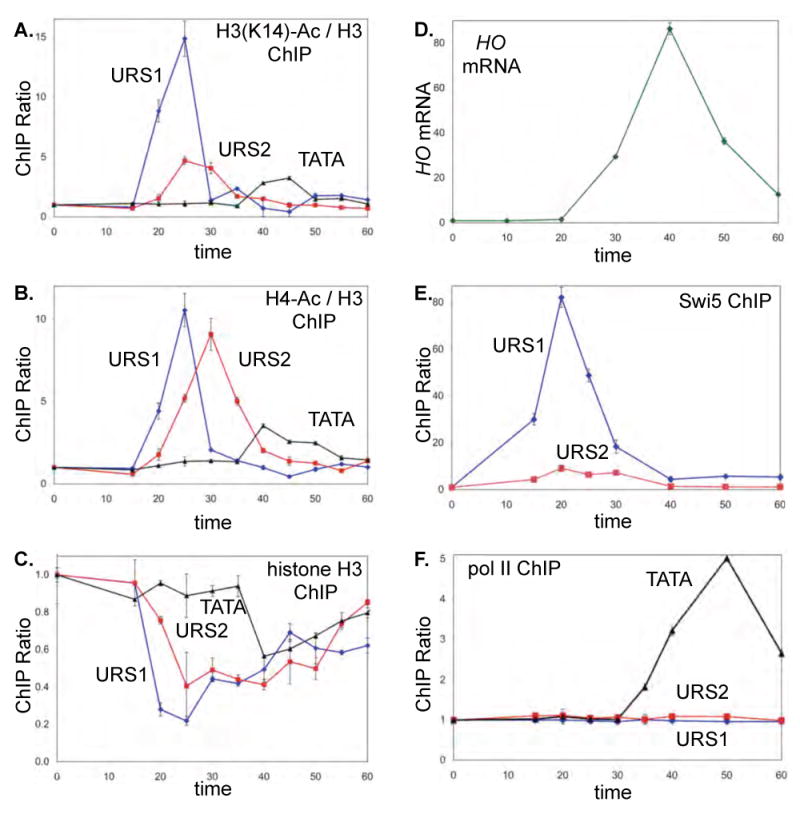
DY6669 cells (GALp::CDC20) were used for ChIPs for H3(K14)-Ac (A), H4-Ac (B), histone H3 (C), and RNA pol II (F). The H3(K14)-Ac and H4-Ac ChIPs are normalized to the ChIP of total H3. D. HO mRNA measured by RT-qPCR from synchronized DY8602 (GALp::CDC20 SWI2-Myc). E. Swi5-Myc ChIP with synchronized DY6542 (GALp::CDC20 SWI5-Myc). Error bars in ChIP or RT-qPCR assays reflect the standard deviation of three replicate PCRs.
Histone acetylation is often associated with promoter activation (Li et al., 2007), and this was examined in a separate time course experiment (Fig 2). Chromatin samples were immunoprecipitated with antibodies that detect acetylation at H3(K14) (Fig 2A) or the H4 tail (Fig 2B), as well as histone H3 (Fig 2C). Histone acetylation occurs progressively, first at URS1 (PCR interval centered at −1293), then URS2 (−666), and finally TATA. Acetylation is then lost quickly, much faster than the repopulation following nucleosome displacement (Fig 2C). These results are substantially different from a previous study that used ash1 mutant strains (Krebs et al., 1999). Notably, we find chromatin disassembly and histone acetylation occur concurrently at each promoter region in our ASH1 strain.
Swi5 recruits coactivators to the URS1 region of the HO promoter
A major question is what factors are responsible for this sequence of changes at the HO promoter. Cosma et al. (1999) showed that Swi5 first recruits Swi/Snf to the promoter, followed by SAGA. These experiments were conducted using ash1 mutant strains, allowing HO expression in both mother and daughter cells. However, we found that an ash1 mutation allows Swi/Snf to be recruited to HO in the absence of the Gcn5 acetyl transferase present in SAGA, while Gcn5 is required for Swi/Snf binding in ASH1 strains (Mitra et al., 2006). We therefore decided to examine the kinetics of Swi/Snf, SAGA, and Mediator binding to HO during the cell cycle in ASH1 strains, and how these coactivators are recruited to the promoter. ChIP experiments with synchronized cells show that the Swi5 DNA-binding protein and Swi/Snf, SAGA, and Mediator coactivators all bind to the HO URS1 region, with peak binding at 20 min following release (Fig 2E, 3A, 3B, 3C). Gcn5 is present in multiple complexes (Eberharter et al., 1999; Pray-Grant et al., 2002; Sterner et al., 2002), and to verify that SAGA is present at HO we used a strain with a Flag tag on the Spt20 subunit which is part of SAGA’s core. A ChIP experiment shows Spt20-Flag binds to HO with the same cell cycle kinetics as Gcn5-Myc (Suppl Fig S2). It is known that Swi5 interacts directly with Swi/Snf (Neely et al., 1999) and Mediator (Bhoite et al., 2001) which can explain their recruitment to sites of Swi5 binding. To address whether SAGA also interacts with Swi5 we used purified recombinant GST-Swi5 in a pulldown experiment (Suppl Fig S3). Extracts were prepared from strains with Myc tags on two SAGA subunits, and Gcn5-Myc and Ada2-Myc both bound to GST-Swi5 but not to GST only; Swi4-Myc did not bind to GST-Swi5, demonstrating specificity of the interaction. An interaction between SAGA and Swi5 is consistent with studies showing interaction of other activators with the Ada2 subunit of SAGA (Berger et al., 1992). Swi5 can therefore recruit each of the three coactivators independently.
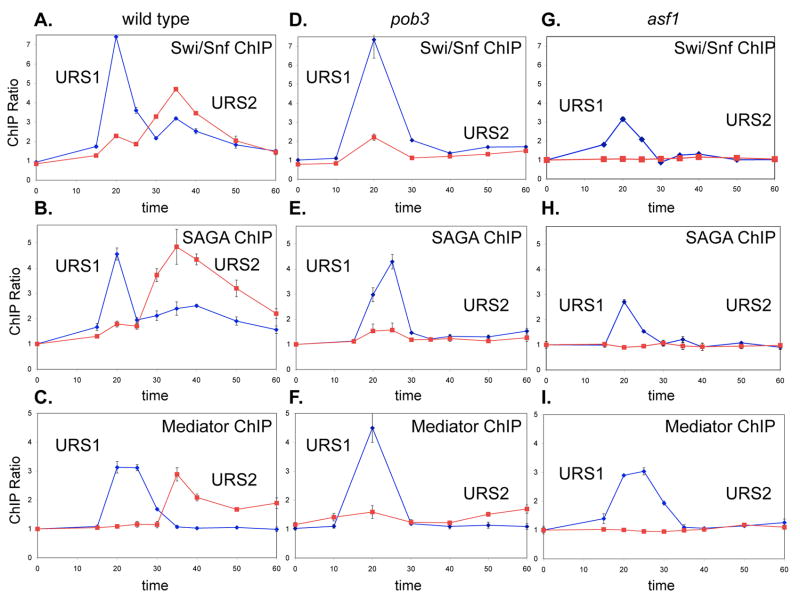
Fig 3. Coactivators bind first to URS1 and then to URS2 at the HO promoter.
Binding of Swi/Snf, SAGA, and Mediator to HO URS1 and URS2 were analyzed by ChIP in synchronized cells. The following cells were used: A. DY8602 (GALp::CDC20 SWI2-Myc), B. DY12752 (GALp::CDC20 GCN5-Myc), C. DY8460 (GALp::CDC20 GAL11-Myc), D. DY10790 (GALp::CDC20 SWI2-Myc pob3), E. DY12750 (GALp::CDC20 GCN5-Myc pob3), F. DY12748 (GALp::CDC20 GAL11-Myc pob3), G. DY12964 (GALp::CDC20 SWI2-Myc asf1), H. DY12927 (GALp::CDC20 GCN5-Myc asf1), I. DY13137 (GALp::CDC20 GAL11-Myc asf1). Error bars in ChIP assays reflect the standard deviation of three replicate PCRs.
To determine the localization of the three coactivators with higher resolution, we used the set of PCR primers tiled across the HO promoter. Swi/Snf binding is found throughout the URS1 region (Suppl Fig S4A) with the peak of binding centered on the Swi5 binding site at −1300 (Suppl Fig S4D). SAGA also binds throughout URS1, but shows two peaks of binding at the two Swi5 binding sites (Suppl Fig S4B). In contrast, Mediator binding is limited to the immediate vicinity of the −1300 Swi5 binding site, with no binding over background seen in the upstream portion of URS1 or at the −1800 Swi5 binding site (Suppl Fig S4C). A swi5 mutation eliminates binding of Swi/Snf, SAGA, and Mediator to HO URS1 (data not shown), consistent with previous work (Bhoite et al., 2001; Cosma et al., 1999). We conclude that Swi5 is responsible for recruiting all three coactivators to HO URS1.
Re-recruitment of coactivators to the URS2 region of HO
Swi5 is unstable after it is imported into the nucleus (Tebb et al., 1993), and Swi5 binding at HO disappears soon after the peak at 20 min after the release (Fig 2E). Coactivator occupancy at URS1 declines concomitant with loss of Swi5 binding (Fig 3A–C). However, binding of the three coactivators, Swi/Snf, SAGA, and Mediator, is seen at the promoter proximal URS2 region with a peak of binding at about 35 min following release (Fig 3A–C). We analyzed ChIP samples from the 35 min time point with the same tiled HO PCR primers to determine where these coactivators are binding. All three coactivators bind throughout URS2 at this time, with highest binding detected with the primer set centered at −729 (Suppl Fig S4).
Thus the Swi5 DNA-binding protein recruits the Swi/Snf, SAGA, and Mediator coactivator complexes to the URS1 region of the HO promoter, then subsequently all three complexes leave URS1 and are found instead at URS2. We refer to this as coactivator re-recruitment, as we do not know whether specific molecules of the Swi/Snf, SAGA, and Mediator move down the promoter, or whether the coactivators dissociate from URS1 and different coactivator molecules subsequently bind to URS2.
It is possible that transcription by RNA polymerase II results in the observed changes in chromatin structure, although there are no known promoters, transcripts, or ORFs in this far upstream region of the HO promoter. To address this question, we performed ChIPs to measure pol II binding to the HO promoter in synchronized cells (Fig 2F). Although pol II is seen binding to the TATA region at the time the gene is transcribed, no binding is seen at URS1 or URS2 at any time during the cell cycle. These results suggest that both the re-recruitment of coactivators and the changes in chromatin structure at HO are independent of transcription by RNA pol II.
FACT is required for coactivator re-recruitment at HO
We hypothesized that other chromatin factors may be required for the changes in chromatin structure at HO. In addition to functioning in DNA replication and transcription elongation, the FACT complex has a role in facilitating initiation of transcription (Formosa, 2003). The SPT16 and POB3 genes encoding FACT subunits are essential for viability, but there are viable mutant alleles that show reduced HO expression (Formosa et al., 2001). A pob3 mutation also reduces expression of the SBF-dependent CLN2 gene, consistent with work with other FACT alleles (Lycan et al., 1994; Rowley et al., 1991), but not expression of PIR1, a Swi5-dependent gene (Fig 4A). We examined binding of the Swi/Snf, SAGA, and Mediator coactivator complexes to HO in a pob3 mutant strain (Fig 3D–F). All three coactivators bind to HO URS1 with kinetics similar to wild type. However, in the pob3 mutant essentially no binding of Swi/Snf, SAGA, or Mediator is seen at URS2, suggesting that FACT is required for re-recruitment of the three coactivators to URS2.
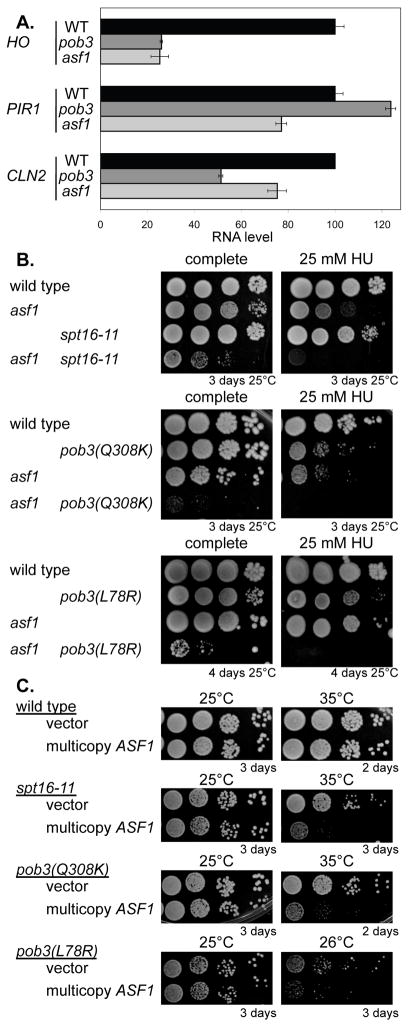
Fig 4. FACT mutants are sensitive to Asf1 levels.
A. HO expression is reduced in pob3 and asf1 mutants. RNA was isolated from log phase wild type (DY150), pob3 (DY7379), and asf1 (DY12869) strains, and HO, PIR1, and CLN2 expression measured by RT-qPCR. Error bars in RT-qPCR assays reflect the standard deviation of three replicate PCRs.
B. Double mutants affecting FACT and Asf1 are additive. Serial ten fold dilutions of strains DY150 (wild type), DY7815 (spt16-11), DY13178 (asf1 spt16-11), DY12554 (pob3-Q308K), DY12869 (asf1), DY13168 (asf1 pob3-Q308K), DY7379 (pob3-L78R), and DY13156 (asf1 pob3-L78R) were grown in plates with or without 25 mM hydroxyurea.
C. Overexpression of ASF1 is toxic in FACT mutants. Strains DY150 (wild type), DY7815 (spt16-11), DY10308 (pob3-Q308K), and DY7379 (pob3-L78R) were transformed with either the pRS426 (YEp-URA3) vector or multicopy YEp-ASF1, and serial dilutions were grown on plates lacking uracil at the indicated temperature.
Asf1 is required for HO expression and for coactivator re-recruitment
Asf1 encodes a histone chaperone, and asf1 mutants are like spt16 and pob3 mutations in FACT in showing sensitivity to the DNA synthesis inhibitor hydroxyurea (Formosa et al., 2001; Schlesinger and Formosa, 2000; Sutton et al., 2001). We find that asf1 spt16 and asf1 pob3 double mutants show a strong additive sensitivity to hydroxyurea (Fig 4B), suggesting that Asf1 and FACT function in distinct ways to promote DNA replication. The asf1 spt16 and asf1 pob3 double mutants also show additive growth defects on complete medium. Because the asf1 mutation is toxic in FACT mutants, we tested whether overexpression of ASF1 could suppress FACT mutants. Instead of suppression, however, we found ASF1 overexpression is toxic in FACT mutants (Fig 4C). Thus FACT mutants are quite sensitive to Asf1 levels, as either too much or too little Asf1 inhibits growth in FACT mutants. Based on these results, we investigated whether an asf1 mutation, like pob3, affects gene expression, and found that asf1 sharply reduces HO expression (Fig 4A). These mutations have only a modest effect on expression of the Swi5-dependent PIR1 and SBF-dependent CLN2 genes, and thus this effect is at least partly specific to HO. We next examined binding of the Swi/Snf, SAGA, and Mediator coactivator complexes to HO in an asf1 mutant strains synchronized by GALp::CDC20 arrest and release (Fig 3G–I). All three coactivators bind to URS1 in the asf1 mutant, although binding is reduced. More importantly, the asf1 mutation blocks coactivators binding to URS2. Thus Asf1 and FACT are both required for coactivator at HO.
FACT and Asf1 bind to the HO promoter
Previous work has shown that FACT is recruited to transcribed regions by elongating RNA polymerase, but little FACT binding to promoters was seen (Mason and Struhl, 2003). We therefore performed ChIP experiments with synchronized cells using polyclonal antibody to the Spt16 subunit of FACT (Fig 5A). No FACT binding is seen at URS1, but strong FACT binding is seen at URS2 at 20 min after release and at TATA at 40 min. FACT binding at TATA is expected when HO is transcribed, as FACT is present at transcribed open reading frames (Mason and Struhl, 2003). Binding at URS2 is unexpected, however, as FACT binding to promoter elements had not been described. The ChIP samples from the 0 and 20 min time points were analyzed with the PCR primers tiled across the HO promoter and the results show that FACT binds across the URS2 region, but primarily at the left end (Suppl Fig S5A). There are several notable observations. FACT binding to URS2 is concurrent with the distant binding of Swi5 and coactivators at URS1. Additionally, the position of peak FACT binding at 20 min approximates the site of peak coactivator binding at later times in the cell cycle. Finally, the nucleosome displacement that occurs at the left end of URS2 starts soon after FACT binds, while the coactivators do not bind until later in the cell cycle.
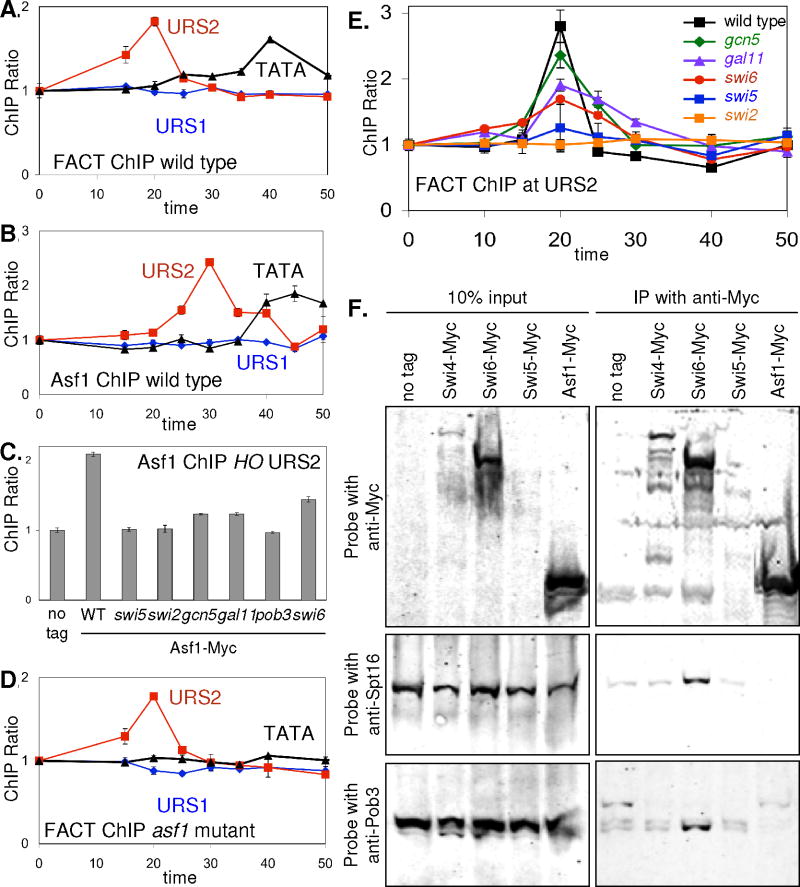
Fig 5. FACT binds to URS2 before Asf1.
ChIPs were performed with synchronized cells in A, B, D, and E.
A. FACT binds to URS2 at 20 min in wild type (DY6669; GALp::CDC20).
B. Asf1 binds to URS2 at 30 min in wild type (DY13010; GALp::CDC20 ASF1-Myc).
C. Using asynchronous cells, Asf1-Myc binds to HO URS2 in wild type (DY12997), but Asf1-Myc binding is lost in swi5 (DY13145), swi2 (DY13176), gcn5 (DY13164), gal11 (DY13162), pob3 (DY13150), and swi6 (DY13147) mutants. DY150 was used as the untagged control.
D. FACT binds to URS2 in an asf1 mutant (DY12914; GALp::CDC20 asf1).
E. FACT binding to URS2 in wild type (DY6669; GALp::CDC20), gcn5 (DY6650; GALp::CDC20 gcn5), gal11 (DY7419; GALp::CDC20 gal11), swi6 (DY13373; GALp::CDC20 swi6), swi5 (DY6570; GALp::CDC20 swi5), and swi2 (DY9718; GALp::CDC20 swi2) strains.
F. FACT interacts with Swi6. Extracts prepared from strains DY150 (no tag), DY8341 (SWI4-Myc[18]) (150 kd), DY8353 (SWI6-Myc[18]) (117 kd), DY5832 (SWI5-Myc[8]) (92 kd), and DY12997 (ASF1-Myc[13]) (51 kd), were immunoprecipitated with anti-Myc antibody and analyzed on western blots, along with controls corresponding to 10% of the input before immunoprecipitation, and the blots were probed with anti-Myc, anti-Spt16, and anti-Pob3 antibodies.
Error bars in ChIP assays reflect the standard deviation of three replicate PCRs.
ChIP experiments were performed using synchronized cells to determine where Asf1 binds to the HO promoter (Fig 5B). Although no binding was seen at URS1, strong binding was seen at URS2 and at TATA. Binding at TATA is expected, as it has been previously shown that Asf1 binds to promoters and ORFs concomitant with transcriptional activation (Schwabish and Struhl, 2006); however, the binding at URS2 is unexpected. Analysis with the PCR primers tiled across the HO promoter shows that Asf1 binds more to the right half of URS2 (Suppl Fig S5B).
Swi/Snf is required for FACT binding to HO
ChIP experiment with various mutants were performed to determine what factors are required for Asf1 binding at HO. As shown in Fig 5C, mutations affecting the Swi5 DNA-binding factor or the Swi/Snf, SAGA, or Mediator coactivator complexes eliminate binding of Asf1 to URS2. A pob3 mutation affecting the FACT complex also eliminates Asf1 binding to URS2, consistent with FACT binding earlier in the cell cycle. A swi6 mutation affecting the SBF factor also reduces Asf1 binding. We conclude that coactivators, FACT, and SBF are required for Asf1 binding to the HO promoter.
We next used synchronized cells to determine what factors are required for FACT binding at HO. FACT binding to HO URS2 is not affected by an asf1 mutation (Fig 5D); FACT binding to TATA is sharply reduced in the asf1 mutant, consistent with the decreased HO transcription. FACT binding to URS2 is lost in a swi5 mutant (Fig 5E), and we conclude that FACT binding to HO is SWI5-dependent. We next determined the effect of mutations affecting coactivators or the SBF DNA-binding factor. FACT binds to URS2 in a gcn5Δ mutant lacking the acetyltransferase normally present in SAGA, in a gal11Δ mutant affecting Mediator, or in a swi6Δ mutant affecting SBF (Fig 5E). However, on these mutants the peak of FACT binding at 20 min is reduced, with binding persisting at 25 min as opposed to the precipitous decline in binding in wild type. Thus the binding peaks are shifted to the right in these mutants, and FACT may dissociate more slowly. Alternatively, integrating under the curves suggests the same amount of FACT binds, but the peak of FACT binding is delayed. Finally, FACT binding is eliminated in a swi2(E834K) mutant affecting Swi/Snf (Fig 5E). We conclude that Swi/Snf is required for FACT binding, although the sites at 20 min where Swi/Snf binds in URS1 and FACT binds in URS2 are separated by some 600 nt.
FACT interacts with SBF
The reduced FACT binding in the swi6 mutant suggested that the SBF DNA-binding factor could help recruit FACT to the promoter. Consistent with this idea, immunoprecipitation of Swi6-Myc from cells also brings down the Spt16 and Pob3 subunits of FACT (Fig 5F). Swi6 nuclear localization is regulated within the cell cycle, and FACT binding at 20 min after CDC20 release occurs when Swi6 enters the nucleus (Geymonat et al., 2004; Sidorova et al., 1995). Asf1 does not bind to HO in a swi6 mutant (Fig 5C), consistent with SBF promoting FACT binding. Finally, the three coactivators do not bind to URS2 in a swi6 mutant, while coactivator binding to URS1 is unaffected (Suppl Fig S6). This results contradict an earlier study that found Swi/Snf and SAGA binding to URS2 was not affected by a swi6 mutation (Cosma et al., 1999); this study used strains with an ash1 mutation affecting a HDAC complex, however. We find that SBF binding to URS2 is markedly reduced by swi5 or swi2 mutations, but only modestly affected by gal11 or gcn5; strong SBF binding is seen in the swi2 ash1 double mutant (Suppl Fig S7). Finally, a pob3 mutation modestly reduces SBF binding (Suppl Fig S7), and thus FACT and SBF mutually promote binding. In summary, SBF interacts with FACT and facilitates its binding, and both factors are required for Asf1 binding and coactivator recruitment at URS2.
Swi/Snf is required for nucleosome displacement at URS1
We next examined what factors are required for the waves of chromatin disassembly that occurs at HO using mutants that had been synchronized during the cell cycle (Fig 6). A swi5 mutation eliminates coactivator recruitment and HO expression, and no nucleosome displacement was seen at any part of the HO promoter in the swi5 mutant. We next examined coactivator mutants. Nucleosome displacement at URS1 in minimal, and very transient, in a swi2(E834K) mutant; nucleosome eviction at URS2 is also lost. In contrast, nucleosome displacement at URS1 is unaffected in gcn5Δ or gal11Δ mutants. The gcn5Δ mutation also limits nucleosome eviction at URS2, while gal11Δ does not affect nucleosome eviction here. We conclude that histone acetylation by Gcn5 is required for efficient nucleosome displacement at URS2. More significantly, Swi/Snf is critically required for nucleosome disassembly at URS1.
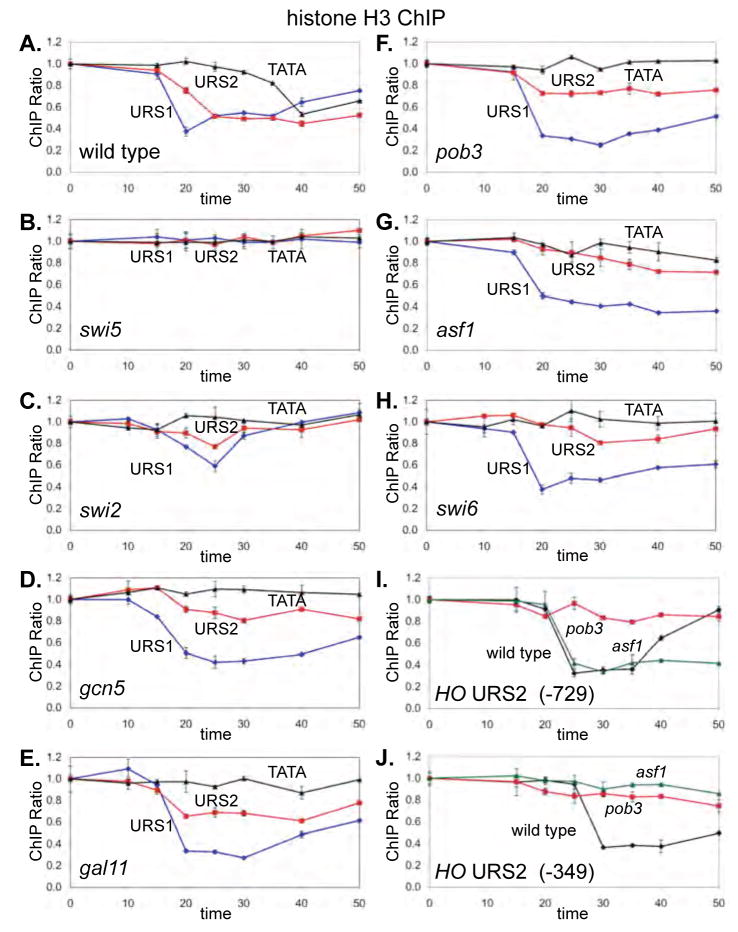
Fig 6. Swi5 and Swi/Snf are required for nucleosome eviction at URS1 and FACT and Asf1 for eviction at URS2.
Histone H3 ChIP was performed using the following synchronized cells and PCR primers for URS1 centered at −1293, URS2 at −666, at TATA at −55: A. DY6669 (GALp::CDC20), B. DY6570 (GALp::CDC20 swi5), C. DY9718 (GALp::CDC20 swi2), D. DY6650 (GALp::CDC20 gcn5), E. DY7419 (GALp::CDC20 gal11). F. DY11246 (GALp::CDC20 pob3), G. DY12914 (GALp::CDC20 asf1), and H. DY13373 (GALp::CDC20 swi6). I. The wild type, pob3 and asf1 H3 ChIPs were probed with URS2 primers centered at −729. J. The wild type, pob3 and asf1 H3 ChIPs were probed with URS2 primers centered at −349. Error bars in ChIP assays reflect the standard deviation of three replicate PCRs.
FACT, SBF, and Asf1 are required for nucleosome displacement at URS2
We next performed H3 ChIPs to examine the effect of pob3 and asf1 mutations on nucleosome eviction (Fig 6F, 6G). In both mutants chromatin disassembly occurs normally at the URS1 region, but eviction is severely reduced at URS2. A similar effect is seen in the swi6 mutant (Fig 6H), consistent with SBF facilitating FACT recruitment. Additionally, the repopulation of URS1 nucleosomes in wild type cells is defective in these mutants. We conclude that mutations in FACT, SBF, and Asf1 have similar effects at HO URS2, eliminating both coactivator re-recruitment and chromatin disassembly.
Nucleosome loss at the left end of URS2 (centered at −729) precedes loss at the right end of URS2 (−349) (Fig 1B), and we therefore examined chromatin changes at these regions in FACT and Asf1 mutants. Nucleosome loss at the left end of URS2 is lost in pob3 mutant but occurs normally in an asf1 mutant (Fig 6I). In contrast, FACT and Asf1 are both required for nucleosome loss at the right end of URS2 (Fig 6J). The timing of events at URS2 is interesting. Binding of FACT and Asf1 precede binding of the coactivators, with FACT binding primarily to the left half of URS2 at 20 min, while Asf1 binds to the right half at 30 min. Chromatin disassembly shows similar kinetics, with chromatin disassembly at the left end of URS2 occurring first and requiring FACT, while nucleosome loss at the right end of URS2 occurs later in the cell cycle and requires Asf1.
DISCUSSION
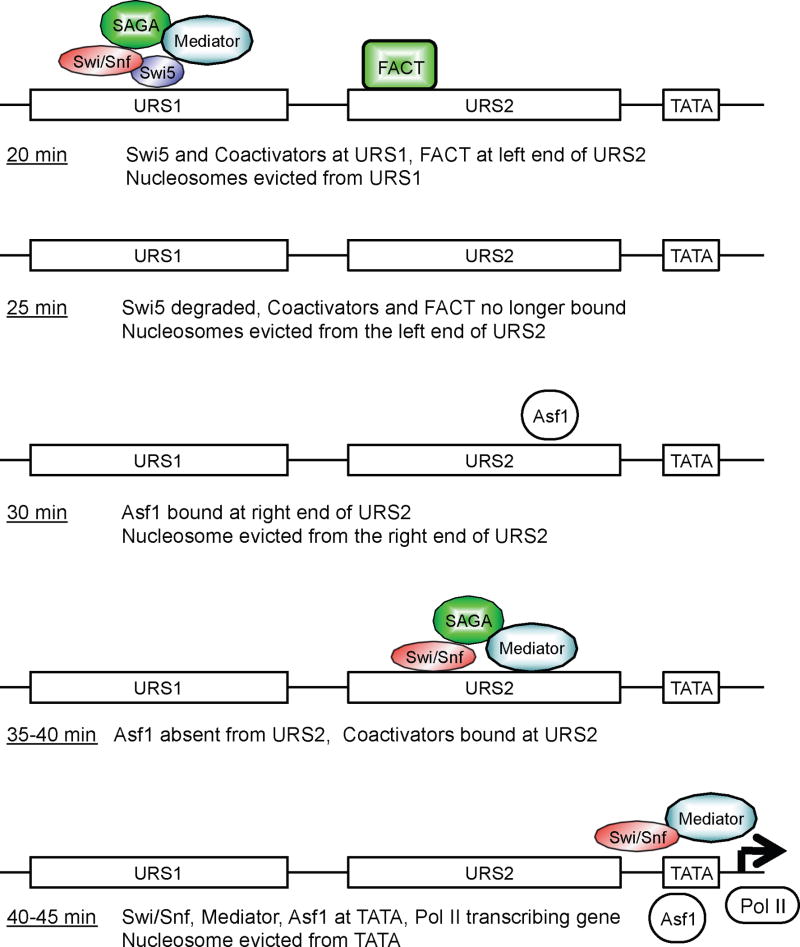
Transcriptional activation of HO involves sequential changes in chromatin structure along the promoter during the cell cycle. Nucleosome displacement at HO occurs stepwise, first at URS1, then in sequence at the left end of URS2, the right end of URS2, and finally at the TATA region when the gene is transcribed (Fig 1B). Concomitant with this chromatin disassembly, acetylation of the residual nucleosomes also occurs in waves along the promoter (Fig 2A,B). ChIP experiments show there is sequential recruitment of three coactivator complexes, Swi/Snf, SAGA, and Mediator, first to URS1 and subsequently to URS2 (Fig 3). Importantly, these are dependent changes: the nucleosome eviction and coactivator binding must occur first at URS1 before chromatin changes and coactivator binding can occur at URS2 later in the cell cycle. Finally, two chromatin factors that have not been previously described as binding to upstream promoter elements, FACT and Asf1, are specifically required for the chromatin disassembly and coactivator binding events at URS2 (Fig 7). FACT binding to URS2 occurs before Asf1 binding, and FACT and Asf1 are each required for nucleosome eviction in distinct regions of URS2 (Fig 6I,J).
Fig 7. Model of events at the HO promoter during the cell cycle.
Times are minutes after release from GALp::CDC20 arrest.
The Swi5 DNA-binding protein is required for all subsequent changes at the HO promoter, and thus can be considered a “pioneer” factor (Cirillo et al., 2002). Swi5 is cell cycle regulated, entering the nucleus after anaphase (Moll et al., 1991). Swi5 interacts directly with the Swi/Snf, SAGA, and Mediator coactivators, and recruits them to the URS1 region of the HO promoter. Swi5 is rapidly degraded by ubiquitin mediated proteolysis (Kishi et al., 2008), and binding of the three coactivators quickly disappears. There are two Swi5 binding sites at URS1, separated by 500 bp. Both binding sites are required for HO activation, and experiments suggest an interaction between the two sites (McBride et al., 1997). Swi5 binds more strongly in vitro to Site B at −1300 (McBride et al., 1997), and ChIP experiments also show stronger Swi5 binding to Site B in vivo (Suppl Fig S4). Swi/Snf and SAGA bind to both Site A and Site B within URS1, with stronger binding to Site B; Mediator appears to bind only to Site B. We suggest that there is an interaction between the Swi5 molecules bound to these two sites, and that it is the interaction of Swi5 and coactivators bound to the two sites that leads to eviction of the nucleosomes in between.
The three coactivators are recruited to URS1 by Swi5, and 10–15 min later they are present at the URS2 region of the promoter, which we call coactivator re-recruitment (Fig 7). FACT and Asf1 are required for this recruitment of coactivators to URS2, and FACT and Asf1 both bind to URS2 before the peak of coactivator binding here (Fig 7). We think of URS1 as the “staging area,” where coactivators are first recruited. Subsequently, the coactivators move from the staging area to the URS2 region where the “traditional” activation occurs. This two part activation scheme allows greater stringency of control over HO expression by imposing additional requirements before activation of expression. The requirement for FACT and Asf1 for coactivator re-recruitment suggests that mobilization of coactivators from the staging area to the activation site is blocked by an inhibitory chromatin structure.
Previous work showed FACT binding to transcribed open reading frames, but not to upstream promoter elements (Mason and Struhl, 2003; Saunders et al., 2003), with FACT recruited by ubiquitylated H2B (Fleming et al., 2008). FACT had not been previously shown to interact with sequence-specific DNA-binding proteins, but here we show that FACT interacts with SBF to facilitate binding to HO (Fig 5E,5F). At the HO URS2 promoter region SBF and FACT are both required for nucleosome eviction and coactivator binding. Importantly, FACT binding precedes the nucleosome eviction which precedes coactivator binding.
Disassembly of chromatin can occur at promoters when a gene is transcriptionally activated (Boeger et al., 2003; Reinke and Horz, 2003; Workman, 2006). Nucleosome displacement at certain promoters requires Asf1 (Adkins et al., 2004), Gcn5 (Verdone et al., 2002), or Swi/Snf (Biddick et al., 2008; Schwabish and Struhl, 2007). Nucleosome eviction is often accompanied by histone acetylation (Boeger et al., 2003; Erkina and Erkine, 2006; Zhao et al., 2005), as we observe at HO. Remarkably, nucleosome eviction at HO occurs stepwise during the cell cycle, moving across the promoter (Fig 1). Swi/Snf is required for efficient nucleosome eviction at URS1, as there is only weak and transient nucleosome loss in the swi2 mutant (Fig 6C). This residual nucleosome eviction may be facilitated by SAGA or Mediator; the swi2 mutation sharply reduces, but does not completely eliminate, binding of these coactivators. This histone eviction at URS1 at 20 min is followed by repopulation of nucleosomes over the next 20–40 min (Fig 1B). Interestingly, this histone loss at URS1 persists in a number of the mutants, including gcn5, gal11, pob3 and asf1 (Fig 6). The mechanism underlying this defect in nucleosome repopulation at URS1 is not clear; the gcn5, pob3, and asf1 mutants are defective in nucleosome loss at URS2 while the gal11 mutant is not.
In contrast to URS1, chromatin disassembly at URS2 occurs before the coactivators bind to this region. Nucleosome loss here requires FACT and Asf1, but these factors are differentially required at two regions of URS2 (Fig 6). Chromatin disassembly at the left end of URS2 requires FACT but not Asf1, and FACT binds primarily to this part of URS2 (Suppl Fig S5A) shortly before the chromatin disassembly. Asf1 binds later to URS2 (Fig 5), primarily at the right end of URS2 (Suppl Fig S5B) when the Asf1-dependent nucleosome eviction occurs here. FACT and Swi/Snf may contribute to this chromatin disassembly, but both factors are required for Asf1 binding to HO. Importantly, FACT and Asf1 are not required for nucleosome displacement at URS1, and we therefore conclude that chromatin disassembly at URS1 and the two regions of URS2 are all mechanistically different. The Asf1 histone chaperone may receive the H3-H4 tetramers evicted from URS1 or the left end of URS2, but stable Asf1 binding is only detected by ChIP slightly later in the cell cycle. Finally, the absence of RNA polymerase II at URS1 or URS2 at any time during the cell cycle (Fig 2F) eliminates the possibility that nucleosome eviction or coactivator recruitment are due to either poised polymerase or expression of a noncoding transcript.
The yeast HO gene encodes an endonuclease that initiates mating type switching. Inappropriate expression of an endonuclease could be disastrous in natural populations, and HO expression is therefore tightly regulated. Multiple activators and coactivators are required to overcome the repression at HO. Here we show that different factors are required to evict nucleosomes from different regions of the HO promoter. Rather than being uniform, the chromatin of the HO promoter appears to contain a series of nucleosomes with individual properties, much like a series of padlocks that each requires a different key and that must be unlocked in a defined order.
EXPERIMENTAL PROCEEDURES
All yeast strains used are listed in Suppl Table 1 and are isogenic in the W303 background (Thomas and Rothstein, 1989). Standard genetic methods were used for strain construction (Rothstein, 1991; Sherman, 1991). Cell cycle synchronization was performed by galactose withdrawal and re-addition with a GALp::CDC20 strain grown at 25°C in YEP medium containing 2% galactose and 2% raffinose (Bhoite et al., 2001). A high degree of synchrony was demonstrated by flow cytometry analysis, budding indices, and analysis of cycle regulated mRNAs (data not shown). In other experiments cells were grown in YEPD medium (Sherman, 1991).
Chromatin immunoprecipitations (ChIPs) were performed as described (Bhoite et al., 2001; Voth et al., 2007) using 9E11 (Abcam) or 4A6 (Upstate) monoclonal antibody to the Myc epitope, anti-Flag (M2, Sigma) monoclonal antibody to the Flag epitope, anti-histone H3 (07–690, Upstate) anti-histone H3(Ac-Lys14) (07–353, Upstate), anti-acetyl-histone H4 (06–598, Upstate), rabbit anti-Spt16 (provided by Tim Formosa), anti-RNA pol II (8WG16, Covance) and antibody coated magnetic beads (Rabbit and Pan Mouse IgG beads, Dynal Biotech). ChIP assays were analyzed by real time PCR as described (Eriksson et al., 2004). PCR primers are listed in Suppl Table 2. Each ChIP sample was first normalized to an input DNA sample to and then to the ChIP signal for a control region on chromosome I. Immunoprecipitations were performed as described previously (Biswas et al., 2008) using anti-Myc antibody and blots were probed with anti-Myc, anti-Spt16, and anti-Pob3 antibodies and scanned using a Odyssey Infrared Imaging System (Li-Cor Biosciences).
GST-Swi5 was expressed and purified from E. coli containing plasmid M1202 (Brazas et al., 1995) and used for GST pull down experiments as described (Barlev et al., 1995). Western blots were performed with anti-Myc antibody and fluorescent secondary antibodies. RT-qPCR was used to measure HO mRNA levels as described (Voth et al., 2007) using primers listed in Suppl Table 2, except that RDN25 RNA was used as the internal control. The YEp-ASF1 plasmid M5382 was constructed by cloning a PCR-generated 2.03 kb EcoRI fragment with the ASF1 gene into pRS426 (Christianson et al., 1992).
Supplementary Material
Acknowledgments
We thank Tim Formosa for FACT antibodies and for many helpful discussions. We thank Tim Formosa, Peter Geiduschek, Emily Parnell, Dean Tantin, and Warren Voth for comments on the manuscript. This work was supported by a grant from the National Institutes of Health
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Agez M, Chen J, Guerois R, van Heijenoort C, Thuret JY, Mann C, Ochsenbein F. Structure of the histone chaperone ASF1 bound to the histone H3 C-terminal helix and functional insights. Structure. 2007;15:191–199. doi: 10.1016/j.str.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Antczak AJ, Tsubota T, Kaufman PD, Berger JM. Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics. BMC Struct Biol. 2006;6:26. doi: 10.1186/1472-6807-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev NA, Candau R, Wang L, Darpino P, Silverman N, Berger SL. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- Berger SL, Pina B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- Bhoite LT, Allen JM, Garcia E, Thomas LR, Gregory ID, Voth WP, Whelihan K, Rolfes RJ, Stillman DJ. Mutations in the Pho2 (Bas2) transcription factor that differentially affect activation with its partner proteins Bas1, Pho4, and Swi5. J Biol Chem. 2002;277:37612–37618. doi: 10.1074/jbc.M206125200. [DOI] [PubMed] [Google Scholar]
- Bhoite LT, Yu Y, Stillman DJ. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 2001;15:2457–2469. doi: 10.1101/gad.921601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddick RK, Law GL, Chin KK, Young ET. The transcriptional coactivators SAGA, SWI/SNF, and mediator make distinct contributions to activation of glucose-repressed genes. J Biol Chem. 2008;283:33101–33109. doi: 10.1074/jbc.M805258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Dutta-Biswas R, Mitra D, Shibata Y, Strahl BD, Formosa T, Stillman DJ. Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J. 2006a;25:4479–4489. doi: 10.1038/sj.emboj.7601333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Dutta-Biswas R, Stillman DJ. Chd1 and yFACT act in opposition in regulating transcription. Mol Cell Biol. 2007;27:6279–6287. doi: 10.1128/MCB.00978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Takahata S, Stillman DJ. Different genetic functions for the Rpd3(L) and Rpd3(S) complexes suggest competition between NuA4 and Rpd3(S) Mol Cell Biol. 2008;28:4445–4458. doi: 10.1128/MCB.00164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Yu Y, Mitra D, Stillman DJ. Genetic Interactions Between Nhp6 and Gcn5 With Mot1 and the Ccr4-Not Complex That Regulate Binding of TATA-Binding Protein in Saccharomyces cerevisiae. Genetics. 2006b;172:837–849. doi: 10.1534/genetics.105.050245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Yu Y, Prall M, Formosa T, Stillman DJ. The Yeast FACT Complex Has a Role in Transcriptional Initiation. Mol Cell Biol. 2005;25:5812–5822. doi: 10.1128/MCB.25.14.5812-5822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- Brazas RM, Bhoite LT, Murphy MD, Yu Y, Chen Y, Neklason DW, Stillman DJ. Determining the requirements for cooperative DNA binding by Swi5p and Pho2p (Grf10p/Bas2p) at the HO promoter. J Biol Chem. 1995;270:29151–29161. doi: 10.1074/jbc.270.49.29151. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Florens L, Swanson SK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim Biophys Acta. 2005;1731:77–87. doi: 10.1016/j.bbaexp.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Panizza S, Nasmyth K. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol Cell. 2001;7:1213–1220. doi: 10.1016/s1097-2765(01)00266-0. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates JR, 3rd, Berger SL, Workman JL. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson P, Biswas D, Yu Y, Stewart JM, Stillman DJ. TATA-binding protein mutants that are lethal in the absence of the Nhp6 high-mobility-group protein. Mol Cell Biol. 2004;24:6419–6429. doi: 10.1128/MCB.24.14.6419-6429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkina TY, Erkine AM. Displacement of histones at promoters of Saccharomyces cerevisiae heat shock genes is differentially associated with histone H3 acetylation. Mol Cell Biol. 2006;26:7587–7600. doi: 10.1128/MCB.00666-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Formosa T. Changing the DNA landscape: putting a SPN on chromatin. Curr Top Microbiol Immunol. 2003;274:171–201. doi: 10.1007/978-3-642-55747-7_7. [DOI] [PubMed] [Google Scholar]
- Formosa T. FACT and the reorganized nucleosome. Mol Biosyst. 2008;4:1085–1093. doi: 10.1039/b812136b. [DOI] [PubMed] [Google Scholar]
- Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman DJ. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 2001;20:3506–3517. doi: 10.1093/emboj/20.13.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Wells GP, Smerdon SJ, Sedgwick SG. Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol Cell Biol. 2004;24:2277–2285. doi: 10.1128/MCB.24.6.2277-2285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Yoon S, Qiu H, Govind S, Hinnebusch AG. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol Cell Biol. 2005;25:5626–5638. doi: 10.1128/MCB.25.13.5626-5638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Seol JH, Han JW, Youn HD, Cho EJ. Histone chaperones regulate histone exchange during transcription. EMBO J. 2007;26:4467–4474. doi: 10.1038/sj.emboj.7601870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi T, Ikeda A, Koyama N, Fukada J, Nagao R. A refined two-hybrid system reveals that SCFCdc4-dependent degradation of Swi5 contributes to the regulatory mechanism of S-phase entry. Proc Natl Acad Sci U S A. 2008;105:14497–14502. doi: 10.1073/pnas.0806253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs JE, Kuo MH, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lycan D, Mikesell G, Bunger M, Breeden L. Differential effects of Cdc68 on cell cycle-regulated promoters in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7455–7465. doi: 10.1128/mcb.14.11.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride HJ, Brazas RM, Yu Y, Nasmyth K, Stillman DJ. Long range interactions at the HO promoter. Mol Cell Biol. 1997;17:2669–2678. doi: 10.1128/mcb.17.5.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra D, Parnell EJ, Landon JW, Yu Y, Stillman DJ. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol Cell Biol. 2006;26:4095–4110. doi: 10.1128/MCB.01849-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- Mousson F, Ochsenbein F, Mann C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma. 2007;116:79–93. doi: 10.1007/s00412-006-0087-z. [DOI] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright AP, Workman JL. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, 3rd, Grant PA. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- Rhoades AR, Ruone S, Formosa T. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol Cell Biol. 2004;24:3907–3917. doi: 10.1128/MCB.24.9.3907-3917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Meth Enzymol. 1991;194:281–302. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rowley A, Singer RA, Johnston JC. CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol Cell Biol. 1991;11:5718–5726. doi: 10.1128/mcb.11.11.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, Reinberg D, Lis JT. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- Schlesinger MB, Formosa T. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics. 2000;155:1593–1606. doi: 10.1093/genetics/155.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J Biol Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol. 2007;27:6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Meth Enzymol. 1991;194:1–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sidorova JM, Mikesell GE, Breeden LL. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol Biol Cell. 1995;6:1641–1658. doi: 10.1091/mbc.6.12.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil A, Herskowitz I. Identification of an asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. Embo J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Belotserkovskaya R, Berger SL. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc Natl Acad Sci U S A. 2002;99:11622–11627. doi: 10.1073/pnas.182021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A, Bucaria J, Osley MA, Sternglanz R. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebb G, Moll T, Dowser C, Nasmyth K. SWI5 instability may be necessary but is not sufficient for asymmetric HO expression in yeast. Genes Dev. 1993;7:517–528. doi: 10.1101/gad.7.3.517. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark AP, Blanksma M, Ferris E, Heroux A, Hill CP, Formosa T. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell. 2006;22:363–374. doi: 10.1016/j.molcel.2006.03.025. [DOI] [PubMed] [Google Scholar]
- Verdone L, Wu J, van Riper K, Kacherovsky N, Vogelauer M, Young ET, Grunstein M, Di Mauro E, Caserta M. Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions. EMBO J. 2002;21:1101–1111. doi: 10.1093/emboj/21.5.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth WP, Yu Y, Takahata S, Kretschmann KL, Lieb JD, Parker RL, Milash B, Stillman DJ. Forkhead proteins control the outcome of transcription factor binding by antiactivation. EMBO J. 2007;26:4324–4334. doi: 10.1038/sj.emboj.7601859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SK, Tyler JK. Transcriptional regulation by chromatin disassembly and reassembly. Curr Opin Genet Dev. 2007;17:88–93. doi: 10.1016/j.gde.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Wittmeyer J, Joss L, Formosa T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry. 1999;38:8961–8971. doi: 10.1021/bi982851d. [DOI] [PubMed] [Google Scholar]
- Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- Zhao J, Herrera-Diaz J, Gross DS. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol Cell Biol. 2005;25:8985–8999. doi: 10.1128/MCB.25.20.8985-8999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.