Summary
Cvt19 is specifically required for the transport of resident vacuolar hydrolases that utilize the cytoplasm-to-vacuole targeting (Cvt) pathway. Autophagy (Apg) and pexophagy, processes that use the majority of the same protein components as the Cvt pathway, do not require Cvt19. Cvt19GFP is localized to punctate structures on or near the vacuole surface. Cvt19 is a peripheral membrane protein that binds to the precursor form of the Cvt cargo protein aminopeptidase I (prAPI) and travels to the vacuole with prAPI. These results suggest that Cvt19 is a receptor protein for prAPI that allows for the selective transport of this protein by both the Cvt and Apg pathways.
Introduction
Degradation by the lysosomal/vacuolar compartment is the primary cellular mechanism for the turnover of cytoplasmic organelles, and facilitates extensive cellular remodeling as developmental cues or environmental conditions dictate. When cells undergo nutrient limitation, organelles and cytosolic proteins are taken into the vacuole nonspecifically by a process called macroautophagy (Kim et al., 2000). A related constitutive process, the cytoplasm-to-vacuole targeting (Cvt) pathway, is used for the specific delivery of cytosolic proteins to the vacuole in yeast (Scott et al., 1996). In certain conditions, some organelles and proteins are selectively transported to the vacuole by the autophagy (Apg) pathway (Baba et al., 1997; Hutchins et al., 1999). Aminopeptidase I (API) is one of the specific cargo proteins delivered to the vacuole by the Cvt pathway in vegetative conditions. However, API can also be specifically transported to the vacuole by autophagy when that pathway is induced (Baba et al., 1997; Scott et al., 1996). The fact that prAPI is transported selectively by both the Cvt and Apg pathways suggests a receptor-mediated mechanism for API concentration at the site(s) of vesicle formation.
API is synthesized in the cytosol as a precursor protein (prAPI) that contains a propeptide region that is required for its specific vacuole delivery (Oda et al., 1996; Segui-Real et al., 1995). Immediately after synthesis, prAPI assembles into a dodecamer in the cytosol (Kim et al., 1997). Precursor API dodecamers are recruited to cytosolic structures called Cvt complexes. Subsequent membrane enwrapping results in the formation of double-membrane vesicles (Cvt vesicles) that are targeted to the vacuole (Baba et al., 1997). Upon fusion of the outer membrane of the Cvt vesicle with the vacuole, the still intact inner vesicle (called a Cvt body) passes into the vacuole lumen where it is ultimately broken down by resident hydrolases, resulting in the release of prAPI (Scott et al., 1997). The propeptide is subsequently removed, generating the mature-sized protein. The Cvt complex breaks apart in the vacuole lumen but the mature API retains its oligomeric form and exists as a 600 kDa protein made up of identical polypeptides (Kim et al., 1997; Metz et al., 1977).
Many of the molecular components that mediate the Cvt/Apg pathways have been recently identified (for a review, see Klionsky and Emr, 2000). The majority of proteins required for autophagy are also required for the Cvt pathway. Based on morphological and biochemical analysis, the Cvt and Apg pathways are thought to use analogous mechanisms (Baba et al., 1997; Scott et al., 1997). In the Apg pathway, double-membrane vesicles engulf cytosolic cargo into autophagosomes that fuse with the vacuole membrane, releasing autophagic bodies into the vacuole lumen (Baba et al., 1994). These vesicles are then broken down, and their contents are digested and recycled (Takeshige et al., 1992).
Despite overall molecular and mechanistic similarities, there are several fundamental differences between the Cvt and Apg pathways. In the Cvt pathway, only selective cargo is transported, whereas the Apg pathway transports both selective and nonselective cargoes (Baba et al., 1997; Scott and Klionsky, 1998). Cvt vesicles are smaller than autophagosomes (150 nm compared to 300–900 nm; Baba et al., 1997), and the Cvt pathway is active in vegetative conditions, whereas autophagy is induced by starvation (Scott et al., 1996). Recent studies have identified a complex of proteins (Apg1, Apg13, Apg17, Vac8, and Cvt9) that may be involved in switching between the Cvt and Apg pathways (Kamada et al., 2000; Scott et al., 2000).
The transport of prAPI by the Cvt pathway is known to be specific and saturable, further suggesting that a receptor may be involved in specifically recruiting prAPI to the site of vesicle formation (Klionsky et al., 1992; Scott et al., 1996). To date, none of the characterized proteins involved in the targeting of prAPI appear to fit the criteria expected for a receptor for this protein. Recently, YOL082w was identified as a potential API-interacting protein by a genome-wide two-hybrid screen carried out in S. cerevisiae (Uetz et al., 2000). Our analysis suggests that the YOL082w gene product, renamed Cvt19, has the basic properties that would be predicted for a prAPI receptor. Precursor API binding is destabilized in cells lacking Cvt19, and prAPI binds to Cvt19 in a propeptide-dependent manner. In addition, Cvt19 appears to be localized at a site consistent with vesicle formation, and travels to the vacuole along with prAPI. Cvt19 is not required for autophagy or for the specific vacuolar delivery of peroxisomes by pexophagy. Together, these results suggest that Cvt19 is a receptor for the targeting of resident hydrolases by the Cvt pathway.
Results
cvt19Δ Cells Are Specifically Defective in the Cvt Pathway
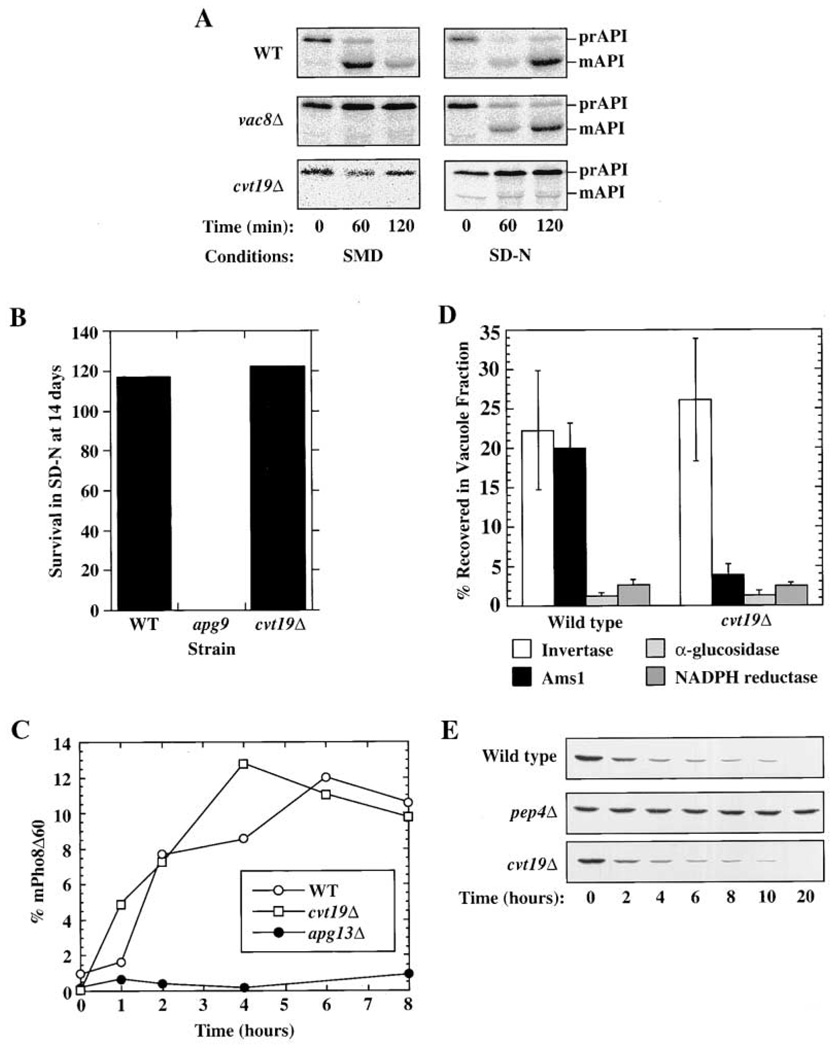
Recently, a systematic analysis of protein-protein interactions in Saccharomyces cerevisiae was performed using a yeast two-hybrid screen (Uetz et al., 2000). In these studies, the Cvt pathway cargo protein prAPI was found to interact with the unknown yeast open reading frame YOL082w. To examine whether YOL082w has a role in the Cvt pathway, the transport of prAPI was examined in a YOL082w deletion strain. In wild-type cells, prAPI is delivered to the vacuole with a half-time of approximately 30 min. In contrast, very little of the vacuolar mature form of API was detected in YOL082wΔ cells (Figure 1A, left panels), suggesting that this protein is required for the vacuole delivery of prAPI. Because of its role in the Cvt pathway, YOL082w was renamed CVT19.
Figure 1. Cvt19 Is Specifically Required for the Cvt Pathway.
(A) Cells from the indicated strains were pulse labeled for 5 min in SMD medium, washed, and resuspended in either SMD or SD-N and subjected to a nonradioactive chase for the amount of time indicated. Samples were immunoprecipitated with anti-API antiserum. The positions of prAPI and mature API (mAPI) are shown. Note that there is a background band that migrates just below the position of mAPI in the cvt19Δ cells chased in SD-N.
(B) The indicated strains were incubated for 14 days in SD-N and then plated. The number of surviving colonies was compared with the number of colonies on day 0.
(C) Cells lacking PHO8 and carrying sequences encoding Pho8Δ60 on a plasmid were subjected to pulse-chase analysis followed by immunoprecipitation with antiserum against ALP. The percent of Pho8Δ60 present in the vacuolar mature form at the indicated time points is plotted. Note that the parental strain for these cells, SEY6210, displays a maximal level of autophagy corresponding to approximately 15% uptake of Pho8Δ60.
(D) Enzyme assays were performed on isolated vacuoles and whole-cell lysates. The percent recovered in the purified vacuole fraction is plotted. Cells carrying a vacuole-delivered CPY-invertase fusion protein and lacking wild-type invertase were used. Invertase recovery (units in vacuole fraction divided by total units loaded on gradient) is a measure of vacuole recovery, and α-glucosidase and NADPH cytochrome c reductase measure contamination by cytosol and ER, respectively.
(E) Cells from the indicated strains were grown under conditions that induce peroxisomes (see Experimental Procedures), washed, and resuspended in SD-N for the time indicated. The presence of the peroxisomal marker protein Fox3 was detected by immunoblotting.
The majority of proteins required for Cvt transport are also required for autophagy, and prAPI is delivered to the vacuole by both pathways (Scott et al., 1996). Accordingly, we decided to examine whether cvt19Δ cells are blocked in autophagy. Yeast mutants defective for autophagy, such as apg9, die rapidly upon shifting to a nitrogen starvation medium (SD-N; Figure 1B). When cvt19Δ cells were grown in SD-N, however, their viability was the same as wild-type cells (Figure 1B). To examine autophagy in cvt19Δ cells more quantitatively, we followed the vacuolar delivery of the marker protein Pho8Δ60. Pho8Δ60 is a construct that consists of the vacuole protein alkaline phosphatase (encoded by PHO8) in which the transmembrane domain that acts as an internal uncleaved signal sequence has been removed, resulting in a cytosolic version of the enzyme. This protein is delivered to the vacuole under nutrient conditions that promote autophagy, and is cleaved to a lower molecular weight form by vacuolar proteinase B (Noda et al., 1995). In cvt19Δ cells, the rate of vacuole delivery of Pho8Δ60 in SD-N was the same as in wild-type cells (Figure 1C). By contrast, in the autophagy mutant apg13Δ, no mature Pho8Δ60 was detected. These results suggest that Cvt19 is required for Cvt transport, but is not necessary for autophagy.
While most components of the Cvt and Apg pathways overlap, some are specific for one or the other pathway. cvt19Δ cells are blocked in prAPI import by the Cvt pathway and are not defective in autophagy. For this reason, we decided to determine whether Cvt19 was specific for the Cvt pathway by examining prAPI import under nutrient-rich and starvation conditions. Wild-type cells import and mature prAPI in both nutrient-rich (SMD, Cvt pathway) and starvation (SD-N, Apg pathway) conditions (Figure 1A). Vac8 is a previously characterized protein that is required for Cvt transport, but has a more limited role in autophagy (Scott et al., 2000). Because prAPI is selectively transported to the vacuole by autophagosomes in starvation conditions, the prAPI delivery block observed in rich media in the vac8Δ strain was overcome when autophagy was induced in these cells. By contrast, the prAPI delivery block was not overcome in cvt19Δ cells when autophagy was induced (Figure 1A, right panels). These results could be explained if prAPI is not selectively localized to the site of autophagosome formation in cvt19Δ cells.
Recently, our lab has characterized a second marker protein for Cvt transport. The vacuole protein α-mannosidase (Ams1) also travels to the vacuole by the Cvt pathway (Hutchins and Klionsky, 2001). Vacuole delivery of Ams1 is blocked in cvt and apg mutant cells. Delivery of Ams1 to the vacuole was examined in cvt19Δ cells by isolating vacuoles and performing enzyme assays to detect the presence of Ams1 and various marker proteins. In wild-type cells, recovery of the vacuole marker carboxypeptidase Y (CPY)-invertase and Ams1 in the purified vacuole fraction was similar (22% compared to 20%). In contrast, in cvt19Δ cells, 25% of CPY-invertase was recovered compared with 5% of Ams1 (Figure 1D), suggesting that Ams1 was not delivered to the vacuole in cvt19Δ cells.
While peroxisomes and other organelles can be degraded nonspecifically by macroautophagy, the uptake of peroxisomes by pexophagy is a specific process (Kim and Klionsky, 2000). All cvt and apg mutants characterized thus far also display defects in the degradation of peroxisomes by pexophagy (Hutchins et al., 1999). We examined pexophagy in cvt19Δ cells by growing the cells in an oleic acid-containing medium to induce peroxisomes, switching them to SD-N, and monitoring the level of the peroxisome enzyme Fox3 (thiolase) by immunoblotting. In both wild-type and cvt19Δ cells, Fox3 was rapidly turned over, indicating that the uptake of peroxisomes by pexophagy is not defective in cvt19Δ cells (Figure 1E). When degradation in the vacuole was halted by deleting proteinase A (encoded by PEP4), Fox3 was stabilized, confirming that Fox3 turnover was vacuole dependent. These results indicate that the cvt19Δ strain was not defective in the process of specific peroxisome degradation. This is in contrast to other mutants such as cvt9Δ that are not defective for autophagy but are still blocked in pexophagy (Kim et al., 2001).
Precursor API Has a Binding Defect in cvt19Δ Cells
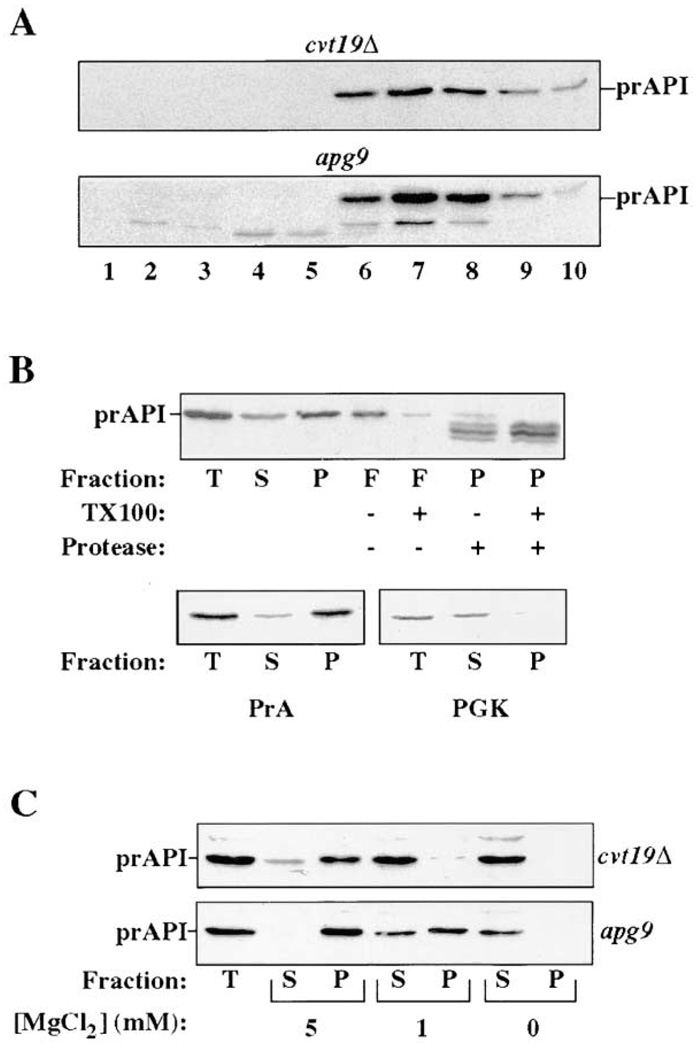
To determine the site of action of Cvt19, we examined the biochemical state of prAPI trapped in cvt19Δ cells. After synthesis, prAPI is rapidly oligomerized into a homododecameric protein complex (Kim et al., 1997). Accordingly, we examined the oligomeric state of prAPI in cvt19Δ cells by glycerol gradient. In wild-type cells or in characterized apg/cvt mutants, both mature API and transiting prAPI peak in fraction 7 (Kim et al., 1997). Similarly, accumulated prAPI in cvt19Δ cells peaked at fraction 7, suggesting that prAPI oligomerization is not blocked in this mutant (Figure 2A).
Figure 2. Precursor API Binding Is Destabilized in cvt19Δ Cells.
(A) Glycerol gradient fractionation of cell lysates isolated from the indicated strains. The resulting fractions were immunoblotted with anti-API antiserum. The position of prAPI is indicated. A small amount of mAPI is present in the apg9 cells, and also peaks in fraction 7.
(B) Subcellular fractionation of spheroplasts isolated from cvt19Δ cells (see Experimental Procedures). T, total lysate; S, supernatant fraction; P, low-speed pellet; and F, fraction that floats through a Ficoll step gradient. Samples were solubilized with 0.2% Triton X-100 or treated with 50 µg/ml proteinase K as indicated. The resulting fractions were subjected to immunoblotting with antisera against the indicated proteins.
(C) Subcellular fractionation was performed as in (B) except that either 5 mM, 1 mM, or 0 mM MgCl2 was added to the lysis buffer, as indicated.
Further analysis of prAPI in cvt19Δ cells was performed by subcellular fractionation and protease treatment. When cvt19Δ cells were lysed osmotically and fractionated using previously characterized conditions, the majority of prAPI was recovered in the 5000 g pellet fraction (Figure 2B). Marker proteins proteinase A (PrA, vacuole) and phosphoglycerate kinase (PGK, cytosol) indicate that the majority of vacuoles were intact and were effectively separated from cytosolic proteins in this experiment. Precursor API located in the pellet fraction was protease accessible and floated through a Ficoll step gradient in the absence, but not the presence, of detergent. These results suggest that prAPI was associated with a cytosolic face of a membrane or with lipid, but was not present within an enclosed membrane compartment in the cvt19Δ strain.
In examining the fractionation pattern of API in many mutant backgrounds, we have observed that wild-type prAPI is typically recovered completely in the pellet fraction when spheroplasts are fractionated by osmotic lysis in buffer containing 5 mM MgCl2 (as in Figure 2B). However, when MgCl2 is omitted from the lysis buffer, prAPI is stripped off into the supernatant fraction (Oda et al., 1996). In the experiment in Figure 2B, a fraction of the prAPI was recovered in the supernatant fraction in the presence of 5 mM MgCl2 in cvt19Δ cells, suggesting that prAPI binding may be destabilized in this strain. To examine this more carefully, MgCl2 was titrated in subcellular fractionation experiments with cvt19Δ cells and a control mutant strain, apg9, which displays typical binding interactions (Figure 2C). As before, in the presence of 5 mM MgCl2, a small fraction of prAPI was observed in the supernatant fraction in cvt19Δ cells. When 1 mM MgCl2 was used, all of the prAPI was recovered in the supernatant fraction. This was in contrast to the control strain, apg9, in which no prAPI was detected in the supernatant fraction after lysis with 5 mM MgCl2, and the majority remained associated with the pellet fraction in 1 mM MgCl2. In agreement with previous results, in the absence of MgCl2, prAPI was stripped off the membrane and recovered in the supernatant fraction in both strains. These results indicate that prAPI is more easily stripped off the membrane in cvt19Δ cells compared with the more typical apg9 cells.
Cvt19 Is a Peripheral Membrane Protein
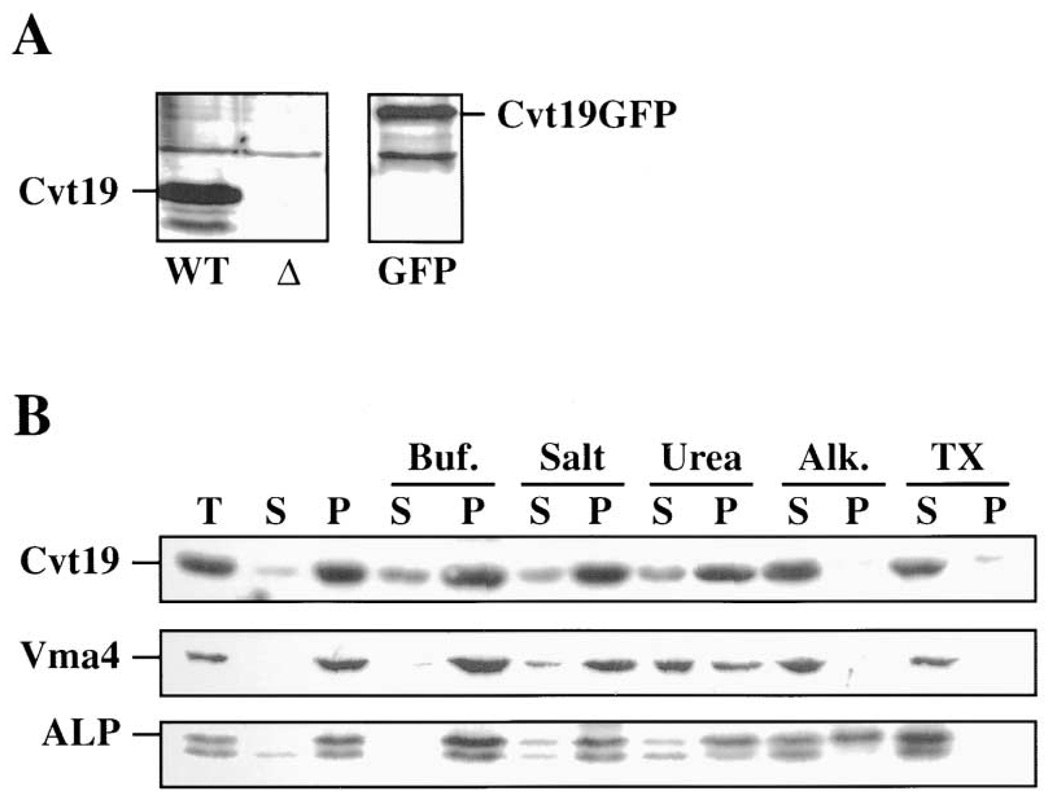
To examine Cvt19 directly, antiserum was generated against the protein (see Experimental Procedures). Cvt19 is predicted to be 48 kDa. In wild-type cells, a band that migrates at about 55 kDa was detected that was not present in cvt19Δ cells (Figure 3A). Subcellular fractionation experiments were performed to determine whether Cvt19 is associated with membranes. After permeabilizing spheroplasts using the conditions described in Figure 2, Cvt19 was recovered in the 13,000 g pellet fraction (Figure 3B), suggesting that it was membrane associated. The nature of the interaction of Cvt19 with the membrane was analyzed by treating the pellet fraction with various reagents. The treated pellets were then subjected to centrifugation at 13,000 g and the resulting supernatant and pellet fractions collected. Washing the Cvt19 pellet with lysis buffer, high salt buffer, or urea resulted in limited stripping of Cvt19 into the supernatant fraction. In contrast, Cvt19 was completely removed from the pellet fraction by alkaline extraction or detergent treatment (Figure 3B). Interestingly, another peripheral membrane protein required for the Cvt and Apg pathways, Apg5, is also extracted from the membrane by high pH, but not by urea (George et al., 2000). As a control, we examined the distribution of Vma4, a peripheral membrane subunit of the vacuolar membrane ATPase, and ALP, a vacuolar integral membrane protein. Vma4 was also stripped off of the membrane by alkaline extraction, and was more sensitive to extraction by urea than Cvt19. In contrast, ALP remained largely in the pellet fraction after alkaline extraction, and was only completely released from the membrane by detergent treatment. These results suggest that Cvt19 is a peripheral membrane protein associated with a low-speed pellet fraction.
Figure 3. Cvt19 Is a Peripheral Membrane Protein.
(A) Cells from the indicated strains were subjected to immunoblotting with antiserum against Cvt19. WT, SEY6210; Δ, SSY31 (cvt19Δ); and GFP, SSY32 (cvt19Δ) containing pCVT19GFP. The positions of Cvt19 and Cvt19GFP are indicated.
(B) Spheroplasts from SEY6210 cells were lysed osmotically as described in Experimental Procedures and fractionated into total (T), supernatant (S), and pellet (P) fractions. The resulting pellet was subjected to the following treatments: Buf., lysis buffer; Salt, 0.66 M KOAc, 0.34 M KCl; Urea, 3 M urea; Alk., 50 mM Na2CO3 (pH 10.5); and TX, 0.5% Triton X-100. After centrifugation, S and P fractions were collected and subjected to immunoblotting. The positions of Cvt19, Vma4 (peripheral membrane), and ALP (integral membrane) are indicated.
Cvt19 Is Delivered to the Vacuole by the Cvt Pathway
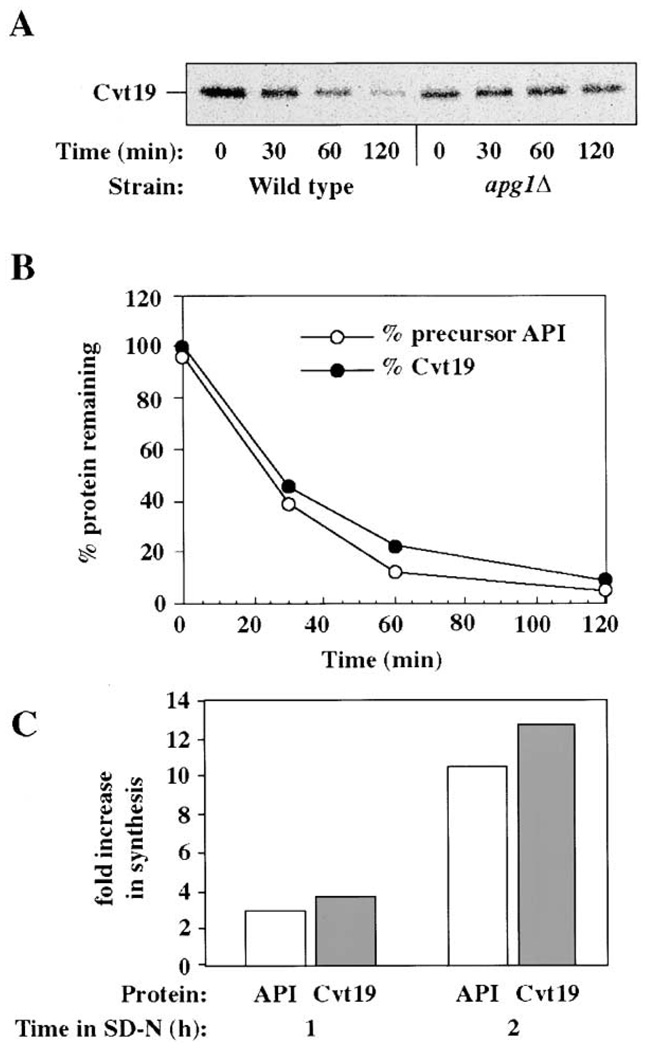
To extend our analysis, we examined the biogenesis of Cvt19 with a pulse-chase experiment. After pulse labeling wild-type cells, a single Cvt19 band was detected by immunoprecipitation analysis. When the cells were subjected to a nonradioactive chase reaction, the Cvt19 band was rapidly degraded with a half-life of about 30 min in both rich and starvation conditions (Figure 4A and data not shown). Because this rate is similar to the rate of delivery of prAPI by the Cvt pathway (Klionsky et al., 1992), we examined the kinetics of prAPI maturation in the same extracts. The rate of degradation of Cvt19 and the rate of maturation of prAPI were nearly the same (Figure 4B), suggesting that these two proteins may be traveling to the vacuole together. To examine whether the degradation of Cvt19 required the components needed for the Cvt pathway, we examined Cvt19 in apg1Δ cells that are defective for both the Cvt and Apg pathways (Scott et al., 2000). Cvt19 was stabilized in this mutant (Figure 4A) as well as in pep4Δ cells (data not shown). These results indicate that Cvt19 travels to the vacuole by the Cvt and Apg pathways, and that it is degraded by vacuole resident proteases.
Figure 4. Cvt19 Is Delivered to the Vacuole by the Cvt Pathway.
(A) Wild-type (SEY6210) or apg1Δ (NNY20) cells were pulse labeled for 10 min, chased for the indicated times in SMD, immunoprecipitated with antiserum against Cvt19, and analyzed by SDS-PAGE.
(B) The extracts from the wild-type cells in (A) were also immunoprecipitated with anti-API antiserum. The kinetics of prAPI maturation and Cvt19 degradation are depicted.
(C) Wild-type cells (SEY6210) were pulse labeled for 10 min, in SMD or in SD-N after growth in SD-N for either 1 or 2 hr as indicated. API and Cvt19 were recovered by immunoprecipitation, and the change in the level of synthesis in SD-N compared to SMD is plotted.
Cvt19 appears to utilize either the Cvt or the Apg pathway for vacuole delivery. Like vacuole delivery of prAPI, the rapid kinetics of degradation suggest that this transport event is specific and must require recruitment into the forming vesicle. To examine whether prAPI itself is involved in this recruitment step, degradation of Cvt19 was examined in ape1Δ cells. We found that the rate of Cvt19 degradation was reduced in the absence of prAPI. The half-life of Cvt19 in rich medium increased approximately 4-fold in the ape1Δ mutant compared to wild-type cells (data not shown). These results suggest that the presence of prAPI triggers the efficient inclusion of Cvt19 in the forming autophagosomes and Cvt vesicles.
The synthesis of prAPI is highly induced in nitrogen starvation conditions (Scott et al., 1996). We examined the synthesis of Cvt19 to determine whether production of this protein is also increased during nitrogen starvation. Cells were pulse labeled after growth in SMD media or after incubation in starvation medium for either 1 or 2 hr, and the levels of Cvt19 and prAPI were examined by immunoprecipitation. The induction of Cvt19 paralleled the induction of prAPI (Figure 4C), consistent with a role for Cvt19 in the correct targeting of prAPI in both rich and starvation conditions.
Overproduction of prAPI in rich medium leads to cytosolic accumulation of the precursor form of the protein (Klionsky et al., 1992). To investigate whether this is the result of titrating out a limited pool of Cvt19, API sorting was examined in cells overproducing both prAPI and Cvt19. We found that the overproduced prAPI was still not efficiently targeted even in the presence of overproduced Cvt19 (data not shown). This result suggests that an additional factor(s) is limiting when prAPI is expressed at high levels. This component(s) could be a membrane protein that serves as an anchor for the peripheral membrane protein Cvt19.
Cvt19GFP Localizes to Punctate Structures near the Vacuole
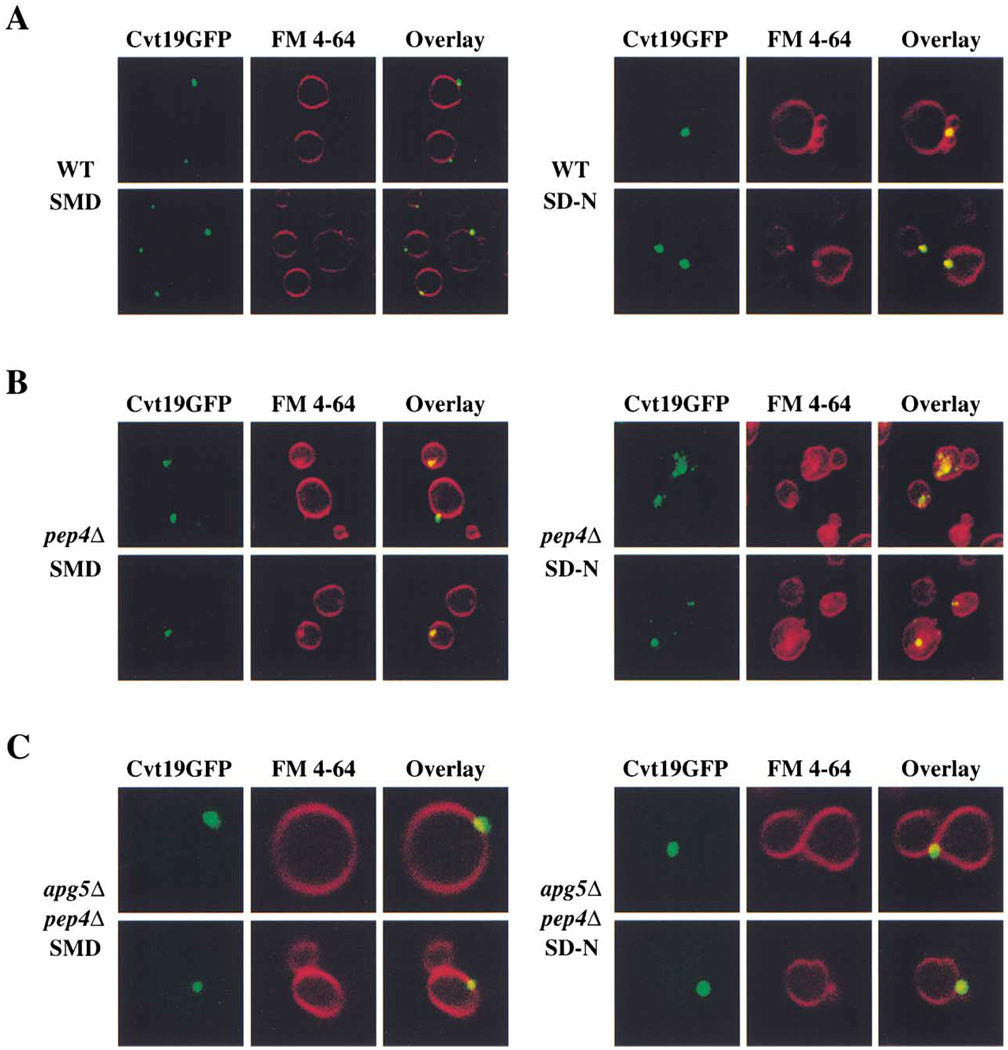
To examine the localization of Cvt19 in intact cells, we constructed a fusion protein consisting of Cvt19 with GFP at its C terminus. The resulting Cvt19GFP was detected as a band at about 85 kDa on immunoblots ( Figure 3A), and complemented the prAPI accumulation phenotype of cvt19Δ cells (data not shown). Cells bearing the Cvt19GFP fusion protein on a centromeric plasmid were incubated with the dye FM 4–64 to label vacuoles. When wild-type cells were examined by fluorescence microscopy after growth in rich medium, Cvt19GFP was observed in punctate structures that appeared to be near or on the vacuole surface (Figure 5A). In starvation conditions, the overall staining pattern was similar, but the fluorescent dots appeared to be slightly larger than in rich medium.
Figure 5. Cvt19GFP Is Localized to Punctate Structures Near the Vacuole.
(A) Wild-type (SEY6210) cells expressing Cvt19GFP were grown in SMD or shifted to SD-N for 2 hr. Cells were treated with FM 4–64 to label vacuoles and analyzed using a fluorescent microscope as described in Experimental Procedures.
(B) Cvt19GFP is trapped in subvacuolar vesicles in pep4Δ cells. pep4Δ (YMTA) cells expressing Cvt19GFP were prepared and examined as in (A).
(C) Delivery of Cvt19GFP to the vacuole is dependent on the machinery of the Cvt and Apg pathways. apg5Δ pep4Δ (SSY101) cells expressing Cvt19GFP were prepared and examined as in (A).
Cvt bodies and autophagic bodies are stabilized and thus easily detected within the vacuoles of cells that lack PrA, one of the major processing enzymes (Klionsky et al., 1990). If Cvt19GFP travels to the vacuole within Cvt vesicles and autophagosomes, it should be present within these subvacuolar structures when PrA is absent. When the staining pattern of Cvt19GFP was examined in pep4Δ cells grown in nonstarvation conditions, punctate structures consistent with the expected abundance of Cvt vesicles were observed (Figure 5B). In starvation conditions, subvacuolar vesicular structures containing Cvt19GFP were even more apparent in the pep4Δ cells. To confirm that the observed structures were Cvt bodies and autophagic bodies, Cvt19GFP was examined in pep4Δ apg5Δ cells. In the absence of Apg5, a protein required for autophagy and Cvt transport, only perivacuolar Cvt19GFP structures were observed (Figure 5C). These studies support the biochemical data (Figure 4) that indicated that Cvt19 travels to the vacuole inside Cvt vesicles and autophagosomes.
Because the vacuolar delivery of Cvt19 is delayed in cells lacking prAPI, we also examined the localization of Cvt19GFP in ape1Δ cells and in ape1Δ pep4Δ cells. In ape1Δ cells, punctate structures indistinguishable from those seen in wild-type cells were observed (data not shown). In the ape1Δ pep4Δ double-mutant strain, punctate structures both outside and inside the vacuole were observed (data not shown). Compared with pep4Δ cells, more punctate structures were present outside the vacuole, and fewer subvacuolar vesicles were observed. These results are consistent with the reduced degradation rate of Cvt19 seen in ape1Δ cells, and suggest that Cvt19 is not as efficiently delivered to the vacuole in the absence of prAPI as in wild-type cells.
Cvt19 Physically Associates with Precursor API
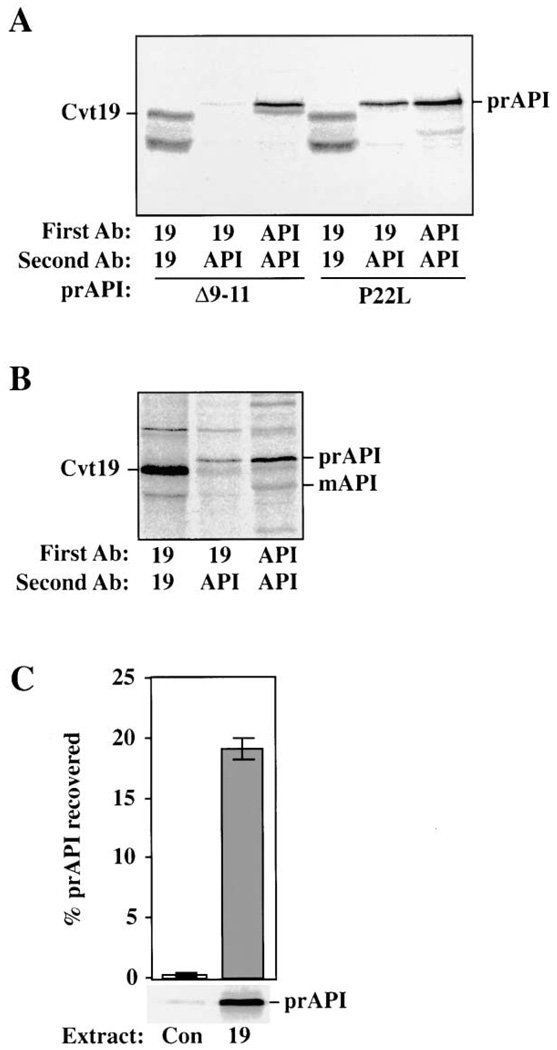
Our experiments, together with two-hybrid data implicating Cvt19 and prAPI as interacting proteins, suggest that Cvt19 may be a receptor protein for prAPI targeting. If this were the case, we would expect that Cvt19 would interact with prAPI during the import process. We examined the interaction of Cvt19 and prAPI in cell lysates by coimmunoprecipitation analysis. For these experiments, we used ape1Δ cells that carried plasmids encoding mutant API precursor proteins that have been previously characterized (Kim et al., 1997; Oda et al., 1996; Scott et al., 1997). Δ9–11 API oligomerizes normally, but does not bind membrane. In contrast, P22L API is tightly associated with the membrane, but is transported to the vacuole inefficiently. Spheroplasts were pulse labeled, lysed in the presence of detergent, and subjected to native immunoprecipitation with antisera against either Cvt19 or API. After the washing of the immune complexes, a second denaturing immunoprecipitation was performed. When either anti-Cvt19 or anti-API was used in both immunoprecipitation reactions, the respective proteins were successfully immunopurified (Figure 6A). In lysates precipitated first with Cvt19 antiserum and then with API antiserum, a strong prAPI band was recovered from cells bearing P22L API. Under the same conditions, only an extremely weak band was recovered from the Δ9–11 API cells.
Figure 6. Coimmunoprecipitation of Cvt19 and prAPI.
(A) Spheroplasts isolated from ape1Δ (THY101) cells containing either Δ 9–11 API or P22L API on a plasmid were pulse labeled for 20 min, chased for 30 min, and lysed in native immunoprecipitation buffer. The resulting extract was cleared by centrifugation at top speed in a microcentrifuge for 5 min before immunoprecipitating with antisera as indicated by “First Ab.” A second, nonnative immunoprecipitation reaction was then performed using the antisera denoted as “Second Ab.”
(B) As in (A) except that wild-type (SEY6210) cells were used. The positions of Cvt19, prAPI, and mAPI are indicated. The lower molecular weight band in the Cvt19 immunoprecipitations is a Cvt19 degradation product.
(C) In vitro binding of Cvt19 and prAPI. Bacterial lysate from cells containing either the control vector alone (Con), or expressing Cvt19 (19) were incubated with in vitro-translated prAPI. API was then recovered by native immunoprecipitation with Cvt19 antiserum. The prAPI recovered is plotted as a percent of the total added to the binding reaction. Error bars represent the standard deviation from three separate experiments.
We also examined whether native API precursor or mature-sized proteins interact with Cvt19. Spheroplasts from wild-type cells were pulse labeled and then subjected to a nonradioactive chase to allow for the presence of both mature API and transiting prAPI in the sample. After coimmunoprecipitation with Cvt19 and then API antisera, the precursor form, but not the mature form of API, was found to be associated with Cvt19 (Figure 6B). These results indicate that Cvt19 interacts with prAPI in a manner that is dependent on the prAPI propeptide.
To investigate whether additional factors might be required for Cvt19-prAPI interaction, an in vitro binding assay was utilized. Protein extracts were isolated from E. coli transformed with either a plasmid encoding Cvt19 or the expression vector alone. The extracts were incubated with radiolabeled prAPI synthesized in vitro in a rabbit reticulocyte lysate-coupled transcription/translation system. The extracts were then subjected to a native immunoprecipitation with Cvt19 antiserum. Precursor API was highly enriched in an immunoprecipitation reaction containing Cvt19 extract compared to a protein extract isolated from bacteria expressing the vector alone (Figure 6C). This result indicates that an additional yeast protein is not necessary to act as a bridge between Cvt19 and prAPI. It is very unlikely that a heterologous protein factor could be provided by the reticulocyte lysate translation system because the level of such a factor would be substoichiometric in our binding assay. Furthermore, there is no evidence for either a Cvt19 homolog or a Cvt pathway equivalent in mammalian cells.
Discussion
Cvt19 Is Required for prAPI Import through the Cvt and Apg Pathways
Precursor API is delivered to the vacuole by the related Cvt and Apg pathways (Baba et al., 1997; Scott et al., 1996). The particular pathway that is utilized depends on nutrient conditions. In contrast to bulk cytosol that is taken up nonselectively by the Apg pathway, transport of prAPI is always selective. However, despite the characterization of numerous proteins required for the Cvt and Apg pathways, identification of a prAPI receptor protein has remained elusive. Here, we report the char acterization of Cvt19, a protein that binds to the precursor form of API and is specifically required for biosynthetic transport from the cytoplasm to the vacuole. Cvt19 is necessary for vacuole delivery of both known Cvt pathway substrates, prAPI and Ams1 (Figure 1). Although the delivery of bulk cytosol by the Apg pathway does not require Cvt19, prAPI cannot be efficiently delivered to the vacuole under autophagic conditions in cvt19Δ cells. These results suggest that Cvt19 may lend specificity to the targeting of prAPI by the Apg pathway. Interestingly, although peroxisomes are also specifically degraded by a mechanism that utilizes the Cvt and autophagic machinery (Hutchins et al., 1999; Kim et al., 2001), this process is independent of Cvt19 (Figure 1E). Thus, Cvt19 is unusual among the characterized Apg/Cvt proteins in that it is required for Cvt transport but is not required for pexophagy.
In cvt19Δ cells, the early steps of the Cvt pathway were not disrupted. Precursor API was found to oligomerize and bind to a membrane fraction (Figure 2). However, when the prAPI binding interaction was probed more rigorously, it became apparent that the binding interaction was destabilized in cvt19Δ cells. It may be that a second receptor protein that has still not been identified also participates in Cvt transport. Precursor API may first bind to this unknown protein as part of the Cvt complex assembly process, and then be passed on to Cvt19 for enclosure into vesicles. Alternatively, the prAPI propeptide forms two α helices, the first being amphipathic, which may have lipid-seeking properties that allow for the membrane binding interaction detected in cvt19Δ cells.
Cvt19 Is a Peripheral Membrane Protein that Localizes near the Vacuole
Biochemical analysis of Cvt19 indicates that it is a peripheral membrane protein that is present in a low-speed membrane fraction (Figure 3). This is consistent with a role in binding prAPI, because the bound form of prAPI is also known to pellet in a low-speed membrane fraction (Oda et al., 1996; Scott and Klionsky, 1995; Figure 2). Localization of Cvt19GFP suggests that Cvt19 is concentrated in punctate structures that are on or near the surface of the vacuole (Figure 5). These may represent the cellular sites of Cvt complex formation or the site of the donor membrane for the forming vesicles.
Cvt19 is a very short-lived protein with a half-life of about 30 min, similar to the rate of vacuole delivery of prAPI (Figures 4A and 4B). Cvt19 degradation is dependent on PrA, suggesting that Cvt19 is delivered to the vacuole (data not shown). In addition, Cvt19 is stabilizedin apg1 cells that are defective in transport by both the Apg and Cvt pathways, indicating that it is delivered to the vacuole within Cvt vesicles or autophagosomes. Cvt19 is also stabilized in ape1Δ cells (data not shown), suggesting that interaction between Cvt19 and prAPI facilitates the inclusion of both proteins into forming vesicles. API synthesis is induced in starvation conditions. As expected for a specific receptor, Cvt19 is similarly induced at levels that are stoichiometric with prAPI (Figure 4C).
Cvt19 Interacts with the Precursor API Propeptide
Coimmunoprecipitation studies were carried out to demonstrate the interaction between Cvt19 and prAPI directly, and to identify the region of prAPI required for the interaction. In lysates taken from cells containing chromosomal levels of both prAPI and Cvt19, antibodies against Cvt19 precipitated not only Cvt19, but also prAPI in a native immunoprecipitation reaction (Figure 6B). Mature API present in the same reaction did not coprecipitate in these conditions, suggesting that it is the precursor form of API that is bound to Cvt19. Additional immunoprecipitation reactions indicate that the previously characterized propeptide mutant that is defective in membrane binding, Δ9–11 API, did not coimmunoprecipitate efficiently with Cvt19 (Figure 6A). Conversely, a propeptide mutant that displays a strong membrane binding interaction but that is blocked in a subsequent stage of import, P22L API, was coimmunoprecipitated with Cvt19. These results suggest that a binding-competent propeptide region is necessary for the interaction between API and Cvt19. An in vitro binding analysis was carried out to determine whether additional protein components are required for prAPI and Cvt19 to interact. Cvt19 expressed in E. coli bound to in vitro-synthesized prAPI, suggesting that Cvt19 and prAPI bind to each other directly and do not require an additional yeast protein to form a stable complex (Figure 6C).
Cvt19 Is Required for Ams1 Transport
Additional genome-wide two-hybrid studies have uncovered an interaction between Cvt19 and Ams1 (Ito et al., 2001). This result is consistent with our finding that like prAPI, Ams1 requires Cvt19 for correct vacuolar delivery (Figure 1D). We were unable to demonstrate an interaction between Cvt19 and Ams1 by coimmunoprecipitation (data not shown); however, this may be due to the fact that Ams1 is a much less abundant protein than API, and thus more difficult to detect.
Cvt19 Is Not a Classical Receptor Protein
Many characterized vacuole/lysosome receptors such as Vps10 and the mannose 6-phosphate receptor are integral membrane proteins that are recycled so that they can mediate multiple rounds of protein sorting. In contrast, Cvt19 is a peripheral membrane protein that is delivered to the vacuole with its cargo and is degraded. Autophagosomes appear to contain few integral membrane proteins (Baba et al., 1995). There may be no benefit to Cvt19 or other proteins in spanning the inner membrane of these double-membrane vesicles. These proteins would only reach the lumenal space between the two membranes rather than the inside or cytosolic face of the vesicle where they could directly transmit a signal or act to recruit additional factors. Long-lived recycling receptors such as Vps10 use their transmembrane domains and cytosolic sequences for maintaining position in the secretory pathway and for recycling. The fact that Cvt19 is turned over rather than recycled is an outcome of the fact that complexes of Cvt19 and prAPI are enclosed in the inner vesicle of Cvt vesicles and autophagosomes. As a result, Cvt19 ends up within the vacuole lumen following fusion, and is degraded.
These studies demonstrate that Cvt19 is an atypical receptor protein for the Cvt pathway. How this peripheral membrane protein localizes to the correct membrane for inclusion in Cvt vesicles and autophagosomes remains to be determined. We are currently attempting to identify a protein that is required for Cvt19 localization. Continued analysis of cytoplasm-to-vacuole targeting will provide further insight into these dynamic membrane and protein transport processes, and the molecular mechanism of Cvt19 action.
Experimental Procedures
Materials
Vent DNA polymerase, DNA restriction and modifying enzymes, and vector pMAL-c2 were purchased from New England Biolabs. T4DNA ligase and protease inhibitor tablets were from Roche Molecular Biochemicals. The S. cerevisiae strain S288C genomic DNA was from Research Genetics. Oligonucleotides were synthesized by Operon Technologies and Invitrogen. N-(triethlyammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide (FM 4–64) was from Molecular Probes. Expre35S35S label was from NEN Life Science Products. All other reagents were from Sigma-Aldrich.
Antiserum Preparation
A maltose binding protein-Cvt19 fusion protein (described below) was expressed from E. coli strain BL21 (Amersham Pharmacia) and purified through amylose resin (New England Biolabs). Antiserum was generated by the procedure described (Harlow and Lane, 1999). The preparation of antiserum to 3-ketoacyl-CoA thiolase (Fox3; Hutchins et al., 1999), API (Klionsky et al., 1992), ALP (Klionsky and Emr, 1989), PrA (Klionsky et al., 1988), and Vma4 (Morano and Klionsky, 1994) were described previously. Mouse monoclonal anti-ALP was from Molecular Probes, and antiserum to PGK was generously provided by Dr. Jeremy Thorner (University of California at Berkeley).
Strains and Media
The cvt19Δ strain was generated by PCR-mediated disruption of the YOL082w locus, using the amplified sequence of the HIS5 gene from Schizosaccharomyces pombe flanked by sequences homologous to the CVT19 coding sequence: 5′-GCGGCGGCACTTGCTTCAGTAACGCCCAAAGGAGAGTTCTGGCCCGGGCTGCAGGAATTC-3′ and 5′-AAGAGCATGATGCTAGGTGATAAGTAATGAGAGGCCTTAG GATCGACGGTATCGATAAGC-3′. Strains used are described in Table 1.
Table 1.
Yeast Strains Used in this Study
| Strain | Genotype | Source or reference |
|---|---|---|
| SEY6210 | MATαleu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL | Robinson et al., 1988 |
| TN124 | MATαleu2-3,112 trp1 ura3-52 pho8∷pho8Δ60 pho13Δ∷LEU2 | Noda et al., 1995 |
| D3Y103 | TN124 apg13Δ∷URA3 | Scott et al., 2000 |
| NNY20 | MATαura3 trp1 leu2 apg1Δ∷LEU2 | Matsuura et al., 1997 |
| D3Y102 | SEY6210 vac8Δ∷TRP1 | Scott et al., 2000 |
| THY101 | SEY6210 ape1Δ∷LEU2 | Oda et al., 1996 |
| THY154 | SEY6210 apg9 | Harding et al., 1995 |
| DKY6281 | SEY6210 pho8Δ∷TRP1 | Klionsky and Emr, 1989 |
| WCG4a | MATαhis3-11,15 leu2-3,112 ura3 | Heinemeyer et al., 1993 |
| YMTA | WCG4a pep4Δ∷HIS3 | Thumm et al., 1994 |
| SSY101 | MATα ura3-52 trp1-901 apg5Δ∷LEU2 pep4Δ∷HIS3 | George et al., 2000 |
| SSY31 | SEY6210 cvt19Δ∷HIS5 S.p. | This study |
| SSY32 | WCG4a cvt19Δ∷HIS5 S.p. | This study |
| SSY33 | DKY6281 cvt19Δ∷HIS5 S.p. | This study |
| SSY34 | THY101 cvt19Δ∷HIS5 S.p. | This study |
Media: SGd, 0.67 %YNB without amino acids and brought to pH 5.5 with 50 mM each of 2-(N-morpholino) ethanesulfonic acid and 3-(N-morpholino) propanesulfonic acid, 3% glycerol, and 0.1% glucose; YTO, 0.67% YNB without amino acids, 0.1% Tween 40, and 0.1% oleic acid; and SMD, 0.67% YNB and2% glucose. Auxotrophic amino acids and vitamins were added to the above media as needed; SD-N, 0.17% YNB without ammonium sulfate or amino acids containing 2% glucose; and YPD, 1% bacto yeast extract, 2% bacto peptone, and 2% dextrose.
Plasmid Construction
The open reading frame of CVT19 was amplified by PCR from S. cerevisiae S288C genomic DNA using primers 5′-GTTCGGATCCATGAACAACTCAAAGACTAACC-3′ and 5′-TCTGGGGATCCGTATATCGTTTGAAAGTAG-3′. The PCR product was digested with PstI and BamHI, and ligated into pMAL-c2. The resulting plasmid carries the in-frame fusion between maltose binding protein and CVT19 coding sequence (1–1093). To generate an in-frame fusion of Cvt19 with GFP, the CVT19 open reading frame together with part of the 5′-untranslated region was amplified by PCR from genomic DNA. Both the forward primer 5′-TCTGGGGATCCGTATATCGTTTGAAAG TAG-3′ and the reverse primer 5′-TCGGATCCATAGACCTCTGTAAATGTTGG-3′ incorporated BamHI sites. The GFP vector pCAPG5-GFP(416) (George et al., 2000) was digested with BamHI to remove the APG5 coding region, and ligated with the above PCR product digested with BamHI to give rise to the plasmid pCVT19GFP(416). The Pho8Δ60 plasmid, pMUH9, was generated from the BamHI/KpnI fragment of pCC5 (Campbell and Thorsness, 1998) ligated into the same sites of pRS416 (Sikorski and Hieter, 1989). The full-length CVT19 ORF was PCR amplified from the pCVT19(426) plasmid using the following 5′ and 3′ oligonucleotides, respectively: 5′-GCTTCAG TAACGCCCATGGGAGAGTTCTGGTAAATGAAC-3′ and 5′-CATTG CTGTATAAAACTCGAGTTTGACCTAGAGTTCTTCCC-3′. The 5′ and 3′ primers contain NcoI and XhoI sites flanking the CVT19 ORF, respectively. The PCR product was digested with NcoI/XhoI and subcloned into the bacterial expression vector pET-14b (Novagen), resulting in pET-CVT19. The full-length APE1 gene was PCR-amplified from pfAPIXhoI (Oda et al., 1996). The following 5′ and 3′ primers containing NcoI and SalI sites flanking the APE1 ORF, respectively, were used for the PCR reaction: 5′-GTAGAAACCTGCACAACCATGGAAATTAAG-3′ and 5′-GAAATAAAAAGAGGTCGACAAAATCACAAC-3′. The PCR product was digested with NcoI and SalI and subcloned into pET-14b prepared by NcoI and XhoI digests, resulting in pET-API.
Pulse-Chase, Nitrogen Starvation, and Pexophagy
Pulse-chase, nitrogen starvation, and pexophagy experiments were performed as described (Hutchins et al., 1999; Scott et al., 1996).
Vacuole Preparations and Enzyme Assays
Vacuoles were isolated as described previously (Haas et al., 1995; Hutchins and Klionsky, 2001). Enzyme assays for marker proteins were performed on material loaded onto the gradient and the recovered vacuole float fraction. Invertase, α-glucosidase, and NADPH cytochrome c reductase assays were performed as described (Johnson et al., 1987). Ams1 activity was determined based on established protocol (Opheim, 1978). Results from assays for each strain were tabulated from four independent vacuole preparations.
Subcellular Fractionation and Protease Treatment
Cells were grown in SMD until 1 A600/ml and spheroplasted as described (Harding et al., 1995). Differential osmotic lysis was carried out by resuspending in PS200 (20 mM PIPES [pH 6.8] and 200 mM sorbitol) containing 5 mM MgCl2 and gently pipeting up and down ten times. Supernatant and pellet fractions were generated by centrifugation at 5000 g for 5 min. The membrane-associated float fraction was generated as described (Scott and Klionsky, 1995). Protease digestion was performed by incubation with 50 µg/ml proteinase K for 15 min on ice in the presence or absence of 0.2% Triton X-100. All samples were collected by TCA precipitation and subjected to cimmunoblotting as described (Harding et al., 1995). Glycerol gradient analysis was as described (Kim et al., 1997).
Cvt19 Membrane Binding
Fractionation of Cvt19 was as above except that spheroplasts were isolated in the presence of 2% glucose to maintain the membrane association of the peripheral subunits of the vacuolar ATPase. In addition, 1 × protease inhibitor tablets, 1 mM PMSF, and 2 µg/ml pepstatin A were added to all buffers, and the pellet fraction was collected after centrifugation at 13,000 g. Membrane treatments were performed by resuspending the pellet fraction in the following buffers and incubating for 10 min at room temperature: 0.66 M KOAc, 0.34 M KCl; 3 M urea, 20 mM PIPES [pH 6.8]; 50 mM Na2CO3 [pH 10.5]; 0.5% Triton X-100, 20mMPIPES [pH 6.8]. The supernatant and pellet fractions were collected after centrifugation at 13,000 g for 5 min.
Coimmunoprecipitation
Spheroplasts were pulse labeled for 20 min, chased for 30 min, pelleted, and lysed by resuspension in native immunoprecipitation buffer (1 × phosphate buffered saline, [pH 7.5], 5 mM MgCl2, 0.5% Tween 20, 1 × protease inhibitor tablet, 1 mM PMSF, 2 µg/ml pepstatin A). The resulting lysate was cleared by centrifugation at 13,000 g for 3 min. The supernatant was removed, added to antiserum against either Cvt19 or API and protein A-sepharose beads, and incubated overnight at 4°C with rocking. The beads were then pelleted, washed once with native immunoprecipitation buffer, and eluted and subjected to a second nonnative immunoprecipitation reaction as described (Scott et al., 1996), except that protease inhibitors were added. The resulting supernatant was resolved by SDS-PAGE and detected using a Molecular Dynamics Storm phosphorImager.
In Vitro Binding
BL21(DE3) E. coli cells (Novagen) transformed with pET-CVT19 were grown to A600 = 0.6 and induced with 400 µM IPTG for 4 hr at 37°C. The cells were harvested, washed, and then resuspended in 1/10 volume of native immunoprecipitation buffer. Bacterial lysis was performed by treating the resuspended cells with three successive rounds of freeze/thaw in an ethanol/dry ice bath, followed by probe sonication at 30% duty using a Branson Digital Sonifier (Branson Ultrasonics). The lysed bacteria were centrifuged at 14,000 g for 10 min at 4°C to generate the supernatant lysate. Fifty microliters of lysate (115 µg of protein) was incubated with 1.6 µl of radiolabeled prAPI synthesized using the TNT Quick Coupled Transcription/Translation rabbit reticulocyte lysate system (Promega) in a final volume of 500 µl of native immunoprecipitation buffer for 2 hr on ice. Radiolabeled prAPI was recovered by native immunoprecipitation with Cvt19 antiserum (see above).
Fluorescent Microscopy
Cultures were grown in SMD until early log phase. To label the vacuolar membrane, the cells were pelleted and resuspended in fresh SMD at 1 A600/ml. FM 4–64 was added to a final concentration of 8 µM, and the culture was incubated at 30°C for 30 min. The cells were washed and resuspended in either SMD or SD-N at 1 A600/ml. After two hr of incubation, the samples were examined on a Leica TCS-SP Laser Scanning Confocal Microscope (Leica), utilizing a 510–525 nm band pass filter for GFP in combination with a long pass 585 nm filter to observe the FM 4–64.
Acknowledgments
The authors would like to thank Dr. Sarah Teter for helpful comments. This work was supported by Public Health Service Grant GM53396 from the National Institutes of Health (to D.J.K.).
References
- Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct. Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Thorsness PE. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J. Cell Sci. 1998;111:2455–2464. doi: 10.1242/jcs.111.16.2455. [DOI] [PubMed] [Google Scholar]
- George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol. Biol. Cell. 2000;11:969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A, Scheglmann D, Lazar T, Gallwitz D, Wickner W. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 1995;14:5258–5270. doi: 10.1002/j.1460-2075.1995.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J. Biol. Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy components in Saccharomyces cerevisiae. J. Biol. Chem. 2001 doi: 10.1074/jbc.M101150200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 1999;112:4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bankaitis VA, Emr SD. Distinct sequence determinants direct intracellular sorting and modification of a yeast vacuolar protease. Cell. 1987;48:875–885. doi: 10.1016/0092-8674(87)90084-5. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Klionsky DJ. Autophagy, Cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu. Rev. Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- Kim J, Scott SV, Oda MN, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Scott SV, Klionsky DJ. Alternative protein sorting pathways. Int. Rev. Cytol. 2000;198:153–201. doi: 10.1016/s0074-7696(00)98005-7. [DOI] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: bio-synthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Banta LM, Emr SD. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol. Cell. Biol. 1988;8:2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 1990;54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Metz G, Marx R, Röhm KH. The quaternary structure of yeast aminopeptidase I. 1. Molecular forms and subunit size. Z. Naturforsch. 1977;32C:929–937. doi: 10.1515/znc-1977-11-1209. [DOI] [PubMed] [Google Scholar]
- Morano KA, Klionsky DJ. Differential effects of compartment deacidification on the targeting of membrane and soluble proteins to the vacuole in yeast. J. Cell Sci. 1994;107:2813–2824. doi: 10.1242/jcs.107.10.2813. [DOI] [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD, Klionsky DJ. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J. Cell Biol. 1996;132:999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opheim DJ. α-D-Mannosidase of Saccharomyces cerevisiae. Characterization and modulation of activity. Biochim. Biophys. Acta. 1978;524:121–130. doi: 10.1016/0005-2744(78)90110-9. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Klionsky DJ. In vitro reconstitution of cytoplasm to vacuole protein targeting in yeast. J. Cell Biol. 1995;131:1727–1735. doi: 10.1083/jcb.131.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Klionsky DJ. Delivery of proteins and organelles to the vacuole from the cytoplasm. Curr. Opin. Cell Biol. 1998;10:523–529. doi: 10.1016/s0955-0674(98)80068-9. [DOI] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Amino-peptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- Segui-Real B, Martinez M, Sandoval IV. Yeast amino-peptidase I is post-translationally sorted from the cytosol to the vacuole by a mechanism mediated by its bipartite N-terminal extension. EMBO J. 1995;14:5476–5484. doi: 10.1002/j.1460-2075.1995.tb00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]