Abstract
Nanostructures constructed from metal and semiconductor nanocrystals conjugated to, and organized by DNA are an emerging class of material with collective optical properties. We created discrete pyramids of DNA with gold nanocrystals at the tips. By taking small angle X-ray scattering (SAXS) measurments from solutions of these pyramids we confirmed that this pyramidal geometry creates structures which are more rigid in solution than linear DNA. We then took advantage of the tetrahedral symmetry to demonstrate construction of chiral nanostructures.
Introduction
Double-stranded DNA (dsDNA) is an attractive material for building nanoparticle assemblies. It has been shown previously that single-stranded DNA (ssDNA) can be attached to gold nanoparticles via a thiol linker. Then, the specific, complementary hydrogen-bonding of the base pairs allows the nanoparticles to be assembled by the hybridization of the DNA into a double helix1, 2, 3. Several researchers have also designed discrete DNA nanostructures in a variety of nonlinear geometries. By careful sequence design triangles, pyramids, cubes, and more complicated polyhedra have all been made from dsDNA4, 5, 6, 7, 8, 9. Additionally, much work has been done separately to build large and infinite structures from DNA building blocks, with and without nanocrystal decoration10, 11. Other researchers have begun to combine nanoparticle conjugation and novel DNA structures12.
In this work, at the intersection of these fields, we built discrete, pyramidal nanostructures in which dsDNA is used as a scaffold to to control the placement of gold nanocrystals. This opens up many possibilities to enhance the functionality of previously designed DNA nanostructures. Using noble metal nanocrystals in these assemblies can allow the construction of plasmonic nanostructures, a kind of artificial molecule in which the surface plasmons of the nanocrystals hybridize in a manner analogous to atomic orbitals13. This may provide a more sensitive plasmon ruler than in previous experiments because conformation changes in such a multi-particle assembly create a symmetry breaking apparent in the optical spectrum14, not just a shift in resonance wavelength and intensity as with pairs of particles15.
DNA conjugation techniques are not limited to metal nanocrystals however. Semiconductor nanocrystals are also a potential building block and have been assembled by DNA with metal nanoparticles in previous studies16. The use of three-dimensional DNA structures as a scaffold for these nanocrystals may open up further opportunities for tuning the optical properties of the assemblies, at the very least, providing greater control over the relative positions of the nanocrystals.
Finally, the tetrahedral symmetry of DNA pyramids suggests the possibility of creating chiral nanostructures. In our pyramids, a nanocrystal is conjugated to each tip. Creating chiral pyramids can be accomplished by using four different nanocrystals and building a structure with inverted symmetry is then a matter of switching the placement of any two nanocrystals. We did this, specifically, by using four different sizes of gold nanocrystals.
The DNA itself need not be though of as a passive structural element. Previous studies on dsDNA pyramids used sequences that specifically included hairpins17. Additional of complementary ‘fuel’ strands lengthened the side with the hairpin by opening these secondary structures up. This is just one possibility for altering the dimensions and symmetry of an artificial plasmonic molecule.
Experimental Methods
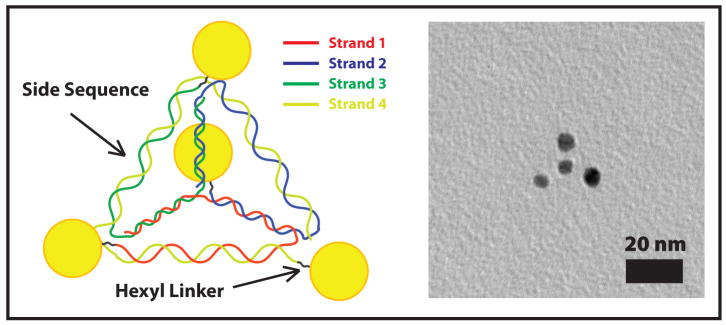
In our most simple pyramid design, referred to as G1, each strand of DNA travels through three pyramid sides and traces out one face of the pyramid. Thus, each third of each strand must be complementary to a third of each of the other strands. In between each of these side sequences, three thymine bases are used to add sufficient flexibility to bend without straining the structure. In each of these side sequences the terminal three base pairs are all guanine-cytosine (GC) pairs. Their higher melting point18 reduces fraying at the corners. The length of each face is 26 bp (base pairs). Given that dsDNA makes a twist every 10.5 bp, this length accommodates 2.5 helical repeats and allows each strand to enter and exit a double helix from the same side, and thus reduces torque and strain at the corners. We designed our sequences with a custom C++ program (Code listing available in Supporting Information) that takes these constraints into consideration as well as rejecting sequences that form stable hairpins, self dimers, or additional unwanted complementary interactions. (Fig 1)
Figure 1.
Schematic and TEM image of DNA-nanocrystal pyramids.
To make larger pyramids, referred to as G2, we used a side length of 37 base pairs, an additional helical repeat. However, the resulting sequences were too long to obtain as synthetic oligonucleotides. Instead, we ordered each sequence in two pieces, broken in the middle of the second side sequence. This results in nicks in the middle of four sides, but we minimized this disturbance by restricting the sequences to GC pairs only for the six bases around the nicks. We used both sets of DNA with 5 nm diameter gold nanocrystals to produce symmetric pyramids. (See Supporting Information for Sequences)
The tetrahedral symmetry of the DNA pyramids allowed us to create chiral nanostructures. This required that the nanocrystals at each tip be different. While this could in principal be achieved with a variety of metal and semiconductor particles, as a proof of concept we used gold nanocrystals in four sizes: 5 nm, 10 nm, 15 nm, and 20 nm. There has been recent work in building chiral structures from magnetic particles which is well suited to long-range structures 19, but using DNA strands gives more direct control over nanoparticle placement and allows a wider range of materials to be used.
The building blocks for synthesizing our materials are DNA-nanocrystal monoconjugates: nanocrystals bearing a single strand of ssDNA. We ordered synthetic oligonucleotides with hexyl-thiol linkers attached to the 5′ phosphate ends (Integrated DNA Technologies Inc., Coralville, IA, 52241). After rehydrating the DNA in 0.5 X tris-borate EDTA buffer (TBE), we prepared monoconjugates using established gel electrophoresis methods for the 5 nm and 10 nm diameter particles20, and recently developed HPLC methods21 for the 15 nm and 20 nm particles. We quantified the monoconjugate concentration with optical absorption at 520 nm22.
We constructed the pyramids by mixing all four monoconjugates in equal measure and adding 1 M sodium chloride in TBE to raise the ionic strength to 100 mM. For pyramids consisting solely of 5 nm diameter particles the concentration of each conjugate was 0.5 μM. For pyramids which included the larger particles the total concentration of each conjugate was 1 nM. The structures were allowed to hybridize overnight at room temperature. We also explored a two-step process in which we mixed the conjugates in pairs, then later combined the pairs with each other after the first hybridization step. This did not affect the yield, as observed by optical density in gel electrophoresis bands.
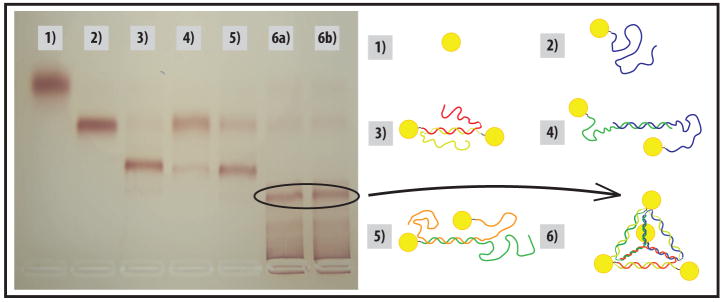
We purified the pyramids built from 5 nm particles using gel electrophoresis. We prepared agarose/TBE gels with an agarose content of 3% by weight. The gels were run for 1 hour at 100 V. We ran the pyramid mixture next to bare gold, monoconjugates, and two-strand products made from combining various pairs of monoconjugates (with different relative positions of the double-stranded section) to verify the contents of each band. The bands were extracted with equal success either by cutting them out and crushing them into TBE, or by cutting the gel ahead of the band, inserting filter paper, and then running the gel an additional 10 minutes to push the band into the paper for extraction. (Fig. 2)
Figure 2.
Gel electrophoresis purification of pyramids, next to incomplete structures as standards. The direction of migration is bottom to top. 1) Bare gold; 2) monoconjugate; 3–5) All possible two-strand products; 6a) Pyramids formed by mixing all strands at once; 6b) Pyramids formed by mixing strands in pairs, then later combining the two-strand products. No difference in yield was observed by this change in protocol.
For transmission electron microscopy (TEM) measurements we pipetted our extracted pyramids onto carbon film and formvar TEM grids and wicked excess liquid away with paper wipes after several minutes. We collected images with a Tecnai 12 electron microscope. For SAXS measurements we pipetted the samples into 2.0 mm quartz capillaries and then flame sealed the ends. In addition to pyramids extracted from gels by both methods, we also prepared samples of monoconjugate (strand 1), and dimers of particles, effectively isolated sides of pyramids (strands 1 and 4 hybridized). (Fig 1) We collected our SAXS data for 5 hours with a Bruker Nanostar. Our illumination source was a copper tube (λ= 1.54 Å). We collected scattered photons up to 2θ= 3 with a twodimensional gas-filled detector (Bruker HiStar) positioned at 105 cm, calibrated with silver behenate. We report our data in terms of scattering vector q, where . Our lowest scattering angles are occluded by a beam stop and we collect data in the range 0.008 Å−1 < q < 0.213 Å−1.
Experimental Results
We took numerous TEM images of the products extracted from our gels to determine the contents of the bands. Lanes 1 and 2 each contain a single product, and thus show the position of bare gold and unhybridized monoconjugate in the other lanes. The additional band in lanes 3 – 5 carries groups of two particles, with the mobility apparently unaffected by the position of the double-stranded portion. The variation in yield for the three different two-particle groupings may result from the fact that in each case a different sequence is hybridized to form the dsDNA portion. The distinct band in lanes 6a and 6b is our pyramid product. Running immediately ahead of it are groups of three particles, unformed pyramids, and following this band back to the well are aggregates from partially-formed structures hybridizing with each other. The contents of the single-particle and two-particle bands were determined by comparison with the standards in lanes 1–5 and the three-particle products were observed by TEM. (See Supporting Information for Additional Images) In 6a the conjugates were mixed all at once while in 6b the conjugates were prehybridized in pairs, then later combined. No difference was observed indicating that cooperation between all four strands is not necessary and that mixing partially formed structures can still cause formation of higher order aggregates. (Fig. 2) From optical density measurements, a typical yield prior to purification is 20%, with the greatest apparent loss coming from higher order structures and aggregates. A gentle thermal annealing is expected to improve this yield, although in this first study we avoided this procedure due to the potential complication of gold-thiol bond cleavage.
The images of the four particle groups show several structures suggestive of tetrahedral geometry in solution. However, many structures are stretched and distorted on the grid due to the high capillary forces upon drying23. It has been shown for pyramids constructed of DNA only though that the structural difference between an open and a closed structure affects the electrophoretic mobility and creates separate bands17. We observe a single distinct (four-particle) band in our gels so we rule out the presence of different populations of four-particle structures, open and closed, in solution.
Electron microscopy allows us to view the nanoparticle assemblies directly, but with a loss of structural information. An indirect technique to study their behavior in solution is SAXS. In previous work we have shown that SAXS may be used to distinguish between dimers of particles tethered by dsDNA and ssDNA24. Here we use it to measure our complete pyramids, and dimers made from a side of the pyramid. Specifically, we performed our SAXS experiments using the G1 pyramids, and dimers made from strands 1 and 4, effectively an isolated side consisting solely of dsDNA. (See structure 3 in Fig. 2) We expect greater flexibility in larger pyramids and in two-particle products which include ssDNA segments and so we deliberately chose the structures in which the observed discrepancy would be most elusive.
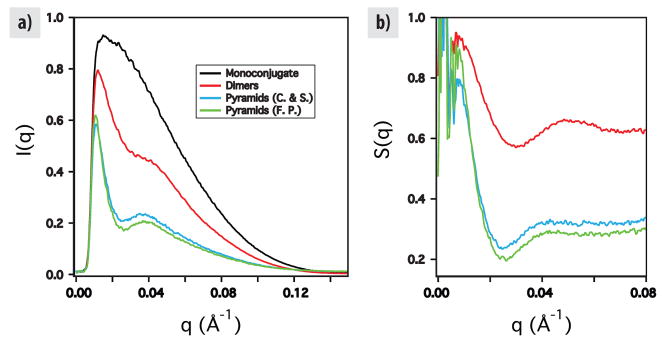
For isolated particles the scattering intensity I(q) is proportional to the scattering form factor P(q). For particles in proximity to each other I(q) is proportional to the product of P(q) and the structure factor S(q), the Fourier transformation of the radial distribution function g(r). We used the program GNOM25 to estimate I(0) and normalize our data such that I(0) = 1 for each scan. (Fig. 4a) Then, we calculated S(q) for our dimers and pyramids by dividing their scans by that of the monoconjugates. (Fig. 4b)
Figure 4.
Small-angle X-ray scattering results. Ordinates are in arbitrary units. Abscissa are scattering vector q, in terms of inverse angstroms. a) Scattered intensity from monoconjugates; dimers made from one side of the pyramid; pyramids extracted from gels by crushing and soaking the appropriate band; pyramids extracted by running the band onto filter paper. b) Structure factors calculated by dividing I(q) for each ordered structure by I(q) for the monoconjugates.
The normalized structure factor for a pair of particles with centers of mass separated by D is given by . We attempted to fit this simple formula to the data from dimers, and to the data from full pyramids extracted both by crush and soak methods, and by running the pyramid band onto filter paper. For the dimers we measured a distance D = 14.3 ± 1.0, while for pyramids purified by these two methods we measured D = 16.1 ± 1.6 nm and D = 16.0 ± 1.6 nm respectively.
It is difficult however to derive much quantitative information from these measurements as they refer simply to the center-to-center distances of the particles. The nanoparticles are tethered with flexible alkyl linkers, and the DNA itself is flexible, even at these length scales. D measured by this simple fit in reciprocal space is not mathematically equivalent to the ensemble average, 〈D〉 in real space. It can be stated qualitatively though that the particles are farther apart in the pyramids than in the dimers. This indicates that the pyramidal structure of the DNA enhances the rigidity of each side, as consistent with previous experiments carried out using atomic force microscopy26.
Building Chiral Nanostructures
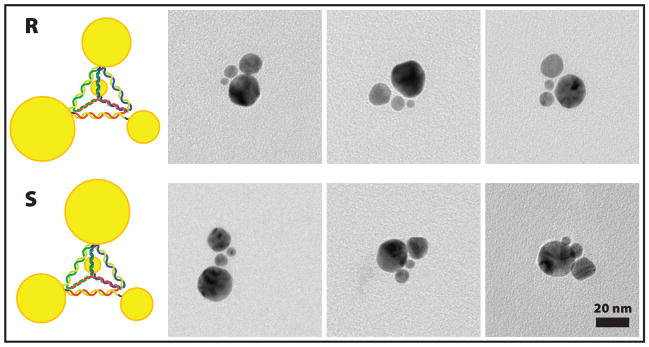
We were able to construct pyramids of DNA bearing a small (5 nm) nanocrystal at each corner. However, at the stage of preparing monoconjugates we can choose to put any type of nanocrystal on each strand. The tetrahedral symmetry of our DNA scaffold then allows us to construct chiral nanoparticles. For a simple proof of concept we used four different sizes of gold colloid. For the ‘R’ enantiomer we used a 5 nm particle on strand 1; 10 nm on strand 2; 15 nm on strand 3; and 20 nm on strand 4. To construct the ‘S’ enantiomer we simply switched the position of 5 nm and 10 nm particles by conjugating them with the opposite strands.
The variety of nanocrystal sizes, especially the inclusion of the largest particles, made purification with electrophoresis impractical. However, noting the high yield of the 5 nm pyramids we simply took TEM images of unpurified chiral pyramid solutions and we were able to observe our products, although some incomplete structures were found as well. (Fig. 5) We can assign the structures to particular enantiomers based on our known choice of monoconjugates. Studies of the optical properties of these chiral nanostructures are underway.
Figure 5.
Examples of chiral pyramids. Top row: ‘R’; Bottom row: ‘S’
Conclusions
We have demonstrated the capability to construct discrete nanostructures decorated with metal nanocrystals. Because each strand has a unique sequence, it is possible to specifically place a different type of nanocrystal at each tip. As a proof of concept, we built chiral nanostructures by using four sizes of gold nanocrystals, but we are not limited just to gold. A wide variety of linker moieties are readily available for synthetic DNA allowing the use of many nanoparticle materials. Additionally, the 3′ ends of our pyramids are available for further functionalization either with additional chemical linkers, or simply with trailing ‘sticky ends’ of ssDNA. This will allow these assemblies to be immobilized on surfaces or incorporated into larger structures, such as self-assembled DNA tiles.
Use of these DNA scaffolds will allow us to create artificial molecules with controllable optical properties. Simply combining metal and semiconductor particles around a DNA pyramid can take previous works a step further by better controlling the placement of the nanocrystals. While multi-particle assemblies have been made using nanocrystals conjugated to several strands of ssDNA, each of these assemblies likely differs in the relative position of the DNA strands around the central nanoparticle. Simple immobilization of the particles at the tips of a pyramid avoids this complication. In our pyramid geometry, using the G1 (shorter) strands with 20 nm gold on each conjugate will bring the surfaces of the particles within 8 nm, assuming the greatest possible extension of the DNA. This should be sufficient for plasmonic coupling. Monoconjugates with 30 nm and 40 nm gold can also be purified using HPLC techniques and can be accommodated by the longer G2 strands.
Through the use of these larger nanoparticles we can envision a threedimensional plasmon ruler. This will build on previous work with plasmon rulers and switchable nanostructures15, 27. We will be able to tune the degree of optical coupling in the structure by adjusting the relative sizes of the DNA scaffold and the nanocrystals. The bending or cleavage of a specific strand will break the symmetry of such a structure and so by incorporating various sequences into the DNA we can make the structure, and thus the optical properties of these artificial molecules sensitive to the action of enzymes, perhaps ultimately in vivo.
Supplementary Material
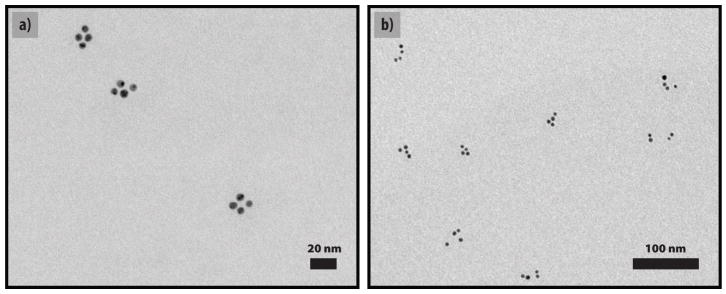
Figure 3.
Typical TEM images of DNA-nanocrystal pyramids. a) G1 pyramids made from four strands of DNA. b) G2 pyramids made from eight strands of DNA.
Acknowledgments
This work was supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contact No. DEAC02- 05CH11231 and by NIH-NCI Grant No. 5U54CA112970
Footnotes
Supporting Information Available
Additional TEM Images; DNA Sequences; SequenceDesign Program. This materials is available free of charge via the Internet at http://pubs.acs.
References
- 1.Mirkin C, Letsinger R, Mucic R, Storhoff J. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 2.Alivisatos AP, Johnsson K, Peng X, Wilson T, Loweth C, Bruchez M, Jr, Schultz P. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell W, Peng X, Alivisatos AP, Schultz P. Angew Chem Int Ed. 1999;38:1808–1812. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1808::AID-ANIE1808>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Seeman N. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Seeman N. J Am Chem Soc. 1994;116:1661–1669. [Google Scholar]
- 6.Yang X, Wenzler L, Qi J, LI X, Seeman N. J Am Chem Soc. 1998;120:9779–9786. [Google Scholar]
- 7.Seeman N. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 8.Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 9.Erben C, Goodman R, Tuberfield A. J Am Chem Soc. 2007;129:6992–6993. doi: 10.1021/ja071493b. [DOI] [PubMed] [Google Scholar]
- 10.Rothemund P, Papadakis N, Winfree E. PLoS Biol. 2004;2:2041–2053. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng J, Constantinou P, Micheel C, Alivisatos AP, Kiehl R, Seeman N. Nano Lett. 2006;6:1502–1504. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aldaye F, Sleiman H. Angew Chem Int Ed. 2006;45:2204–2209. doi: 10.1002/anie.200502481. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Brandl D, Nordlander P, Halas N. Acc Chem Res. 2007;40:53–62. doi: 10.1021/ar0401045. [DOI] [PubMed] [Google Scholar]
- 14.Brandl D, Mirin N, Nordlander P. J Phys Chem B. 2006;110:12302–12310. doi: 10.1021/jp0613485. [DOI] [PubMed] [Google Scholar]
- 15.Reinhard B, Sheikholeslami S, Mastroianni A, Alivisatos AP, Liphardt J. Proc Natl Acad Sci U S A. 2007;104:26672672. doi: 10.1073/pnas.0607826104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu A, Micheel C, Cha J, Chang H, Yang H, Alivisatos AP. J Am Chem Soc. 2004;126:10832–10833. doi: 10.1021/ja046747x. [DOI] [PubMed] [Google Scholar]
- 17.Goodman R, Heilemann M, Doose S, Erben C, Kapanidis A, Turberfield R. Nat Nanotechnol. 2008;3:93–96. doi: 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- 18.SantaLucia J., Jr Proc Natl Acad Sci U S A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zerrouki D, Baudry J, Pine D, Chaikin P, Bibette J. Nature. 2008;455:380–382. doi: 10.1038/nature07237. [DOI] [PubMed] [Google Scholar]
- 20.Zanchet D, Micheel C, Parak W, Gerion D, Alivisatos AP. Nano Lett. 2001;1:32–35. [Google Scholar]
- 21.Claridge S, Liang HW, Basu SR, Frechet JMJ, Alivisatos AP. Nano Lett. 2008;8:1202–1206. doi: 10.1021/nl0802032. [DOI] [PubMed] [Google Scholar]
- 22.Claridge S, Goh S, Frechet JMJ, Williams S, Micheel C, Alivisatos AP. Chem Mater. 2005;17:1628–1635. [Google Scholar]
- 23.Liddle J, Cui Y, Alivisatos AP. J Vac Sci Technol B. 2004;22:3409–3414. [Google Scholar]
- 24.Claridge S, Mastroianni A, Au YB, Liang HW, Micheel C, Frechet JMJ, Alivisatos AP. J Am Chem Soc. 2008;130:95989605. doi: 10.1021/ja8026746. [DOI] [PubMed] [Google Scholar]
- 25.Svergun D. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 26.Goodman R, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 27.Sebba DS, Mock JJ, Smith DR, La Bean TH, Lazarides AA. Nano Lett. 2008;8:1803–1808. doi: 10.1021/nl080029h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.