Abstract
Several cytosolic sulfotransferase enzyme isoforms are functional in placenta but there is limited information available on the utility of cultured trophoblast cells for studying sulfation. The trophoblast cell layer constitutes the rate-determining barrier for trans-placental transfer. The objective of this work was to examine the mRNA expression and enzyme activities of four sulfotransferase isoforms reported to be functional in human placenta (SULT1A1, SULT1A3, SULT1E1, and SULT2A1) in primary cytotrophoblast cells and the trophoblast-like BeWo cell line. Reverse transcription polymerase chain reaction (RT-PCR) was performed to determine mRNA expression. Enzyme activities were assessed using the following substrates: 4-nitrophenol for SULT1A1, dopamine for SULT1A3, 17β-estradiol for SULT1E1, and dehydroepiandrosterone for SULT2A1. For 4-nitrophenol and dopamine sulfation, apparent Km values, response to inhibitors (2,6-dichloro-4-nitrophenol and sodium chloride), and thermal stability profiles indicated that 4-nitrophenol and dopamine sulfation in BeWo cells were being mediated by SULT1A1 and SULT1A3, respectively. SULT1A1 and SULT1A3 were also functional in the cytotrophoblast cells. Both at the protein and at the mRNA levels, SULT1A1 was more abundant in BeWo cells in comparison to the primary cytotrophoblast cells. SULT1E1 and SULT2A1 mRNA were not detected in the cytotrophoblasts. SULT1E1 mRNA was weakly expressed in BeWo but there was negligible functional activity. Although SULT2A1 mRNA was abundantly expressed in BeWo, Western blot and enzyme activities revealed that the protein is not expressed in BeWo cells. The results suggest that the BeWo cells and the cytotrophoblast cells can be used to examine the roles of SULT1A1 and SULT1A3 in placental metabolism.
1. Introduction
Sulfotransferase (SULT) enzymes catalyze the transfer of a sulfuryl group (SO3-1) from a physiological donor substrate 3′-phosphoadenosine-5′-phosphosulfate (PAPS) to acceptor substrates containing hydroxyl, amine, N-hydroxyl, or sulfhydryl groups in a process known as sulfonation. Sulfoconjugates are generally inactive but in certain cases sulfated metabolites can be pharmacologically active or potentially carcinogenic [1, 2].
Sulfotransferase enzymes can be either cytosolic or membrane-associated. In human tissues, 13 cytosolic SULT isoforms have been identified so far [3] out of which four isoforms have been very well characterized. These are the phenolic sulfotransferase isoforms SULT1A1, SULT1A3, and SULT1E1, and the hydroxysteroid sulfotransferase SULT2A1. SULT1A1 sulfates small, planar phenolic compounds with a high affinity while SULT1A3 sulfates monoamines with a high affinity. Endogenous substrates of SULT1A1 include 17β-estradiol and thyroid hormones (T3 and T4), while endogenous substrates of SULT1A3 include catecholamines (dopamine, epinephrine, and norepinephrine)[3]. Both SULT1A1 and SULT1A3 also sulfate several medicinal drugs and environmental chemicals which suggests a major role of the enzymes in xenobiotic elimination[3]. SULT1E1 (also referred to as estrogen sulfotransferase), exhibits the highest affinity for estrogenic substances among all the sulfotransferase enzymes [4]. This enzyme sulfates 17β-estradiol at physiological concentrations. Its physiological functions are most likely concerned with maintaining estrogen balance, and protection of tissues from the untoward effects of estrogen [5, 6]. SULT2A1 sulfates steroid hormones (dehydroepiandrosterone), cholesterol, bile acids, and is predominantly functional in the adrenal gland [3]. Sulfated dehydroepiandrosterone serves as the source of estrogens and androgens in peripheral tissues. In the liver, SULT2A1 is responsible for majority of the bile acid sulfation [4, 7]. All four isoforms are functional in the liver and the intestine [8-10] and tissues such as brain and lung contain several of these isoforms [11]. The tissue localization supports the hypothesis that these sulfotransferase isoforms play a major role in modulating xenobiotic exposure.
Sulfation of medicinal drugs has been detected, and subsequently several sulfotransferase isoforms have been identified in term and mid-gestation human placenta [12, 13]. Examination of placental tissue sections (of both maternal and fetal origin) revealed that the phenolic sulfotransferase isoforms SULT1A1 and SULT1A3 were functional in all sections. Highest activity was reported in sections containing syncytiotrophoblast cells [13]. The trophoblast cell layer consisting of multinucleated syncytiotrophoblast cells and mononucleated cytotrophoblast cells, is the rate-limiting barrier in the placental transfer of substances[14]. Dehydroepiandrosterone sulfation was attributed to SULT2A1 activity [13]. Although this study, that examined placental sections, reported low SULT1E1 protein expression in syncytiotrophoblast fractions, in another study SULT1E1 was found to be localized to the syncytiotrophoblasts [15]. In addition, other sulfotransferase isoforms are also functional in syncytiotrophoblasts [16].
Several metabolic enzymes and efflux transporters are functional in trophoblast cells, which individually as well as in concert, have the ability to modify the extent of fetal exposure to substances in the maternal circulation [14, 17, 18]. The process of sulfation introduces a sulfonate moiety into a molecule. In other tissues, the negatively charged sulfate metabolites are eliminated by efflux transporters [19, 20]. It is likely that a similar concerted pathway for sulfate metabolite elimination exists in placental tissue as well but this has not yet been examined. The purpose of this study was to determine whether selected sulfotransferase enzymes are functional in trophoblast cells, in an effort to comprehend if trophoblast cells can ultimately be utilized as a suitable model to examine how the placenta inactivates and eliminates substances in the maternal circulation by sulfation.
Although, the above-mentioned sulfotransferase enzymes have been extensively characterized in other human tissue, there is only limited information available on whether cultured trophoblast cells may serve as suitable in vitro models for studying placental sulfation [21]. The objective of this work was to determine the expression and activities of SULT1A1, SULT1A3, SULT1E1, and SULT2A1 in trophoblast cells. We used primary cytotrophoblast cells and the BeWo cell line as trophoblast models. The BeWo cell line is derived from a methotrexate-resistant malignant choriocarcinoma, and in culture consists predominantly of cytotrophoblast-like cells with a smaller proportion of syncytiotrophoblast-like cells[22]. BeWo cells have also been shown to be adequately representative of placental phase I metabolism [17]. This study significantly extends the work of Tamura et al. who demonstrated sulfation activity in a wild type BeWo cell culture system [21]. Also, we utilized a subclone of the BeWo cells (b30) that possesses some functional differences with respect to wild type BeWo cells[23].
2. Materials and methods
2.1. Cell culture materials
The BeWo cell line (clone b30) was obtained from Dr. Alan Schwartz (Washington University, St. Louis, MO). Penicillin-streptomycin, L-glutamine, and minimal essential medium nonessential amino acid (MEM-NEAA) solutions were purchased from Invitrogen (Carlsbad, CA). Heat inactivated fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Norcross, GA). All other materials and cell culture reagents were from Sigma (St. Louis, MO).
2.2. Cell culture
The BeWo cell line was cultured as described previously [24]. Passages 29 through 45 were used in this study. Cytotrophoblast cells were obtained from Dr. Joan S. Hunt (University of Kansas Medical Center, Kansas City, KS). They were isolated from placenta following an uncomplicated caesarean delivery as per the procedure of Petroff et al. [25] and stored in liquid nitrogen until further use.
2.3. Homogenate preparation for sulfotransferase assays
BeWo cell homogenate was prepared as described previously with some modifications [26]. Cells at confluence were washed and scraped in phosphate buffered saline (PBS) containing 0.5 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM phenylmethylsulfonylfluoride (PMSF). Cells were then centrifuged and PBS was aspirated off. Cells were then lysed on ice in 5 mM potassium phosphate buffer (pH 7.4) containing 1.25 mM EDTA, 2.5 mM dithiothreitol (DTT), and Complete Mini Protease Inhibitor cocktail tablets (Roche, Indianapolis, IN) for 20 strokes using a Dounce homogenizer. The lysates were centrifuged at 13,000 × g for 20 minutes at 4°C. The supernatant was further centrifuged at 100,000 × g for 1 hour at 4°C. The resulting high speed supernatant was used for determination of total protein content. Following this, the high-speed supernatant was diluted in a BSA solution (5 mM potassium phosphate buffer, pH 7.4, containing 5 mg/mL BSA) at a 1:1 ratio and stored at -80°C until further use. Cytotrophoblast cell homogenate was prepared in a similar manner.
2.4. Determination of sulfotransferase enzyme activities
Sulfotransferase enzyme assays were performed with the appropriate diagnostic substrates: 4-nitrophenol for SULT1A1, dopamine for SULT1A3, dehydroepiandrosterone (DHEA) for SULT2A1, and 17β-estradiol for SULT1E1. Preliminary experiments were optimized to ensure that product formation was linear with respect to time and protein concentration.
Enzyme assays for SULT1A1 and SULT1A3 were a modification of the method of Foldes and Meeks [27]. The complete reaction mixture consisted of 0.4 μM [35S]-3′-phosphoadenosine-5′-phosphosulfate (PAPS, Perkin Elmer, Waltham, MA), varying concentrations of the acceptor substrates, and BeWo/primary cytotrophoblast protein (20 μg) in 20 mM potassium phosphate buffer (pH 7.4). The final reaction volume was 150 μL. Controls contained vehicle in place of the acceptor substrate. Reactions were started by the addition of PAPS to the other components and incubated at 37°C for 20 minutes. The reaction was terminated by sequential additions of 200 μL of 1:1 0.1 M barium hydroxide:0.1 M barium acetate and 100 μL of 0.1 M zinc sulfate, and centrifugation of the reaction mixture at 10,000 r.p.m. for 3 minutes. Following this 100 μL each of 0.1M barium hydroxide and 0.1 M zinc sulfate were added, and the reaction mixture was centrifuged at 10,000 r.p.m. for 10 minutes. The supernatant was analyzed for the sulfated product by liquid scintillation counting. Each assay was performed at least in duplicate.
Estradiol (E2) sulfation was measured using a method based on that of Harris et al. with a few modifications [28]. The reaction mixture consisted of 10 μM PAPS, BeWo cytosolic protein (50 μg) in a final volume of 200 μL in 100 mM Tris buffer containing 10 mM magnesium acetate and 0.1 mM EDTA (pH 7.9). Reactions were started by the addition of [2,4,6,7-3H(N)]-estradiol (0.5 Ci/mmol, Perkin Elmer, Waltham, MA). The rest of the procedure was performed as outlined previously [28].
Sulfation of 3 μM or 10 μM [1,2,6,7-3H(N)]-dehydroepiandrosterone (diluted to 0.4 Ci/mmol, Perkin Elmer, Waltham, MA) was measured according to the protocol of Chen et al. [8]. The reaction mixtures contained BeWo cytosolic protein in the range 50-600 μg.
2.5. Reverse transcription polymerase chain reaction (RT-PCR)
BeWo cells at confluence were washed, scraped in PBS, and then centrifuged to remove the PBS. Cytotrophoblast cells were retrieved from liquid nitrogen, thawed, and centrifuged to remove the storage medium. Total RNA was isolated from either BeWo cells or cytotrophoblast cells, using a commercially available phenol-guanidine isothiocyanate solution (TRIZOL reagent, Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. An Oligotex mRNA Mini Kit (Qiagen, Valencia, CA) was used to purify mRNA from total RNA. To concentrate mRNA prior to the experiment, volume containing 200 ng mRNA was mixed with 1/10th the volume of 3M diethylpyrocarbonate treated sodium acetate (pH 5.2), 2 μL glycogen (Roche Diagnostics, Indianapolis, IN), and ethanol (at a volume 2.5 fold more than the volume of mRNA). The mixture was stored overnight at -20°C to precipitate the mRNA. The mixture was centrifuged at 14,000 rpm for 15 minutes at 4°C. The mRNA pellet was washed in 75% ethanol and most of the supernatant was aspirated off. The remainder was allowed to evaporate in room air and the mRNA pellet was reconstituted in RNAse free water. Reverse transcription and subsequent cDNA amplification were performed with an Access RT-PCR Introductory System (Promega, Madison, WI) in an Eppendorf Mastercycler. PCR cycle parameters were according to manufacturer's protocol. The primer sequences for amplifying SULT1A1, SULT1A3, SULT2A1 and SULT1E1 (Table I) have previously been published [21, 29, 30]. All primers were synthesized by the Biotechnology Support Facility at the University of Kansas Medical Center (Kansas City, KS). Primers for β-actin were obtained from Promega (Madison, WI).
Table I. Primer sequences used for reverse transcribing sulfotransferase mRNA and expected base pair sizes of the reaction products.
| Target sequence | Genbank accession ID | Primers (Forward, Reverse) | Expected size (Expected size after digestion) |
|---|---|---|---|
| SULT1A1 (39-1025) |
L10819 | 5′-ATGGAGCTGATCCAGGACAC-3′ 5′-TGACCTACCGTCCCAGGCCC-3′ |
987 |
| SULT1A3 (139-1125) |
L19956 | 5′-ATGGAGCTGATCCAGGACAC-3′ 5′-TGAGCCACTGTGCCTGACTC-3′ |
987 |
| SULT1E1 (389-852) |
Y11195 | 5′-ACCTGAACTTCTTCCTGCC-3′ 5′-TCCAGTCTCCTGTAATTCCC-3′ |
464 |
| SULT2A1 (21-875) |
U08024 | 5′-ATGTCGGACGATTTCTTATG-3′ 5′-AATCCCATGGGAACAGCTC-3′ |
856 |
Electrophoresis of all RT-PCR products was performed at 80 V on a 2% agarose gel. The gel was post-stained with 0.5 μg/mL ethidium bromide for 45 minutes, washed four times in distilled water at 10 minute intervals, and viewed with a UV transilluminator.
2.6. Data analyses
Sulfation of both 4-nitrophenol and dopamine exhibited substrate inhibition at high substrate concentrations. Kinetic constants were determined from a modification of the Michaelis-Menten equation that incorporates substrate inhibition, which has been outlined as follows.
where, v, Vmax, Km, and [S] represent the sulfation rate, maximal sulfation rate, the apparent Michaelis-Menten constant, and the utilized substrate concentration respectively. For many enzymes, a second substrate molecule binds to the active site of the enzyme at high substrate concentrations to form an inactive enzyme-substrate complex. Ki represents the dissociation constant of the substrate from this inhibitory complex. Regression analysis was performed using the GraphPad Prism software, version 5 (La Jolla, CA).
The IC50 values (inhibitor concentration at which sulfation rate is 50% of the untreated controls) for 2,6,dichloro-4-nitrophenol were calculated from the Four-Parameter Logistic Function as follows:
where, y is the sulfation rate, x is the log concentration of the inhibitor and C is the IC50. Regression analysis was performed using the Sigma Plot software (SPSS, Chicago, IL).
The IC50 values in the presence of sodium chloride and the temperatures at which enzyme activities were inhibited by 50% were calculated from the Four-Parameter Logistic Function (Linear) as follows:
where, y is the sulfation rate, x is the concentration of the inhibitor and C is the IC50. Regression analysis was performed using the Sigma Plot software (SPSS, Chicago, IL).
3. Results
3.1. Sulfotransferase mRNA expression in trophoblast cells
SULT1A1 and SULT1A3 mRNA were expressed in both BeWo cells and cytotrophoblast cells (Figures 1 and 2). Although semi-quantitative, the results illustrate that SULT1A1 mRNA is expressed more abundantly in BeWo in comparison to the cytotrophoblast cells. SULT1A3 was equivalently expressed in the two cell types. SULT2A1 mRNA was abundantly expressed in BeWo but not in the primary cytotrophoblast cells (Figure 2). In contrast, SULT1E1 mRNA was faintly expressed in BeWo cells and was undetectable in the cytotrophoblast cells (data not shown).
Figure 1. Reverse transcription polymerase chain reaction (RT-PCR) analysis of SULT1A1 in BeWo cells and in primary cytotrophoblast cells.
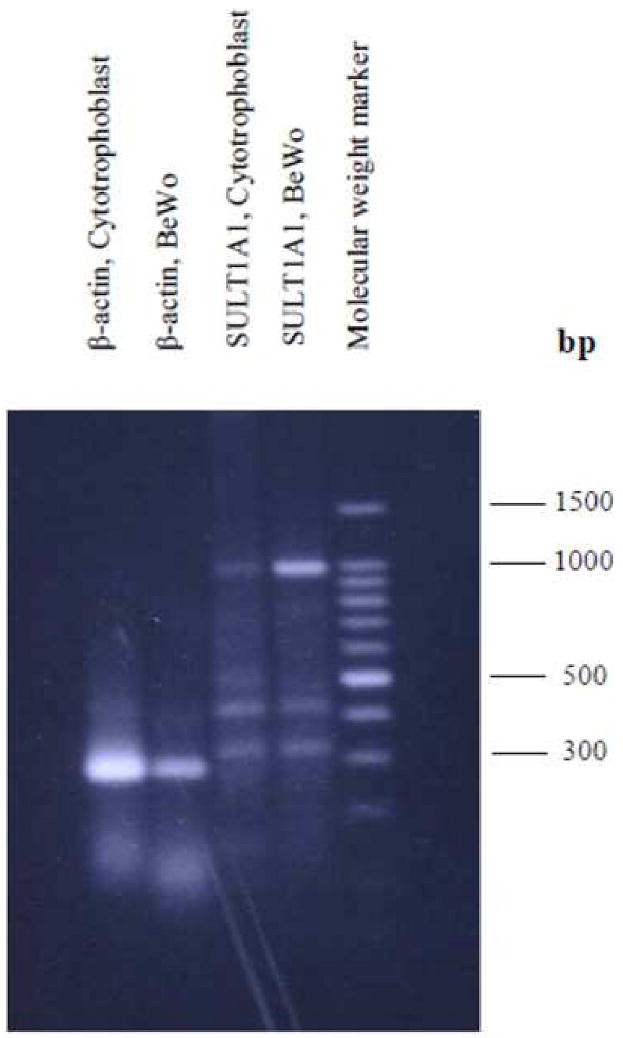
200 ng mRNA was used for reverse transcription and polymerase chain reaction amplification of SULT1A1. Expected size: SULT1A1 (987 bp).
Figure 2. Reverse transcription polymerase chain reaction (RT-PCR) analyses of SULT1A3 and SULT2A1 in BeWo cells and in primary cytotrophoblast cells.
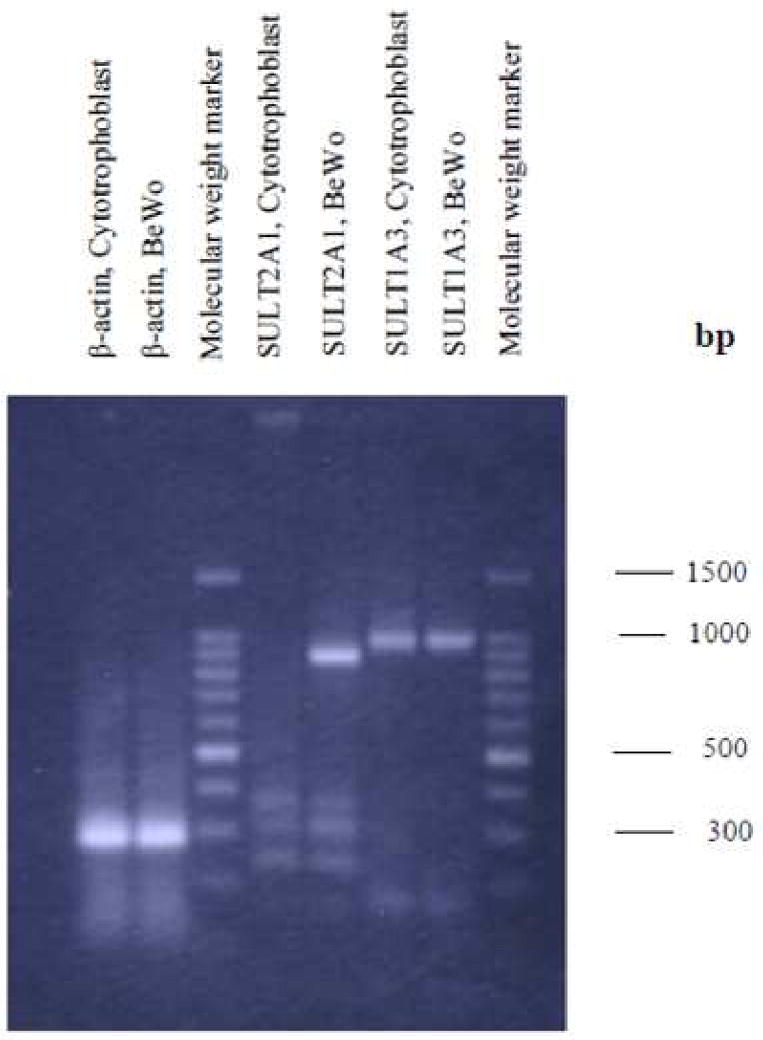
200 ng mRNA was used for reverse transcription and polymerase chain reaction amplification of SULT1A3 and SULT2A1. Expected sizes: SULT1A3 (987 bp), SULT2A1 (872 bp).
3.2. Sulfation activities in BeWo cells
The enzyme activities of SULT1A1 and SULT1A3 were tested with 4-nitrophenol and dopamine respectively. Sulfation of both 4-nitrophenol (0.1-160 μM) and dopamine (0.1-80 μM) exhibited substrate inhibition at higher concentrations (Figures 3 and 4), which is typical of SULT1A1 and SULT1A3 mediated sulfation respectively [31].
Figure 3. Sulfation of 4-nitrophenol (pmol/min/mg protein) in BeWo cells as a function of 4-nitrophenol concentration.
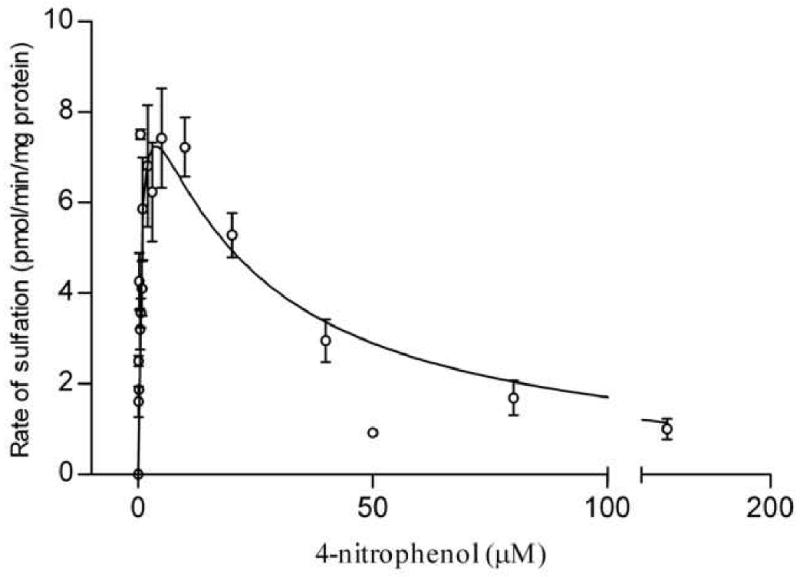
The figure represents a fit to the substrate inhibition equation (section 2.6). Each assay was repeated at least twice and each point represents the mean ± S.E.M. of three or more determinations. Kinetic parameters were as follows: Km = 0.63 ± 0.21 μM, Ki = 21.2 ± 8.1 μM.
Figure 4. Sulfation of dopamine (pmol/min/mg protein) in BeWo cells as a function of dopamine concentration.
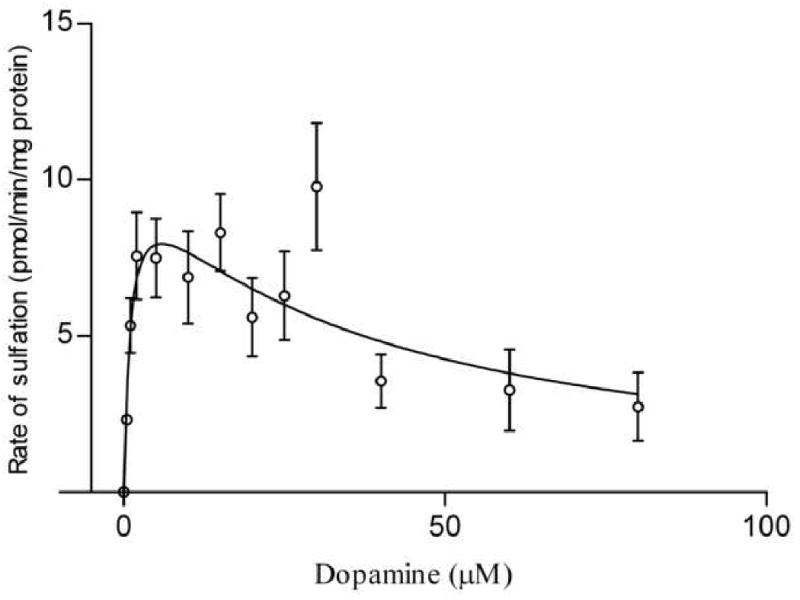
The figure represents a fit to the substrate inhibition equation (section 2.6). Each assay was repeated at least twice and each point represents the mean ± S.E.M. of three or more determinations. Kinetic parameters were as follows: Km = 1.1 ± 0.66 μM, Ki = 32.9 ± 17.5 μM.
For sulfation of 4-nitrophenol, the apparent Km and Ki values were 0.63 ± 0.21 μM and 21.2 ± 8.1 μM, respectively. The apparent Km value in BeWo cells compared well with SULT1A1-mediated 4-nitrophenol sulfation either in other tissues or with the recombinant protein which report the Km value to be in the range 0.5-1 μM [26, 32-34]. In BeWo, the Km obtained for dopamine sulfation was 1.1 ± 0.66 μM, while the Ki was 32.9 ± 17.5 μM. Though multiple articles cite that SULT1A3-mediated sulfation of dopamine exhibits higher apparent Km values (3-10 μM) [2, 26], much lower Km values have also been observed (0.65 μM) [35]. Maximal sulfation rates (Vmax) were similar for 4-nitrophenol (9.8 ± 1.3 pmol/min/mg) and dopamine (10.8 ± 2.4 pmol/min/mg).
It should be mentioned here that at fixed concentrations of 4-nitrophenol or dopamine (2 μM), biphasic Eadie-Hofstee plots were observed when PAPS concentration was varied. High and low affinity Km of PAPS in the presence of 4-nitrophenol were (0.034 ± 0.036 μM) and (18.3 ± 6.5 μM) respectively. In the presence of dopamine, the Km values for PAPS were (0.029 ± 0.016 μM) and (17.7 ± 4.1 μM) respectively. Control reactions lacked PAPS.
For measuring SULT1A1 and SULT1A3 activities, control reactions contained vehicle in place of 4-nitrophenol or dopamine. SULT1A1 and SULT1A3 activities were not observed at 20 μM PAPS, which is commonly utilized in sulfotransferase assays. One of the products of the sulfation reaction, PAP (3′-phosphoadenosine-5′-phosphate), is a potent inhibitor of SULT1A1 and SULT1A3 (Ki= 0.1 ± 0.01 μM and 0.1 ± 0.02 μM respectively)[36]. Accumulation of this reaction product could be a likely cause of the enzyme inhibition observed at higher PAPS concentrations. This necessitated performing the enzyme assays at a lower PAPS concentration (0.4 μM), which is also frequently cited in the literature [27, 37, 38].
SULT2A1 is reported to exhibit maximal activity towards DHEA in the range 3-10 μM [4]. In BeWo cells, there was negligible sulfation of DHEA (3 μM and 10 μM). Under the same assay conditions, purified SULT2A1 readily sulfated DHEA. This indicated that SULT2A1 is not functional in BeWo cells. Western blots confirmed that SULT2A1 protein is not expressed in BeWo cells (data not shown). Sulfation of 17β-estradiol (10-30 nM) was negligible (0.02-0.05 pmol/min/mg). This combined with the low mRNA levels of SULT1E1 indicated that SULT1E1 is not functional in BeWo cells (data not shown).
3.3. Effect of inhibitors 2,6-dichloro-4-nitrophenol (DCNP) and NaCl, and temperature on 4-nitrophenol and dopamine sulfation in BeWo cells
As there can be overlapping substrate specificities between sulfotransferase isoforms, 4-nitrophenol and dopamine sulfation activities were further characterized in the presence of inhibitors 2,6-dichloro-4-nitrophenol (DCNP) and sodium chloride. Typically SULT1A1 is more sensitive to both DCNP and sodium chloride, than SULT1A3. In BeWo cells, 4-nitrophenol sulfation was more sensitive to inhibition by DCNP than dopamine sulfation, whereas both exhibited similar sensitivities to sodium chloride inhibition (Figures 5 and 6). The IC50 value for 4-nitrophenol sulfation obtained with DCNP (0.052 μM) is similar to previously cited values (0.4–1.5 μM) [26, 34, 39, 40]. Similarly, the IC50 obtained with sodium chloride (368 ± 1 mM) was comparable to what has been reported (80-100 mM) [26, 34]. The IC50 values with DCNP (830 μM) and sodium chloride (303 ± 1.4 mM) for dopamine sulfation agreed very well with references of SULT1A3-mediated sulfation of dopamine, which put the values in the range 0.4-0.6 mM and 180-340 mM respectively [2, 26]. Contrary to previous reports [26, 37], in BeWo cells dopamine sulfation was more sensitive to sodium chloride than 4-nitrophenol sulfation.
Figure 5. Effect of 2,6,-dichloro-4-nitrophenol on sulfotransferase activities in BeWo cells.
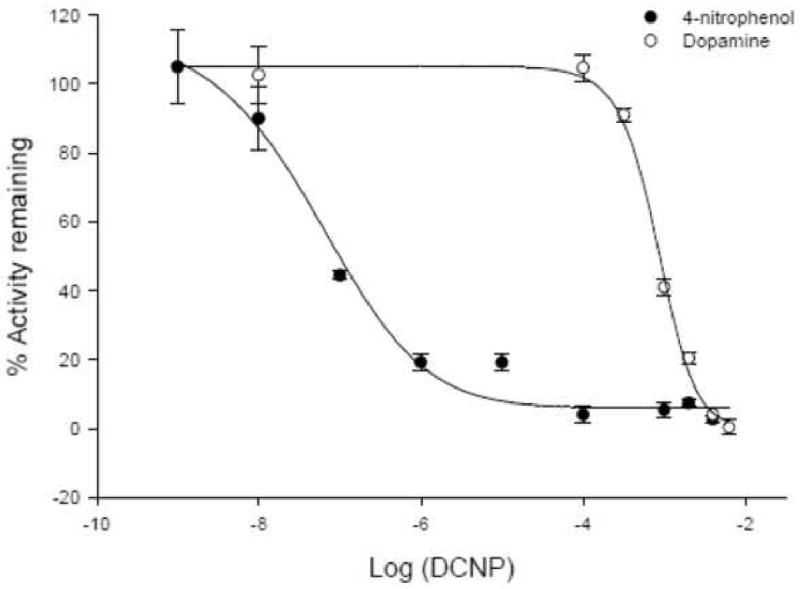
The effect of 2,6-dichloro-4-nitrophenol (DCNP) on 4-nitrophenol (solid circles) and dopamine (open circles) sulfation activities was determined in BeWo cells. Enzyme activity in the presence of the inhibitor was expressed as a percentage of the activity in the absence of the inhibitor. The IC50 values for 4-nitrophenol and dopamine were 0.05 μM and 830 μM respectively. Each assay was repeated twice and each point represents mean ± S.E.M of three or more determinations.
Figure 6. Effects of sodium chloride on sulfotransferase activities.
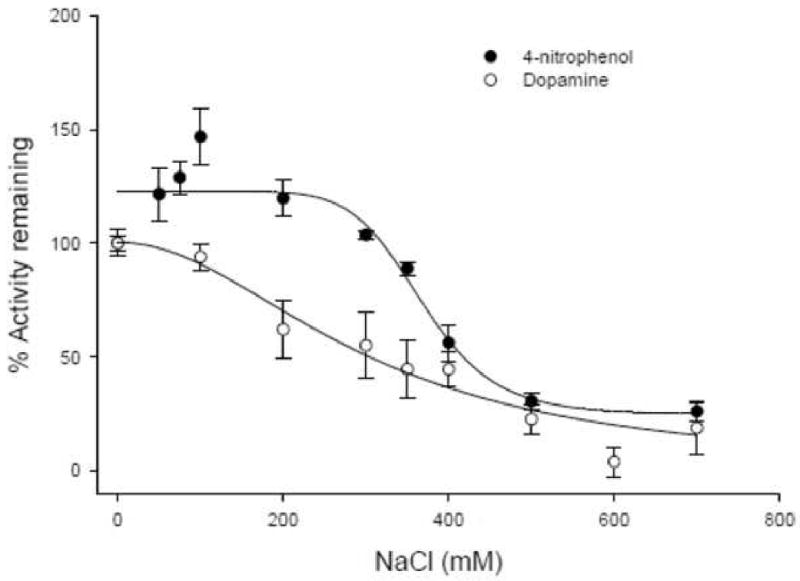
The effect of NaCl on 4-nitrophenol (solid circles) and dopamine (open circles) sulfation activities was determined in BeWo cells. Enzyme activity in the presence of the inhibitor was expressed as a percentage of the activity in the absence of the inhibitor. The IC50 values for 4-nitrophenol and dopamine were 368 ± 1 mM and 303 ± 1.4 mM respectively. Each assay was repeated twice and each point represents mean ± S.E.M of three or more determinations.
SULT1A1 and SULT1A3 exhibit differential thermal stability, and this has led to their nomenclature as thermostable and thermolabile phenolic sulfotransferase isoforms respectively. SULT1A1 has been reported to be 50% inactivated between 43-44°C while SULT1A3 is 50% inactivated between 38.5-40.5°C [2, 26, 40]. Though there are discrepancies between studies with regards to the absolute values reported, SULT1A1 is relatively more thermostable than SULT1A3 [39]. In BeWo cells, 4-nitrophenol and dopamine sulfation were 50% inactivated at 41.5 ± 1.0 °C and at 39.1 ± 1.0 °C, respectively (Figure 7). These observations matched well with previously reported values and indicated that in BeWo the enzymes retain their distinctive thermal stability properties.
Figure 7. Thermal stability of 4-nitrophenol (solid circles and broken line) and dopamine (open circles and solid line) sulfation activities in BeWo cells.

The net sulfotransferase activity was expressed as a percentage of the basal (unheated) activities. The 50% inactivation temperatures (T50) were 41.5 ± 1.0 °C and at 39.1 ± 1.0 °C for 4-nitrophenol and dopamine respectively. Each point represents mean ± S.E.M of three or more determinations.
Overall, IC50 values obtained in the presence of 2,6,dichloro-4-nitrophenol and sodium chloride, as well as the temperatures at which enzyme activities were 50% inhibited, agreed well with previous reports to confirm that 4-nitrophenol and dopamine sulfation in BeWo cells were being mediated by SULT1A1 and SULT1A3 respectively [2, 26, 34, 39, 40]. Also, in BeWo the biochemical properties of SULT1A1 and SULT1A3 are similar to their properties observed in other tissues such as platelets, liver, thyroid glands, and bone [2, 26, 37, 40].
3.4. SULT1A1 and SULT1A3 activities in primary cytotrophoblast cells
Among the four sulfotransferase isoforms examined in this study, only the phenolic sulfotransferase isoforms SULT1A1 and SULT1A3 were functional in BeWo cells. Thus the enzyme activities of only these two isoforms were determined in the primary cytotrophoblast cells. Again, 4-nitrophenol and dopamine were used to test for SULT1A1 and SULT1A3 respectively. Sulfation was examined in the presence of 2,6,dichloro-4-nitrophenol (DCNP) concentrations that previously completely inhibited enzyme activities in BeWo cells (Figure 8). At 1 mM and 4 mM DCNP, sulfation of 4-nitrophenol and dopamine were also completely inhibited in cytotrophoblast cells. This indicated that 4-nitrophenol sulfation and dopamine sulfation were being mediated by SULT1A1 and SULT1A3 respectively in the primary cytotrophoblast cells. SULT1A3 activity was similar (6.9±0.6 pmol/min/mg in BeWo cells vs. 3.8 ± 0.8 pmol/min/mg in primary cytotrophoblast cells) whereas SULT1A1 activity was much higher in BeWo cells (21.6 ±2 pmol/min/mg in BeWo cells vs. 2.8 ± 2.4 pmol/min/mg in the primary cytotrophoblast cells).
Figure 8. Sulfation activities of 4-nitrophenol (4 μM) and dopamine (1 μM) in primary cytotrophoblast cells.
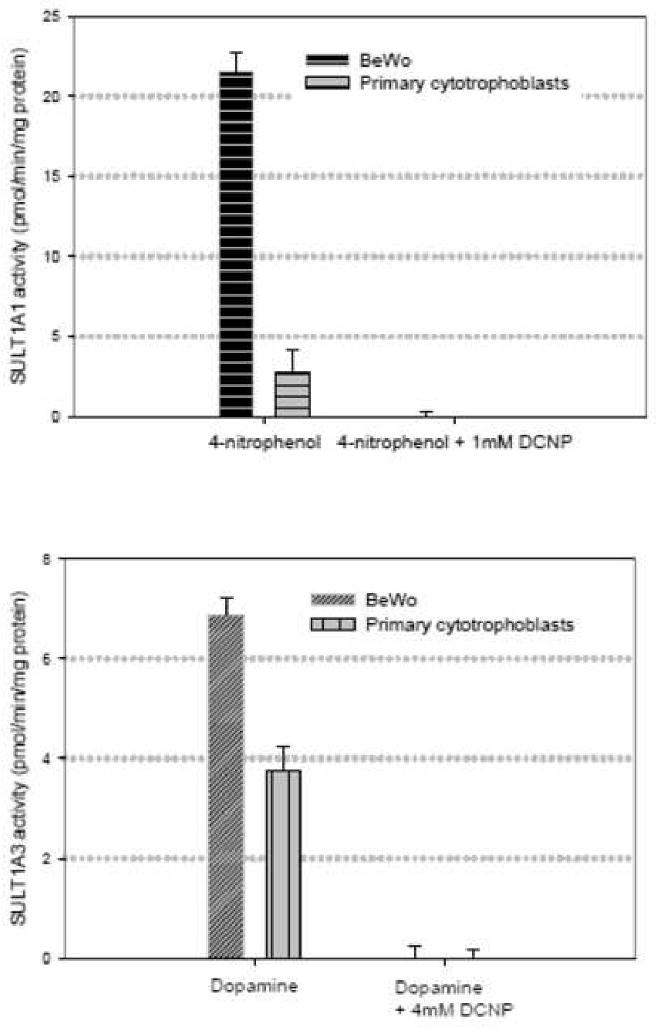
Sulfotransferase activities were measured in the presence and in the absence of 1 mM and 4 mM 2,6,-dichloro-4-nitrophenol (DCNP). At these concentrations, DCNP previously completely abolished 4-nitrophenol and dopamine sulfation in BeWo cells. DCNP also completely inhibited 4-nitrophenol and dopamine sulfation in the primary trophoblast cells to indicate that 4-nitrophenol and dopamine sulfation are being mediated by SULT1A1 and SULT1A3 respectively.
4. Discussion
Cytosolic sulfotransferase enzymes are functional in whole placental tissue but not much work has focused on their presence in trophoblast cells which constitute the rate-limiting barrier to trans-placental transfer [13]. Tamura et al. determined SULT1A1 and SULT1A3 enzyme activities in BeWo cells using single concentrations of 4-nitrophenol and dopamine [21]. However, there are often overlapping substrate specificities between the sulfotransferase enzymes. For instance, SULT1A1, SULT1A2, and SULT1B1, all sulfate 4-nitrophenol at low micromolar concentrations, and thus single concentration determinations can lead to erroneous identification of sulfotransferase isoforms [32, 41]. Our work examines 4-nitrophenol and dopamine sulfation activities in the presence of inhibitors of these enzymes and extends the work of Tamura et al [21]. For 4-nitrophenol and dopamine sulfation, apparent Km values, response to inhibitors and thermal stability profiles agreed well with previously reported values, to lead to the conclusion that 4-nitrophenol and dopamine sulfation in BeWo cells are being mediated by SULT1A1 and SULT1A3, respectively. SULT1A1 and SULT1A3 were also functional in the primary trophoblast cells. Both at the protein and mRNA levels (a semi-quantitative determination for the latter), SULT1A1 was more abundant in BeWo in comparison to the primary cytotrophoblast cells, whereas SULT1A3 was equivalently expressed in the two cell types. SULT1E1 and SULT2A1 mRNA were not detectable in the primary trophoblast cells and the enzymes were not functional in BeWo cells. There appears to be some contradiction about the expression of SULT2A1 in placenta. While one study reports negligible SULT2A1 mRNA in the placenta, Stanley et al reported high DHEA (10 μM) sulfation rates in fractions rich in syncytiotrophoblasts and attributed this to SULT2A1 [13, 42]. However, as mentioned above sulfotransferases enzymes often exhibit overlapping substrate specificities, and SULT2A1 is not the only isoform that sulfates DHEA at such concentrations. Another sulfotransferase isoform SULT2b1b, which also sulfates hydroxysteroids, is localized primarily to the nuclei of syncytiotrophoblasts [16]. One of the physiological substrates of SULT2b1b is cholesterol [7] which is the precursor for progesterone synthesis. SULT2b1b and SULT2A1 exhibit similar affinities for DHEA [16]. It is possible that the DHEA sulfation activity reported by Stanley et al. [13] was mediated by SULT2b1b. Another possible reason for the discrepancy observed in our work and the work of Stanley et al. [13] with respect to presence of SULT2A1 protein in trophoblast could be differential enzyme expression between the cytotrophoblasts and syncytiotrophoblasts as has been observed with several transport proteins [43]. Our results indicated that SULT1E1 is not functional in BeWo cells. Similar to SULT2A1 there are conflicting reports on placental/trophoblast SULT1E1 expression. While some studies report much lower activity/mRNA expression in comparison to SULT1A1/SULT1A3 [13, 42], others have demonstrated SULT1E1 to be localized to syncytiotrophoblasts [15]. One possible reason for observing low SULT1E1 activity in BeWo cells in this study could be the presence of steroid sulfatase enzymes, which hydrolyze sulfate molecules. These enzymes exhibit particularly high activity in placental tissue and have been localized to the syncytiotrophoblasts [15, 44].
SULT1A1 sulfates a wide range of substrates that includes steroid hormones, medicinal drugs, as well as procarcinogens. The physiological functions of this enzyme in the placenta have not been elucidated, although it has been reported that it is not the source of the high levels of thyroid hormone sulfates detected in the fetal circulation [13]. One of the endogenous substrates of SULT1A1 is 17β-estradiol, although SULT1A1 sulfates the hormone at much higher concentrations than SULT1E1 [45]. As the syncytiotrophoblast secretes increasing amounts of the 17β-estradiol with advancing gestation [46], it is tempting to speculate that a possible physiological function of the enzyme is maintaining tissue estrogen balance, at least under conditions of high local concentrations. Endogenous substrates of SULT1A3 include catecholamines and catecholestrogens. The majority of plasma catecholamines exist in the sulfated form [47] and plasma levels of catecholamine sulfates are elevated in disease conditions such as hypertension and pheochromocytoma [48-50]. The physiological significance of catecholamine sulfation is not very well understood. However, catecholamine sulfates are inactive at the adrenergic and dopaminergic receptors and it has been suggested that conjugation serves as a protective mechanism against the adverse responses that may be produced from the high circulating levels of catecholamines [7, 51-54]. While studies have not dealt with the specific role of SULT1A3 in the placenta, similar to its function in other tissues, physiological functions of SULT1A3 in the placenta may be catecholamine inactivation.
Fetal sulfation develops as early as the 14th week of gestation and there are no correlations between gestation age (14-35 weeks) and fetal sulfotransferase activity [55]. Several studies report fetal SULT1A1 and SULT1A3 activities to be higher or similar to placental activity [55, 56]. Nevertheless, the SULT1A family also appears to be well developed in the placenta and likely acts as a support of fetal metabolism for fetal protection.
In conclusion our work establishes the phenolic sulfotransferase isoforms SULT1A1 and SULT1A3 to be functional in BeWo cells and in primary cultures of cytotrophoblast cells. A lot of the work on placental metabolism has focused on the effects of cigarette smoke, medicinal drugs, and environmental chemicals on placental enzyme activity, as a way of determining if this may be related to fetal toxicity. There has not been a lot of focus on the effects of foreign chemicals on placental sulfotransferase activity. SULT1A1 catalyses the sulfation of several procarcinogens such as N-hydroxylated aromatic amines, nitroalkanes, and benzylic alcohols to produce nucleophilic DNA adducts [1, 57]. The functional activities of SULT1A1 and SULT1A3 in BeWo cells suggests that this cell line can be used as a suitable model for studying placental sulfotransferase induction/inhibition as a basis for developing an understanding of the role of these enzymes in physiological and disease processes. Also, the BeWo cells express several efflux transporters and it is likely that one or more of these eliminate sulfate metabolites. Thus the BeWo cell line can be used to demonstrate how firstly sulfation, and secondly, elimination of sulfate metabolites, serves to protect the fetus.
Acknowledgments
The authors are grateful to Peter Silverstein, Ph.D., and Claudia Bode, Ph.D., for their instructions on RT-PCR and cloning studies respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Williams JA, Stone EM, Fakis G, Johnson N, Cordell JA, Meinl W, et al. N-Acetyltransferases, sulfotransferases and heterocyclic amine activation in the breast. Pharmacogenetics. 2001;11:373–88. doi: 10.1097/00008571-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Kudlacek PE, Clemens DL, Anderson RJ. Characterization of recombinant human liver thermolabile phenol sulfotransferase with minoxidil as the substrate. Biochem Biophys Res Commun. 1995;210:363–9. doi: 10.1006/bbrc.1995.1670. [DOI] [PubMed] [Google Scholar]
- 3.Lindsay J, Wang LL, Li Y, Zhou SF. Structure, function and polymorphism of human cytosolic sulfotransferases. Curr Drug Metab. 2008;9:99–105. doi: 10.2174/138920008783571819. [DOI] [PubMed] [Google Scholar]
- 4.Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–16. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 5.Falany JL, Macrina N, Falany CN. Regulation of MCF-7 breast cancer cell growth by beta-estradiol sulfation. Breast Cancer Res Treat. 2002;74:167–76. doi: 10.1023/a:1016147004188. [DOI] [PubMed] [Google Scholar]
- 6.Kotov A, Falany JL, Wang J, Falany CN. Regulation of estrogen activity by sulfation in human Ishikawa endometrial adenocarcinoma cells. J Steroid Biochem Mol Biol. 1999;68:137–44. doi: 10.1016/s0960-0760(99)00022-9. [DOI] [PubMed] [Google Scholar]
- 7.Strott CA. Sulfonation and molecular action. Endocr Rev. 2002;23:703–32. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- 8.Chen G, Zhang D, Jing N, Yin S, Falany CN, Radominska-Pandya A. Human gastrointestinal sulfotransferases: identification and distribution. Toxicol Appl Pharmacol. 2003;187:186–97. doi: 10.1016/s0041-008x(02)00073-x. [DOI] [PubMed] [Google Scholar]
- 9.Cappiello M, Giuliani L, Pacifici GM. Differential distribution of phenol and catechol sulphotransferases in human liver and intestinal mucosa. Pharmacology. 1990;40:69–76. doi: 10.1159/000138643. [DOI] [PubMed] [Google Scholar]
- 10.Furimsky AM, Green CE, Sharp LE, Catz P, Adjei AA, Parman T, et al. Effect of resveratrol on 17beta-estradiol sulfation by human hepatic and jejunal S9 and recombinant sulfotransferase 1E1. Drug Metab Dispos. 2008;36:129–36. doi: 10.1124/dmd.107.016725. [DOI] [PubMed] [Google Scholar]
- 11.Baranczyk-Kuzma A, Audus KL, Borchardt RT. Catecholamine-metabolizing enzymes of bovine brain microvessel endothelial cell monolayers. J Neurochem. 1986;46:1956–60. doi: 10.1111/j.1471-4159.1986.tb08519.x. [DOI] [PubMed] [Google Scholar]
- 12.Sodha RJ, Schneider H. Sulphate conjugation of beta 2-adrenoceptor stimulating drugs by platelet and placental phenol sulphotransferase. Br J Clin Pharmacol. 1984;17:106–8. doi: 10.1111/j.1365-2125.1984.tb05009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanley EL, Hume R, Visser TJ, Coughtrie MW. Differential expression of sulfotransferase enzymes involved in thyroid hormone metabolism during human placental development. J Clin Endocrinol Metab. 2001;86:5944–55. doi: 10.1210/jcem.86.12.8081. [DOI] [PubMed] [Google Scholar]
- 14.Young AM, Allen CE, Audus KL. Efflux transporters of the human placenta. Adv Drug Deliv Rev. 2003;55:125–32. doi: 10.1016/s0169-409x(02)00174-6. [DOI] [PubMed] [Google Scholar]
- 15.Miki Y, Nakata T, Suzuki T, Darnel AD, Moriya T, Kaneko C, et al. Systemic distribution of steroid sulfatase and estrogen sulfotransferase in human adult and fetal tissues. J Clin Endocrinol Metab. 2002;87:5760–8. doi: 10.1210/jc.2002-020670. [DOI] [PubMed] [Google Scholar]
- 16.Meloche CA, Falany CN. Expression and characterization of the human 3 beta-hydroxysteroid sulfotransferases (SULT2B1a and SULT2B1b) J Steroid Biochem Mol Biol. 2001;77:261–9. doi: 10.1016/s0960-0760(01)00064-4. [DOI] [PubMed] [Google Scholar]
- 17.Avery ML, Meek CE, Audus KL. The presence of inducible cytochrome P450 types 1A1 and 1A2 in the BeWo cell line. Placenta. 2003;24:45–52. doi: 10.1053/plac.2002.0876. [DOI] [PubMed] [Google Scholar]
- 18.Vaidya SS, Walsh SW, Gerk PM. Formation and Efflux of ATP-Binding Cassette Transporter Substrate 2,4-Dinitrophenyl-S-Glutathione from Cultured Human Term Placental Villous Tissue Fragments. Mol Pharm. 2009 doi: 10.1021/mp900019z. [DOI] [PubMed] [Google Scholar]
- 19.Brand W, van der Wel PA, Rein MJ, Barron D, Williamson G, van Bladeren PJ, et al. Metabolism and transport of the citrus flavonoid hesperetin in Caco-2 cell monolayers. Drug Metab Dispos. 2008;36:1794–802. doi: 10.1124/dmd.107.019943. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Lin H, Hu M. Absorption and metabolism of genistein and its five isoflavone analogs in the human intestinal Caco-2 model. Cancer Chemother Pharmacol. 2005;55:159–69. doi: 10.1007/s00280-004-0842-x. [DOI] [PubMed] [Google Scholar]
- 21.Tamura HO, Taniguchi K, Hayashi E, Hiyoshi Y, Nagai F. Expression profiling of sulfotransferases in human cell lines derived from extra-hepatic tissues. Biol Pharm Bull. 2001;24:1258–62. doi: 10.1248/bpb.24.1258. [DOI] [PubMed] [Google Scholar]
- 22.Friedman SJ, Skehan P. Morphological differentiation of human choriocarcinoma cells induced by methotrexate. Cancer Res. 1979;39:1960–7. [PubMed] [Google Scholar]
- 23.Liu F, Soares MJ, Audus KL. Permeability properties of monolayers of the human trophoblast cell line BeWo. Am J Physiol. 1997;273:C1596–604. doi: 10.1152/ajpcell.1997.273.5.C1596. [DOI] [PubMed] [Google Scholar]
- 24.Bode CJ, Jin H, Rytting E, Silverstein PS, Young AM, Audus KL. In vitro models for studying trophoblast transcellular transport. Methods Mol Med. 2006;122:225–39. doi: 10.1385/1-59259-989-3:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petroff MG, Phillips TA, Ka H, Pace JL, Hunt JS. Isolation and culture of term human trophoblast cells. Methods Mol Med. 2006;121:203–17. doi: 10.1385/1-59259-983-4:201. [DOI] [PubMed] [Google Scholar]
- 26.Dubin RL, Hall CM, Pileri CL, Kudlacek PE, Li XY, Yee JA, et al. Thermostable (SULT1A1) and thermolabile (SULT1A3) phenol sulfotransferases in human osteosarcoma and osteoblast cells. Bone. 2001;28:617–24. doi: 10.1016/s8756-3282(01)00463-x. [DOI] [PubMed] [Google Scholar]
- 27.Foldes A, Meek JL. Rat brain phenolsulfotransferase: partial purification and some properties. Biochim Biophys Acta. 1973;327:365–74. doi: 10.1016/0005-2744(73)90419-1. [DOI] [PubMed] [Google Scholar]
- 28.Harris RM, Wood DM, Bottomley L, Blagg S, Owen K, Hughes PJ, et al. Phytoestrogens are potent inhibitors of estrogen sulfation: implications for breast cancer risk and treatment. J Clin Endocrinol Metab. 2004;89:1779–87. doi: 10.1210/jc.2003-031631. [DOI] [PubMed] [Google Scholar]
- 29.Spink BC, Katz BH, Hussain MM, Pang S, Connor SP, Aldous KM, et al. SULT1A1 catalyzes 2-methoxyestradiol sulfonation in MCF-7 breast cancer cells. Carcinogenesis. 2000;21:1947–57. doi: 10.1093/carcin/21.11.1947. [DOI] [PubMed] [Google Scholar]
- 30.Taskinen J, Ethell BT, Pihlavisto P, Hood AM, Burchell B, Coughtrie MW. Conjugation of catechols by recombinant human sulfotransferases, UDP-glucuronosyltransferases, and soluble catechol O-methyltransferase: structure-conjugation relationships and predictive models. Drug Metab Dispos. 2003;31:1187–97. doi: 10.1124/dmd.31.9.1187. [DOI] [PubMed] [Google Scholar]
- 31.Barnett AC, Tsvetanov S, Gamage N, Martin JL, Duggleby RG, McManus ME. Active site mutations and substrate inhibition in human sulfotransferase 1A1 and 1A3. J Biol Chem. 2004;279:18799–805. doi: 10.1074/jbc.M312253200. [DOI] [PubMed] [Google Scholar]
- 32.Chapman E, Best MD, Hanson SR, Wong CH. Sulfotransferases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl. 2004;43:3526–48. doi: 10.1002/anie.200300631. [DOI] [PubMed] [Google Scholar]
- 33.Gamage NU, Duggleby RG, Barnett AC, Tresillian M, Latham CF, Liyou NE, et al. Structure of a human carcinogen-converting enzyme, SULT1A1. Structural and kinetic implications of substrate inhibition. J Biol Chem. 2003;278:7655–62. doi: 10.1074/jbc.M207246200. [DOI] [PubMed] [Google Scholar]
- 34.Anderson RJ, Jackson BL, Liebentritt DK. Human platelet thermostable phenol sulfotransferase from blacks and whites: biochemical properties and variations in thermal stability. J Lab Clin Med. 1988;112:773–83. [PubMed] [Google Scholar]
- 35.Dajani R, Sharp S, Graham S, Bethell SS, Cooke RM, Jamieson DJ, et al. Kinetic properties of human dopamine sulfotransferase (SULT1A3) expressed in prokaryotic and eukaryotic systems: comparison with the recombinant enzyme purified from Escherichia coli. Protein Expr Purif. 1999;16:11–8. doi: 10.1006/prep.1999.1030. [DOI] [PubMed] [Google Scholar]
- 36.Rens-Domiano SS, Roth JA. Inhibition of M and P phenol sulfotransferase by analogues of 3′-phosphoadenosine-5′-phosphosulfate. J Neurochem. 1987;48:1411–5. doi: 10.1111/j.1471-4159.1987.tb05679.x. [DOI] [PubMed] [Google Scholar]
- 37.Ebmeier CC, Anderson RJ. Human thyroid phenol sulfotransferase enzymes 1A1 and 1A3: activities in normal and diseased thyroid glands, and inhibition by thyroid hormones and phytoestrogens. J Clin Endocrinol Metab. 2004;89:5597–605. doi: 10.1210/jc.2003-031939. [DOI] [PubMed] [Google Scholar]
- 38.Anderson RJ, Weinshilboum RM. Phenolsulphotransferase in human tissue: radiochemical enzymatic assay and biochemical properties. Clin Chim Acta. 1980;103:79–90. doi: 10.1016/0009-8981(80)90233-8. [DOI] [PubMed] [Google Scholar]
- 39.Raftogianis RB, Wood TC, Weinshilboum RM. Human phenol sulfotransferases SULT1A2 and SULT1A1: genetic polymorphisms, allozyme properties, and human liver genotype-phenotype correlations. Biochem Pharmacol. 1999;58:605–16. doi: 10.1016/s0006-2952(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 40.Anderson RJ, Liebentritt DK. Human platelet thermostable phenol sulfotransferase: assay of frozen samples and correlation between frozen and fresh activities. Clin Chim Acta. 1990;189:221–9. doi: 10.1016/0009-8981(90)90095-a. [DOI] [PubMed] [Google Scholar]
- 41.Tabrett CA, Coughtrie MW. Phenol sulfotransferase 1A1 activity in human liver: kinetic properties, interindividual variation and re-evaluation of the suitability of 4-nitrophenol as a probe substrate. Biochem Pharmacol. 2003;66:2089–97. doi: 10.1016/s0006-2952(03)00582-3. [DOI] [PubMed] [Google Scholar]
- 42.Nishimura M, Naito S. Tissue-specific mRNA expression profiles of human phase I metabolizing enzymes except for cytochrome P450 and phase II metabolizing enzymes. Drug Metab Pharmacokinet. 2006;21:357–74. doi: 10.2133/dmpk.21.357. [DOI] [PubMed] [Google Scholar]
- 43.Evseenko DA, Paxton JW, Keelan JA. ABC drug transporter expression and functional activity in trophoblast-like cell lines and differentiating primary trophoblast. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1357–65. doi: 10.1152/ajpregu.00630.2005. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki T, Miki Y, Nakata T, Shiotsu Y, Akinaga S, Inoue K, et al. Steroid sulfatase and estrogen sulfotransferase in normal human tissue and breast carcinoma. J Steroid Biochem Mol Biol. 2003;86:449–54. doi: 10.1016/s0960-0760(03)00356-x. [DOI] [PubMed] [Google Scholar]
- 45.Yasuda S, Suiko M, Liu MC. Oral contraceptives as substrates and inhibitors for human cytosolic SULTs. J Biochem. 2005;137:401–6. doi: 10.1093/jb/mvi047. [DOI] [PubMed] [Google Scholar]
- 46.Kallen CB. Steroid hormone synthesis in pregnancy. Obstet Gynecol Clin North Am. 2004;31:795–816. doi: 10.1016/j.ogc.2004.08.009. x. [DOI] [PubMed] [Google Scholar]
- 47.Kuchel O, Buu NT. Conjugated catecholamines and their measurement: some pharmacokinetic aspects. Curr Med Res Opin. 1983;8 3:3–8. doi: 10.1185/03007998309109831. [DOI] [PubMed] [Google Scholar]
- 48.Yoshizumi M, Kitagawa T, Hori T, Katoh I, Houchi H, Ohuchi T, et al. Physiological significance of plasma sulfoconjugated dopamine in patients with hypertension--clinical and experimental studies. Life Sci. 1996;59:324–30. doi: 10.1016/0024-3205(96)00301-3. [DOI] [PubMed] [Google Scholar]
- 49.Kuchel O. Clinical implications of genetic and acquired defects in catecholamine synthesis and metabolism. Clin Invest Med. 1994;17:354–73. [PubMed] [Google Scholar]
- 50.Kuchel O, Buu NT, Fontaine A, Hamet P, Beroniade V, Larochelle P, et al. Free and conjugated plasma catecholamines in hypertensive patients with and without pheochromocytoma. Hypertension. 1980;2:177–86. doi: 10.1161/01.hyp.2.2.177. [DOI] [PubMed] [Google Scholar]
- 51.Michel GL, Lenz T, Lernhardt U, Weicker H, Bieger WP, Werle E. Sulfoconjugated catecholamines: lack of beta-adrenoceptor binding and adenylate cyclase stimulation in human mononuclear leukocytes. Eur J Pharmacol. 1987;143:179–88. doi: 10.1016/0014-2999(87)90531-0. [DOI] [PubMed] [Google Scholar]
- 52.Kyncl JJ, Buckner SA, Brondyk H, Kerkman DJ, DeBernardis JF, Bush EN, et al. Adrenergic and dopaminergic properties of dopamine sulfoconjugates. J Cardiovasc Pharmacol. 1985;7:1198–204. doi: 10.1097/00005344-198511000-00030. [DOI] [PubMed] [Google Scholar]
- 53.Lenz T, Werle E, Strobel G, Weicker H. O-Methylated and sulfoconjugated catecholamines: differential activities at human platelet alpha 2-adrenoceptors. Can J Physiol Pharmacol. 1991;69:929–37. doi: 10.1139/y91-141. [DOI] [PubMed] [Google Scholar]
- 54.Stokes AH, Hastings TG, Vrana KE. Cytotoxic and genotoxic potential of dopamine. J Neurosci Res. 1999;55:659–65. doi: 10.1002/(SICI)1097-4547(19990315)55:6<659::AID-JNR1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 55.Pacifici GM. Sulfation of drugs and hormones in mid-gestation human fetus. Early Hum Dev. 2005;81:573–81. doi: 10.1016/j.earlhumdev.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 56.Cappiello M, Giuliani L, Rane A, Pacifici GM. Dopamine sulphotransferase is better developed than p-nitrophenol sulphotransferase in the human fetus. Dev Pharmacol Ther. 1991;16:83–8. [PubMed] [Google Scholar]
- 57.Kreis P, Brandner S, Coughtrie MW, Pabel U, Meinl W, Glatt H, et al. Human phenol sulfotransferases hP-PST and hM-PST activate propane 2-nitronate to a genotoxicant. Carcinogenesis. 2000;21:295–9. doi: 10.1093/carcin/21.2.295. [DOI] [PubMed] [Google Scholar]


