Abstract
Some genes cannot be cloned by conventional methods because in most cases the genes or gene products are toxic to E. coli. CCH1 is a high-affinity Ca2+ channel present in the plasma membrane of Cryptococcus neoformans and other fungi. Like many toxic genes, the molecular cloning of CCH1 has been a major challenge and consequently direct studies of CCH1 channel activity in heterologous expression systems have been impossible. We devised a straightforward approach that resulted in the molecular cloning and functional expression of CCH1 by exploiting homologous recombination both in vitro and in vivo. This approach precluded the standard enzyme digestion-mediated ligation reactions and the subsequent isolation of plasmids from E. coli. The shuttle plasmid carrying CCH1-GFP, which was prepared in vitro and propagated in yeast, was successfully expressed in a mammalian cell line (HEK293). CCH1 transcripts were detected only in HEK293 cells transfected with the plasmid DNA. Fluorescence microscopy studies revealed the expression of CCH1-GFP fusion protein on the cell surface of HEK293 cells, similar to the localization pattern of a well-characterized plasma membrane-associated K+ channel. This approach will be particularly useful for genes that encode ion channels and transporters that cannot be cloned by conventional techniques requiring E. coli.
Keywords: calcium channel, toxic gene, Cryptococcus neoformans, homologous recombination, E. coli
Introduction
It is practically impossible to perform functional expression studies of genes that cannot be cloned and maintained by the standard molecular biological techniques that use E. coli as a host. Cch1 is a high-affinity Ca2+ channel that is expressed in the plasma membrane of different fungi including Cryptococcus neoformans [1, 2, 3, 4, 5]. This basidiomycete is a pathogen that causes life-threatening meningitis primarily in individuals that lack an adequate T-cell dependent immune response [6, 7, 8]. Its ability to survive in environments of low Ca2+ is dependent on Cch1 activity and Mid1, a binding partner of Cch1 and possibly a co-regulator [1, 2, 9]. Genetic analysis has shown that a strain of C. neoformans that lacked CCH1 was not viable in conditions of low extracellular Ca2+ consistent with its role as the only high-affinity Ca2+ channel in the plasma membrane of C. neoformans [1, 2, 9]. Among the mechanisms that regulate Cch1 activity is the highly organized trafficking of Cch1 to the plasma membrane by EF3, an elongation factor unique to fungal cells [10, 11, 12, 13, 14]. This is supported by evidence demonstrating that Cch1 is mislocalized in an EF3 repression strain of C. neoformans. As expected, this strain is not viable during conditions where extracellular Ca2+ is limiting [10]. This is consistent with the phenotype of the cch1Δ null mutant strain and suggests that Cch1 is nonfunctional when it is not expressed in the plasma membrane [1, 10].
Despite the gains made in resolving the regulation of Cch1, direct analysis of Cch1 channel activity by electrophysiological techniques has been elusive because the inability to clone Cch1 precluded all structure-function studies in heterologous expression systems. This problem is not unique to CCH1, as examples of other genes encoding ion channels that could not be cloned and maintained by standard molecular biology techniques involving E. coli have been documented [16, 17, 18, 19]. Here we have devised a simple and reliable approach that successfully led to the functional expression of Cch1. This approach is amenable to other problematic genes, particularly those encoding ion channels and transporters.
In this study we bypassed standard enzyme-mediated ligation reactions and the need to propagate plasmids in E. coli and instead exploited the idea of homologous recombination to generate plasmids containing the gene of interest. The cloning strategy involved preparing a linearized plasmid with homologous ends to the DNA fragment intended for insertion into the plasmid. As long as the end sequences of both the plasmid and the DNA insert share sequence identity, two targeted double crossover events can be achieved to integrate the DNA insert into the plasmid [20, 21, 22]. These crossover events are dependent on the actions of the homologous recombination machinery in yeast [20]. The idea of producing new plasmids by this mechanism in yeast has been well documented and it has served as a powerful molecular biological tool [21, 22]. However we found that if an in vitro recombination reaction is performed prior to transforming yeast, then the number of positive clones increases dramatically. This is probably because the in vitro conditions can be easily optimized to generate the best possible yield. In addition, incomplete in vitro reactions can be more readily completed by the in vivo homologous recombination machinery once the DNA is transformed into yeast. The recovery of plasmids from yeast is usually accomplished by using E. coli as the host. However, in the case of CCH1 this was not an option. Instead we devised a protocol for recovering high yields of CCH1-plasmid DNA directly from yeast sufficient for transfection and expression in a human embryonic kidney (HEK293) cell line where we could measure Cch1 channel activity directly.
HEK293 cells are routinely used for functional studies of ion channels. With this approach CCH1 transcripts were detected in HEK293 cells and localization studies in HEK293 cells revealed a plasma membrane distribution of Cch1. Taken together, the results clearly demonstrate that the approach presented in this study has successfully facilitated the molecular cloning of CCH1 and this has allowed a direct functional analysis of Cch1. This same approach will be extremely useful for other transporters and ion channels that cannot be cloned by conventional methods that are dependent on E. coli.
Material and Methods
Media and growth of yeast cultures and HEK293 cells
The wild type Saccharomyces cerevisiae strain (AGSc29, W303 MATa) used in this study was recovered from a 15% glycerol stock stored at –80ºC. The strain was maintained on YPD (1% yeast extract, 2% peptone and 2% dextrose) or synthetic medium lacking leucine and cultured in YPD medium at 30ºC for 24 h. Cells from the human embryonic kidney cell line (HEK293) were cultured in Dulbecco’s modified Eagle’s medium with 10% calf serum and antibiotics in a 5% CO2 incubator at 37ºC. HEK293 cells were purchased from ATCC (CRL-1573).
Preparation of vector and CCH1 amplicon for in vitro homologous recombination
The aim was to clone CCH1 into the pcDNA3.1/CT-GFP-topo expression plasmid under the control of the constitutive mammalian CMV promoter (Invitrogen, Carlsbad CA). To circumvent propagating the expression plasmid in E. coli, the essential features of pcDNA3.1/CT-GFP-topo that were needed for the expression of CCH1 in HEK293 cells were PCR-amplified and cloned into a yeast expression plasmid, pRS425. These features included the DNA fragment containing the CMV promoter, the GFP tag, the unique multiple cloning sites between CMV and GFP, and the polyadenylation sequence. The entire DNA fragment (~2 Kb) was amplified from pcDNA3.1/CT-GFT-topo with primer 19 and 20 inserted into a SacII site on pRS425 using standard molecular techniques. The pcDNA3.1/CT-GFP-topo was supplied in a linearized form for topo-cloning and as a result recircularization of the vector was needed prior to using it as a template in the PCR reaction. Although the ends of the linearized pcDNA3.1/CT-GFP-topo were not compatible, a ligated, circular form of the plasmid was obtained by transforming the topo vector into E. coli. Even during standard topo cloning reactions, a small percentage of the topo vector will re-ligate to form an empty circular plasmid.
The CCH1 amplicon was generated from cDNA that was synthesized by the SuperScript III First-Strand synthesis system for RT-PCR (Invitrogen) using total RNA as template. RNA was isolated from a culture (~5 × 107 cells) of a wild type strain of Cryptococcus neoformans var. grubii (H99). Cells were lysed with acid washed glass beads and total RNA was isolated and purified according to the manufacturers instructions (RNAeasy Kit, Qiagen).
In vitro recombination reaction
In order to avoid an enzyme digestion-mediated ligation reaction in E. coli an in vitro recombination reaction was performed with the In-Fusion 2.0 Dry-Down PCR kit (Clontech). The PCR-generated CCH1 amplicon (strategy 1, primers 1 & 2; strategy 2, primers 3 & 4) was purified using the QIAquick PCR purification kit (Qiagen). Next, the linearized vector and the CCH1 fragment were added to the In-Fusion (Dry-down mix) ligation mixture at a 1:5 vector to fragment ratio and incubated at 37ºC for 15 min followed by a 15 min incubation at 50ºC.
Yeast Transformation by lithium acetate
A wild type strain of Saccharomyces cerevisiae (AGSc29) was transformed by the lithium acetate method according to published protocols. Briefly, yeast cells were grown in YP media with 2% glucose to an OD600 of 0.4. Cells were washed and resuspended in a 100µl LiTE (1M LiOAc, 100mM TrisCl pH 7.5, 10mM EDTA) to which 10)µl of the In-Fusion mix and 10µl of sonicated salmon sperm (Stratagene) were added. The mixture was added to 600)µl of PEG/LiTE (44% PEG 3300), mixed well by repetitive pipeting and incubated in a rotating wheel for 30min at 30ºC followed by a 15 min incubation in a 42ºC water bath. Cells were washed once in TE, resuspened in 200)µl of TE and 200)µl was plated onto agar plates lacking leucine. Agar plates were incubated at 30ºC for two days at which point colonies were selected and screened by PCR using Choice Taq Blue DNA polymerase (Denville Scientific) with DNA primers 6 and 12 (Table 1). PCR conditions were as follows: 95ºC for 1 min, 95ºC for 40s and 55ºC for 40s and 68ºC for 5:30 min repeated 35 cycles, followed by an additional extension step at 68ºC for 10 min.
Table 1.
A list of all primers used in this study.
| Name | Sequence of Primer | |
|---|---|---|
| 1. | fusion- CCH1-Fa−- strategy 1 | gtctagaatggctagccccactcccgcaa |
| 2. | fusion- CCH1-Rb- strategy 1 | cttctcctttgctagccataccctgttcat |
| 3. | fusion- CCH1-F- strategy 2 | atggcccactcccgcaaccacagcctcgcaatc |
| 4. | fusion- CCH1-R- strategy 2 | same as primer #7 |
| 5. | CMV-F | cgatgtacgggccagatatac |
| 6. | GFP-R | tagtgcgttcctgtacataaccttc |
| 7. | CCH1-R | accctgttcattttcaatgccttcttgatctac |
| 8. | CCH1 seq1 | cagagagaagaggtggatcacggccgacta |
| 9. | CCH1 seq2 | tactttcttgtttttgctgcgtgcata |
| 10. | CCH1 seq3 | ggaactgttttgacctttttct |
| 11. | CCH1 seq4 | acggacgagcatatggatgcatttag |
| 12. | CCH1 seq5 | tactgttgacgaaagcatttggt |
| 13. | CCH1 seq6 | atctttttcttctctgggctaccat |
| 14. | CCH1 seq7 | actattttcaagactatataacga |
| 15. | CCH1 3521-R | caggcccagagccacttctgtg |
| 16. | CCH1 5’440-R | tccctgtcgtctgatgatct |
| 17. | CCH1-CMV-R | ggctgtggttgcgggagtgggccataatttcgataagccagtaagcag |
| 18. | CCH1-GFP-F | agaaggcattgaaaatgaacagggtatggctagcaaaggagaagaact |
| 19. | CMV-SacII-F | ccgcgggttaggcgttttgcgctgcttcgcgatgtacg |
| 20. | PolyA-SacII-R | ccgcggcactacgtgaaccatcaccctaatcaag |
Primers used in the 5’ forward direction are denoted by F.
Primers used in the 3’ reverse direction are denoted by R.
Extraction of CCH1- plasmid DNA from yeast
The yeast colonies that were identified as positive transformants were cultured in leucine drop-out media containing 2% glucose overnight at 30ºC. Yeast cells were processed according to the E.Z.N.A. Yeast plasmid mini spin kit (Omega Bio-Tek) with some noted changes. Approximately 1 × 10 cells (50 times more cells than that recommended by the manufacturer) were resuspended in 24 ml of buffer SE containing 240 µl of β-mercaptoethanol and 2 ml of lyticase cocktail and incubated at 30ºC for 30 min in order to digest cell walls. The resulting spheroplasts were lysed and centrifuged at 10 000 × g for 10min until a clear lysate was obtained according the manufacturer recommendation. Clear lysates were added to spin columns in order to isolate pure plasmid DNA. However despite scaling up the number of cells, the yield of plasmid DNA was very low (~80ng/µl) and not sufficient for transfection studies in HEK293 cells.
In order to improve the yield of plasmid DNA other types of spin columns were tested. Approximately 2×109 cells were resuspended in 4ml Buffer P1 from Qiagen Plasmid Midi Kit and 4ml of lyticase from E.Z.N.A. Yeast Plasmid Kit was added and incubated at 30ºC for 30 min [32]. Glass beads were added to the sample and cells were lysed by vortexing. The supernatant was removed and added to 4ml of Buffer P2 from the Midi kit. At this point the instructions provided by the manufacturer were followed as described in the Qiagen Plasmid Midi Kit protocol. The yield of plasmid DNA obtained with the Midi kit was on the order of ~ 3 µg/µl The recovered CCH1-plasmid DNA was sequenced with DNA primers 8, 9, 10, 11, 12, 13, 14, 15 and 16 (Table 1). It is important to note that none of the available commercial kits for isolating plasmid DNA from yeast provided enough yield for further study of CCH1. We have found that the Midi-DNA plasmid kit from Qiagen, which is commercially available for isolating plasmids from bacteria, was also very good for isolating plasmid DNA from yeast. This proved to be a key step in the successful cloning of CCH1 since sufficient yields of plasmid DNA were required for further study of CCH1.
Functional expression of CCH1-plasmid in HEK293 cells
HEK293 cells were counted and trypsinized 24 h before transfection. Approximated 1.25 × 105 in 1 ml of complete medium were plated per well such that the cell density was ~50–80% confluent on the day of transfection. Transient transfection of HEK 293 with CCH1-GFP plasmid DNA was performed with Lipofectamine 2000 and Plus reagents according to the manufacturer’s instructions (Invitrogen). Approximately 1µg of CCH1-GFP plasmid DNA was added to 100 µl, of Opti-MEM I reduced serum media and mixed with 1.75 µl of Lipotectamine 2000 and the mixture was incubated for 30 min at room temperature. The mixture was added directly to HEK293 cells that had been grown in a culture dish and maintained in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 4 mM L-Glutamine, 10% fetal bovine serum. HEK293 cells were maintained at 37ºC with 5% CO2 18–24 h post-transfection prior to assaying cells for transgene expression. HEK293 cells were fixed in 4% paraformaldehyde for 20 min at room temperature, washed in PBS and subsequently visualized with a fluorescent microscope. Immunofluorescence was examined using a Leica DMR series fluorescent microscope equipped with a chroma 86013 filter set (Chroma Technology, Rockingham VT) and CoolSNAP-HQ (Roper Scientific, Tucson AZ). The fluorescence label (GFP) was visualized by using filters S484/15 for excitation and S517/30 for emission.
Results and Discussion
A straightforward approach that permits the molecular cloning and functional expression of genes that cannot be cloned by conventional methods
We had previously determined that the CCH1 gene from Cryptococcus neformans could not be cloned by standard ligation-mediated techniques that used E. coli as a host because the gene encoding Cch1 or possibly the gene product is toxic to E. coli [1, 2]. This was particularly frustrating because it precluded a direct functional study of Cch1 channel activity in a heterologous expression system.
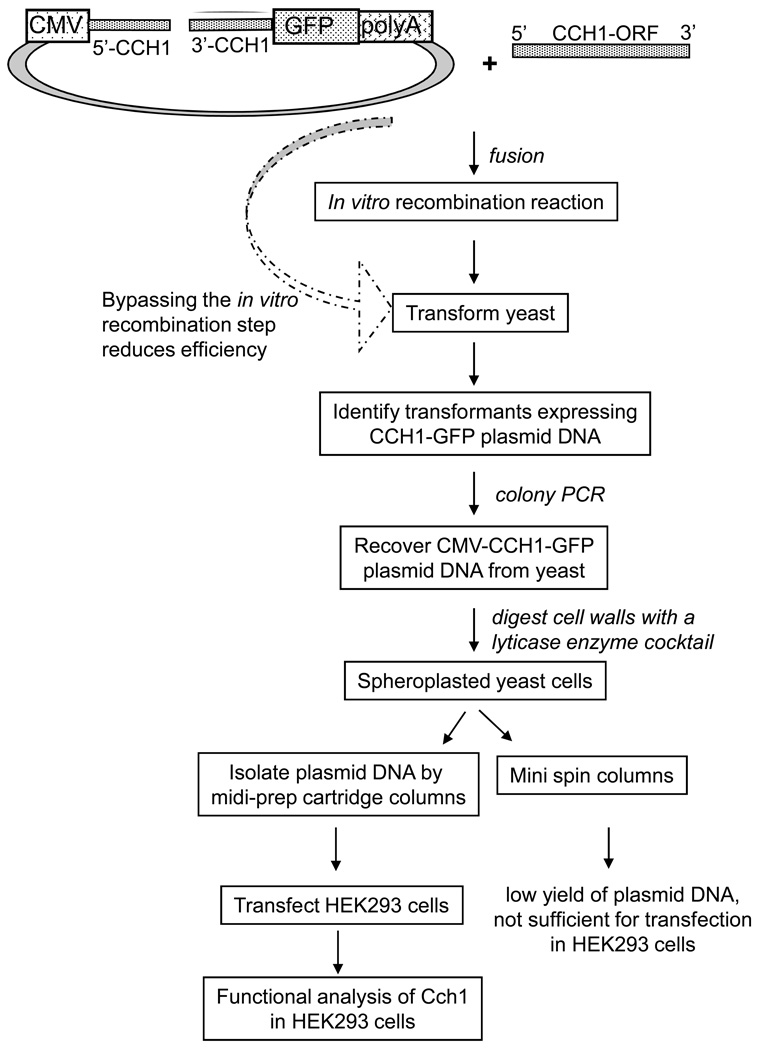
Our goal was to expeditiously clone CCH1 into an expression vector that could be efficiently isolated from S. cerevisiae and transfected into a mammalian cell line so that a functional study of the CCH1 channel could be done (Figure 1). Firstly, in order for this approach to be successful, it required a yeast plasmid that also expressed specific elements for functional expression in mammalian cells. Secondly, it was necessary to bypass the use of E. coli since it was clear that CCH1 could not be cloned using the standard protocols that utilized E. coli as a host. Instead we made use of an in vitro homologous recombination reaction to target CCH1 to a specific region of the shuttle vector used in our study. Thirdly, yeast was used as a vehicle to further promote homologous recombination between vector and insert and to propagate the new CCH1-expressing plasmid. Finally the approach required the recovery of a large quantity of plasmid DNA that could be simply and directly extracted from yeast and effectively transfected into a heterologous expression system, like a mammalian cell line where functional activity of CCH1 could be resolved (Figure 1).
Figure 1.
A schematic representation of the approach devised in order to clone CCH1 independently of conventional methods that require E. coli and to ultimately functionally express CCH1 in a mammalian cell line (human embryonic kidney cells, HEK293). The details of the methods listed are explained in the Materials and Methods section. Large arrow represents an alternative approach that bypassed the in vitro recombination reaction step. The construction of the vector is described in Figure 2.
Generating a vector that can be propagated in yeast and expressed in mammalian cells
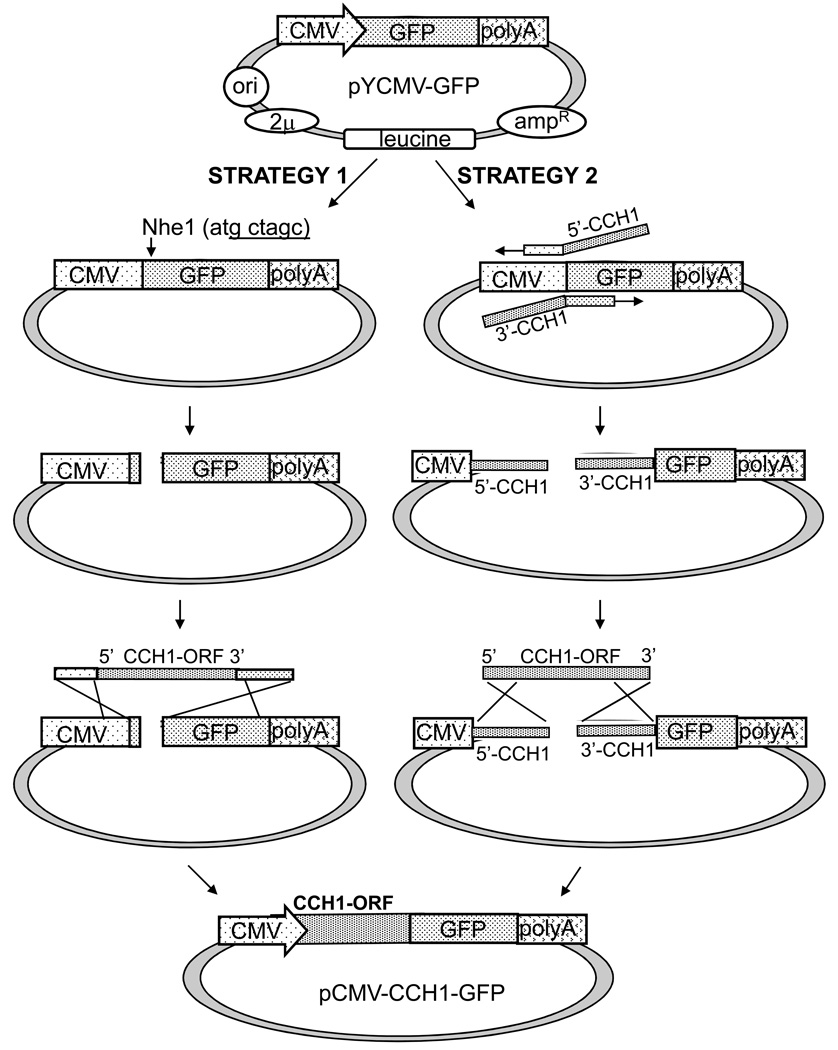
In order to construct a vector that could be propagated in yeast and expressed in mammalian cells, we began with the yeast pRS425 shuttle vector (a 2μplasmid with leucine as the nutritional marker) as the backbone. A stretch of DNA consisting of a CMV promoter, a GFP tag and a polyadenylation sequence was PCR- amplified from a mammalian expression vector (pcDNA3.1/CT-GFP) and cloned into the multiple cloning site of pRS425 generating the new vector, pYCMV-GFP (Figure 2). The polyadenylation sequence was added to the vector since it is known that this sequence is required to stabilize newly generated mature messenger RNA for translation in mammalian cells.
Figure 2.
A diagram illustrating the steps involved in constructing the CCH1 expression vector (pCMV-CCH1-GFP). Strategy 1) A fragment consisting of a CMV promoter, a GFP tag and a polyadenylation sequence was PCR-amplified from pcDNA3.1/CT-GFP-topo and cloned into a SacII site of a yeast shuttle vector (pRS425) generating the new vector, pYCMV-GFP. This vector was linearized with restriction enzyme Nhe1 and homologous end sequences of CMV and GFP were placed at the 5’ and 3’ ends of the CCH1 open reading frame, respectively. Strategy 2) Homologous end sequences of CCH1 were located on the pYCMV-GFP vector - at the end of CMV and at the beginning of GFP. To target CCH1 to pYCMV-GFP, primers were designed to amplify pYCMV-GFP with overhangs that corresponded to the 5’ end and the 3’ end of CCH1. Recombination between the ends of CCH1 and the vector was promoted by in vitro and in vivo homologous recombination reactions resulting in pCMV-CCH1-GFP (bottom of Figure 2).
Prior to initiating the in vitro reaction, pYCMV-GFP was linearized between the CMV promoter and the GFP tag so that the ends of the vector could facilitate the process of recombination. Two different versions of pYCMV-GFP were generated and used in two separate approaches. Both approaches positioned CCH1 after the CMV promoter in frame with the GFP tag but each approach used a different set of sequences at the homologous ends (Figure 2). In the first strategy, homologous end sequences of CMV and GFP were placed at the 5’ and 3’ ends of the CCH1 open reading frame, respectively (Figure 2, Strategy 1). To accomplish this, primers were designed to amplify the CCH1 allele with overhangs that corresponded to the 3’ end of CMV and the 5’ end of GFP (Table 1, primers 1 & 2). In the second strategy, the two homologous end sequences were located on the pYCMV-GFP vector - at the end of CMV and at the beginning of GFP (Figure 2, Strategy 2). To target CCH1 to pYCMV-GFP, primers were designed to amplify pYCMV-GFP with overhangs that corresponded to the 5’ end and the 3’ end of CCH1 (Table 1, primers 17 & 18). Primers were positioned such that CCH1 would be cloned after the CMV promoter and in frame with the GFP tag. This second strategy turned out to be the best approach for efficiently cloning CCH1. A limitation of the first approach was the difficulty in obtaining sufficient amounts of CCH1 cDNA with primers that had more than 15 nucleotides of homologous end sequences. In contrast, PCR amplification of pYCMV-GFP to generate a linearized plasmid with homologous ends was achieved with far less effort and greater yield (Figure 2, Strategy 2). A second limitation of the initial strategy was due to the limited number of restriction sites in the multiple cloning site of pYCMV-GFP. The only restriction site that did not cut within the sequence of CCH1 (NheI) resulted in a break in the open reading frame of the GFP tag which had to be corrected with the CCH1 reverse primer in order to ultimately create the CCH1-GFP fusion protein from pCMV-CCH1-GFP.
An in vitro recombination reaction and propagation of the CCH1-GFP plasmid in yeast negated the use of E. coli
Since the gene encoding CCH1 has 13 introns it was not possible to PCR amplify CCH1 from genomic DNA isolated from C. neoformans [1]. Instead, CCH1 was amplified from total RNA and the resulting 6.0 Kb PCR fragment was sequenced to ensure that the cDNA reflected the entire open reading frame of CCH1 and that it was free of all introns and any PCR-induced errors (Table 1). To avoid restriction enzyme-mediated ligation reactions in E. coli, an in vitro recombination system was used (In-Fusion 2.0 Dry-down cloning kit, Clontech). This system does not involve DNA digestion with restriction enzymes but rather it promotes the in vitro recombination between a PCR-generated sequence, such as the CCH1 cDNA, to a linearized vector by recognizing a 15 bp overlap at its ends - similar to the mechanism of homologous recombination in yeast. The instructions provided by the manufacturer were slightly modified because we found a more favorable outcome when we incorporated the following changes. Firstly, prior to combining the PCR-amplified CCH1 product with the linearized vector, the product was purified and concentrated with the QIAquick PCR purification kit (Qiagen). Secondly, the vector to insert ratio was increased from 1:2 to 1:5 in the fusion mixture.
Transformation of the fusion mixture into a wild type strain of S. cerevisiae was achieved by the standard lithium acetate method, and yeast colonies were selected on agar plates lacking leucine [24, 25]. It should be noted that an alternative approach was initially tried such that the in-fusion recombination step was bypassed and yeast was directly transformed with the linearized vector and CCH1 fragment; however excluding the in vitro fusion step resulted in not only fewer transformants but also many more transformants carrying empty plasmids. This was in stark contrast to the results reported by another group which found that in vivo homologous recombination in yeast was sufficient for cloning the CCH1 gene from S. cerevisiae (ScCCH1) [22]. They were able to express ScCCH1 in yeast without the need for the in vitro recombination reaction [22]. However we have found that ScCCH1 and the CCH1 gene from C. neoformans (CnCCH1) have significant sequence dissimilarity and this difference may result in the formation of secondary structures of CnCCH1 that could pose significant challenges for its cloning in S. cerevisiae, in addition to its toxicity to E. coli [1].
It is also likely that our in vitro recombination reaction was more efficient than the in vivo homologous recombination reactions in yeast partly because the in vitro conditions were optimized until the best yield was obtained. In our approach, yeast served merely as a means to propagate the recombinant plasmids generated in vitro. However, yeast may have provided an additional opportunity for any in vitro fusion reactions that did not go to completion. For example, an incomplete in vitro reaction may have yielded one double crossover event whereas two double crossover events would be required for the successful integration of CCH1 into the plasmid [20]. This second double crossover event can be achieved in vivo by the yeast homologous recombination machinery [20]. It is conceivable that incomplete in vitro recombination reactions may be more readily completed in vivo since the insert is already tethered to the plasmid and this close proximity could facilitate the recombination process.
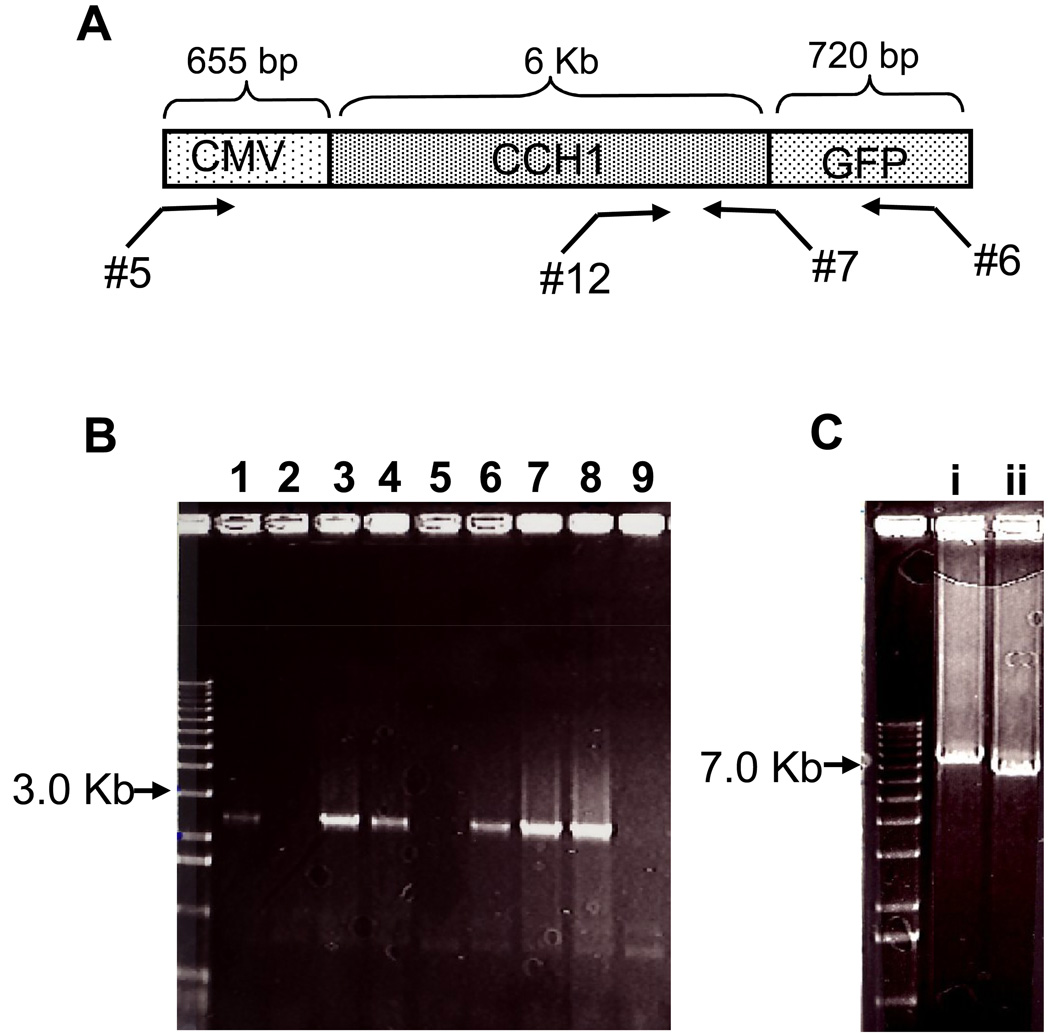
To identify the transformants that expressed the plasmid carrying CCH1-GFP, yeast colony PCR was used (Figure 3). Here individual yeast colonies were added to a buffer and boiled and the resulting supernatant was added to a PCR reaction with appropriate primers (DNA primers 6 and 12) (Figure 3A, Table 1). Of the ~ 33 colonies that were screened by PCR, 19 colonies expressed the plasmid carrying the CCH1-GFP fusion (Figure 3B). This suggests that roughly 58% of all colonies screened contained the CCH1-GFP insert. It is likely that further optimization of the reactions discussed above may have resulted in a larger number of colonies carrying the CCH1-GFP DNA sequence. Further analysis of the positive clones revealed the presence of the intact CMV-CCH-GFP sequence within the plasmid (Figure 3B).
Figure 3.
Yeast colony PCR identified DNA plasmids containing the CMV-CCH1-GFP sequence. A) A schematic diagram of the PCR strategy used for screening yeast colonies for the presence of the CCH1-GFP plasmid. Primers 5, 6, 7 and 12 are listed in Table 1. B) A representative agarose gel showing the amplified DNA products from the colony PCR reaction. Yeast colony PCR (Table 1, primers 6 & 12) was used to rapidly screen colonies and identify transformants carrying the CCH1-GFP plasmid. Amplified DNA was separated by agarose electorphoresis and stained with ethidium bromide to visualize bands. Lanes 1, 3, 4, 6 7 and 8 represent positive clones carrying the CCH1-GFP plasmid (~ 2.5 Kb). Shown are 9 representatives of the total 33 colonies screened. C) Further analysis revealed the presence of the CCH1 open reading frame within the plasmid. PCR-amplified DNA using primers 5 & 6 (Ci) and 5 & 7 (Cii) that targeted two different regions of the CMV-CCH1-GFP plasmid was separated by agarose electrophoresis and the DNA bands were visualized with ethidium bromide. Lanes (i) and (ii) revealed two DNA bands from pCMV-CCH1-GFP that corresponded to the expected size (Ci ~ 7.4 Kb and Cii ~ 6.6 Kb).
Isolating and purifying sufficient amounts of the CCH1-GFP plasmid DNA from yeast
The next step involved the recovery of a significantly large amount of pCMV-CCH1-GFP from yeast. Normally, plasmid DNA is extracted from yeast by using E. coli as a host. However in the case of CCH1 we could not use this approach and instead we devised a method to extract high-yields of plasmid DNA directly from yeast. Because yeast have a cell wall that must be removed before plasmid DNA can be isolated, yeast cells were treated with lyticase. We found that the lyticase cocktail provided by the E.Z.N.A. Yeast Plasmid Kit (Omega Bio-Tek) was very effective in generating large quantities of spheroplasts (yeast cells lacking the cell wall). The spheroplasts were disrupted and the resulting supernatant was added to a spin column in order to isolate and purify plasmid DNA. However the yield of plasmid DNA that resulted from using the mini spin columns provided by this kit was very low (~ 80ng/µl) probably because the DNA-binding capacity of these columns is very low. When large cartridge columns from a midi-plasmid DNA preparation kit (Qiagen) were used, the yield of plasmid DNA improved significantly (~ 3µg/µl). Sequencing analysis confirmed the presence of the intact allele of CCH1 in frame with the GFP tag.
Functional Expression of the CCH1 channel
Patch-clamp techniques represent the most direct approach for resolving channel activity because these techniques can effectively determine specific kinetic parameters of ion channels such as channel selectivity and channel conductance [31]. We had previously tried to apply these techniques directly to isolated spheroplasts of C. neoformans in order to analyze CCH1 channel function by measuring CCH1 channel activity straightaway. Unfortunately this approach was fraught with technical difficulties that led to inconsistencies in CCH1 channel measurements. For this reason the expression of CCH1 in a heterologous expression system appeared to be the only approach that would permit direct functional analysis of the CCH1 channel.
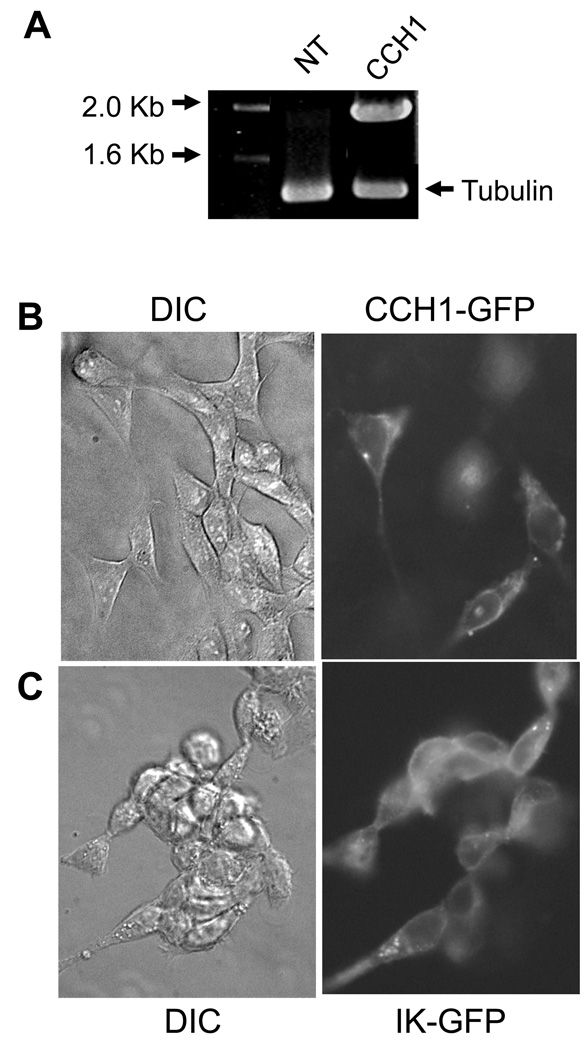
To confirm the functional expression of CCH1, pCCH1-GFP was transiently transfected into a human embryonic kidney cell line (HEK293). We chose this cell line because it is routinely used for functional expression studies of ion channels in part because these cells are easily maintained and transfected. The high yield of the CCH1-GFP plasmid that was recovered directly from yeast using the approach described in this study was key to the successful transfection of HEK293 cells. In order to confirm the expression of mRNA transcripts of CCH1 in the HEK293 cell line, RT (reverse transcriptase)-PCR was used. Here total RNA isolated from transfected HEK293 cells was used as template in the RT-PCR reaction and the amplified products were separated by agarose gel electrophoresis. A band of DNA corresponding to CCH1 was detected only in transfected HEK293 cells and not in untransfected HEK293 cells (Figure 4A).
Figure 4.
The functional expression of CCH1 in HEK293 cells. A) Reverse transcriptase (RT) PCR analysis revealed the presence of CCH1 transcripts in HEK293 cells that had been transfected with CCH1-GFP plasmid DNA. CCH1 transcripts were not detected in non-transfected HEK293 cells. Tubulin is shown as a loading control. Total RNA was isolated from HEK293 cells by standard protocols and used as template in the RT-PCR reaction. PCR amplified products were separated by agarose electrophoresis and visualized by ethidium bromide. HEK293 cells were transiently transfected with CCH1-GFP plasmid DNA as follows: approximately 1µg plasmid DNA was added to 100 µl, of Opti-MEM I reduced serum media and mixed with 1.75 µl of Lipotectamine 2000 and the mixture was incubated for 30 min at room temperature. HEK293 cells were maintained in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 4 mM L-Glutamine, 10% fetal bovine serum at 37ºC and 5% CO2. β) Localization studies revealed a predominant plasma membrane distribution of Cch1-GFP in HEK293 cells, similar to the localization pattern of a well-characterized plasma membrane-bound IK+ channel shown here as a plasma membrane marker, (DIC, differential interference contrast) (C). HEK293 cells were fixed in 4% paraformaldehyde for 20 min at room temperature and washed in PBS. The fluorescence label (GFP) was visualized by using filters S484/15 for excitation and S517/30 for emission.
The expression of the CCH1-GFP fusion protein in HEK293 cells was monitored by fluorescence microscopy 48 hr following transfection. The fluorescence images revealed the expression of CCH1 along the surface of the HEK293 cells (Figure 4B). This localization pattern was similar to that of the IK+ channel that was used here as a marker of the plasma membrane (Figure 4C). The IK-type channel represents a Ca2+-activated K+ channel with intermediate conductance [30]. These data demonstrate that CCH1 was localized to the plasma membrane of HEK293 cells and they support the functional viability of CCH1. Moreover we applied patch-clamp techniques to HEK293 cells and successfully measured CCH1 channel activity in these cells (data not shown). This provides definitive support for the functional activity of CCH1 in HEK293 cells. Collectively, the results demonstrate that the CCH1-GFP plasmid was successfully constructed and expressed in a mammalian cell line and they attest to the feasibility of the approach put forward in this study.
The limitations with other approaches
Originally the molecular cloning of CCH1 was tried using several different strains of E. coli as hosts. Despite having tried almost all commercially available E. coli strains capable of withstanding toxic genes, our extensive screens of thousands of colonies only yielded clones carrying empty plasmid and in some cases plasmids that contained small fragments of CCH1 (data not shown). Moreover we identified one potential positive transformant, which was later found to have carried a near complete CCH1 cDNA lacking a start codon (data not shown). These findings further underscore the necessity of sequencing every single positive clone to ensure that the open reading frame of any toxic gene is not altered by E. coli in their effort to cope with the toxicity of the gene. An additional disadvantage to using these specialized E. coli strains was that they often yielded a very low number of plasmids. Because the successful transfection of HEK293 cells depends on a sufficient concentration of plasmid DNA this requirement made it difficult to isolate enough plasmid using this method.
Once we were able to successfully clone CCH1 using both in vitro and in vivo recombination reactions, isolating sufficient amounts of CCH1 plasmid DNA from yeast for transfection in HEK293 cells remained a challenge. Since we could not use E. coli as a host to propagate large numbers of CCH1 plasmid DNA, we reasoned that perhaps E. coli might tolerate the CCH1 allele if it was expressed in a low-copy plasmid. These types of plasmids can be useful because they contain an origin of replication that maintains a very low-copy number (~ 5 to 20) per cell. Unfortunately this approach proved to be inefficient since it appeared that E. coli could not consistently tolerate even a low-copy plasmid carrying the CCH1 gene (data not shown). This is consistent with the results of a similar approach used to reconstitute ScCCH1 expression in S. cerevisiae [22]. Our results confirm that CCH1 is a highly toxic gene and any effort to clone it using either specialized E. coli strains or low-copy plasmids appears to be futile.
Another approach that was attempted involved a PCR based strategy known as PCR overlap where we generated a linear expression construct consisting of a CMV promoter, the CCH1-GFP sequence and a polyadenylation signal. This construct was then transfected directly into HEK 293 cells and ectopically integrated into the genome for functional analysis of Cch1 channel activity. This technique has been used extensively to generate targeting or disruption alleles in S. cerevisiae, C. neoformans and C. albicans because it eliminates the need for cloning and thereby increases high-throughput genetic analysis [26, 27, 28, 29]. In our case, PCR overlap was used to generate three fragments (CMV promoter, the CCH1-GFP sequence and a polyadenylation signal), each with short flanking regions homologous to one of the other fragments that were then used to generate the complete CCH1 expression allele. Regrettably, generating a sufficient amount of the CCH1-GFP linear expression construct for transfection into HEK293 cells proved to be extremely inefficient, especially for a large gene like CCH1 and therefore this approach was abandoned (data not shown).
In summary, we have devised a method that has led to the successful molecular cloning of CCH1 and for the first time the functional expression of CCH1 in a heterologous expression system. As a result of this method, a full-scale structure-function analysis of the CCH1 channel is now possible. The strategy described in this study will be particularly useful for the cloning and functional expression of other transporters and ion channels that cannot tolerate conventional methods requiring E. coli.
Acknowledgements
We are grateful to E. Blumwald for providing assistance with the fluorescence study of CCH1 expression in HEK293 cells. This study was supported by a National Institutes of Health grant awarded to A.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu M, Du P, Heinrich G, Cox GM, Gelli A. Cch1 mediates calcium entry in Cryptococcus neoformans and is essential in low-calcium environments. Eukaryot Cell. 2006;5:1788–1796. doi: 10.1128/EC.00158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Locke EG, Bonilla M, Liang L, Takita Y, Cunningham KW. A homolog of voltage-gated Ca2+ channels is stimulated by depletion of secretory Ca2+ in yeast. Mol Cell Biol. 200;20:6686–6694. doi: 10.1128/mcb.20.18.6686-6694.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hallen HE, Trail F. The L-type calcium ion channel Cch1 affects ascospore discharge and mycelial growth in the filamentous fungus Gibberella zeae (Anamorph Fusarium graminearum) Eukaryot. Cell. 2008;7:415–424. doi: 10.1128/EC.00248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer MN, Schnell N, Chattaway J, Davies P, Dixon G, Saunders D. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Lett. 1997;419:259–262. doi: 10.1016/s0014-5793(97)01466-x. [DOI] [PubMed] [Google Scholar]
- 5.Peiter E, Fischer M, Sidaway K, Roberts SK, Saunders S. The Saccharomyces cerevisiae Ca2+ channel Cch1Mid1 is essential for tolerance to cold stress and ion toxicity. FEBS Lett. 2005;579:5697–5703. doi: 10.1016/j.febslet.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 6.Casadevall A, Perfect JR. Cryptococcus neoformans. Washington, DC: American Society for Microbiology; 1998. [Google Scholar]
- 7.Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS – 100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bicanic T, Harrison TS. Cryptococcal meningitis. British Med. Bulletin. 2004;72:99–118. doi: 10.1093/bmb/ldh043. [DOI] [PubMed] [Google Scholar]
- 9.Iida H, Nakamura H, Ono T, Okumura MS, Anraku Y. Mid1a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein is required for Ca2+ influx and mating. Mol. Cell Biol. 1994;14:8259–8271. doi: 10.1128/mcb.14.12.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu M, Gelli A. Elongation factor 3, EF3, associates with the calcium channel Cch1 and targets Cch1 to the plasma membrane in Cryptococcus neoformans. Eukaryot Cell. 2008;7:1118–1126. doi: 10.1128/EC.00116-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen CBF, Becker T, Blau M, Anand M, Halic M, Balar B, Mielke T, Boesen T, Pedersen JS, Spahn CMT, Kinzy TG, Andersen GR, Beckmann R. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- 12.Blakely G, Hekman J, Chakraburtty K, Williamson PR. Evolutionary divergence of an elongation factor 3 from Cryptococcus neoformans. J Bacteriol. 2001;183:2241–2248. doi: 10.1128/JB.183.7.2241-2248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakraburtty K, Triana-Alonso FJ. Yeast elongation factor 3: structure and function. J. Biol. Chem. 1998;379:831–840. doi: 10.1515/bchm.1998.379.7.831. [DOI] [PubMed] [Google Scholar]
- 14.Colthurst DR, Schauder BS, Hayes MV, Tuite MF. Elongation factor 3 (EF3) from Candida albicans shows both structural and functional similarity to EF-3 from Saccharomyces cerevisiae. Mol. Microbiol. 1992;6:1025–1033. doi: 10.1111/j.1365-2958.1992.tb02168.x. [DOI] [PubMed] [Google Scholar]
- 15.Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 16.Lee SH, Soderlund DM. Cloning and maintenance of the housefly sodium channel gene using low copy number vector and two sequential host strains. J Asia-Pacific Entomology. 2008;12:51–53. [Google Scholar]
- 17.Mjk1, a K+ channel from M. jannaschii, mediates K+ uptake and K+ sensitivity in E. coli. 2003. FEBS Letters. 2003;547:165–169. doi: 10.1016/s0014-5793(03)00706-3. [DOI] [PubMed] [Google Scholar]
- 18.Sun S, Gan JH, Paynter JJ, Tucker SJ. Cloning and functional characterization of a superfamily of microbial inwardly rectifying potassium channels. Phsiol. Genomics. 2006;26:1–7. doi: 10.1152/physiolgenomics.00026.2006. [DOI] [PubMed] [Google Scholar]
- 19.Paidhungat M, Garrett S. A homolog of mammalian, voltage-gated calcium channels mediates yeast pheromone-stimulated Ca2+ uptake and exacerbates the cdc1(Ts) growth defect. Mol Cell Biol. 1997;17:6339–6347. doi: 10.1128/mcb.17.11.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: A model system for the study of recombination. Proc Natl Acad Sci. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–206. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 22.Iida K, Tada T, Iida H. Molecular cloning in yeast by in vivo homologous recombination of the yeast putative a1 subunit of the voltage-gated calcium channel. FEBS Letters. 2004;576:291–296. doi: 10.1016/j.febslet.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 23.Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, D’Souza C, Wang P, Heitman J. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- 24.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterlogous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 25.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 26.Lorenz MC, Muir RS, Lim E, McElver J, Weber SC, Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- 27.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eberhardt I, Hohman S. Strategy for deletion of complete open reading frames in Saccharomyces cerevisiae. Curr Genet. 1995;27:306–308. doi: 10.1007/BF00352097. [DOI] [PubMed] [Google Scholar]
- 30.Vergara C, Latorre R, Marrion NV, Adelman JP. Calcium-activated potassium channels. Curr Opin Neurobiol. 1998;8:321–329. doi: 10.1016/s0959-4388(98)80056-1. [DOI] [PubMed] [Google Scholar]
- 31.Hamil OP, Marty A, Neher E, Sakman B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 32.Singh MV, Weil PA. A method for plasmid purification directly from yeast. Analyt Biochem. 2002;307:13–17. doi: 10.1016/s0003-2697(02)00018-0. [DOI] [PubMed] [Google Scholar]