SUMMARY
Pluripotent stem cell lines can be derived from blastocyst embryos, which yield embryonic stem cell lines (ES cells), as well as the post-implantation epiblast, which gives rise to epiblast stem cell lines (EpiSCs). Remarkably, ES cells and EpiSCs display profound differences in the combination of growth factors that maintain their pluripotent state. Molecular and functional differences between these two stem cell types demonstrate that the tissue of origin and/or the growth factor milieu may be important determinants of the stem cell identity. We explored how developmental stage of the tissue of origin and culture growth factor conditions affect the stem cell pluripotent state. Our findings reveal that novel stem cell lines can be generated from blastocyst embryos with unique functional and molecular properties. We demonstrate that the culture growth factor environment and cell-cell interaction play a critical role in defining several unique and stable stem cell ground states.
INTRODUCTION
Following fertilization, the totipotent zygote undergoes rapid cleavage divisions to form a preimplanation blastocyst embryo, a hollow sphere in which two different cell types can be identified. An outer layer of trophectoderm cells enclosing a small group of pluripotent cells known as the inner cell mass (ICM), from which the embryo proper will develop. At implantation, the ICM forms the extraembryonic endoderm and the epiblast, consisting of pluripotent cells that give rise to all embryonic germ layers. It was the pioneering work of Martin and Evans that demonstrated that cells in the ICM can be propagated indefinitely in vitro in a stable pluripotent state as embryonic stem (ES) cell, while maintaining the ability to generate all tissues of the adult body (Evans and Kaufman, 1981; Martin, 1981; Martin and Evans, 1975). The subsequent derivation of human ES cells sparked the hope that the unique properties of these cells could be harnessed to enable regenerative therapies and advance our understanding of human development (Thomson et al., 1998). Like their murine counterparts, human ES cells can be propagated indefinitely in vitro, and demonstrate the ability to generate derivatives of all three germ layers. Human ES cells were long thought to be the homo sapiens equivalent to murine ES (mES) cells, despite clear morphological differences and different growth factor requirements between these two ES cell types.
The recent derivation of Epiblast Stem Cells (EpiSCs) from post-implantation epiblasts provides a new perspective on the nature of human ES cells (Brons et al., 2007; Tesar et al., 2007). At the molecular level EpiSCs are much more similar to human ES cells than mES cells. EpiSCs display a flattened 2-D colony morphology, which is also characteristic for human ES cells, and are maintained under similar growth factor conditions. The close match between EpiSCs and human ES cells suggests a functional similarity between these cells. EpiSCs display many characteristic hallmarks of pluripotent stem cells such as the expression of Oct4, Sox2, Nanog and the ability to generate derivatives of all three germ layers both during in vitro differentiation and in vivo teratoma formation. But interestingly, EpiSCs fail to contribute to chimera formation when injected into recipient blastocysts.
The above comparisons of mES cells, human ES cells and EpiSCs illustrate that stem cell pluripotency is not a fixed ground state, but is strongly influenced by developmental-and environmental context. Distinct pluripotent stem cell lines with unique functional characteristics can be derived from different parts of the embryo and under different growth factor conditions. For example, the functional differences in developmental potential between mES cells and EpiSCs may reflect the tissue of origin from which the stem cell line is initially derived; inner cell mass vs. epiblast, or may be a consequence of their different culture conditions. After all, mES cells require a combination of Leukemia Inhibitory Factor (LIF) and Bone Morphogenetic Protein 4 (BMP4) to maintain their undifferentiated state (Ying et al., 2003), while the factors that support murine EpiSC or human ES cell self-renewal are a combination of bFGF, ActivinA or TGFβ and activation of the Wnt signaling pathway (Brons et al., 2007; Carpenter et al., 2004; Denning et al., 2006; Mallon et al., 2006; Rosler et al., 2004; Tesar et al., 2007; Xu et al., 2005).
To dissect the effect of the growth factor milieu and the developmental age of the tissue of origin on the stem cell pluripotent state, we derived novel stem cell lines from murine blastocyst embryos in culture conditions previously applied to derivation of EpiSCs from epiblast stage embryos. We designated these cells FAB-SCs for bFGF, Activin and BIO-derived stem cells. We demonstrate that FAB-SCs are molecularly and functionally distinct from both ES cells and EpiSCs. FAB-SCs express common molecular markers of stem cell pluripotency, Oct4, Nanog and Sox2, but unexpectedly, the cells fail to pass hallmark tests of pluripotent differentiation such as in vitro embryoid body formation, teratoma formation or contribution to embryonic development upon blastocyst transplantation. However, brief (transient) stimulation of FAB-SCs with LIF and BMP4 induces the potential to generate teratomas, and give germline contribution in chimeric mice. Our study provides new insights into the role of growth factor environment in reprogramming of the stem cell pluripotent state and identifies an unexpected role for cell-cell adhesion in this process.
RESULTS
Derivation and characterization of blastocyst stem cells
To analyze the role of the developmental stage of the embryo on the developmental potential of embryo-derived stem cells, we explored whether novel cell lines could be derived from blastocysts under similar growth factor conditions as previously described for EpiSC- and human ES cell cultures (bFGF, ActivinA and BIO). Initial experiments using matrigel as substrate were unsuccessful, but when instead, blastocysts were hatched on MEFs in the presence of bFGF, ActivinA and BIO and a blocking antibody against murine LIF we were able to derive stable cell lines with novel properties, which we designated FAB-SCs, for bFGF, Activin and BIO-derived stem cells. If the zona pellucida was left intact, only 10% of the blastocysts hatched and upon trypsinization and passaging of the ICM outgrowths, a third of the hatched blastocysts yielded stable cultures that were homogenous in appearance (n=184). Removal of the zona pellucida prior to plating of the embryos improved the derivation frequency significantly, as 80% of the embryos demonstrated robust ICM outgrowth and upon passaging 30% of the original blastocysts yielded stable FAB-SC lines (n=99). It is interesting to note that under FAB-SC conditions ICM expansion was noticeable within 2 days after plating. In contrast, when blastocysts are plated under mES cell conditions, ICM expansion is delayed and occurs several days later. The difference in ICM outgrowth is not due to differences in cell proliferation rates, since FAB-SCs and mES cell proliferation rates are similar (not shown). Instead, the delay in ICM outgrowth under ES cell conditions may indicate that ES cell lines are derived from a small subpopulation of cells within the ICM while FAB-SC conditions allow the entire ICM to expand. Alternatively, the delayed ICM outgrowth in ES cell conditions may reflect a pause in cell proliferation associated with epigenetic reprogramming events that are essential for the derivation of mES cells (Kaji et al., 2006). While speculative, this latter option would imply that such reprogramming does not occur under FAB-SC conditions.
FAB-SCs share features with EpiSCs and mES cells, yet are distinct from both
Unlike mES cell colonies, which have a characteristic 3-dimensional appearance of tight shiny colonies, FAB-SCs grew as monolayer colonies reminiscent of EpiSCs (Figure 1A). Q-PCR analysis of Oct4, Sox2 and Nanog demonstrated that all three pluripotency transcription factors were expressed in FAB-SC lines as well as in traditional mES cells (Figure 1B). We further confirmed the homogeneous expression and nuclear localization of these transcription factors using immunohistochemistry (Figure 1C). In addition, we confirmed the expression of the cell surface marker SSEA1 on the FAB-SCs (not shown) further demonstrating that the FAB-SC cultures homogeneously express molecular hallmarks of pluripotent cells. Although FAB-SCs are derived on a feeder layer of MEFs, in the presence of a blocking antibody to LIF, we cannot formally exclude that very low residual levels of LIF signaling are required for FAB-SC derivation. However, established FAB-SC lines can be maintained on gelatin or Matrigel coated dishes in serum-free media in the absence of LIF or BMP4, with sustained expression of pluripotent markers, demonstrating that these growth factors are not required for the maintenance of these cells. Finally, as we will demonstrate below, stimulation of FAB-SCs with LIF and BMP4 induces profound permanent phenotypic changes in these cells, arguing that FAB-SCs do not experience these growth factors during their derivation.
Figure 1. Blastocyst-derived stem cells (FAB-SC).
FAB-SCs were derived as described in the results section. A: Brightfield image of FAB-SCs (Top panel) and mES cells (bottom panel) B: Q-PCR expression analysis of Oct4, Sox2 and Nanog expression on FAB-SC and ES cell as indicated. C: Left panels: Immunofluorescence staining of FAB-SCs for Oct4, Sox2 and Nanog as indicated. Right panels: DAPI nuclear staining. D. Hierarchy clustering of MicroRNA profiles of MEF, mES, EpiSC and FAB-SC cell lines. E. Normalized expression intensity values (scaled median ratio) were obtained from Agilent whole-genome microarrays. Three biological replicates were used for all three cell types.
FAB-SCs are derived under culture conditions similar to those recently reported for the derivation of murine EpiSCs (Brons et al., 2007; Tesar et al., 2007), but FAB-SCs and EpiSCs originate from different developmental stages of the embryo. To chart similarities and differences among FAB-SCs, ES cells and EpiSCs, we performed global gene- and microRNA (miRNA) expression analysis on these cells.
Using a Luminex bead platform (Lu et al., 2005), we analyzed the expression levels of >430 miRNAs in independent ES, EpiSC and FAB-SC clones. Heatmap analysis of these samples revealed clear differences in the global miRNA profiles of these three cell lines (Figure 1D). Interestingly, FAB-SCs express miRNAs recently shown to be ES cell specific, such as the miR-290 cluster (Houbaviy et al., 2003), or enriched in self-renewing ES cells, such as miR-18a, miR-19a and miR- 20a (Hayashi et al., 2008). In contrast, EpiSCs express low levels of these miRNAs. These data demonstrate that FAB-SCs express miRNAs typical of ES cells, reflecting the blastocyst origin of these cells. EpiSCs on the other hand, express several miRNAs associated with post-implantation development including miR-1 and miR-206 (associated with muscle development), and miR-150 and miR-142 (hematopoietic differentiation) (Chen et al., 2004; Xiao et al., 2007). The microRNA expression profiling of these pluripotent stem cell lines underscores the unique character of FAB-SCs and reflects their early developmental origin. EpiSCs on the other hand demonstrate the expression of miRNAs associated with early lineage commitment, in line with the post-implantation epiblast origin of these cells.
To further interrogate the similarities and differences between FAB-SCs, mES cells and EpiSCs, we performed microarray analysis of the transcriptional profile of these different cell lines. FAB-SCs express several known pluripotency factors, including Oct4, Sox2 and Nanog, at similar levels as mES and EpiSCs (Figure 1E). Yet, the expression of epiblast markers is absent or low in FAB-SCs compared to EpiSCs, demonstrating that while these cells are propagated under similar growth factor conditions, they are not the same (Figure 1E). FAB-SCs distinguish themselves from ES cells as well, as they do not express many of the genes associated with germ cell differentiation, such as Stella, Blimp1 or Dazl, which are commonly expressed in mES cells (Figure 1E). The mRNA expression analysis further demonstrates that FAB-SCs represent an alternative stable stem cell state that is distinct from both mES cells and EpiSCs.
FAB-SCs fail to demonstrate pluripotency in assays of development
We next tested the ability of FAB-SCs to generate derivatives of all three germ layers in in vitro and in vivo assays of development. Embryoid body (EB) formation is a simple and widely used method in which aggregates of pluripotent cells initiate a differentiation program that is reminiscent of early embryonic development (Doetschman et al., 1985; Leahy et al., 1999). In the context of the EB, molecular interactions that drive early embryonic development are recapitulated and cells differentiate to form ectoderm, endoderm and mesoderm derivatives.
Surprisingly, and in stark contrast to ES cells, EBs made from FAB-SCs remained small and failed to expand. We next interrogated the ability of FAB-SCs to form teratomas when injected into immunodeficient mice. FAB-SCs failed to form any teratomas at 3 months after injection (n=20) whereas teratomas formed within 1 month in all mice injected with ES cells (n=5).
Pluripotency is characterized by the ability of a stem cell to self renew indefinitely while maintaining the capacity to differentiate into derivatives of all three germ layers. While FAB-SC cultures display sustained expression of hallmark molecular markers of pluripotency >30 passages (Figure 1C), the cells fail to pass standard in vitro and in vivo tests of pluripotency such as EB differentiation or teratoma formation. As such, FAB-SCs are also molecularly and functionally distinct from primitive epiblast-like cells (EPL cells) that are derived when ES cells are cultured in the presence of HEPG2 conditioned medium (Rathjen et al., 1999). Epiblast marker genes, which are expressed in EPL cells are low or absent in FAB-SCs and in contrast, FAB-SCs express the ICM marker Gbx2 and EPL cells do not. In addition, EPL cells are capable of forming teratomas, while FAB-SCs are not, excluding the possibility that FAB-SCs are EPL cells.
Growth factor stimulation induces FAB-SC pluripotency
To examine the influence of the growth factor milieu on FAB-SC pluripotency, we explored the effect of LIF and BMP4 stimulation on the ability of FAB-SCs to generate teratomas (Figure 2A).
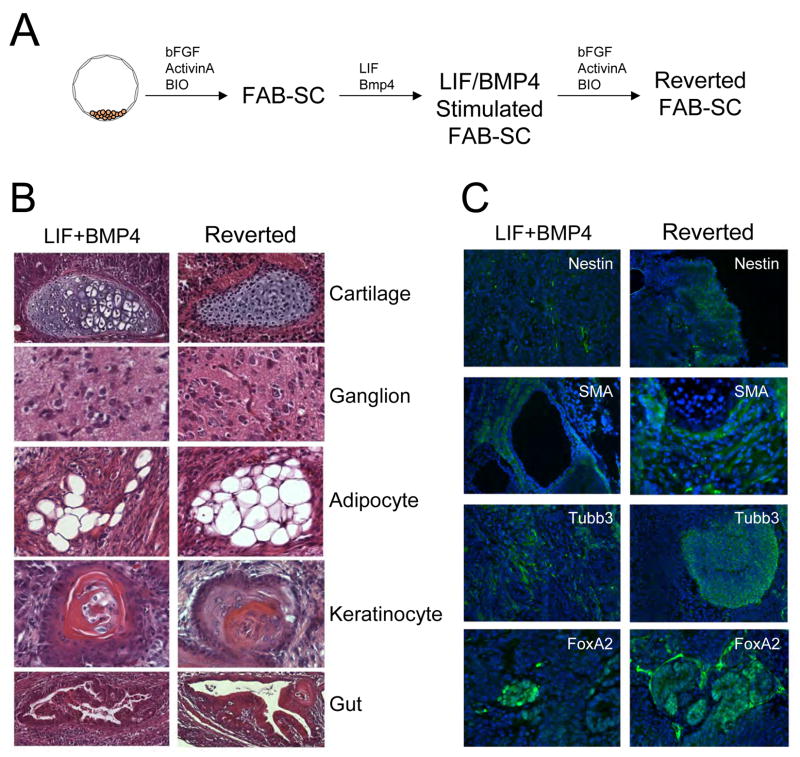
Figure 2. LIF and BMP4 stimulate FAB-SC teratoma formation.
A: Schematic representation of FAB-SC derivation (with bFGF/ActivinA and BIO), LIF/BMP4 stimulated FAB-SCs cultured in the presence of LIF and BMP4 and Reverted FAB-SCs which are again maintained in bFGF/ActivinA and BIO B. H&E staining of teratomas generated from a clonal FAB-SC line stimulated with LIF/BMP4 (Left panels) or stimulated and subsequently cultured for 7 days in FAB-SC conditions (Right panels). Derivatives of all three germ layers are observed as indicated. C: Immunohistochemistry analysis of markers of ectoderm (Nestin, Tubulin β3 (Tubb3)), mesoderm (Smooth Muscle Actin, (SMA) or endoderm (FoxA2).
To ensure a homogeneous FAB-SC population, we used flow-cytometry to sort single cell clones of FAB-SCs containing a GFP transgene into 96-well plates, and visually confirmed the presence of a single cell in each well. Clonal FAB-SCs were stimulated with LIF (100ng/ml) and BMP4 (40 ng/ml) for 1 week and injected subcutaneously into NOD-SCID mice (1×106 cells, n=7) to assess their teratoma-forming potential. While none of the native FAB-SC clones gave rise to teratomas, injection of LIF/BMP4 stimulated FAB-SC clones resulted in the formation of teratomas in all recipients (3 independent clones, n=7 for each clone). H&E staining and immunofluorescent detection of markers of germlayer differentiation demonstrated that the teratomas generated by the LIF/BMP4 stimulated FAB-SCs displayed derivatives of all three embryonic germlayers (Figure 2B and C, left panels), demonstrating that brief culture in LIF/BMP4 induces FAB-SC developmental potential. Interestingly, when the LIF/BMP4 stimulated FAB-SCs were returned back to the original FAB-SC growth factor conditions and cultured for another week, these “reverted” FAB-SCs retained the ability to generate teratomas (Figure 2, B and C, right panels). Thus, the FAB-SC pluripotent state, induced by transient LIF/BMP4 stimulation, is retained even when the LIF/BMP4 stimulus is subsequently removed.
Blastocyst contribution by FAB-SCs
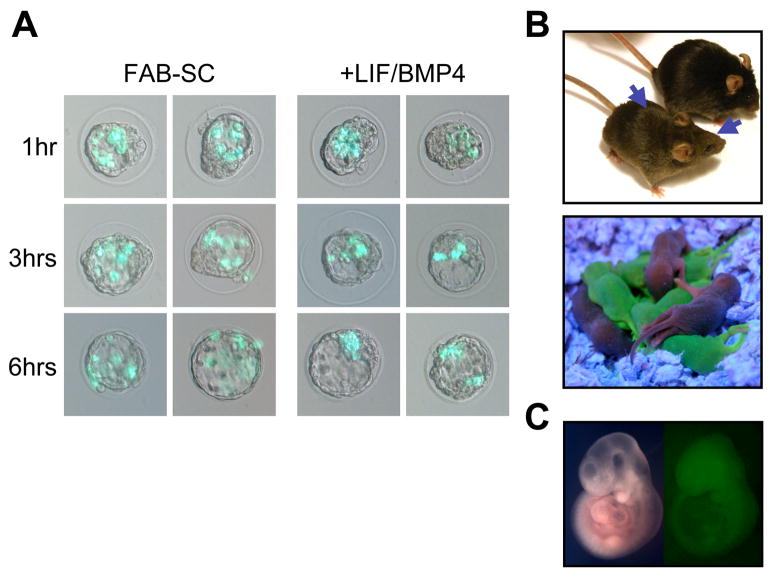
To further explore our observation that LIF/BMP4 stimulation induced FAB-SC pluripotency, we examined the effect of growth factor stimulation on the ability of FAB-SCs to form chimeras upon transfer into recipient blastocysts. Control- and LIF/BMP4 stimulated FAB-SCs, expressing the GFP transgene described above, were injected into blastocyst embryos and their integration into the recipient blastocyst was monitored over time. While the LIF/BMP4-exposed FAB-SCs integrated with the cells of the ICM within 6 hours after injection, unstimulated FAB-SCs remained dispersed in the blastocyst cavity, revealing that integration of FAB-SCs into the recipient embryos was impaired (Figure 3A).
Figure 3. Growth factor stimulation induces FAB-SC chimera formation and germline contribution.
A Temporal analysis of the integration of GFP-transgenic FAB-SC before and after LIF/BMP4 stimulation into recipient blastocysts. Integration of FAB-SCs and LIF/BMP4 stimulated FAB-SCs was monitored at 1, 3 and 6 hours after injection as indicated. B: Top panel, high contribution chimera derived from LIF/BMP4 stimulated FAB-SC (48 hr stimulation). Middle panel: GFP+ offspring of the FAB-SC chimera, demonstrating germline transmission. Bottom panel: Chimera from clonal FAB-SCs stimulated for 7 days with LIF/BMP4; arrows indicate agouti coat color chimerism.
To further analyze the developmental potential of FAB-SCs before and after growth factor stimulation, we transferred the embryos into pseudopregnant females and analyzed chimerism by the expression of the GFP transgene and/or coat color. No chimerism was observed in any of the >320 pups from blastocysts injected with 10–12 FAB-SCs each. Sectioning and immunohistochemistry staining of the embryos using an anti-GFP antibody revealed no GFP contribution to the recipient embryos at mid-gestation (E9.5–E11.5). In contrast, even brief 48 hour stimulation of FAB-SCs with LIF and BMP4 induced the ability of these cells to form chimeras. Although the chimeric frequency was low (7 out of 254 pups), the chimeras demonstrated high (40–90%) FAB-SC contribution (Figure 3B, top panel). Mating with a wild-type female showed transmission of the FAB-SC-derived GFP transgene to the offspring, demonstrating that LIF/BMP4 stimulated FAB-SCs are capable of germline contribution as well (Figure 3B, middle panel). These embryo chimerism experiments were repeated with single cell-derived clonal FAB-SC lines. Again, no chimerism was observed upon blastocyst transfer of clonal FAB-SCs (n=160). However, LIF/BMP4 stimulation of clonal FAB-SCs for 7 days induced the ability of these cells to contribute to recipient embryos with a frequency similar to the parental cell line (not shown).
Finally, we analyzed whether FAB-SCs exposed to LIF/BMP4 for 1 week would contribute to chimera formation after further culture in bFGF, ActivinA and BIO. Clonal LIF/BMP4 stimulated FAB-SCs were cultured for an additional 7 days in FAB-SC media containing bFGF, ActivinA, BIO and anti-LIF antibody, and subsequently injected into recipient blastocyst embryos. GFP contribution by the injected reverted FAB-SCs was detected in 4 out of 123 embryos analyzed, with chimerism ranging from 20–80% (Figure 3C), demonstrating that pluripotency is retained even after removal of the LIF/BMP4 signal.
The above data demonstrate that LIF/BMP4 stimulation of FAB-SCs induces their ability to contribute to chimera formation. The relatively low number of chimeras obtained suggests that only a fraction of the FAB-SCs undergoes full growth-factor mediated conversion to the pluripotent state. Robust contribution by cells that do successfully undergo pluripotent conversion, including contribution to the germline, indicates however that the induced cells are truly pluripotent. Importantly, the ability of FAB-SCs to display this effect at the clonal level demonstrates that the LIF/BMP4 stimulated chimera formation is the result of an induction of FAB-SC pluripotency rather than clonal selection of “competent” cells from a heterogeneous starter population.
E-Cadherin is induced by transient LIF/BMP4 stimulation of FAB-SCs
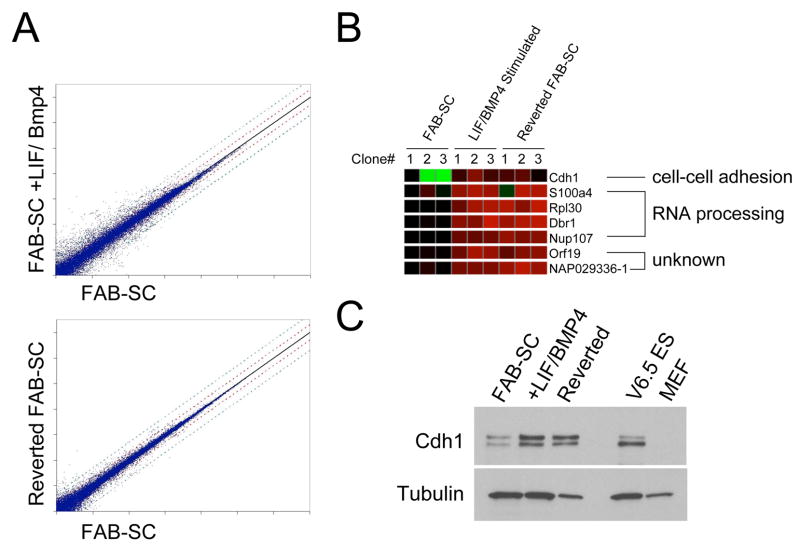
The unique properties of FAB-SCs allowed us to further probe the molecular mechanism behind the induction of FAB-SC pluripotency by growth factor stimulation. Microarray comparison of the FAB-SC gene expression profile of three independent clonal FAB-SC lines, with the expression profile of LIF/BMP4 stimulated FAB-SCs demonstrated profound changes in gene expression (Figure 4A, upper panel). Comparison of FAB-SCs to cells that were transiently stimulated with LIF/BMP4 and subsequently reverted to the original growth conditions of bFGF, ActivinA and BIO enabled us to focus on gene expression changes linked to the induction of pluripotency (Figure 4A, lower panel). We compared the gene expression profiles of FAB-SCs, the LIF/BMP4 stimulated FAB-SC and the growth factor reversed FAB-SCs and searched for genes that were up- or downregulated at least 3-fold by LIF/BMP4 stimulation, and remained altered in the reverted FAB-SCs. Only a handful of genes displayed this expression profile consistently in 3 independent FAB-SC clones (Figure 4B). Four “hits” were genes involved in RNA translation and two were genes with unknown function. Cdh1 (E-Cadherin) displayed the most profound induction, as it demonstrated a 4–6 fold upregulation on all seven features of the microarray. Western analysis of E-Cadherin expression in FAB-SCs, LIF/BMP4 stimulated FAB-SCs and reverted FAB-SCs demonstrated that while FAB-SCs express low levels of E-Cadherin, LIF/BMP4 stimulation resulted in upregulation of E-Cadherin expression to comparable levels as those observed in mES cells, which was sustained in the reverted FAB-SCs (Figure 4C).
Figure 4. E-Cadherin (Cdh1) is induced by LIF/BMP4 stimulation of FAB-SC.
A: microarray comparison of gene expression of FAB-SC and LIF/BMP4 stimulated FAB-SC (Upper panel) or FAB-SC and growth factor reversed FAB-SC (Lower panel). B. Heatmap of genes permanently upregulated by LIF/BMP4 stimulation of FAB-SC. C: Western blot analysis of E-Cadherin expression in FAB-SC, LIF/BMP4 stimulated FAB-SC and growth factor reverted FAB-SC.
E-Cadherin is a critical regulator of FAB-SC pluripotency
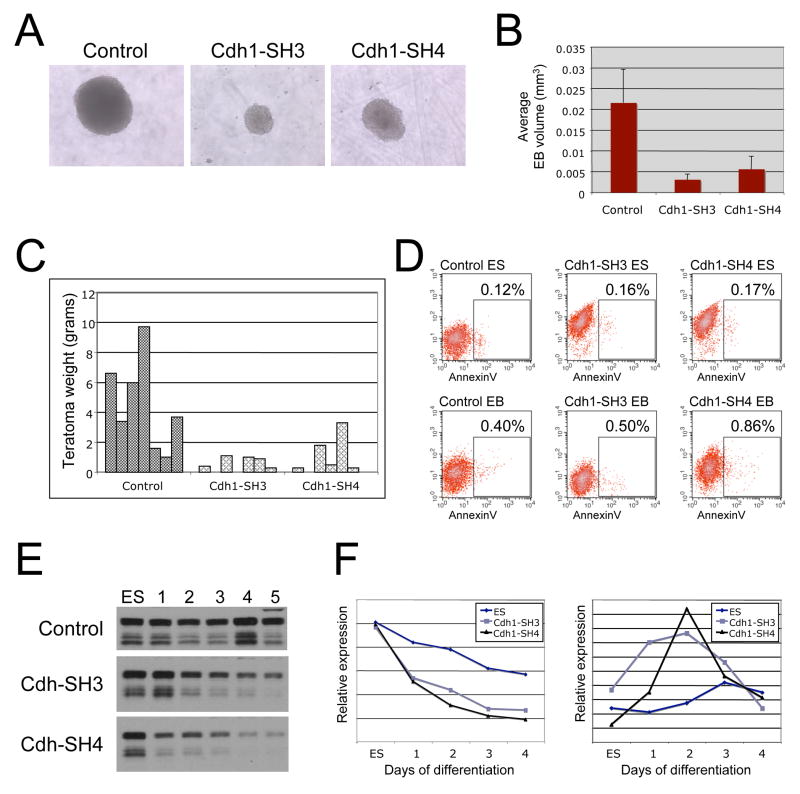
To further analyze the function of E-Cadherin in pluripotent stem cells, we generated lentiviral shRNA hairpins to examine the effect of downregulation of E-Cadherin expression on the ability of FAB-SCs to generate teratomas. shRNA hairpins were tested to functionally downregulate E-Cadherin expression in mES cells (Supplemental figure 1A). While E-Cadherin expression was unaffected in the control hairpin, two E-Cadherin hairpins (Cdh1-SH3 and Cdh1-SH4) demonstrated >90% downregulation of E-Cadherin expression and were selected for further experiments. Functional knockdown of E-Cadherin expression was tested in a cell aggregation assay. Cadherins mediate cell-cell adhesion through homotypic interactions. Two cell populations expressing similar levels of the same cadherin will form homogeneous aggregates, while dissimilarities in the nature or level of cadherin expression will result in segregation of the two cell types (Gibralter and Turner, 1985; Takeichi et al., 1981). While ES cells transduced with the control shRNA hairpin formed homogeneous aggregates when mixed with wild-type ES cells, ES cells expressing the E-Cadherin hairpins did not mix with the wild-type cells, but instead aggregated in spatially separate domains, demonstrating functional consequences of E-Cadherin knock-down in these cell lines (Supplemental figure 1B). Next we explored the effect of knockdown of E-Cadherin expression on LIF/BMP4 stimulated FAB-SCs, but we were unable to establish stable clones using the E-Cadherin knockdown hairpins. Transduction of LIF/BMP4 stimulated FAB-SCs with the control vector yielded stable clones expressing the tdTomato reporter gene present in the lentiviral shRNA vector (Figure 5A). Loss of E-Cadherin has been reported to induce anoikis mediated apoptosis in certain cancer cell lines, yet no difference in apoptosis was observed between the control and E-Cadherin knockdown samples (not shown). When we analyzed the transduced cells by fluorescent microscopy however, we observed a striking difference in the morphology of the E-Cadherin knockdown cells compared to control. In the control sample, tdTomato-positive (Lentiviral transduced) cells proliferated as undifferentiated colonies (Figure 5A, lower panels). In contrast, stimulated FAB-SC cells transduced with the E-Cadherin hairpins had a fibroblast-like morphology, demonstrating that downregulation of E-Cadherin resulted in rapid FAB-SC differentiation (Figure 5A, upper panels, arrows).
Figure 5. E-Cadherin regulates FAB-SC pluripotency.
A FAB-SCs constitutively expressing a GFP transgene were transduced with control vector or shRNA to knock down E-Cadherin. A tdTomato reporter gene was co-expressed from the lentiviral shRNA vector to allow identification of knockdown cells. Cdh1-knockdown results in FAB-SC differentiation, middle top panel, arrowhead. B. FAB-SCs were transduced with either control vector or Cdh1 expression vector and 1 × 106 cells were injected subcutaneously into NOD-SCID mice. Tumors were analyzed for germlayer differentiation 1 month after injection of the cells. Top panels: H&E staining of teratomas generated from Cdh1-FAB-SCs. I: Keratinocyte, II: Adipocyte, III: Gut. Lower panels: Immunohistochemistry analysis of markers of IV: Nestin, ectoderm, V: Smooth Muscle Actin, mesoderm and VI: FoxA2, endoderm.
Induction of FAB-SC pluripotency by ectopic expression of E-Cadherin
The upregulation of E-Cadherin expression in FAB-SCs following LIF/BMP4 stimulation correlates with the induction of the ability of these cells to form teratomas containing derivatives of all three germ layers. We next examined whether this upregulation of E-Cadherin expression is sufficient to induce the teratoma forming potential of FAB-SCs. FAB-SCs transduced with a lentiviral vector expressing E-Cadherin formed teratomas in immunodeficient mice after one month (5 out of 7), whereas none of 7 mice injected with control FAB-SCs developed teratomas. H&E staining and immunofluorescent detection of markers of germlayer differentiation demonstrated that the teratomas generated by the E-Cadherin FAB-SCs displayed derivatives of all three embryonic germ layers (Figure 5B). These data demonstrate that overexpression of E-Cadherin alone is sufficient to induce robust teratoma-forming potential in FAB-SCs and suggest that a key target of LIF/BMP4 stimulation is upregulation of E-Cadherin expression.
Accelerated ES cell differentiation in the absence of E-Cadherin expression
Above data demonstrate that E-Cadherin plays an important role in regulating the FAB-SC pluripotent state. To test whether abrogation of E-Cadherin expression would compromise the pluripotency of mES cells, we generated stable mES cell lines expressing the control or Cdh1 knockdown hairpins. Downregulation of E-Cadherin expression in mES cells changes the morphology of the cells from the tight, 3-dimensional colony shape to a more flattened colonies of loosely connected cells, very much resembling FAB-SCs (Supplemental Figure 2A). ES cells stably expressing the E-Cadherin hairpins could be propagated for over 20 passages and the proliferation and apoptotic rates between the control and E-cadherin knockdown ES cell lines were comparable (see Figure 6D below) suggesting knockdown of E-cadherin did not impair ES self-renewal, proliferation or apoptosis. Using immunohistochemistry we analyzed the expression of Oct4 in the control and E-Cadherin knockdown ES cell lines (Supplemental Figure 2B). No differences in the level, localization or percentage of Oct4 expression were observed between control and E-Cadherin knockdown ES cells. Furthermore, even cells that demonstrated a rounded-up clustered colony morphology were expressing Oct4 (Arrowheads, supplemental Figure 2B). Thus, while knockdown of E-Cadherin expression in traditional mES cells results in a change in morphology and loss of tight adhesion of the ES cells, loss of E-Cadherin does not impair ES cell self-renewal. Like FAB-SCs, the E-Cadherin knock-down ES cells formed small EBs (Figure 6A and B). Moreover, whereas E-Cadherin knock-down ES cells form teratomas upon subcutaneous injection into NOD-SCID mice, loss of E-Cadherin expression results in a profound reduction in teratoma size, as measured by the weight of the teratomas (Figure 6C). Analysis of the teratomas revealed multilineage differentiation however, indicating that differentiation of the Cdh1 knockdown ES cells per se was not impaired (not shown). The reduced size of the E-Cadherin knock-down EBs was not due to decreased proliferation or increased apoptosis of these cells (Figure 6D). Finally we analyzed the expression of pluripotency markers Oct4, Sox2 and Nanog before and during differentiation of control ES cells or the E-Cadherin knockdown cell lines. While we observed no difference in the downregulation of Oct4 and Sox2 expression, Western blot analysis revealed an accelerated loss of Nanog protein expression in the absence of E-Cadherin expression as compared to control cells (Figure 6E). The rapid loss of Nanog expression in the E-Cadherin knockdown cells was also observed when we monitored Nanog RNA levels by Q-PCR (Figure 6F). Oct4, Sox2 and Nanog have been shown to bind to promoter elements of genes involved in early cell fate decisions (Boyer et al., 2005; Boyer et al., 2006). The concerted binding of these transcription factors mediates the recruitment of the polycomb silencing complex, thereby suppressing gene expression. Thus part of the role Oct4, Sox2 and Nanog play in ES cell self-renewal is to prevent the expression of genes associated with differentiation, such as genes in the Hox-cluster. Since loss of E-Cadherin results in early downregulation of Nanog upon cell differentiation, we analyzed the expression of HoxA1 and HoxB1 in these cells. Upregulation of HoxA1 and HoxB1 is a hallmark sign of cell differentiation toward the somatic lineages and distinguishes pluripotent cells from early somatic cells in the primitive epiblast (Saitou et al., 2002; Yabuta et al., 2006). While we observed no upregulation of HoxA1 expression during early differentiation of control, or E-Cadherin knockdown ES cells, HoxB1 expression was upregulated prematurely in both E-Cadherin knockdown cell lines compared to control (Figure 6F). Together our data demonstrate that loss of E-Cadherin expression results in rapid differentiation of FAB-SCs, and while mES cells can maintain their pluripotent state in the absence of E-Cadherin expression, they demonstrate an accelerated loss of Nanog expression and premature upregulation of HoxB1 expression upon induction of differentiation.
Figure 6. Loss of E-Cadherin expression compromises EB- and teratoma formation by accelerating ES cell differentiation.
A: Representative images demonstrating EB differentiation of wild type and E-Cadherin knockdown ES cells. Loss of Cadherin expression results in small EB size compared to the wild-type EBs. B: Bar-graph analysis of the size of Wild-type or E-Cadherin knockdown EBs C: The effect of loss of E-Cadherin expression on teratoma formation. Plotted is the weight (in grams) of seven teratomas of wild-type and seven each of two independent E-Cadherin knockdown ES cell lines. Loss of E-Cadherin results in a profound reduction in teratoma size. D: Analysis of apoptotic frequency in control and E-Cadherin knockdown cells. Stable ES cell lines transduced with control vector or two independent E-Cadherin knockdown hairpins were analyzed by flow cytometry for AnnexinV staining. The ES cells expressed a tdTomato fluorescent reporter gene to distinguish them from the MEF feeder cells. Top panels: AnnexinV-FITC staining of control and two E-Cadherin knockdown ES cell lines. The percentage of AnnexinV-positive cells in indicated. Bottom panels: AnnexinV staining of the same cell lines after 7 days of monolayer differentiation of the cells. E: E-Cadherin downregulation results in early loss of Nanog expression and premature upregulation of HoxB1. Western-blot analysis of Nanog protein expression during differentiation of control ES cells (top) and two independent E-Cadherin knockdown ES cell lines (middle and lower panels). Numbers indicate days of differentiation. F: Left: Q-PCR analysis of the Nanog RNA levels in the same samples. Right: Q-PCR analysis of HoxB1 expression in the same samples.
DISCUSSION
We have derived a novel stem cell line from murine blastocyst embryos using growth factor conditions previously reported for murine EpiSCs and human ES cells (Brons et al., 2007; Ginis et al., 2004; Tesar et al., 2007). Transcriptome and miRNA expression analyses demonstrate that EpiSCs, mES cells and now FAB-SCs represent distinctive stem cell lines derived from early murine embryos. Our data demonstrate that stem cells can exist in a number of distinct metastable epigenetic states, that each display unique phenotypic properties which are determined by the growth factor environment and the developmental stage of the embryo from which the cell line is derived (Table 1). FAB-SCs may be trapped in a state of “partial pluripotency”, akin to the partially reprogrammed states identified in cultures of induced pluripotent stem cells (iPS cells). In a seminal paper Takahashi and Yamanaka demonstrated that introducing four transcription factors, Oct4, Sox2, c-Myc and Klf4, is sufficient to reprogram fibroblasts into pluripotent stem cells (Takahashi and Yamanaka, 2006). The authors selected successfully reprogrammed iPS cells by their re-expression of an ES cell specific Fbx15 reporter. While these initial iPS cells displayed the ability to generate teratomas with derivatives of all three germ layers and contributed to somatic lineages in chimera experiments, they failed to contribute to the germline, indicating that full pluripotency was not achieved. Subsequent application of alternative reporter genes demonstrated that the pluripotent potential of the derived iPS cell lines is determined by the method of iPS colony selection, and germline competent iPS cells can be obtained if cells are selected for the re-expression of Nanog or Oct4 transcription factors (Maherali et al., 2007; Meissner et al., 2007; Okita et al., 2007) or when selection is omitted entirely (Stadtfeld et al., 2008). These data suggested that iPS cells can exist in a stable, partially-reprogrammed state. Indeed, a recent report demonstrates that incomplete reprogramming of chromatin marks associated with somatic lineages in some iPS lines results in failure of these iPS cells to repress lineage-specifying transcription factors and hampers their pluripotent potential (Mikkelsen et al., 2008). Similarly, our FAB-SCs may be in a stable ground-state of “near pluripotency” that can be reprogrammed to full pluripotency by altering the growth factor milieu. The role and mechanism of growth factor signaling in the reprogramming process has thus far not been recognized and our FAB-SC system uniquely allows the dissection of this process at the molecular level. We identified E-Cadherin as a candidate gene which is induced upon transient LIF/BMP4 stimulation of FAB-SCs. E-cadherin plays an essential role during pre-implantation embryonic development. In its absence, proper cell polarization is absent and trophectoderm formation is impaired (Larue et al., 1994; Riethmacher et al., 1995). Remarkably, embryo compaction is unaffected in E-Cadherin null embryos, but the cells fail to sustain the compacted state and do not form a blastocyst. E-Cadherin mutant embryos fail to hatch and do not demonstrate the typical compacted ICM morphology but instead form clusters of rounded cells, much like bunches of grapes (Larue et al., 1994), and shRNA knockdown of E-Cadherin expression in mES cells resulted in similar colony morphology. In addition, E-Cadherin mutant ES cells fail to generate organized tissue structures in teratoma assays (Larue et al., 1996). Ectopic expression of E-Cadherin or N-Cadherin in E-Cadherin mutant ES cells restored their ability to generate defined teratomas and revealed an unexpected differential effect on the type of tissue formed by the expression of these two cadherins, demonstrating that cell-cell adhesion plays an important role in directing tissue differentiation. Our data extend these observations by demonstrating that E-Cadherin plays an active role in regulating the stem cell pluripotent state itself.
Table 1.
Origin, Culture Conditions and Functional Properties of Different Pluripotent Stem Cell Lines
| Cell line | Origin | Growth factor conditions | Tertoma formation | Chimera formation | Reference |
|---|---|---|---|---|---|
| Murine ES | Blastocyst | LIF, BMP4 | All germ layers | Somatic and germline contribution | (Evans and Kaufman, 1981; Martin, 1981; Martin and Evans, 1975) |
| Murine EpiSC | Epiblast | bFGF, Activin | All germ layers | No | (Brons et al., 2007; Tesar et al., 2007) |
| Human ES | Blastocyst | bFGF, Activin, (BIO), MEF conditioned media | All germ layers | Not Tested | (Thomson et al., 1998) |
| FAB-SC | Blastocyst | bFGF, Activin, BIO | No | No | This report |
| LIF/BMP4 stimulated FAB-SC | Blastocyst | LIF, BMP4 | All germ layers | Somatic and germline contribution | This Report |
| Reverted FAB-SC | Blastocyst | bFGF, Activin, BIO | All germ layers | Somatic contribution, germline contribution not tested | This Report |
| Murine IPS cells | Somatic cells | LIF, BMP4 | All germ layers | Somatic and germline contribution | (Maherali et al., 2007; Meissner et al., 2007; Okita et al., 2007; Takahashi and Yamanaka, 2006) |
| Human IPS cells | Somatic cells | bFGF, serum, MEF conditioned media | All germ layers | Not tested | (Park et al., 2008; Takahashi et al., 2007; Yu et al., 2007) |
| Murine EG cells | Embryonic gonad | Derivation: bFGF, LIF and SCF Maintenance: LIF and fetal bovine serum |
All germ layers | Somatic and germline contribution | (Matsui et al., 1992; Resnick et al., 1992) |
| Human EG cells | Embryonic gonad | LIF, bFGF and Forskolin | All germ layers | Not Tested | (Shamblott et al., 1998) |
| Murine mGS cells | Post-natal testis | Derivation: GDNF, LIF Maintenance: LIF and fetal bovine serum |
All germ layers | Somatic contribution | (Guan et al., 2006; Kanatsu-Shinohara et al., 2004; Seandel et al., 2007) |
The differential need for E-Cadherin expression for the maintenance of pluripotency in FAB-SCs and mES cells may be due to redundant effects of N-Cadherin expression in mES cells. Indeed, a knock-in ES cell line in which N-Cadherin cDNA was inserted into the E-Cadherin locus demonstrated that while N-Cadherin failed to correct the trophectoderm phenotype of the E-Cadherin mutant embryos, it did rescue ES cell colony morphology (Kan et al., 2007). Since FAB-SCs only express E-Cadherin, downregulation of E-Cadherin expression results in rapid differentiation. In contrast, mES cells express low levels of N-Cadherin which may act redundantly to rescue stem cell self-renewal. Upon knockdown of E-Cadherin expression, differentiating mES cells display premature downregulation of Nanog expression concomitant with an early upregulation of HoxB1 expression. We conclude that E-Cadherin plays a critical role in regulating the stem cell pluripotent state and prevents premature differentiation by regulating Nanog expression.
The molecular link between E-Cadherin and Nanog expression is unknown and may be founded in the close cell-cell contact mediated by E-Cadherin. For example, DE-cadherin is an important component of the Drosophila germ cell niche, where it anchors germline stem cells to the somatic component of the niche (Song and Xie, 2002). Close contact between the stem cells and the soma assures that the stem cells receive high levels of growth factor signals that are required for their self-renewal (Song and Xie, 2002; Tulina and Matunis, 2001; Yamashita et al., 2003). It is interesting to hypothesize that pluripotent stem cells, such as FAB-SCs and ES cells, in a similar fashion require Cadherin-mediated cell-cell contacts for optimal self-renewal, possibly by an unknown paracrine mechanism. Alternatively, E-Cadherin itself could provide essential downstream signals that mediate the regulation of Nanog expression and maintenance of pluripotency. E-Cadherin associates with β-catenin, which serves dual roles as mediator of cell adhesion and transcriptional regulator. β-catenin activation through the Wnt signaling pathway has been implicated in ES cell self-renewal (Sato et al., 2004). In addition, TCF binding sites were recently found to co-localize with many Oct4 and Nanog binding sites in the genome, indicating that the Wnt-β-catenin signaling pathway is integral to the pluripotency circuitry mediated by these transcription factors (Cole et al., 2008). E-cadherin may fine-tune this pathway by modulating intracellular β-catenin levels. Additional signaling pathways are known to be activated by Cadherin stimulation and dissection of the molecular associations between the growth factor environment, The expression of E-Cadherin by epithelial cells has been postulated as a reason for why transcription factor induced reprogramming of epithelia is more efficient than the generation of iPS cells from fibroblasts, which lack E-Cadherin expression (Aoi et al., 2008). Thus, unraveling the role of the growth factor environment and cell-cell interactions in the induction and maintenance of FAB-SC pluripotency may serve as a paradigm for other systems of epigenetic reprogramming.
EXPERIMENTAL PROCEDURES
Derivation and maintenance of FAB-SC cell lines
E3.5 blastocysts were obtained from C57/BL6 female X 129SvEv-EGFP male mice. Briefly, blastocysts were flushed from uterus with M2 media (Chemicon) and placed on MEFs in media that consisted of DMEM, 15% KOSR, 2mM L-glutamine, 1% non-essential amino acids, 100 U of penicillin, 100 μg of streptomycin, 1 mM Sodium Pyruvate (all from Invitrogen), 0.1 mM β-mercaptoethanol, 50 μg/ml ascorbic acid, 100 μg/ml FE-saturated transferring (all from Sigma). For the maintenance of FAB-SCs and reverted FAB-SCs growth factors were added: 1 ng/ml bFGF (R&D systems), 50 ng/ml human ActivinA (Peprotech), 0.5 μM BIO (Sigma) and 100 ng/ml LIF-blocking antibody (R&D systems). 7 days after initiation of explant culture, blastocyst outgrowths were dissociated with Trypsin-EDTA (Invitrogen) and transferred to new wells containing MEFs. The established FAB-SC lines were passaged every other day by at a subculture ratio of 1:10. The LIF/BMP4 stimulated FAB-SCs were cultured in the same basal media containing 100 ng/ml rmLIF and 50 ng/ml rhBMP4 (R&D systems).
miRNA expression profiling
MicroRNA expression profiling was performed as described (Lu, 2008; Mi et al., 2007). One microgram total RNA was used for profiling using the plate-capture method of labeling and using the bead-based platform for detection. MicroRNAs were captured using plates coupled with oligonucleotides antisense to microRNAs, ligated with adaptors, reverse-transcribed and amplified through PCR, incorporating biotin as a label. Labeled microRNAs were hybridized to detecting oligonucleotides on colored beads, before detection using flow cytometry on a Luminex 100IS machine. Median fluorescence intensity was used for each microRNA as a measure of expression. Data were preprocessed as described (Lu et al., 2005). Briefly, samples were normalized assuming the same total fluorescence intensity. Data were then log2-transformed and thresholded at 5. Hierarchical clustering was performed in matlab after filtering out microRNAs that were detected in the noise range (with maximum expression in any sample of less than 7.25).
Supplementary Material
Acknowledgments
The authors thank Dr. George Daley for his review of our manuscript and help with the blastocyst injections. Dr. Paul Tesar for the EpiSC image, Dr. Roger Tsien for the use of the tdTomato fluorescent reporter, Dr. Keith Orford for critical review of the manuscript, Kelly Shea for providing technical support and the HSCI/MGH Flow Cytometry Core Facility for help with cell purifications. This work was supported by grants from the National Institutes of Health and the Harvard Stem Cell Institute. M.E. was supported by the Dutch Cancer Society (Koningin Wilhelmina Fonds)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science. 2008 doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Carpenter MK, Rosler ES, Fisk GJ, Brandenberger R, Ares X, Miura T, Lucero M, Rao MS. Properties of four human embryonic stem cell lines maintained in a feeder-free culture system. Dev Dyn. 2004;229:243–258. doi: 10.1002/dvdy.10431. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning C, Allegrucci C, Priddle H, Barbadillo-Munoz MD, Anderson D, Self T, Smith NM, Parkin CT, Young LE. Common culture conditions for maintenance and cardiomyocyte differentiation of the human embryonic stem cell lines, BG01 and HUES-7. Int J Dev Biol. 2006;50:27–37. doi: 10.1387/ijdb.052107cd. [DOI] [PubMed] [Google Scholar]
- Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Gibralter D, Turner DC. Dual adhesion systems of chick myoblasts. Dev Biol. 1985;112:292–307. doi: 10.1016/0012-1606(85)90400-2. [DOI] [PubMed] [Google Scholar]
- Ginis I, Luo Y, Miura T, Thies S, Brandenberger R, Gerecht-Nir S, Amit M, Hoke A, Carpenter MK, Itskovitz-Eldor J, et al. Differences between human and mouse embryonic stem cells. Dev Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- Guan K, Nayernia K, Maier LS, Wagner S, Dressel R, Lee JH, Nolte J, Wolf F, Li M, Engel W, et al. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199–1203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, O’Carroll D, Das PP, Tarakhovsky A, Miska EA, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kan NG, Stemmler MP, Junghans D, Kanzler B, de Vries WN, Dominis M, Kemler R. Gene replacement reveals a specific role for E-cadherin in the formation of a functional trophectoderm. Development. 2007;134:31–41. doi: 10.1242/dev.02722. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, et al. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 2004;119:1001–1012. doi: 10.1016/j.cell.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Larue L, Antos C, Butz S, Huber O, Delmas V, Dominis M, Kemler R. A role for cadherins in tissue formation. Development. 1996;122:3185–3194. doi: 10.1242/dev.122.10.3185. [DOI] [PubMed] [Google Scholar]
- Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci U S A. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy A, Xiong JW, Kuhnert F, Stuhlmann H. Use of developmental marker genes to define temporal and spatial patterns of differentiation during embryoid body formation. J Exp Zool. 1999;284:67–81. doi: 10.1002/(sici)1097-010x(19990615)284:1<67::aid-jez10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Lu J. MicroRNA-Mediated Control of Cell Fate in Megakaryocyte-Erythrocyte Progenitors. Developmental Cell. 2008 doi: 10.1016/j.devcel.2008.03.012. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly Reprogrammed Fibroblasts Show Global Epigenetic Remodeling and Widespread Tissue Contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mallon BS, Park KY, Chen KG, Hamilton RS, McKay RD. Toward xeno-free culture of human embryonic stem cells. Int J Biochem Cell Biol. 2006;38:1063–1075. doi: 10.1016/j.biocel.2005.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR, Evans MJ. Differentiation of clonal lines of teratocarcinoma cells: formation of embryoid bodies in vitro. Proc Natl Acad Sci U S A. 1975;72:1441–1445. doi: 10.1073/pnas.72.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang H, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008 doi: 10.1038/nature07056. Advance online publication, May 28 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Rathjen J, Lake JA, Bettess MD, Washington JM, Chapman G, Rathjen PD. Formation of a primitive ectoderm like cell population, EPL cells, from ES cells in response to biologically derived factors. J Cell Sci. 1999;112(Pt 5):601–612. doi: 10.1242/jcs.112.5.601. [DOI] [PubMed] [Google Scholar]
- Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- Riethmacher D, Brinkmann V, Birchmeier C. A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc Natl Acad Sci U S A. 1995;92:855–859. doi: 10.1073/pnas.92.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosler ES, Fisk GJ, Ares X, Irving J, Miura T, Rao MS, Carpenter MK. Long-term culture of human embryonic stem cells in feeder-free conditions. Dev Dyn. 2004;229:259–274. doi: 10.1002/dvdy.10430. [DOI] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Seandel M, James D, Shmelkov SV, Falciatori I, Kim J, Chavala S, Scherr DS, Zhang F, Torres R, Gale NW, et al. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449:346–350. doi: 10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamblott MJ, Axelman J, Wang S, Bugg EM, Littlefield JW, Donovan PJ, Blumenthal PD, Huggins GR, Gearhart JD. Derivation of pluripotent stem cells from cultured human primordial germ cells. Proc Natl Acad Sci U S A. 1998;95:13726–13731. doi: 10.1073/pnas.95.23.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Xie T. DE-cadherin-mediated cell adhesion is essential for maintaining somatic stem cells in the Drosophila ovary. Proc Natl Acad Sci U S A. 2002;99:14813–14818. doi: 10.1073/pnas.232389399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takeichi M, Atsumi T, Yoshida C, Uno K, Okada TS. Selective adhesion of embryonal carcinoma cells and differentiated cells by Ca2+-dependent sites. Dev Biol. 1981;87:340–350. doi: 10.1016/0012-1606(81)90157-3. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Xu RH, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. 2005;2:185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod. 2006;75:705–716. doi: 10.1095/biolreprod.106.053686. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.