Abstract
Background
Analysis of 23 published transcriptome studies allowed us to identify nine genes displaying frequent alterations in HNSCC (FN1, MMP1, PLAU, SPARC, IL1RN, KRT4, KRT13, MAL, and TGM3). We aimed to independently confirm these dysregulations and to identify potential relationships with clinical data for diagnostic, staging and prognostic purposes either at the tissue level or in saliva rinse.
Methods
For a period of two years, we systematically collected tumor tissue, normal matched mucosa and saliva of patients diagnosed with primary untreated HNSCC. Expression levels of the nine genes of interest were measured by RT-qPCR in tumor and healthy matched mucosa from 46 patients. MMP1 expression level was measured by RT-qPCR in the salivary rinse of 51 HNSCC patients and 18 control cases.
Results
Dysregulation of the nine genes was confirmed by the Wilcoxon test. IL1RN, MAL and MMP1 were the most efficient diagnostic markers of HNSCC, with ROC AUC > 0.95 and both sensitivity and specificity above 91%. No clinically relevant correlation was found between gene expression level in tumor and T stage, N stage, tumor grade, global survival or disease-free survival. Our preliminary results suggests that with 100% specificity, MMP1 detection in saliva rinse is potentially useful for non invasive diagnosis of HNSCC of the oral cavity or oropharynx, but technical improvement is needed since sensitivity was only 20%.
Conclusion
IL1RN, MAL and MMP1 are prospective tumor diagnostic markers for HNSCC. MMP1 overexpression is the most promising marker, and its detection could help identify tumor cells in tissue or saliva.
Background
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignancy worldwide and every year an estimated 600,000 people are newly diagnosed. HNSCC accounts for about 10% of the total cancer burden in men [1]. It is often diagnosed at late stages in heavy alcohol and tobacco users and patients presenting advanced disease have a short overall survival with major post-therapeutic side effects. Despite ongoing efforts, no molecular markers have yet been validated as useful clinical tools for the early detection and management of this disease. Recently, the transcriptome of HNSCC was extensively probed by several groups using expression microarray technology. The goals were to identify the genes potentially involved in this pathology and to identify diagnostic and/or prognostic gene-expression signatures [2-27]. Although the heterogeneous designs of these studies (number of sample, tumor sites, tumor stages, microdissection of tissue samples, RNA extraction and RT protocols, microarray technologies or biostatistics analyses) probably explain the diversity of the results, there is nevertheless strong motivation to develop robust molecular diagnostic tests [28].
We reviewed 23 studies published between March 2000 and November 2007, in which expression profiles of HNSCC tumors versus their normal matched mucosa were compared. Despite the important methodological heterogeneity of gene expression studies 9 genes appeared to be repeatedly dysregulated, in at least nine of the 23 studies for each gene [2-27]. Four genes were up-regulated in HNSCC tissues (FN1, MMP1, PLAU and SPARC) and five were down-regulated (IL1RN, KRT4, KRT13, MAL and TGM3). Because these genes were recurrently found to be dysregulated whatever the tumor location and despite the important methodological heterogeneity of the gene expression studies, we hypothesized that these nine genes could be candidate as HNSCC-specific molecular markers. We therefore evaluated by RT-qPCR their expression level in 46 HNSCC samples and 46 healthy matched mucosa taken from an independent cohort of 74 patients consecutively diagnosed with primary untreated HNSCC in our institution. We assessed their tumor diagnostic values and looked at possible correlations with clinical data. The need for early detection in high-risk patients led several groups to develop techniques for identifying molecular markers in bodily fluid rather than in an invasive biopsy unsuitable for cancer screening. Saliva is an easily accessible diagnostic fluid for screening HNSCC [29]. In this study, we assessed the capacity of MMP1, one of the most relevant markers, to detect HNSCC tumor cells in saliva cells; this provided information on its utility as a non invasive diagnostic marker.
Methods
Patients and sample collection
All normal and tumor tissues, as well as saliva samples, were obtained from the Institutional Tumor Bank of the University Hospital of Nîmes, France. All samples were collected with the informed consent of the patients. This study was approved by the local ethics committee and the scientific board of the Tumor Bank.
Tumor tissue, normal matched mucosa and saliva were obtained from 74 patients consecutively diagnosed with primary untreated HNSCC between April 2005 and April 2007 in our institution. All HNSCC patients were Caucasian heavy smokers and drinkers. Additional saliva samples from 18 healthy controls were collected, between February and April 2008. This cohort of control cases was composed of two populations: 9 consecutive patients refereed to our institution by their physician for HNSCC screening, motivated by heavy drinking and smoking addictions; 9 consecutive patients without alcohol or tobacco addiction in their past history that consulted for otologic or rhinologic mechanical disorders. All patients underwent a head and neck examination to determine the absence or presence of HNSCC and to rule out any significant inflammatory lesion of the upper aero-digestive tract. Patients in the control group were statistically a little younger and with more women than in the HNSCC group (average age = 52.8 years for controls vs 58.8 years for HNSCC [p = 0.037, student test]; sex ratio = 0.38 for controls vs 0.11 for HNSCC [p = 0.029 2-sample test for equality of proportions]).
Tissue samples were collected by biopsy during diagnostic endoscopy and were immediately snap frozen and stored in liquid nitrogen. The matched non-malignant tissue was collected from the same anatomical site, as far as possible from the primary lesion for tumors crossing the midline or on the opposite side for well-lateralized tumors.
For saliva collection both HNSCC patients and control cases followed the same protocole. Subjects were asked to refrain from eating, drinking, smoking, or oral hygiene procedures for at least 1 hour before collection and carried out a 30-second oral rinse with 50 ml of NaCl 0.9% solution that was immediately centrifuged at 2600 rpm for 15 minutes at 4°C. The supernatant was discarded and the pellet was diluted into 1 ml of NaCl 0.9% and stored at -80°C before RNA extraction. The clinical and histopathological features of the populations are presented in Table 1.
Table 1.
Clinical and histopathological features of the population: A) tissue sample population; B) saliva sample populations
| Table 1-A | Table 1-B | ||||||
|---|---|---|---|---|---|---|---|
| Tissu samples (n = 46) | Saliva samples: Control population (n = 18) | ||||||
| Median | Min | Max | Median | Min | Max | ||
| Age | 56.7 | 41.2 | 77.7 | Age | 53 | 32 | 71 |
| Qt | % | Qt | % | ||||
| Sex | Woman | 4 | 8.7 | Sex | Woman | 6 | 33.3 |
| Men | 42 | 91.3 | Men | 12 | 66.6 | ||
| Site | Larynx | 8 | 17.4 | ||||
| Oral cavity | 8 | 17.4 | Saliva samples: HNSCC population (n = 51) | ||||
| Hypopharynx | 7 | 15.2 | Median | Min | Max | ||
| Oropharynx | 23 | 50 | Age | 59 | 41 | 83 | |
| T Stage | 1 | 1 | 2.2 | Qt | % | ||
| 2 | 11 | 23.9 | Sex | Woman | 6 | 11.8 | |
| 3 | 19 | 41.3 | Men | 45 | 88.2 | ||
| 4 | 15 | 32,6 | |||||
| N Stage | 0 | 18 | 39.1 | Site | Larynx | 5 | 9.8 |
| 1 | 4 | 8.7 | Oral cavity | 11 | 21.7 | ||
| 2 | 23 | 50 | Hypopharynx | 8 | 15.7 | ||
| 3 | 1 | 2.2 | Oropharynx | 27 | 52.8 | ||
| M Stage | 0 | 46 | 100 | T Stage | 1 | 6 | 11.8 |
| 1 | 0 | 0 | 2 | 12 | 23.5 | ||
| 3 | 14 | 27.4 | |||||
| Grade | 1 | 13 | 28.3 | 4 | 19 | 37.3 | |
| 2 | 27 | 58.7 | |||||
| 3 | 6 | 13 | N Stage | 0 | 17 | 33.2 | |
| + | 34 | 66.7 | |||||
| Death | Yes | 21 | 45.7 | ||||
| No | 25 | 54.3 | M Stage | 0 | 50 | 1.9 | |
| 1 | 1 | 98.1 | |||||
| Follow-up | Controlled | 25 | 54.3 | ||||
| Progression | 10 | 21.7 | |||||
| Recurrence | 11 | 23.9 | |||||
Qt: quantity
%: percentage
RNA isolation, quality control and cDNA synthesis
To obtain homogeneous and histologically well-characterized samples for RNA analyses, tissue samples were cut with a cryo-microtome into 50-200 slices of 9-μm thickness in RNase-free conditions. At least three frozen slices taken from the sample core were mounted on glass slides and briefly stained with eosin-hematoxylin for histopathological examination by an experienced pathologist (H.C.). Tumor tissue versus normal surrounding tissue percentage (T/N %) was determined for malignant samples. HNSCC samples with less than 30% tumor cells were excluded from the study. Tissue samples were not microdissected in order to include in the qPCR analysis not only the tumor cells, but also the surrounding stromal cells, which are known to have altered transcriptional activity during the carcinogenetic process [30]. Normal tissues had to be composed of both stroma and its surrounding normal epithelial layer, without any tumor cells, to be included in the study. Total RNA was extracted from the remaining tissue slices using the RNeasy Mini Kit (Qiagen, Courtaboeuf, France). Saliva rinse RNA extraction was carried out using the RNA Isolation Kit on a MagNa Pure Compact instrument (Roche, Meylan, France).
RNA quality control and quantification were carried out on an Agilent 2100 Bioanalyser using Total RNA Nano II Chips (Agilent Technologies, Massy, France). To limit the impact of possible RNA degradation on our results, we selected for further analysis, only tissue samples with total RNA concentrations >85 ng/μl and values of RNA integrity number (RIN) >6. and saliva samples with total RNA concentration >40 ng/μl and RIN value >4. These criteria were never responsible of sample removal as satisfying qualities could be obtained by a second or third extraction procedure when required.
After quality control of tissues and saliva of the initial cohort of 74 HNSCC patients, 46 matched pairs of tumor tissue and normal mucosa and 51 saliva samples were useful. For 23 patients we analyzed both tissues and saliva. For 23 patients we analysed tissues only and for 28 patients saliva only. From our initial cohort of 74 HNSCC patients, 46 matched pairs of tumor tissue and normal mucosa as well as 51 saliva samples were useful. The amount of tissue, either tumor or normal mucosa, was insufficient for 28 patients to carry out pathological analysis and mRNA extraction. Saliva collection missed for 18 patients and saliva collection procedures were inadequate for 5 other patients. Finally only 23 patients were analyzed both at the tissue and saliva level. No relation was found between sample removal and tumor stage.
After quality control, 1 μg of total RNA was reverse transcribed using M-MLV reverse transcriptase and oligo dT14-16 primers (Applied Biosystems, Courtaboeuf, France). Samples were incubated for 10 minutes at 65°C, cooled on ice for 5 minutes, and incubated with reverse transcriptase for 1 hour at 37°C. Reverse transcriptase was then inactivated by heating at 95°C for 5 minutes. The resulting cDNA were diluted 1:10 for tissue samples and 1:4 for saliva samples before being used as PCR template.
Quantitative real-time PCR (qPCR
We quantified the mRNA expression of three housekeeping genes and the nine genes of interest by real-time RT-qPCR using the Light Cycler Fast DNA Masterplus SYBR green kit on LightCycler 480 (Roche, Meylan, France). Stringent primers sets were designed using Oligo 6 Software (MBI, Cascade, CO, USA). To avoid false detection of genomic DNA, although DNase was used for the extraction procedure, amplification was done on spliced regions of the genes. Gene references and primer characteristics are listed in Table 2. For each qPCR reaction we used 2 μl of the diluted cDNA, 1 μl of 10 μmol.l-1 forward primer, 1 μl of 10 μmol.l-1 reverse primer, 5 μl Light Cycler Fast DNA Masterplus SYBR green I and 11 μl PCR water, for a final volume of 20 μl. The PCR cycle conditions were set as follow: a preincubation step for 10 minutes at 95°C followed by 40 cycles for tissue cDNA and 53 cycles for saliva cell cDNA; each cycle included 15 seconds at 95°C, 15 seconds at 60°C, and 15 seconds at 72°C. The temperature transition rate was 20°C/second. A melting curve was generated by linear heating from 50°C to 95°C in 20 minutes with 10 fluorescence measures every 1°C. A negative control, with no template, and a positive inter-run control were included for each gene in each qPCR run. All measurements were performed in triplicate. Standard curve assays showed an efficient amplification >1.8 for all genes and the specificity was shown by a single peak at the expected temperature on melting curve analyses (Table 2). For each gene the inter-assay coefficient of variation of crossing point values was <10% (data not shown).
Table 2.
Characteristics of the gene-specific qPCR assays
| Gene name (synonym) | Access n° Gene ID | Gene location | Primer sequence 5'-3' | Amplicon size Melting T°c | PCR efficiency |
|---|---|---|---|---|---|
| KRT4 | NM_002272 | 12q12-q13 | f: tcaacaacaagtttgcctc | 185 | 1.94 |
| keratin 4 | 3851 | r: gtcattgcccaaggtatcta | 90 | ||
| IL1RN | NM_173841 | 2q14.2 | f: cctgtcctgtgtcaagtctg | 257 | 1.80 |
| interleukin 1 receptor antagonist | 3557 | r: cgtcctcctggaagtagaat | 90 | ||
| KRT13 | NM_153490 | 17q12-q21.2 | f: tctctgtcttgctggtctga | 234 | 1.88 |
| keratin 13 | 3860 | r: atgaagaggagatgaaggaa | 89 | ||
| MMP1 | NM_002421 | 11q22.3 | f: aaagacagattctacatgcg | 237 | 1.92 |
| matrix metallopeptidase 1 | 4312 | r: tgcttcacagttctaggga | 85 | ||
| PLAU | NM_002658 | 10q24 | f: ggactacatcgtctacctgg | 230 | 1.90 |
| plasminogen activator, urokinase | 5328 | r: caaactggggatcgttatac | 88 | ||
| SPARC | NM_003118 | 5q31.3-q32 | f: ggtgactgaggtatctgtgg | 245 | 1.85 |
| secreted protein acidic cysteie-rich | 6678 | r: aggtcttgttgtcattgctg | 90 | ||
| TGM3 | NM_003245 | 20q11.2 | f: cactctccaatggcagtagt | 215 | 1.93 |
| transglutaminase 3 | 7053 | r: cataaagacgctatccacat | 88 | ||
| FN1 | NM_212482 | 2q34 | f: tgacacttatgagcgtcct | 234 | 1.81 |
| fibronectin 1 | 2335 | r: aaacacttctcagctatggg | 86 | ||
| MAL | NM_002371 | 2cen-q13 | f: ataaagccgcagtagaactt | 181 | 1.95 |
| mal, T-cell differentiation protein | 4118 | r: agagtaaacacagcacccac | 84 | ||
| ACTB | NM_001101 | 7p15-p12 | f: tggctggggtgttgaaggtct | 238 | 1,89 |
| actin, beta | 60 | r: agcacggcatcgtcaccaact | 90 | ||
| B2M | NM_004048 | 15q21-q22.2 | f: cagcgtactccaaagattca | 240 | 1.99 |
| beta-2-microglobulin | 567 | r: gaatgctccactttttcaat | 90 | ||
| RPS18 | NM_022551 | 6p21.3 | f: agcttgttgtccagaccatt | 187 | 1.84 |
| ribosomal protein S18 | 6222 | r: tgaggaaagcagacattgac | 87 |
T°c: Temperature
For the salivary assay, 50 cycles of the above-described qPCR procedure were carried out in order to detect very low concentrations of mRNA. As this number of cycle was extremely high in order to detect very low concentration of mRNA, the PCR specificity was controlled with care by melting curve analysis of the PCR products and by negative controls. Whatever, the highest Cp value observed was 44.2 cycles. The whole RT-qPCR procedure was carried out with and without RT enzyme, confirming the absence of contamination by genomic DNA during the automated saliva rinse RNA extraction process (data not shown).
qPCR data analysis
values were automatically calculated by LightCycler 480® Software using the second derivative method and were imported into qBase, version 1.3.5, a free software program for the management and automated analysis of qPCR data, for quantitative analyses [31]. Normal tissue cDNA of patient 5 was arbitrarily chosen as a calibrator for each gene, and for this sample the expression level was set at 1 for each gene. For each gene qPCR amplification efficiencies were calculated by qBase from standard curves and were applied in the quantification algorithm. Relative quantities were normalized by qBase to a geometric mean of the three housekeeping genes ACT, B2M and RPS18. Concerning the housekeeping gene set stability assessment, raw relative quantities were tested with geNorm software, version 3.4, for the combination of the three genes. geNorm algorithm calculates a gene expression stability measure M for a reference gene based on the average pairwise variation for that gene with all other tested reference genes [32].
Statistical analysis
S-Plus 2000 software (TIBCO Software, Inc., Palo Alto, CA, USA) was used to perform the statistical analyses. The quantitative variables were described by median, minimum and maximum values and the qualitative variables were described by frequencies and percentages. Genetic markers were compared between HNSCC tissues and matched normal mucosa using a Wilcoxon test for paired data. Comparisons were considered significant when p < 0.05.
The ability of each gene to diagnose the tumor tissue (HNSCC diagnostic ability) was represented by a Receiver Operating Characteristic (ROC) curve and the corresponding Area Under the Curve (AUC). The ROC curves and AUC were assessed by a non-parametric method [33]. The optimal cut-off used to calculate both sensitivity and specificity, was defined as the cut-off minimizing the number of misclassified tissues.
The clinical relevance of HNSCC markers was analyzed by correlating the expression level and clinical-pathological parameters. For each gene, the expression level in HNSCC was compared according to stage T, stage N, histological grade and tumor tissue versus normal surrounding tissue percentage (T/N %) using a Kruskal-Wallis test. The Cox model was used to analyze global survival and disease-free survival. For survival tests, the median follow-up was 1 year and 3 months.
Since no data were available concerning the diagnosis values of our candidate markers, it was not possible to determine a statistically based number of sample. Then, we decided to include all patients with exploitable samples, available at our Institutional Tumor Bank at the time of analysis.
Results
Housekeeping gene stability
To date there is no published evidence to guide the selection of suitable housekeeping genes for the normalization of HNSCC RT-qPCR studies. Hence, we chose to normalize our qPCR relative ratio by the geometric mean of three commonly used housekeeping genes in cancer studies: ACT, B2M and RPS18. We validated this approach using geNorm software, version 3.4, which gave an expression stability measure based on the average pairwise variation of the three genes [32]. In our assay the stability value (M) of the association of ACT, B2M and RPS18 was M = 1.2. This M value, below the 1.5 arbitrary cut-off recommended by the software, means that our set of three housekeeping genes was appropriate for normalization.
Differential gene expression between HNSCC and normal matched mucosa
Normalized relative expression levels of the nine selected genes were calculated by qBase for each sample. These expression levels were compared between HNSCC and normal matched tissues for the 46 patients (92 samples). As presented in Table 3, relative mRNA expression levels were significantly higher in tumor than in healthy samples for FN1, MMP1, PLAU and SPARC (Wilcoxon test for paired data, p ≤ 0.002). These levels were lower in tumor than in normal samples for KRT4, KRT13, IL1RN, MAL and TGM3 (Wilcoxon test for paired data, p < 0.001). For each gene, the median expression level ratio between tumor tissues and their matched normal mucosa are presented in Figure 1. Among the overexpressed genes in tumor, MMP1 showed the highest order of magnitude (510-fold) and FN1 the lowest (2.6-fold). Among down-regulated genes in tumor, KRT4 showed the highest order of magnitude (530-fold) and IL1RN the lowest (9-fold).
Table 3.
Expression levels of the 9 genes of interest in HNSCC and normal matched mucosa
| Normal Tissue | HNSCC Tissue | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | median | St Dev | min | max | median | St Dev | min | max | p-values |
| FN1 | 0.076 | 0.280 | 0.004 | 1.577 | 0.268 | 0.471 | 0.007 | 2.185 | 0.002 |
| IL1RN | 1.801 | 1.851 | 0.304 | 9.149 | 0.204 | 0.304 | 0.010 | 1.675 | <0.001 |
| KTR13 | 9.110 | 105.525 | 0.217 | 564.113 | 0.156 | 5.193 | <0.001 | 28.358 | <0.001 |
| KTR4 | 2.647 | 75.657 | 0.080 | 479.146 | 0.004 | 1.120 | <0.001 | 6.003 | <0.001 |
| MAL | 2.319 | 3.378 | 0.262 | 19.968 | 0.033 | 0.266 | <0.001 | 1.182 | <0.001 |
| MMP1 | 0.755 | 7.395 | 0.033 | 43.727 | 319.480 | 1515.1 | 3.038 | 7510.565 | <0.001 |
| PLAU | 0.995 | 15.426 | 0.304 | 105.707 | 6.831 | 18.192 | 0.274 | 109.053 | <0.001 |
| SPARC | 0.302 | 0.198 | 0.065 | 1.000 | 0.985 | 0.996 | 0.140 | 4.791 | <0.001 |
| TGM3 | 1.399 | 17.775 | 0.064 | 121.378 | 0.035 | 13.594 | 0.001 | 92.326 | <0.001 |
The median, minimum and maximum values of relative normalized ratios for each gene of interest in normal tissue versus HNSCC tissue, with corresponding p-value (Wilcoxon test for paired data).
Figure 1.
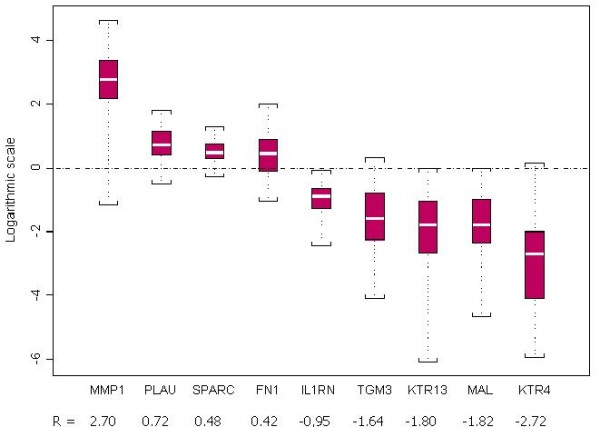
Differential mRNA expression of the nine genes of interest in macroscopically healthy mucosa and HNSCC tissue. For estimation of the individual expression of each gene, the expression ratios of paired tissue specimens were calculated as R = HNSCC/normal. The distribution of the log of these ratios is represented for each gene by a box-plot. The central box represents the interquartile interval, the white line inside the box is the median value, and the minimum and the maximum values are indicated with square brackets.
HNSCC diagnostic ability of the nine genes
The ability of the nine selected genes to diagnose HNSCC was assessed by generating ROC curves. Corresponding AUC were calculated for each gene in order to find the best single marker to differentiate between normal mucosa and HNSCC tissue. Apart from FN1, which showed a poor AUC value (0.693), all genes demonstrated good diagnostic abilities with AUC > 0.86. Three genes were considered remarkable (IL1RN, MAL and MMP1) with AUC > 0.95. As presented in Table 4, when optimized cut-off values were selected, the single expression level of IL1RN, MAL or MMP1 correctly predicted the tumor or normal nature of tissue samples in, respectively, 87, 86 and 88 cases out of the 92 tested samples. Corresponding sensitivities ranged from 93.7 to 95.7% and specificities ranged from 91.3 to 97.8%. Interestingly, the association of these three genes in a multivariate model did not enhance diagnostic efficacy (data not shown). For each gene detailed statistics are listed in Table 4 (AUC of ROC curves, cut-off values and corresponding sensitivity, specificity, true positive value, false positive value, true negative value and false negative value).
Table 4.
Diagnostic values of the 9 gene of interest
| Gene | AUC | Cut-off value | False negative | False positive | True negative | True positive | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|---|
| FN1 | 0,693 | 0.20 | 19 | 11 | 35 | 27 | 58.7 | 76.1 |
| IL1NR | 0,974 | 0.51 | 3 | 2 | 44 | 43 | 93.5 | 95.7 |
| KTR13 | 0,921 | 4.80 | 1 | 11 | 33 | 42 | 75.0 | 95.5 |
| KTR4 | 0,947 | 0.23 | 5 | 4 | 42 | 41 | 89.1 | 91.3 |
| MAL | 0,986 | 0.72 | 2 | 4 | 42 | 44 | 95.7 | 91.3 |
| MMP1 | 0,989 | 24.00 | 3 | 1 | 45 | 43 | 93.5 | 97.8 |
| PLAU | 0,897 | 2.30 | 9 | 5 | 41 | 37 | 80.5 | 89.1 |
| SPARC | 0,864 | 0.67 | 13 | 3 | 43 | 33 | 71.7 | 93.5 |
| TGM3 | 0,939 | 0.31 | 7 | 4 | 42 | 39 | 84.8 | 91.3 |
Calculation of AUC values for the relative expression levels of the nine genes of interest by ROC analyses and the corresponding diagnostic values with optimum cut-off value.
Clinical relevance of the nine genes
We investigated a possible association between the expression levels of the nine genes measured in tumor and T stage, N stage and histological grade. No statistical difference was found between expression levels in tumor of patients with stage N0 versus stage N+. The p-values ranged from 0.26 to 0.98 except for KTR4 (p = 0.08, Kruskal-Wallis test). Statistical differences were found between patients with histological grade 1 versus grade 2-3 for IL1RN (p = 0.01) and SPARC (p = 0.03). For the other genes, p-values ranged from 0.27 to 0.99. Statistical differences were found between patients with stage T2 versus stage T3-T4 for SPARC (p = 0.01). For the other genes, p-values ranged from 0.11 to 0.96.
A univariate survival analysis did not detect genes significantly correlated to global survival. For three genes, hazard ratios (HR) were low but with large confidence intervals: TGM3 (HR = 0.23 [0.02-2.94]), MAL (HR = 0.23 [0.02-2.24]) and IL1RN (HR = 0.38 [0.06-2.46]). Similar results were found when disease-free survival was analyzed.
MMP1 mRNA detection in salivary rinse
Among the three best markers for HNSCC at the diagnostic level, MMP1 was the only one to be overexpressed in tumors. We hypothesized that its expression could be detected in the saliva rinses of patients with HNSCC, where tumor cells are known to desquamate. We carried out a preliminary assay to tested this gene in saliva but not the other 8 candidate genes. Among the 51 HNSCC patients, only 10 (20%) exhibited MMP1 mRNA at a measurable concentration in their saliva rinse. For these positive samples, MMP1 signal was detected at a mean Cp value of 36.4 cycles (31.7-44.2). As presented table 5, among the 10 HNSCC tumors detected by MMP1 salivary test, six aroused from the oropharynx, 3 from the oral cavity and 1 from the hypopharynx. MMP1 expression level in tissues was available for 3 out of the 10 patients with MMP1 positives salivary rinse. These three patients had 1000-fold overexpression of MMP1 in HNSCC comparing to normal matched mucosa. No MMP1 signal was detectable in the control group of 18 patients. ACT, B2M and RPS18 were used as positive internal control in this assay. Contrary to MMP1, their mRNA expression was detectable in all saliva samples at mean Cp values of 27.7, 28.8 and 31.7 cycles for ACT, B2M and RPS18, respectively.
Table 5.
Characteristics of patients with positive MMP1 salivary test
| Patient | Tumor site | T stage | N stage | M stage |
|---|---|---|---|---|
| 1 | Oral cavity | 1 | 0 | 0 |
| 2 | Oral cavity | 4a | 2b | 0 |
| 3 | Oral cavity | 4a | 2c | 0 |
| 4 | Oropharynx | 2 | 0 | 0 |
| 5 | Oropharynx | 2 | 2b | 0 |
| 6 | Oropharynx | 3 | 2b | 0 |
| 7 | Oropharynx | 4a | 0 | 0 |
| 8 | Oropharynx | 4a | 2c | 0 |
| 9 | Oropharynx | 4a | 2c | 1 |
| 10 | Hypopharynx | 3 | 2b | 0 |
Tumor sites and TNM stages of the 10 patients with detectable levels of MMP1 in their saliva rinse.
Discussion
Transcriptome profiling of tumor is a promising approach to identifying gene dysregulations potentially useful at the clinical level to detect or diagnose tumors and predict outcome, as well as to identify the gene pathways involved in carcinogenic processes. In the field of HNSCC, numerous studies have been performed but all have failed to find clinical applications, essentially because of the great diversity in their designs and the lack of a confirmatory step in an independent cohort of patients [2-27]. When we looked closely at the HNSCC transcriptome analyses some genes emerged as frequently dysregulated and therefore as specific candidates as HNSCC molecular markers.
This original study is the first to validate independently the gene dysregulation observed in various HNSCC transcriptome assays. Our study, based on RT-qPCR analysis, is not a definitive transcriptome validation assay, but it clearly proves that reliable biological information with potential clinical applications, can be obtained by pooling the results of several transcriptome data despite heterogeneous designs and small patient cohorts.
Using this gene selection and validation approach, we identified eight efficient transcriptional markers to predict the presence of HNSCC cells in tissue samples, with three remarkable markers: IL1RN, MAL and MMP1. When tested individually, these three markers presented specificity above 91% and sensitivity above 93% in a cohort of 46 patients with various stages, grades and sites of the disease. Surprisingly, there was no additional gain when these three top markers were evaluated together in a multivariate model rather than each separately. This finding implies that above 90% of both specificity and sensitivity little additional information can be expected by associating several transcriptional markers. It also indicates that the biological information associated with these three dysregulated genes should be somehow similar as far as diagnosis is concerned. It is worth mentioning that in clinical routine the use of only one well-characterized diagnostic marker has the advantage of being simple.
No clinically relevant correlations were identified between gene expression level measured in tumor and clinical or pathological parameters. This absence of correlation could signify that these dysregulations are common early events in HNSCC carcinogenesis, making these genes useful as diagnostic markers but useless as staging or prognostic markers. The proportion of early-stage tumor was low in our population, with 12 stage T2 tumors (26%); nonetheless, these nine dysregulations remained statistically significant in this subgroup of patient. This finding confirmed that these markers could be used for the detection of early-stage as well as later-stage tumors.
Among these three most remarkable dysregulated genes for HNSCC diagnosis, MMP1 was highly overexpressed. We thus hypothesized that it could be detected in the saliva rinses of patients with HNSCC for diagnosis or screening purposes. Indeed, several studies recently focused on the detection of such cells, mainly by studying DNA alterations (e.g., mutation, hypermethylation) but also by RNA detection [34]. Through a transcriptome salivary approach, Li et al. identified a set of seven overexpressed genes in saliva from patients presenting oral squamous cell carcinoma; their association in a multivariate model yielded a sensitivity of 91% and a specificity of 91% [35-40]. Li et al used the cell-free saliva for their transcriptome study. On the contrary, we choose to extract RNA from salivary floating-cells. In our opinion mRNA overexpression due to tumour alterations is more likely to be detected in the salivary floating-cells pellets than in the more RNA-diluted free cell saliva. Kim et al reported an elevation of MMP1 in saliva related to refractory periodontitis in microarray study of oral subepithelial connective tissues [41]. We do not think that periodontal health changes were responsible of MMP1 elevation in saliva of our patients since no clear differences of periodontal status were noticed between HNSCC patients and control cases or between MMP1 positive and MMP1 negative HNSCC patients.
As most of the patients likely to develop HNSCC are reluctant to consult physicians when the first symptoms occur, a noninvasive screening method based on the detection of tumor cells among epithelial oral cells could be of interest. In this preliminary salivary study, we confirmed that the expression of MMP1 was confined to HNSCC tumor cells as no expression was detected within the healthy population. Unfortunately, the sensitivity was low as only 20% of the patients presenting the disease were detected. Given that the salivary donors in our study presented advanced tumors, mostly symptomatic or easily detectable by standard clinical examination, this salivary approach could be considered as inefficient. On the other hand, the 100% specificity seems encouraging and HNSCC screening by a non-invasive salivary/oral screening test remains a promising field of research. In our study, the lack of sensitivity to detect salivary expression of MMP1 could have been due to suboptimal procedures for saliva collection, RNA extraction and retro-transcription. Sensitized procedures would increase the quantity of tumor cells and mRNA in samples and therefore improve the sensitivity of this technique [29].
The nine dysregulated genes have very different biological functions and some of them are already known to be implicated in cancer. From a clinical point of view, IL1RN, MAL and TGM3 were not clearly identified as potential HNSCC markers, while few data have been published concerning the implication of FN1, KRT, MMP1, PLAU and SPARC in this type of cancer. From a fundamental point of view, further studies are required to assess the functional mechanisms implicated in the dysregulations we observed.
Conclusion
We confirmed in an independent study the dysregulation of FN1, MMP1, PLAU SPARC, IL1RN, KRT4, KRT13, MAL and TGM3 in HNSCC. Three of them, MMP1, MAL and IL1RN, were remarkable at identifying HNSCC in comparison with normal mucosa. They thus present an interesting potential as screening/diagnostic markers, still to be evaluated at the clinical level. MMP1 was the most promising gene of this study because it was overexpressed in tumor, highly dyregulated (500-fold) and tumor-specific. In addition to salivary detection, MMP1 could be tested at the RNA level for its ability to identify small tumor clusters in diagnostic biopsies, surgical margins or lymph nodes in the setting of primary tumors. This would be a valuable complement to histopathological analyses which efficacy for the diagnosis of microscopic tumors is low.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BL was responsible for the conception, design and experimental work of the study and for drafting the manuscript. AE, CR, DJ, FH and SL made substantial contributions to the study conception and design and greatly enhanced the intellectual content of the manuscript. CC contributed substantially to the study design and carried out the statistical analysis. HC was responsible for pathological examination and quality control. GC, CR, OS and JGL carried out the tissue harvesting. JPB supervised the study and approved the final version of the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Benjamin Lallemant, Email: benjamin.lallemant@chu-nimes.fr.
Alexandre Evrard, Email: alexandre.evrard@univ-montp1.fr.
Christophe Combescure, Email: christophe.combescure@hcuge.ch.
Heliette Chapuis, Email: heliette.chapuis@chu-nimes.fr.
Guillaume Chambon, Email: guillaume.chambon@chu-nimes.fr.
Caroline Raynal, Email: caroline.raynal@univ-montp1.fr.
Christophe Reynaud, Email: christophe.reynaud@chu-nimes.fr.
Omar Sabra, Email: sabraomar@hotmail.com.
Dominique Joubert, Email: djoubert@igf.cnrs.fr.
Frédéric Hollande, Email: frederic.hollande@univ-montp1.fr.
Jean-Gabriel Lallemant, Email: jean.gabriel.lallemant@chu-nimes.fr.
Serge Lumbroso, Email: serge.lumbroso@chu-nimes.fr.
Jean-Paul Brouillet, Email: jean.paul.brouillet@chu-nimes.fr.
Acknowledgements
The authors would like to thank Gislaine Tuech and Michel Canipel (Laboratoire de biochimie, CHU de Nîmes, France) for their excellent technical support and Catherine Carmeni for assistance in manuscript preparation.
References
- Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol. 2007;18(3):581–592. doi: 10.1093/annonc/mdl498. [DOI] [PubMed] [Google Scholar]
- Alevizos I, Mahadevappa M, Zhang X, Ohyama H, Kohno Y, Posner M, Gallagher GT, Varvares M, Cohen D, Kim D. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene. 2001;20(43):6196–6204. doi: 10.1038/sj.onc.1204685. [DOI] [PubMed] [Google Scholar]
- Belbin TJ, Singh B, Smith RV, Socci ND, Wreesmann VB, Sanchez-Carbayo M, Masterson J, Patel S, Cordon-Cardo C, Prystowsky MB. Molecular profiling of tumor progression in head and neck cancer. Archives of otolaryngology--head & neck surgery. 2005;131(1):10–18. doi: 10.1001/archotol.131.1.10. [DOI] [PubMed] [Google Scholar]
- Carles A, Millon R, Cromer A, Ganguli G, Lemaire F, Young J, Wasylyk C, Muller D, Schultz I, Rabouel Y. Head and neck squamous cell carcinoma transcriptome analysis by comprehensive validated differential display. Oncogene. 2006;25(12):1821–1831. doi: 10.1038/sj.onc.1209203. [DOI] [PubMed] [Google Scholar]
- Chin D, Boyle GM, Williams RM, Ferguson K, Pandeya N, Pedley J, Campbell CM, Theile DR, Parsons PG, Coman WB. Novel markers for poor prognosis in head and neck cancer. International journal of cancer. 2005;113(5):789–797. doi: 10.1002/ijc.20608. [DOI] [PubMed] [Google Scholar]
- Choi P, Chen C. Genetic expression profiles and biologic pathway alterations in head and neck squamous cell carcinoma. Cancer. 2005;104(6):1113–1128. doi: 10.1002/cncr.21293. [DOI] [PubMed] [Google Scholar]
- Cromer A, Carles A, Millon R, Ganguli G, Chalmel F, Lemaire F, Young J, Dembele D, Thibault C, Muller D. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23(14):2484–2498. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- El-Naggar AK, Kim HW, Clayman GL, Coombes MM, Le B, Lai S, Zhan F, Luna MA, Hong WK, Lee JJ. Differential expression profiling of head and neck squamous carcinoma: significance in their phenotypic and biological classification. Oncogene. 2002;21(53):8206–8219. doi: 10.1038/sj.onc.1206021. [DOI] [PubMed] [Google Scholar]
- Gonzalez HE, Gujrati M, Frederick M, Henderson Y, Arumugam J, Spring PW, Mitsudo K, Kim HW, Clayman GL. Identification of 9 genes differentially expressed in head and neck squamous cell carcinoma. Archives of otolaryngology--head & neck surgery. 2003;129(7):754–759. doi: 10.1001/archotol.129.7.754. [DOI] [PubMed] [Google Scholar]
- Hwang D, Alevizos I, Schmitt WA, Misra J, Ohyama H, Todd R, Mahadevappa M, Warrington JA, Stephanopoulos G, Wong DT. Genomic dissection for characterization of cancerous oral epithelium tissues using transcription profiling. Oral oncology. 2003;39(3):259–268. doi: 10.1016/S1368-8375(02)00108-2. [DOI] [PubMed] [Google Scholar]
- Ibrahim SO, Aarsaether N, Holsve MK, Kross KW, Heimdal JH, Aarstad JH, Liavaag PG, Elgindi OA, Johannessen AC, Lillehaug JR. Gene expression profile in oral squamous cell carcinomas and matching normal oral mucosal tissues from black Africans and white Caucasians: the case of the Sudan vs. Norway. Oral oncology. 2003;39(1):37–48. doi: 10.1016/S1368-8375(02)00018-0. [DOI] [PubMed] [Google Scholar]
- Leethanakul C, Patel V, Gillespie J, Pallente M, Ensley JF, Koontongkaew S, Liotta LA, Emmert-Buck M, Gutkind JS. Distinct pattern of expression of differentiation and growth-related genes in squamous cell carcinomas of the head and neck revealed by the use of laser capture microdissection and cDNA arrays. Oncogene. 2000;19(28):3220–3224. doi: 10.1038/sj.onc.1203703. [DOI] [PubMed] [Google Scholar]
- Leethanakul C, Patel V, Gillespie J, Shillitoe E, Kellman RM, Ensley JF, Limwongse V, Emmert-Buck MR, Krizman DB, Gutkind JS. Gene expression profiles in squamous cell carcinomas of the oral cavity: use of laser capture microdissection for the construction and analysis of stage-specific cDNA libraries. Oral oncology. 2000;36(5):474–483. doi: 10.1016/S1368-8375(00)00039-7. [DOI] [PubMed] [Google Scholar]
- Lemaire F, Millon R, Young J, Cromer A, Wasylyk C, Schultz I, Muller D, Marchal P, Zhao C, Melle D. Differential expression profiling of head and neck squamous cell carcinoma (HNSCC) British journal of cancer. 2003;89(10):1940–1949. doi: 10.1038/sj.bjc.6601373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez E, Cheng C, Farwell DG, Ricks S, Agoff SN, Futran ND, Weymuller EA Jr, Maronian NC, Zhao LP, Chen C. Transcriptional expression profiles of oral squamous cell carcinomas. Cancer. 2002;95(7):1482–1494. doi: 10.1002/cncr.10875. [DOI] [PubMed] [Google Scholar]
- Nagata M, Fujita H, Ida H, Hoshina H, Inoue T, Seki Y, Ohnishi M, Ohyama T, Shingaki S, Kaji M. Identification of potential biomarkers of lymph node metastasis in oral squamous cell carcinoma by cDNA microarray analysis. International journal of cancer. 2003;106(5):683–689. doi: 10.1002/ijc.11283. [DOI] [PubMed] [Google Scholar]
- Schmalbach CE, Chepeha DB, Giordano TJ, Rubin MA, Teknos TN, Bradford CR, Wolf GT, Kuick R, Misek DE, Trask DK. Molecular profiling and the identification of genes associated with metastatic oral cavity/pharynx squamous cell carcinoma. Archives of otolaryngology--head & neck surgery. 2004;130(3):295–302. doi: 10.1001/archotol.130.3.295. [DOI] [PubMed] [Google Scholar]
- Sok JC, Kuriakose MA, Mahajan VB, Pearlman AN, DeLacure MD, Chen FA. Tissue-specific gene expression of head and neck squamous cell carcinoma in vivo by complementary DNA microarray analysis. Archives of otolaryngology--head & neck surgery. 2003;129(7):760–770. doi: 10.1001/archotol.129.7.760. [DOI] [PubMed] [Google Scholar]
- Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet. 2004;154(1):27–35. doi: 10.1016/j.cancergencyto.2004.01.026. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Tsai ST, Ko JY, Jin YT, Li C, Huang W, Young KC, Lai MD, Liu HS, Wu LW. The mRNA profile of genes in betel quid chewing oral cancer patients. Oral oncology. 2004;40(4):418–426. doi: 10.1016/j.oraloncology.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Villaret DB, Wang T, Dillon D, Xu J, Sivam D, Cheever MA, Reed SG. Identification of genes overexpressed in head and neck squamous cell carcinoma using a combination of complementary DNA subtraction and microarray analysis. The Laryngoscope. 2000;110(3 Pt 1):374–381. doi: 10.1097/00005537-200003000-00008. [DOI] [PubMed] [Google Scholar]
- Warner GC, Reis PP, Jurisica I, Sultan M, Arora S, Macmillan C, Makitie AA, Grenman R, Reid N, Sukhai M. Molecular classification of oral cancer by cDNA microarrays identifies overexpressed genes correlated with nodal metastasis. International journal of cancer. 2004;110(6):857–868. doi: 10.1002/ijc.20197. [DOI] [PubMed] [Google Scholar]
- Whipple ME, Mendez E, Farwell DG, Agoff SN, Chen C. A genomic predictor of oral squamous cell carcinoma. The Laryngoscope. 2004;114(8):1346–1354. doi: 10.1097/00005537-200408000-00006. [DOI] [PubMed] [Google Scholar]
- Suhr ML, Dysvik B, Bruland O, Warnakulasuriya S, Amaratunga AN, Jonassen I, Vasstrand EN, Ibrahim SO. Gene expression profile of oral squamous cell carcinomas from Sri Lankan betel quid users. Oncology reports. 2007;18(5):1061–1075. [PubMed] [Google Scholar]
- Ziober AF, Patel KR, Alawi F, Gimotty P, Weber RS, Feldman MM, Chalian AA, Weinstein GS, Hunt J, Ziober BL. Identification of a gene signature for rapid screening of oral squamous cell carcinoma. Clin Cancer Res. 2006;12(20 Pt 1):5960–5971. doi: 10.1158/1078-0432.CCR-06-0535. [DOI] [PubMed] [Google Scholar]
- Chen C, Mendez E, Houck J, Fan W, Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17(8):2152–2162. doi: 10.1158/1055-9965.EPI-07-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palka KT, Slebos RJ, Chung CH. Update on molecular diagnostic tests in head and neck cancer. Seminars in oncology. 2008;35(3):198–210. doi: 10.1053/j.seminoncol.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, Eisele D, Abemayor E, Elashoff D, Park NH. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10(24):8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- De Wever O, Pauwels P, De Craene B, Sabbah M, Emami S, Redeuilh G, Gespach C, Bracke M, Berx G. Molecular and pathological signatures of epithelial-mesenchymal transitions at the cancer invasion front. Histochemistry and cell biology. 2008;130(3):481–494. doi: 10.1007/s00418-008-0464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome biology. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- Mazumder A, Palma AJ, Wang Y. Validation and integration of gene-expression signatures in cancer. Expert review of molecular diagnostics. 2008;8(2):125–128. doi: 10.1586/14737159.8.2.125. [DOI] [PubMed] [Google Scholar]
- Carvalho AL, Jeronimo C, Kim MM, Henrique R, Zhang Z, Hoque MO, Chang S, Brait M, Nayak CS, Jiang WW. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14(1):97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- Chen K, Sawhney R, Khan M, Benninger MS, Hou Z, Sethi S, Stephen JK, Worsham MJ. Methylation of multiple genes as diagnostic and therapeutic markers in primary head and neck squamous cell carcinoma. Archives of otolaryngology--head & neck surgery. 2007;133(11):1131–1138. doi: 10.1001/archotol.133.11.1131. [DOI] [PubMed] [Google Scholar]
- Righini CA, De Fraipont F, Timsit JF, Faure C, Brambilla E, Reyt E, Favrot MC. Tumor-specific methylation in saliva: a promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res. 2007;13(4):1179–1185. doi: 10.1158/1078-0432.CCR-06-2027. [DOI] [PubMed] [Google Scholar]
- Temam S, Trassard M, Leroux G, Bosq J, Luboinski B, Lenoir G, Benard J, Janot F. Cytology vs molecular analysis for the detection of head and neck squamous cell carcinoma in oesopharyngeal brush samples: a prospective study in 56 patients. British journal of cancer. 2003;88(11):1740–1745. doi: 10.1038/sj.bjc.6600953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spafford MF, Koch WM, Reed AL, Califano JA, Xu LH, Eisenberger CF, Yip L, Leong PL, Wu L, Liu SX. Detection of head and neck squamous cell carcinoma among exfoliated oral mucosal cells by microsatellite analysis. Clin Cancer Res. 2001;7(3):607–612. [PubMed] [Google Scholar]
- Mithani SK, Smith IM, Zhou S, Gray A, Koch WM, Maitra A, Califano JA. Mitochondrial resequencing arrays detect tumor-specific mutations in salivary rinses of patients with head and neck cancer. Clin Cancer Res. 2007;13(24):7335–7340. doi: 10.1158/1078-0432.CCR-07-0220. [DOI] [PubMed] [Google Scholar]
- Kim DM, Ramoni MF, Nevins M, Fiorellini JP. The gene expression profile in refractory periodontitis patients. Journal of periodontology. 2006;77(6):1043–1050. doi: 10.1902/jop.2006.050254. [DOI] [PubMed] [Google Scholar]


