Abstract
Background
Contagious bovine pleuropneumonia (CBPP) is a mycoplasmal disease caused by Mycoplasma mycoides subsp. mycoides SC (MmmSC). Since the disease is a serious problem that can affect cattle production in parts of Africa, there is a need for an effective and economical vaccine. Identifying which of the causative agent's proteins trigger potentially protective immune responses is an important step towards developing a subunit vaccine. Accordingly, the purpose of this study was to determine whether phage display combined with bioinformatics could be used to narrow the search for genes that code for potentially immunogenic proteins of MmmSC. Since the production of IgG2 and IgA are associated with a Th1 cellular immune response which is implicated in protection against CBPP, antigens which elicit these immunoglobulin subclasses may be useful in developing a subunit vaccine.
Results
A filamentous phage library displaying a repertoire of peptides expressed by fragments of the genome of MmmSC was constructed. It was subjected to selection using antibodies from naturally- and experimentally-infected cattle. Mycoplasmal genes were identified by matching the nucleotide sequences of DNA from immunoselected phage particles with the mycoplasmal genome. This allowed a catalogue of genes coding for the proteins that elicited an immune response to be compiled. Using this method together with computer algorithms designed to score parameters that influence surface accessibility and hence potential antigenicity, five genes (abc, gapN, glpO, lppB and ptsG) were chosen to be expressed in Escherichia coli. After appropriate site-directed mutagenesis, polypeptides representing portions of each of these proteins were tested for immunoreactivity. Of these five, polypeptides representing expression products of abc and lppB were recognised on immunoblots by sera obtained from cattle during a natural outbreak of the disease.
Conclusion
Since phage display physically couples phenotype with genotype, it was used to compile a list of sequences that code for MmmSC proteins bearing epitopes which were recognised by antibodies in the serum of infected animals. Together with the appropriate bioinformatic analyses, this approach provided several potentially useful vaccine or diagnostic leads. The phage display step empirically identified sequences by their interaction with antibodies which accordingly reduced the number of ORFs that had to be expressed for testing. This is a particular advantage when working with MmmSC since the mycoplasmal codon for tryptophan needs to be mutated to prevent it from being translated as a stop in E. coli.
Background
Contagious bovine pleuropneumonia (CBPP), a pulmonary disease caused by Mycoplasma mycoides subsp. mycoides SC (MmmSC) is a major constraint to cattle production in Africa [1]. The current vaccines are not always fully effective [2] and there remains an urgent need to control or even eradicate the disease. Although the nucleotide sequence of the MmmSC type strain PG1 genome is available, the proteins responsible for protection have not been identified. Accordingly, an important step towards a subunit vaccine would be to identify which of the potentially large number of antigens encoded in its genome [3-5] actually trigger immune responses during infection. Serum antibodies are likely to be involved in immunity since passive transfer of sera from recovered cattle can protect recipient calves [6,7], but Th1 memory lymphocytes and γδ T-cells are also active [8-10]. Identifying which antigens evoke one or more of these immune pathways therefore remains a key step in developing a subunit-based CBPP vaccine [11].
Phage display [12] makes it possible to identify antigenic proteins by using antibodies from an immune source to select binding peptides from a large repertoire of random amino acid sequences [13]. Fragmented-genome or "shotgun" display libraries [14] can directly identify genes that code for the proteins of which the immunoselected peptides form a part. Provided that artefactual binders expressed by frame-shifted or incorrectly orientated inserts are excluded [15], matching the sequence coding for an antigenic peptide with the sequence of the genome from which it is derived locates the encoding gene. The first MmmSC display library was constructed by Persson and co-workers [16] and more recently, the approach was also applied to Mycoplasma hyopneumoniae [17] as a way of identifying immunogenic polypeptides.
To locate genes coding for potentially immunogenic proteins, enzymatically-generated fragments of MmmSC chromosomal DNA were used to construct a genome-specific filamentous phage display library which was subjected to selection using antibodies from a CBPP outbreak in Botswana [18] and an experimentally infected animal from Mali designated C11 [19]. CD4+ T-cell activation and IFNγ release are associated with an IgG2 humoral immune response [20] while IgA is associated with local mucosal immunity. Accordingly, both immunoglobulin classes were used separately to select peptides as well as using total IgG. Using this approach together with computer algorithms designed to identify linear B-cell epitopes [21], five genes were chosen to be expressed for further analysis and testing to establish whether they were recognised by sera from cattle obtained during a natural outbreak of the disease.
Results
Construction of a fragmented-genome library
A pIII fusion protein phage display library of approximately 4 × 105 primary clones displaying peptides derived from the MmmSC genome was constructed by ligating DNA fragments ranging in size from approximately 30 to 900 bp as determined by agarose gel electrophoresis into a filamentous phage display vector. The probability of the genome being represented was 0.97 if the average insert size was 240 bp. DNA sequencing of 16 arbitrarily-chosen clones showed no obvious bias towards any particular region of the mycoplasmal genome. Of the 16, two copies of one of the sequenced DNA inserts were in-frame and in the correct orientation. The largest insert was 178 base pairs and the smallest 52.
To verify that the library was large and diverse enough to identify other unknown MmmSC antigens, it was first screened in a defined model system by panning on immuno-purified IgG prepared from a rabbit immune serum directed against amino acid residues 328-478 of the proline-rich MmmSC glycoprotein which is coded for by ORF5 (EMBL/GenBank accession number CAE77151). Multiple copies of overlapping peptides that mapped to a defined region on the target glycoprotein spanning residues 333 to 445 were selected (Figure 1).
Figure 1.
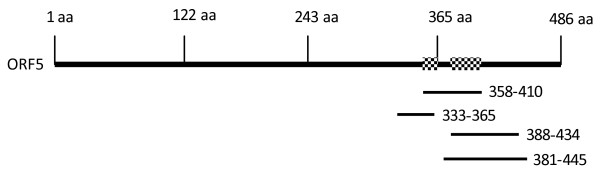
Schematic representation showing alignment of the hypothetical proline-rich glycoprotein ORF5 with selected phage fusion peptides. Antigenic regions suggested by the presence of overlapping sequences located between amino acid residues 358-365 and 388-410 are indicated by shading.
Identification of antigenic peptides
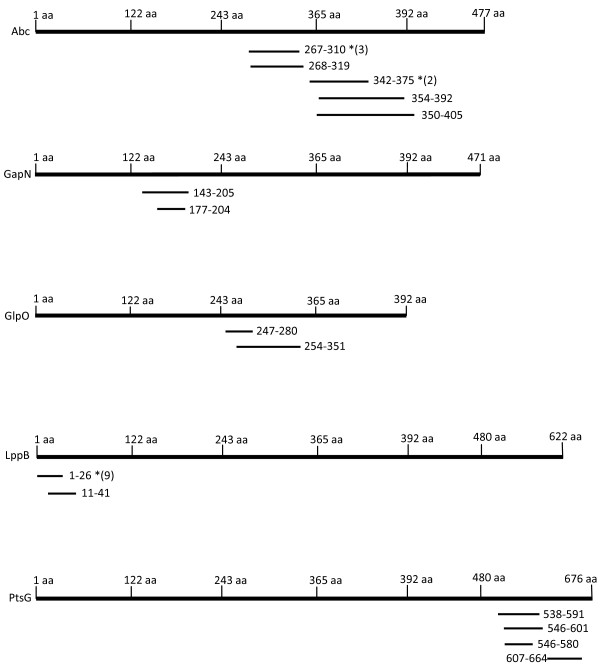
Panning experiments aimed at selecting phage displayed peptides that were recognised by the total serum IgG, the IgG2 component and the IgA subpopulation of animal C11 and pooled serum from seven cattle affected by a CBPP outbreak in Botswana were performed. Clones selected by each immunoglobulin class were randomly chosen for DNA sequencing. After panning, all clones were in-frame and without stops. Fifteen peptides recognised by the total IgG preparation from the serum of animal C11 matched regions in eight different MmmSC proteins in the EMBL/GenBank database, while 29 peptides that had been selected using IgG2 corresponded with regions on 17 different proteins (Table 1). The library was not panned with total serum IgG from Botswana but peptides were selected using IgG2 which matched parts of four of the proteins identified by the C11 antiserum (GyrB, GlpO, LppB and Abc). Bronchoalveolar lavage (BAL) IgA from animal C11 specifically selected four peptides that could be correlated with portions of MmmSC proteins while the Botswana serum selected six peptides that matched three MmmSC proteins. A single C11 IgG2 peptide matched MmmSC glyceraldehyde-3-phosphate dehydrogenase (GapN) which was also identified by one C11 IgA peptide. Four peptides that were isolated with C11 IgG2 matched the ABC transporter, substrate-binding component protein (Abc). This protein was also identified by the IgG from Mali (i.e. animal C11) and IgG2 and IgA from the Botswana serum pool. Glycerol-3-phosphate oxidase (GlpO) was selected by peptides from both C11 and Botswana serum IgG2. Three of the Botswana IgA peptides matched to the mycoplasmal prolipoprotein B (LppB) amino acid sequence. This protein was also selected with both the C11 and Botswana IgG2 serum antibodies. The PTS system, glucose-specific IIBC component (PtsG) was identified by four peptides (Figure 2).
Table 1.
List of phage displayed peptides and their corresponding MmmSC proteins identified after selection with the indicated antibody subpopulations from animal C11 and the Botswana outbreak
| Mmm SC protein match | Identified by serum from: | Accession numbera | Locus Tag | |
|---|---|---|---|---|
| Animal C11c | Botswana outbreakc | |||
| IgG panning | ||||
| Putative variable surface protein | Yes (3) | ND | CAE77433 | MSC_0818 |
| Cell division protein (FtsZ) | Yes (1) | ND | CAE77211 | MSC_0588 |
| ABC transporter, substrate-binding component (Abc) | Yes (1) | ND | CAE77420b | MSC_0804 |
| Prolipoprotein (Lpp) | Yes (1) | ND | CAE77256 | MSC_0635 |
| D-lactate dehydrogenase (Ldh) | Yes (2) | ND | CAE76687 | MSC_0034 |
| DNA gyrase subunit A (GyrA) | Yes (2) | ND | CAE76660 | MSC_0007 |
| DNA gyrase subunit B (GyrB) | Yes (1) | ND | CAE76659 | MSC_0006 |
| PTS system, glucose-specific, IIBC component (PtsG) | Yes (4) | ND | CAE77485b | MSC_0873 |
| IgG2 panning | ||||
| Transposase ISMmy1C | Yes (1) | No | CAE77424 | MSC_0808 |
| 30S ribosomal protein S5 (RpsE) | Yes (1) | No | CAE77346 | MSC_0728 |
| Putative glucose-specific IIABC component (PtsG) | Yes (1) | No | CAE76806 | MSC_0161 |
| Prolipoprotein (Lpp) | Yes (1) | No | CAE77256 | MSC_0635 |
| Prolipoprotein LppC (LppC) | Yes (1) | No | CAE76772 | MSC_0122 |
| Alanine-tRNA ligase (AlaS) | Yes (1) | No | CAE76823 | MSC_0178 |
| DNA gyrase subunit B (GyrB) | Yes (1) | Yes (1) | CAE76659 | MSC_0006 |
| Methionine adenosyltransferase (MetK) | Yes (1) | No | CAE77120 | MSC_0492 |
| Glyceraldehyde-3-phosphate dehydrogenase (GapN) | Yes (1) | No | CAE77137b | MSC_0509 |
| Glycerol-3-phosphate oxidase (GlpO) | Yes (1) | Yes (1) | CAE76900b | MSC_0259 |
| D-lactate dehydrogenase (Ldh) | Yes (1) | No | CAE76687 | MSC_0034 |
| P115-like protein with SMC_C motif | Yes (7) | No | CAE77104 | MSC_0476 |
| Ribose/Galactose ABC transporter, ATP-binding component (RbsA) | Yes (3) | No | CAE76663 | MSC_0010 |
| Prolipoprotein B (LppB) | Yes (2) | Yes (5) | CAE77147b | MSC_0519 |
| Glucose inhibited division protein A (GidA) | Yes (1) | No | CAE77646 | MSC_1042 |
| ABC transporter, substrate-binding component (Abc) | Yes (4) | Yes (1) | CAE77420b | MSC_0804 |
| Fructose-bisphosphate aldolase class II (FbaA2) | Yes (1) | No | CAE76786 | MSC_0139 |
| IgA panning | ||||
| Glyceraldehyde-3-phosphate dehydrogenase (NADP) (GapN) | Yes (1) | No | CAE77137b | MSC_0509 |
| Prolipoprotein B (LppB) | No | Yes (3) | CAE77147b | MSC_0519 |
| ABC transporter, substrate-binding component (Abc) | No | Yes (2) | CAE77420b | MSC_0804 |
| CTP synthase (CtrA) | Yes (1) | No | CAE76781 | MSC_0134 |
| Prolipoprotein LppC (LppC) | No | Yes (1) | CAE76772 | MSC_0122 |
| Thioredoxin reductase (NADPH) (TrxB) | Yes (1) | No | CAE77547 | MSC_0938 |
| Asparagine-tRNA ligase (AsnS) | Yes (1) | No | CAE76732 | MSC_0080 |
a EMBL/GenBank accession numbers for the protein sequences.
b Proteins that were expressed in this study (see Figure 3).
c Figures in parenthesis indicate numbers of peptides selected
ND - not determined
Figure 2.
Schematic representation showing alignment of the five selected proteins with phage displayed peptides obtained with CBPP immune sera. The entire open reading frames that code for the five proteins (Abc, GapN, GlpO, LppB and PtsG) selected for expression aligned with the phage displayed immunogenic polypeptides that were isolated by panning the fragmented MmmSC genome phage display library against CBPP immune sera. Asterisks indicate peptides isolated more than once and the numbers in parentheses specify the number of repeated isolations for such peptides.
Expression and antigenicity of MmmSC polypeptides
Given the difficulty of easily expressing recombinant mycoplasmal proteins due to codon usage, a short list of genes to be expressed in E. coli was compiled. Based on predetermined selection criteria discussed below and using algorithms for antigenicity and surface location together with knowledge of their established characteristics, this list comprised the ABC transporter Abc, GapN, GlpO, LppB and PtsG. Site-directed mutations allowed 62% of the ABC transporter Abc (residues 184 to 477), 96% of GlpO (residues 6 to 380) and 61% of LppB (residues 1 to 376) to be expressed. Fifty percent of GapN (residues 88 to 322) and 38% of the PtsG (residues 421 to 676) polypeptides could be expressed without mutating (Figure 2).
Four of the resulting recombinant polypeptides were found in the E. coli insoluble pellet fraction. Their Mrs were within the range expected from their amino acid sequences. The calculated Mrs were: ABC transporter Abc, Mr ~33 kDa; GapN, Mr ~26 kDa; GlpO, Mr ~41 kDa; and LppB, Mr 43 ~kDa (compare with Figure 3). PtsG was isolated from the soluble fraction using nickel chelation, but it manifested in PAGE as two bands with Mrs ~70 and ~45 kDa (Figure 3; calculated Mr ~28 kDa).
Figure 3.
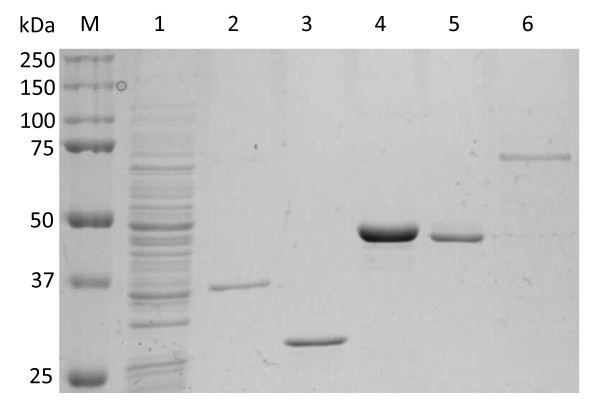
Expression of the five selected proteins in E. coli. SDS-PAGE (10%) showing segments of the protein antigens that were expressed in E. coli. Lanes: M, molecular mass standards; 1, 12 μg of total antigen of MmmSC strain 8740; 2-6, expressed segments of proteins Abc, GapN, GlpO, LppB and PtsG, respectively.
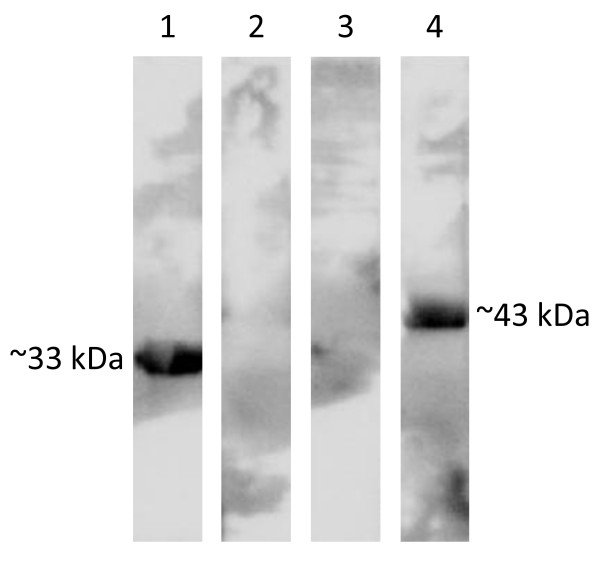
The pool of the seven sera obtained from the Botswana outbreak was also used in immunoblotting. The pool reacted with the expressed Abc and LppB polypeptides (Figure 4). The PtsG polypeptide bands were probed separately with serum obtained from an experimental infection. This immunoblot, however, showed multiple bands that apparently reacted with the pooled sera (not shown).
Figure 4.
Chemiluminescent immunoblot. Recognition of the ABC transporter Abc (lane 1) and lipoprotein LppB (lane 4) polypeptides that were expressed in E. coli by a pool of sera obtained from cattle that were naturally infected with CBPP during the 1995 Botswana outbreak. The GapN (lane 2) and GlpO (lane 3) polypeptides were not recognised in this test format.
Discussion
When a pathogen infects an animal, its epitopes leave an "imprint" in the form of a spectrum of disease-specific antibody paratopes in the serum. Most animals are therefore likely to have antibodies directed against a large number of foreign epitopes. The strategy pursued in this study was to use this complex mixture of antibodies to select binders from a limited repertoire of sequences derived from the genome of MmmSC, thereby focussing the phage display selection process on relevant epitopes. These binders were matched to open reading frames present in the genome. Unlike immunoblotting, this approach also identified the genes that coded for the antigenic proteins.
The fragmented genome library covered approximately 97% of the mycoplasmal genome. While adequate for its purpose, it cannot, however, be considered to have been completely random since among the 1016 proteins encoded in the genome of MmmSC type strain PG1, 797 (78.4%) contain at least one UGAtrp codon, which is read as stop codon in E. coli. Moreover, the frequency of UGAtrp codons in coding sequences of MmmSC genes is relatively high: 1.00% in contrast to 0.05% of UGGtrp codons. This means that epitopes containing such stops could be disrupted. Moreover, in a phage display system, the secreted phages would be unlikely to display large oligopeptides or those that resisted being transported through the bacterial membrane or periplasm. The functionality of the library was therefore first tested by selecting peptides with a polyclonal rabbit antiserum directed against a single recombinant MmmSC polypeptide, i.e., the C-terminal proline-rich portion of ORF5. The ORF5 gene product [22] corresponds to the 486 aa protein having EMBL/GenBank accession number CAE77151 [5]. The ORF5 antiserum selected a series of overlapping peptides thereby identifying a B cell epitope and confirming that polyclonal serum could specifically select antigenic peptides from the phage displayed repertoire. A further important indication that the peptides had been specifically selected was that prior to panning, only 12.5% of the sequenced inserts contained in the library were both in-frame and in the correct orientation for translation as mycoplasmal peptides. In contrast, after panning, all were in-frame and without stops. This finding, together with the way in which immunoselection yielded multiple copies of some peptides (particularly those that overlapped but were not identical), provided additional evidence that the strategy was essentially sound.
While 26 different MmmSC genes matched sequences selected by phage display, those chosen for expression in E. coli were required to have fulfilled criteria which were considered to have a bearing on their usefulness as possible vaccine antigens. Firstly, since the pathogen enters the animal via the nasal passages, preference was given to genes selected by IgA from Mali and Botswana. Secondly, only genes that were identified by multiple overlapping copies of each phage displayed peptide qualified. Thirdly, peptides that fulfilled the first two criteria, but which were selected with a negative bovine serum were excluded. Finally, the protein's likely function or structural position was taken into account with a focus on previously-identified membrane-associated proteins [23] which also fulfilled antigenicity criteria as predicted by bioinformatics analyses. Although not excluded as being potentially useful, any overlapping sequences that coded for internally located proteins e.g. the DNA gyrase subunit B (Table 1) were not investigated in this study. Applying these criteria allowed us to focus on the ABC transporter, substrate-binding component protein (Abc), the glyceraldehyde-3-phosphate dehydrogenase (GapN), the glycerol-3-phosphate oxidase (GlpO), the prolipoprotein B (LppB) and the PTS system, glucose-specific IIBC component (PtsG) for expression in E. coli. By applying these criteria we do not exclude further studies on any of the other apparently antigenic proteins as vaccine or diagnostic targets. Even though the proliporotein LppC fulfilled our criteria, some of the peptides which matched the amino acid sequence included sequences of unknown origins which did not align with the target ORF (not shown).
ABC transporter proteins act on a wide variety of substrates that include sugars, peptides, proteins and toxins [24]. For example, the ATP-binding cassette (ABC) transporter GtsABC together with GlpO forms part of the glycerol catabolism pathway associated with MmmSC virulence [25,26]. GlpO itself has been identified as a significant MmmSC virulence factor [27-29]. GapN, in contrast, may play a role in transcription [30] and apoptosis [31]. Membrane lipoproteins that interact with host cells can stimulate the release of pro-inflammatory cytokines [32] and are major antigens [23,33,34]. The lipoprotein (LppB) identified by the phage display is found in African and Australian strains of MmmSC, but not in the less virulent European strains [22]. The ptsG gene, which occurs in duplicate in many MmmSC strains [35], encodes the permease of the phosphoenolpyruvate:glucose phosphotransferase system. It has also been implicated in intraclonal antigenic variation [36], a possible factor in the evasion of the host immune response. With the exception of GapN, these proteins are likely to be involved in pathogenicity or to be accessible to B cell receptors. They therefore have potential either in vaccine or diagnostics development. Only two of the expressed polypeptides, however, reacted in immunoblots, possibly because their epitopes in the denatured state most faithfully resembled the phage displayed peptides that were originally bound in the selection process. Although phage display of necessity identified B cell epitopes, it is not yet clear whether it is this response, or a cell-mediated one based on CD4 [37], which is a primarily responsible for protection. The proteins identified using phage display will therefore also need to be tested for their ability to cause primed lymphocytes to proliferate and produce IFNγ.
Conclusion
Constructing a phage library that displays peptides derived from the actual organism of interest made it possible to narrow the search for genes that code for antigenic and hence potentially immunogenic proteins of the mycoplasma that causes CBPP. Because of their interaction with antibodies in the serum of infected animals, these proteins may be regarded as potential vaccine targets, in particular those selected using IgG2 and IgA. A model epitope discovery system has shown that many antigenic peptides obtained from such phage libraries have potential as vaccine antigens [38]. It may therefore also be worth examining the actual antigenic MmmSC peptides that were selected from the epitope library as possible components of a subunit vaccine. Knowing which proteins are antigenic may help to identify targets for generating knockout mutants for use as genetically defined vaccines [39]. Lastly, phage display was able to identify polypeptides that were recognised in immunoblotting by serum from animals that were affected by a natural disease outbreak. As well as having potential as vaccine antigens, such peptides may be useful diagnostic targets.
Methods
Strains, growth conditions and vectors
MmmSC strain 8740 from Cameroon, provided by Dr. L. Dedieu, CIRAD-EMVT, Montpellier, France, was cultured in PPLO broth medium (Difco, Detroit, MI, USA) containing thallium acetate (1% w/v), ampicillin (0.5 mg/ml), heat-inactivated horse serum (20%) [40] and supplemented with 0.1 volumes of 25% fresh yeast extract. Mycoplasmas were grown at 37°C in 5% CO2 until stationary growth phase and harvested by centrifugation at 20000 g for 20 min. For genetic manipulation and subcloning, E. coli strains TG1 (Stratagene, La Jolla, CA, USA), DH5α, Top10 (Invitrogen, Carlsbad, CA, USA) and BL21 Star™ (DE3) (Invitrogen) were used. The phage display vector fdtet 8.53 was a gift from Dr. V. K. Chaudhary, University of Delhi, New Delhi, India.
Antisera, antibodies, and immunoblot analysis
Anti-ORF5 immune serum was obtained by injecting rabbits with amino acid residues 328-478 of the 486 aa proline-rich MmmSC ORF5 [22]. Bovine sera and bronchoalveolar lavage (BAL) from animals C11 (recovered from a sub-acute to chronic experimental infection) and T1 (uninfected control) were from Dr. M. Niang, Central Veterinary Laboratory, Bamako, Mali [4,19]. The seven bovine sera used in screening and immunoblotting were a kind gift from the Botswana National Veterinary Laboratory in Gabarone, Botswana [18]. Antibodies were isolated using ImmunoPure® Protein G columns (Pierce, Rockford, IL, USA). Antibody-containing fractions were applied to Excellulose™ GF-5 Desalting columns (Pierce). Before selection by panning, unwanted filamentous phage antibodies were removed from the C11 serum by cross-absorption [41]. BAL IgA from animal C11 and serum IgA from Botswana cattle were used in pannings, but a limited volume was available and the samples were not cross-absorbed. Negative control pannings using BAL IgA and total IgG from the control animal (T1) were also performed.
Immunoblotting was performed according to standard protocols. A volume of 10 μl of each of the seven sera from Botswana were added to 5 ml of 1% milk powder (MP) suspended in PBS, pH 7.4. Blots were incubated overnight in the pool of diluted sera at room temperature. For the detection of bound antibodies, sheep horseradish peroxidise conjugated anti-bovine IgG (catalogue No. PP200; The Binding Site, Birmingham, UK) was diluted 1:10000 and incubated with the blot for an hour at room temperature. Bound antibodies were detected after incubation of the blot with SuperSignal® West Pico chemiluminescent substrate (Pierce) using the Lumi-Imager from Roche Molecular Biochemicals.
Display library construction
Phage library construction using the pIII phage display vector fdtet 8.53 was as described by Gupta and co-workers [42]. This entailed ligating blunt-ended fragments of MmmSC genomic DNA in the presence of the restriction enzyme SrfI and T4 DNA ligase. The extent to which the genome was represented in the primary library with a theoretical probability of 0.99 was calculated using the method of Clarke and Carbon [43]. To deplete the resulting phage repertoire of any peptides that may have been susceptible to binding by irrelevant antibodies present in healthy bovine serum, a 50 μl volume was incubated with 2 mg of naïve bovine IgG at 4°C overnight. Any phage/IgG aggregates were removed by centrifugation for 5 min at 15000 g and the supernatant transferred to a new tube. The depleted library was stored at 4°C.
Affinity selection from the phage library
The peptide display library was subjected to three successive rounds of affinity selection essentially as described [15]. For selection of fusion phages from the library with IgG2a or IgA antibodies, the polystyrene Petri dish (Falcon 1007; Becton Dickinson, Lincoln Park, NJ, USA) used for panning was first coated with antibodies specific for the desired bovine immunoglobulin subclass at a concentration of approximately 20 μg/ml before the blocking step.
Identification of antigens
Sequences of phage displayed peptides were compared with the EMBL/GenBank database using the BLAST programs [44]. Flexibility, hydrophilicity, polarity and surface properties were scored using the programs Bcepred http://www.imtech.res.in/raghava/bcepred/ and BepiPred http://www.cbs.dtu.dk/services/BepiPred/[21,45].
Cloning, site-directed mutagenesis, expression and purification of proteins
For expression, the relevant sequences of the targeted genes were amplified from genomic DNA and cloned in the pET100/D-TOPO® E. coli expression vector (Invitrogen), or in the case of PtsG, in the pQE-TriSystem His·Strep 2 vector (Qiagen). Site-directed mutagenesis (QuikChange Site-Directed mutagenesis kit; Stratagene) was used to change mycoplasmal UGAtrp codons to E. coli UGGtrp codons. Transformed E. coli cells were inoculated into Overnight Express Instant TB medium from Novagen (Madison, WI, USA). Following overnight induction, bacterial cells were lysed using Novagen BugBuster® reagent, after which the supernatant fluids and cell pellets were analysed by SDS-PAGE and immunoblotting on a PVDF membrane using standard protocols. Proteins for PAGE analysis were purified by using ProBond nickel chelate chromatography kits as described by the manufacturer (Invitrogen).
Authors' contributions
The first two authors made equivalent contributions to the study; DRM constructed and screened the epitope library. She was also responsible for mutating and expressing four of the genes as well as testing the resulting polypeptides in immunoblotting. AM identified, mutated and expressed the ptsG gene in E. coli and analysed and correlated the data after DRM left the Onderstepoort Veterinary Institute. EMV identified and expressed ORF5 and raised the rabbit immune serum. DHD conceptualised the study, supervised all facets of the research and is responsible for the manuscript as submitted. The authors have read and approved the final version.
Contributor Information
Dubravka R Miltiadou, Email: miltiadou@bigpond.com.
Arshad Mather, Email: MatherA@arc.agric.za.
Edy M Vilei, Email: edy.vilei@vbi.unibe.ch.
Dion H Du Plessis, Email: duplessisdh@arc.agric.za.
Acknowledgements
We are grateful to Laurence Dedieu, François Thiacourt (CIRAD-EMVT, Montpellier, France) and Joachim Frey (Institute of Veterinary Bacteriology, University of Bern, Switzerland) for stimulating discussions. We thank Jane Banda and Frances Jordaan (Onderstepoort Veterinary Institute, Republic of South Africa) for their technical help. The South African portion of this project was supported by the European Union (FP 6 INCO-DEV, Project CBPPVAC) and the General Directorate for Development and International Cooperation, French Ministry of Foreign and European Affairs (PSF No. 2003-24 LABOVET). The contribution of EMV was funded by the Wellcome Trust, London, UK, grant No. 075804. We thank Dr Philippe Totté of CIRAD for his constructive comments regarding the manuscript.
References
- Tambi NE, Maina WO, Ndi C. An estimation of the economic impact of contagious bovine pleuropneumonia in Africa. Rev Sci Tech. 2006;9:999–1011. [PubMed] [Google Scholar]
- Mbulu RS, Tjipura-Zaire G, Lelli R, Frey J, Pilo P, Vilei EM, Mettler F, Nicholas RA, Huebschle OJ. Contagious bovine pleuropneumonia (CBPP) caused by vaccine strain T1/44 of Mycoplasma mycoides subsp. mycoides SC. Vet Microbiol. 2004;9:229–234. doi: 10.1016/j.vetmic.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Gonçalves R, Regalla J, Nicolet J, Frey J, Nicholas R, Bashiruddin J, De Santis P, Gonçalves AP. Antigen heterogeneity among Mycoplasma mycoides subsp. mycoides SC isolates: discrimination of major surface proteins. Vet Microbiol. 1998;9:13–28. doi: 10.1016/S0378-1135(98)00214-4. [DOI] [PubMed] [Google Scholar]
- Niang M, Diallo M, Cisse O, Kone M, Doucoure M, Roth JA, Balcer-Rodrigues V, Dedieu L. Pulmonary and serum antibody responses elicited in zebu cattle experimentally infected with Mycoplasma mycoides subsp. mycoides SC by contact exposure. Vet Res. 2006;9:733–744. doi: 10.1051/vetres:2006032. [DOI] [PubMed] [Google Scholar]
- Westberg J, Persson A, Holmberg A, Goesmann A, Lundeberg J, Johansson KE, Pettersson B, Uhlen M. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of contagious bovine pleuropneumonia (CBPP) Genome Res. 2004;9:221–227. doi: 10.1101/gr.1673304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiga WN, Roberts DH, Kakoma I, Rurangirwa FR. Passive immunity to contagious bovine pleuropneumonia. Res Vet Sci. 1975;9:330–332. [PubMed] [Google Scholar]
- Masiga WN, Windsor RS. Immunity to contagious bovine pleuropneumonia. Vet Rec. 1975;9:350–351. doi: 10.1136/vr.97.18.350. [DOI] [PubMed] [Google Scholar]
- Dedieu L, Balcer-Rodrigues V, Cisse O, Diallo M, Niang M. Characterisation of the lymph node immune response following Mycoplasma mycoides subsp. mycoides SC infection in cattle. Vet Res. 2006;9:579–591. doi: 10.1051/vetres:2006020. [DOI] [PubMed] [Google Scholar]
- Dedieu L, Balcer-Rodrigues V, Yaya A, Hamadou B, Cisse O, Diallo M, Niang M. Gamma interferon-producing CD4 T-cells correlate with resistance to Mycoplasma mycoides subsp. mycoides S.C. infection in cattle. Vet Immunol Immunopathol. 2005;9:217–233. doi: 10.1016/j.vetimm.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Totté P, Rodrigues V, Yaya A, Hamadou B, Cisse O, Diallo M, Niang M, Thiaucourt F, Dedieu L. Analysis of cellular responses to Mycoplasma mycoides subsp. mycoides small colony biotype associated with control of contagious bovine pleuropneumonia. Vet Res. 2008;9:8. doi: 10.1051/vetres:2007046. [DOI] [PubMed] [Google Scholar]
- Dedieu-Engelmann L. Contagious bovine pleuropneumonia: a rationale for the development of a mucosal sub-unit vaccine. Comp Immunol Microbiol Infect Dis. 2008;9:227–238. doi: 10.1016/j.cimid.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;9:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- Wang LF, Yu M. Epitope identification and discovery using phage display libraries: applications in vaccine development and diagnostics. Curr Drug Targets. 2004;9:1–15. doi: 10.2174/1389450043490668. [DOI] [PubMed] [Google Scholar]
- Wang LF, du Plessis DH, White JR, Hyatt AD, Eaton BT. Use of a gene-targeted phage display random epitope library to map an antigenic determinant on the bluetongue virus outer capsid protein VP5. J Immunol Methods. 1995;9:1–12. doi: 10.1016/0022-1759(94)00235-O. [DOI] [PubMed] [Google Scholar]
- Fehrsen J, du Plessis DH. Cross-reactive epitope mimics in a fragmented-genome phage display library derived from the rickettsia, Cowdria ruminantium. Immunotechnology. 1999;9:175–184. doi: 10.1016/S1380-2933(98)00018-9. [DOI] [PubMed] [Google Scholar]
- Persson A, Jacobsson K, Frykberg L, Johansson KE, Poumarat F. Variable surface protein Vmm of Mycoplasma mycoides subsp. mycoides small colony type. J Bacteriol. 2002;9:3712–3722. doi: 10.1128/JB.184.13.3712-3722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler J, Nieswandt S, Gerlach GF, Meens J, Schirrmann T, Hust M. Identification of immunogenic polypeptides from a Mycoplasma hyopneumoniae genome library by phage display. Appl Microbiol Biotechnol. 2008;9:447–458. doi: 10.1007/s00253-008-1576-1. [DOI] [PubMed] [Google Scholar]
- Amanfu W, Masupu KV, Adom EK, Raborokgwe MV, Bashiruddin JB. An outbreak of contagious bovine pleuropneumonia in Ngamiland district of north-western Botswana. Vet Rec. 1998;9:46–48. doi: 10.1136/vr.143.2.46. [DOI] [PubMed] [Google Scholar]
- Niang M, Diallo M, Cissé O, Koné M, Doucouré M, Le Grand D, Balcer V, Dedieu L. Transmission expérimentale de la péripneumonie contagieuse bovine par contact chez les zébus: étude des aspects cliniques et pathologiques de la maladie. Revue d'Élevage et de Médecine Vétérinaire des Pays Tropicaux. 2004;9:7–14. [Google Scholar]
- Balcer V, Dedieu L. In: COST Action 826-Mycoplasmas of ruminants: pathogenicity, diagnostics, epidemiology and molecular genetics. Bergonier D, Berthelot X, Frey J, editor. Luxembourg: Office for Official Publications of the European Communities; 2000. Cell-mediated immune response induced in cattle by Mycoplasma mycoides subsp. mycoides: comparison between infected and vaccinated animals; pp. 97–100. [Google Scholar]
- Saha S, Raghava GPS. In: ICARIS LNCS 3239. Nicosia G, Cutello V, Bentley PJ, Timis J, editor. Berlin: Springer; 2004. BcePred: Prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties; pp. 197–204. [Google Scholar]
- Vilei EM, Abdo E-M, Nicolet J, Botelho A, Gonçalves R, Frey J. Genomic and antigenic differences between the European and African/Australian clusters of Mycoplasma mycoides subsp. mycoides SC. Microbiology. 2000;9:477–486. doi: 10.1099/00221287-146-2-477. [DOI] [PubMed] [Google Scholar]
- Pilo P, Frey J, Vilei EM. Molecular mechanisms of pathogenicity of Mycoplasma mycoides subsp. mycoides SC. Vet J. 2007;9:513–521. doi: 10.1016/j.tvjl.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;9:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Vilei EM, Frey J. Genetic and biochemical characterization of glycerol uptake in Mycoplasma mycoides subsp. mycoides SC: its impact on H2O2 production and virulence. Clin Diagn Lab Immunol. 2001;9:85–92. doi: 10.1128/CDLI.8.1.85-92.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic SP, Vilei EM, Frey J. Characterization of a chromosomal region of Mycoplasma sp. bovine group 7 strain PG50 encoding a glycerol transport locus (gtsABC) Microbiology. 2003;9:195–204. doi: 10.1099/mic.0.25742-0. [DOI] [PubMed] [Google Scholar]
- Vilei EM, Correia I, Ferronha MH, Bischof DF, Frey J. β-D-Glucoside utilization by Mycoplasma mycoides subsp. mycoides SC: possible involvement in the control of cytotoxicity towards bovine lung cells. BMC Microbiol. 2007;9:31. doi: 10.1186/1471-2180-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilo P, Vilei EM, Peterhans E, Bonvin-Klotz L, Stoffel MH, Dobbelaere D, Frey J. A metabolic enzyme as a primary virulence factor of Mycoplasma mycoides subsp. mycoides Small Colony. J Bacteriol. 2005;9:6824–6831. doi: 10.1128/JB.187.19.6824-6831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof DF, Janis C, Vilei EM, Bertoni G, Frey J. Cytotoxicity of Mycoplasma mycoides subsp. mycoides small colony type to bovine epithelial cells. Infect Immun. 2008;9:263–269. doi: 10.1128/IAI.00938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Roeder RG, Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;9:255–266. doi: 10.1016/S0092-8674(03)00552-X. [DOI] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD. et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;9:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- Rawadi G, Roman-Roman S. Mycoplasma membrane lipoproteins induce proinflammatory cytokines by a mechanism distinct from that of lipopolysaccharide. Infect Immun. 1996;9:637–643. doi: 10.1128/iai.64.2.637-643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilo P, Martig S, Frey J, Vilei EM. Antigenic and genetic characterisation of lipoprotein LppC from Mycoplasma mycoides subsp. mycoides SC. Vet Res. 2003;9:761–775. doi: 10.1051/vetres:2003035. [DOI] [PubMed] [Google Scholar]
- Bonvin-Klotz L, Vilei EM, Kühni-Boghenbor K, Kapp N, Frey J, Stoffel MH. Domain analysis of lipoprotein LppQ in Mycoplasma mycoides subsp. mycoides SC. Antonie Van Leeuwenhoek. 2008;9:175–183. doi: 10.1007/s10482-007-9191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof DF, Vilei EM, Frey J. Genomic differences between type strain PG1 and field strains of Mycoplasma mycoides subsp. mycoides small-colony type. Genomics. 2006;9:633–641. doi: 10.1016/j.ygeno.2006.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaurivaud P, Persson A, Le Grand D, Westberg J, Solsona M, Johansson KE, Poumarat F. Variability of a glucose phosphotransferase system permease in Mycoplasma mycoides subsp. mycoides Small Colony. Microbiology. 2004;9:4009–4022. doi: 10.1099/mic.0.27247-0. [DOI] [PubMed] [Google Scholar]
- Jores J, Nkando I, Sterner-Kock A, Haider W, Poole J, Unger H, Muriuki C, Wesonga H, Taracha EL. Assessment of in vitro interferon-γ responses from peripheral blood mononuclear cells of cattle infected with Mycoplasma mycoides ssp. mycoides small colony type. Vet Immunol Immunopathol. 2008;9:192–197. doi: 10.1016/j.vetimm.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Matthews LJ, Davis R, Smith GP. Immunogenically fit subunit vaccine components via epitope discovery from natural peptide libraries. J Immunol. 2002;9:837–846. doi: 10.4049/jimmunol.169.2.837. [DOI] [PubMed] [Google Scholar]
- Janis C, Bischof D, Gourgues G, Frey J, Blanchard A, Sirand-Pugnet P. Unmarked insertional mutagenesis in the bovine pathogen Mycoplasma mycoides subsp. mycoides SC: characterization of a lppQ mutant. Microbiology. 2008;9:2427–2436. doi: 10.1099/mic.0.2008/017640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poonia B, Sharma AK. Modulation of lympho-proliferative responses of ovine peripheral blood mononuclear cells by Mycoplasma mycoides ssp. mycoides (LC type) Vet Immunol Immunopathol. 1998;9:323–335. doi: 10.1016/S0165-2427(98)00141-X. [DOI] [PubMed] [Google Scholar]
- Smith GP, Petrenko VA, Matthews LJ. Cross-linked filamentous phage as an affinity matrix. J Immunol Methods. 1998;9:151–161. doi: 10.1016/S0022-1759(98)00087-8. [DOI] [PubMed] [Google Scholar]
- Gupta S, Arora K, Sampath A, Khurana S, Singh SS, Gupta A, Chaudhary VK. Simplified gene-fragment phage display system for epitope mapping. Biotechniques. 1999;9:328–334. doi: 10.2144/99272st04. [DOI] [PubMed] [Google Scholar]
- Clarke L, Carbon J. A colony bank containing synthetic Col E1 hybrid plasmids representative of the entire E. coli genome. Cell. 1976;9:91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;9:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;9:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]