Abstract
The expansion of unstable microsatellites is the cause of a number of inherited neuromuscular and neurological disorders. While these expanded repeats can be located in either the coding or non-coding regions of genes, toxic RNA transcripts have been primarily implicated in the pathogenesis of non-coding expansion diseases. In this review, we briefly summarize studies which support this RNA-mediated toxicity model for several neurologic disorders and highlight how pathogenic RNAs might negatively impact nervous system functions. However, it is important to note that the distinction between coding versus non-coding regions has become muddled by recent observations that the transcribed portion of the genome, or transcriptome, is considerably larger than previously appreciated. Thus, we also explore the possibility that a combination of protein and RNA gain-of-function events underlie some microsatellite expansion diseases.
Keywords: RNA-mediated disease, microsatellite, splicing
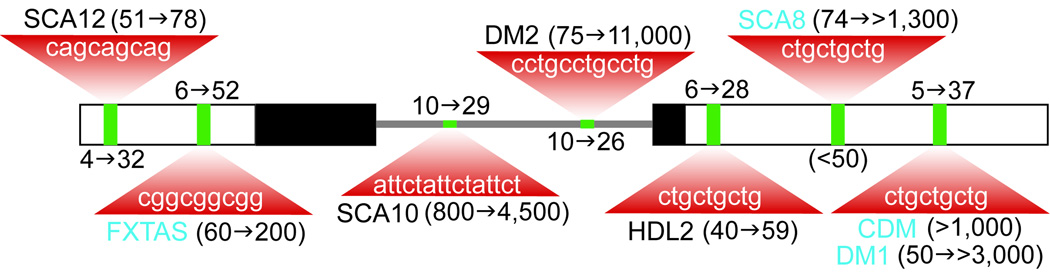
An important class of inherited neurological disorders is distinguished by genetic anticipation, an intergenerational increase in disease severity combined with a decreased age-of-onset. The discovery two decades ago that anticipation resulted from the aberrant expansion of microsatellites led to a flurry of studies designed to clarify the molecular etiology for each disease and the realization that the location of the repeat within an affected gene suggested a pathogenic mechanism. For protein coding region expansions, such as the CAG repeat expansion or (CAG)n in the Huntington disease gene HTT, the production of mutant proteins containing polyglutamine (polyQ) results in toxicity possibly because mutant HTT protein, or polyQ fragments generated by proteolysis, interfere with a variety of cellular pathways [14]. In contrast, microsatellite expansions in the non-coding regions of genes, including introns and untranslated regions (5’- and 3’-UTRs), are associated with three types of spinocerebellar ataxias (SCA8, 10, 12), fragile X-associated tremor/ataxia syndrome (FXTAS), Huntington disease-like 2 (HDL2) and two types of myotonic dystrophy (DM1, DM2) (Fig. 1). The finding that some expansions are pathogenic at the RNA level was first achieved for DM1 [4, 40, 42, 45]. DM1 is a multi-systemic disorder caused by the expansion of a CTG repeat in the 3’ untranslated region of the DMPK gene while DM2 results from the expansion of a related non-coding microsatellite, CCTG, in the first intron of ZNF9 [27]. Surprisingly, these expansions result in a similar adult-onset neuromuscular disorder with such characteristic features as muscle hyperexcitability (myotonia), cardiac conduction defects and insulin resistance. While the DM brain is also profoundly affected, we know very little about the underlying molecular events which lead to dysfunction in the nervous system. These CNS effects can be subdivided into developmental abnormalities, such as mental retardation (DM1 only), and adult-onset degenerative changes, which include executive function deficits, hypersomnia, progressive memory problems and cerebral atrophy (DM1 and DM2) [34–35, 37].
Fig. 1.
Microsatellites in ‘noncoding’ regions. A model two exon gene is illustrated with 5’- and 3’ UTRs (open boxes), ORF (black boxes) and an intron (grey line) together with the relative positions (green boxes) of seven unstable microsatellite diseases, SCA12, FXTAS, SCA10, DM2, HDL2, SCA8 and DM1/CDM. The microsatellite repeat range of mutant alleles (parentheses next to highlighted disease) are indicated by a red triangle together with normal allele repeats (opposite side of green boxes). Bidirectional transcription has been documented for those disease genes highlighted in blue.
Several lines of evidence argue that DM is an RNA-mediated disease [4, 40, 42]. First, the DM inheritance pattern is autosomal dominant but the expansion mutation is located in a non-coding region of the DMPK gene so a gain-of-function at the protein level is unlikely. Second, DMPK and ZNF9 mutant transcripts accumulate in nuclear RNA, or ribonuclear, foci that have been proposed to interfere with normal nuclear functions [6, 50]. Although full-length DMPK mRNA accumulates in ribonuclear foci and this may compromise DMPK protein levels, splicing separates the CCUG repeat from the mRNA so ZNF9 protein levels are unaffected by the DM2 expansion mutation eliminating ZNF9 haploinsufficiency as a cause of this disease [31]. Third, common manifestations of DM disease are observed in several transgenic poly(CUG) mice which have been generated by placing expanded repeats in the non-coding regions of heterologous genes. For example, an early poly(CUG) model was created by inserting either a normal length (CTG)5 or a long mutant (CTG)250 repeat in the 3’ UTR of the human skeletal actin gene (HSA) [28]. Only transgenic mice with the long repeat (HSALR) develop myotonia and myopathy and disease severity correlates with transgene expression indicating that repeats are pathogenic at the RNA level. A limitation of this transgenic model is that the CTG repeat expansion is only expressed in muscle. Restriction of spatial expression patterns is not a problem in DM300-328/DMXL/SXXL transgenic lines, which carry a human DMPK genomic 45 kb fragment with 300–~1,200 CTG repeats although the larger repeats are expressed at low levels [8]. While homozygous mice develop several DM1-relevant disease features, they also show reduced body size which is not a phenotype associated with this disease. Recently, bitransgenic mouse lines have been generated which can be used to inducibly express a non-coding CTG960 repeat (EpA960) in other tissues [41, 53]. Following tamoxifen administration, heart-specific EpA960 mice develop cardiac arrhythmias while muscle-specific lines present with myotonia and severe muscle wasting. Although this is an important new model because it allows for inducible postnatal expression of a toxic RNA in specific tissues, including the brain, induction of the EpA960 transgene results in a burst of high levels of an untranslated RNA with an interrupted repeat, [(CTG)20CTCGA]48.
How do CTG-rich microsatellite expansions in non-coding regions of two unrelated genes result in a similar adult-onset disease? Current evidence argues that transcription of mutant DMPK and ZNF9 alleles produces CUG and CCUG expansion (CUGexp, CCUGexp) RNAs which fold into stable double-stranded (ds) hairpin structures that are toxic to cells because they alter the activities of at least two families of alternative splicing factors [4, 40]. While these CUGexp and CCUGexp RNAs recruit, and subsequently sequester, the muscleblind-like (MBNL) proteins [17, 30, 36], they also trigger the hyperphosphorylation and overexpression of CUGBP1 and ETR3-like factors (CELF) [20]. These two events appear to be tightly coupled in vivo since tamoxifen-inducible expression of the polyCUG transgene EpA960 in mouse heart results in the simultaneous appearance in cardiomyocytes of CUGexp ribonuclear foci, sequestration of Mbnl1 in these foci and increased expression of Cugbp1 [53]. Because the MBNL and CELF proteins regulate the splicing of a specific set of fetal and adult alternative exons during postnatal development, the corresponding fetal protein isoforms persist in the DM adult which is incompatible with normal adult tissue function [10, 44, 47]. For example, expression of the major skeletal muscle chloride channel ClC-1, encoded by the CLCN1 gene, is regulated at the level of alternative splicing [3, 29]. In newborn muscle, CLCN1 pre-mRNA splicing results in the inclusion of exon 7a, which leads to a premature stop codon in exon 7 due to frameshifting, so the resulting mRNA is rapidly degraded via the nonsense-mediated decay (NMD) pathway. CLCN1 exon 7a splicing is promoted by the relatively high levels of CUGBP1 in neonatal myonuclei. During postnatal development, CUGBP1 levels decline while MBNL1 protein translocates from the cytoplasm to the nucleus resulting in exon 7a skipping and synthesis of the full-length ClC-1 protein [21, 26]. Evidence that loss of ClC-1 due to mis-splicing in DM adult muscle results in a channelopathy and myotonia comes from studies using morpholino antisense oligonucleotides (AONs) [54]. Treatment of either HSALR or Mbnl1 knockout mice skeletal muscle with AONs that target the Clcn1 exon 7a 5’ splice site block splicing of this exon and rescues the myotonia observed in these DM mouse models. Although the expression of CUGexp and CCUGexp mutant RNAs also alters alternative splicing in the central nervous system (CNS), only a few target exons have been identified (MAPT exons 2, 6,10, APP exon 7, GRIN1/NMDAR1 exon 5) and unlike the role of CLCN1 in myotonia the relationship between mis-splicing and specific disease manifestations in the nervous system is currently unclear [7, 15, 22–23]. The MBNL and CELF protein families are also involved in a number of other regulatory pathways, including mRNA turnover and localization, so it is likely that CUGexp and CCUGexp mutant RNAs also impact gene expression at several levels [1, 39, 56] [43].
RNA gain-of-function mutations have been implicated in other neurological diseases such as FXTAS (Fig. 1). Fragile X syndrome is a common form of inherited mental retardation and is generally caused by lengthy (>200) CGG expansions in the 5’-UTR of the FMR1 gene. While these elongated repeat tracts repress FMR1 transcription, the shorter FXTAS-associated (CGG)60–200 expansions, originally thought to be premutation alleles, cause a late adult-onset neurodegenerative disease characterized by gait ataxia, progressive intention tremor and parkinsonism [40]. Rather than transcriptional repression, these intermediate-size repeats cause upregulation of FMR1 expression and accumulation of FMR1 RNA transcripts in intranuclear inclusions. Evidence that rCGGexp RNAs are toxic in mammals independent of gene context has been provided recently. Transgenic mice, which selectively express a pathogenic (CGG)90 expansion in the 5’ UTR of either Fmr1 or EGFP in Purkinje cells, develop behavioral deficits, ubiquitin-positive intranuclear inclusions by 8 weeks of age and show enhancement of Purkinje cell axonal swelling and dropout [9]. The structure of rCGG repeats has been analyzed by NMR and, similar to CUGexp, these repeats form stable RNA hairpins which are recognized by several RNA binding proteins, including Purα, hnRNP A2/B1 and CUGBP1 [16, 48, 57]. The RNA gain-of-function model for FXTAS is also supported by observations that Pura knockout mice develop neurological deficits similar to FXTAS and overexpression of Purα in a Drosophila (CGG)90-EGFP transgenic model suppresses disease-associated neurodegeneration [16, 18].
Toxic RNAs may also be involved in the pathogenesis of some autosomal dominant (AD) spinocerebellar ataxias (SCAs), which are late-onset and progressive neurodegenerative diseases [2]. Of the 28 characterized AD SCAs, eight are associated with CAG/CTG (SCA1, 2, 3, 6, 7, 8, 12, 17), and one with ATTCT/AGAAT (SCA10), repeats and these expansions in SCA8 and 12 may occur in either non-coding or coding regions depending on transcription initiation site selection, transcriptional orientation and RNA splicing (Fig. 1). SCA12, a rare disorder except in India where it is the second most common dominant ataxia, is caused by a CAGexp mutation in PPP2R2B, which encodes the Bβ brain-specific regulatory subunit of the ubiquitous heterotrimeric phosphatase PP2A [11, 49]. Action tremor and mild gait ataxia are prominent clinical signs and the CAG repeat is located in several different regions of PPP2R2B transcripts or in the promoter depending on alternative splicing and which transcriptional start sites are utilized [12]. Current evidence indicates that the CAGexp alters transcriptional and/or splicing regulation of PPP2R2B. One disease model proposes that the CAG repeat is in the promoter of the Bβ1 variant and CAGexp causes transcriptional upregulation and dysregulation of phosphatase activity. In addition, use of upstream promoters and alternative splice site selection position the CAGexp in either an alternative 5’-UTR or in intron 6 [11]. When positioned in the intron, the alternative splicing pattern of PPP2R2B may be modified resulting an increase in the Bβ2 isoform that induces neuronal mitochondrial fragmentation and apoptosis [5]. Although these models suggest that SCA12 is not an RNA-mediated disease, a recent study of another SCA, SCA3, highlights the possibility that these relatively short intronic (CAG)51–78 RNAs present in SCA12 brain could contribute to disease pathogenesis when present in PPP2R2B pre-mRNA.
SCA3 is a polyQ-induced disease that is caused by a similarly sized (CAG)61–84 coding region expansion in the ATXN3 gene, which encodes a deubiquitinating enzyme [2, 42]. Surprisingly, a Drosophila modifier screen for ATXN3 polyQ-induced neurodegeneration revealed that upregulation of Mbl, the fly homologue of human MBNL, enhances toxicity suggesting a link between polyQ- and RNA-mediated pathogenesis [24]. In this fly model, Mbl increases the level of both polyQ RNA and protein and substitution of the pure CAG repeat with a CAA/G interrupted repeat, which still encodes a polyQ protein but does not provide an optimal RNA-binding site for Mbl/MBNL proteins, significantly decreased the eye degeneration phenotype. Moreover, expression of a non-coding CAGexp induced neuronal degeneration which suggests that untranslated CAGexp RNAs may be important modifiers of polyQ disease.
While CAGexp RNAs might contribute to polyQ-induced toxicity, the complexity of the transcriptome in the human brain is vast and bidirectional transcription of expanded microsatellites has the potential to generate toxic proteins and RNAs from different strands. SCA8 is a slowly progressive ataxia with reduced penetrance caused by a CTG•CAG expansion on chromosome 13q21 [13, 45]. An early study reported that an SCA8 repeat, (CTA)11CTGCTA(CTG)80, was exclusively transcribed from the ATXN8OS gene in the CTG orientation and produced an alternatively spliced and polyadenylated non-coding CUGexp RNA which led to the conclusion that this ataxia is caused by an RNA gain-of-function mechanism [19]. Subsequent analysis demonstrated that the opposite strand was also transcribed and the CAG expansion in this ATXN8 gene produces a nearly pure polyQ protein which is detectable with the anti-polyglutamine monoclonal antibody 1C2 [38]. The SCA8 repeat may be interrupted with one or multiple CCG, CTA, CTC, CCA or CTT triplets but the influence of these sequence interruptions on pathogenesis has not been addressed. Although there has been some controversy centered on the reduced penetrance associated with SCA8, and the observation that large ATXN8/ATXN8OS CTG•CAG expansions exist in unaffected control individuals, a mouse BAC-transgenic model expressing a human SCA8 (CTG)116 expansion recapitulates several features of this disease, including a progressive neurological phenotype with deficits in cerebellar cortical inhibition as well as 1C2 positive polyQ intranuclear inclusions in cerebellar Purkinje and pontine neurons [38]. Further studies demonstrated that polyQ intranuclear inclusions are present in human SCA8 Purkinje, medullary and dentate neurons but absent in autopsy controls. Although this study is the first to report potentially deleterious gain-of-function effects at both RNA and protein levels in a neurological disease, the relative toxicity of polyQ versus CUGexp RNA in SCA8 requires further investigation.
Another issue is the possibility that toxic proteins are produced from non-trinucleotide repeat expansions, such as the DM2 CCTGexp. No open reading frames exist in the ZNF9 antisense orientation but in the sense direction an initiation codon is located upstream of the expansion region. Alternative splicing, and/or additional transcription start sites proximal to this repeat, could create an open reading frames containing a tetrapeptide repeat. Although proteins containing complex repeat motifs exist in both prokaryotes and eukaryotes [51], the possibility that these types of proteins are toxic to cells has not been tested.
While bidirectional transcription has important implications for microsatellite diseases, it is also possible that toxic RNAs and proteins can be produced from the same transcriptional unit. In addition to SCA3, another example is HDL2 which is an adult-onset progressive neurodegenerative disorder where CTGexp mutations are located in either coding (to produce polyalanine or polyleucine) or noncoding (3’ UTR) regions of the JPH3 gene depending on the splicing pattern (Fig. 1, for simplicity only the 3’-UTR position is shown). Intranuclear ubiquitin and 1C2 positive protein inclusions as well as CUGexp ribonuclear foci are present in HDL2 brain [46] [32]. Together with SCA8, these observations call into question pathogenic mechanisms based solely on the location of unstable microsatellites in coding versus non-coding regions and suggest that disease phenotype and penetrance may be influenced by an interplay between toxic RNAs and proteins.
Other types of repeats that do not form stable dsRNA structures may also be toxic to cells. This is exemplified by the ATTCT pentanucleotide expansion in intron 9 of the ATXN10 gene which causes SCA10, an AD neurodegenerative disorder characterized by cerebellar ataxia, seizures and anticipation [33]. Several results argue that SCA10 is not caused by ATXN10 loss-of-function [52]. Instead, SCA10 is another candidate for an RNA-mediated disease since ribonuclear foci can be detected by RNA fluorescence in situ hybridization following transfection of ATTCT repeats into cultured cells [25].
Currently there are no effective treatments for these neurological disorders. However, understanding the pathogenic mechanisms underlying these diseases should accelerate the development of effective therapeutic approaches. For example, pathogenic RNAs can be targeted using morpholino antisense oligonucleotides that bind to specific repeat expansions to block the access of RNA-binding proteins that are sequestered in disease [55]. Alternatively, screening of small molecules that promote overexpression of the affected RNA-binding proteins can also be considered. Indeed, the possibility that some of these microsatellite expansion diseases result from bidirectional transcription and the production of both toxic proteins and RNAs suggests that multifaceted treatment strategies should be considered for this group of disorders.
Acknowledgements
Our research is supported by grants from the NIH (AR046799, NS058901, NS048843) and the MDA. The authors thank L. Ranum for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adereth Y, Dammai V, Kose N, Li R, Hsu T. RNA-dependent integrin alpha3 protein localization regulated by the Muscleblind-like protein MLP1. Nat. Cell Biol. 2005;7:1140–1147. doi: 10.1038/ncb1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson KM, Andresen JM, Orr HT. Emerging pathogenic pathways in the spinocerebellar ataxias. Curr. Opin. Genet. Dev. 2009 doi: 10.1016/j.gde.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlet BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 4.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dagda RK, Merrill RA, Cribbs JT, Chen Y, Hell JW, Usachev YM, Strack S. The spinocerebellar ataxia 12 gene product and protein phosphatase 2A regulatory subunit Bbeta2 antagonizes neuronal survival by promoting mitochondrial fission. J. Biol. Chem. 2008;283:36241–36248. doi: 10.1074/jbc.M800989200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3' untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhaenens CM, Schraen-Maschke S, Tran H, Vingtdeux V, Ghanem D, Leroy O, Delplanque J, Vanbrussel E, Delacourte A, Vermersch P, Maurage CA, Gruffat H, Sergeant A, Mahadevan MS, Ishiura S, Buee L, Cooper TA, Caillet-Boudin ML, Charlet-Berguerand N, Sablonniere B, Sergeant N. Overexpression of MBNL1 fetal isoforms and modified splicing of Tau in the DM1 brain: two individual consequences of CUG trinucleotide repeats. Expt. Neurol. 2008;210:467–478. doi: 10.1016/j.expneurol.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 8.Gomes-Pereira M, Foiry L, Nicole A, Huguet A, Junien C, Munnich A, Gourdon G. CTG trinucleotide repeat "big jumps": large expansions, small mice. PLoS Genet. 2007;3:e52. doi: 10.1371/journal.pgen.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashem V, Galloway JN, Mori M, Willemsen R, Oostra BA, Paylor R, Nelson DL. Ectopic expression of CGG containing mRNA is neurotoxic in mammals. Hum. Mol. Genet. 2009;18:2443–2451. doi: 10.1093/hmg/ddp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. Embo J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes SE, O'Hearn EE, McInnis MG, Gorelick-Feldman DA, Kleiderlein JJ, Callahan C, Kwak NG, Ingersoll-Ashworth RG, Sherr M, Sumner AJ, Sharp AH, Ananth U, Seltzer WK, Boss MA, Vieria-Saecker AM, Epplen JT, Riess O, Ross CA, Margolis RL. Expansion of a novel CAG trinucleotide repeat in the 5' region of PPP2R2B is associated with SCA12. Nat. Genet. 1999;23:391–392. doi: 10.1038/70493. [DOI] [PubMed] [Google Scholar]
- 12.Holmes SE, O'Hearn E, Cortez-Apreza N, Hwang HS, Ross CA, Strack S, Margolis RL. Spinocerebellar ataxia type 12. In: Wells RD, Ashizawa T, editors. Genetic Instabilities and Hereditary Neurological Diseases. San Diego: Elsevier-Academic Press; 2006. pp. 461–473. [Google Scholar]
- 13.Ikeda Y, Daughters RS, Ranum LP. Bidirectional expression of the SCA8 expansion mutation: one mutation, two genes. Cerebellum. 2008;7:150–158. doi: 10.1007/s12311-008-0010-7. [DOI] [PubMed] [Google Scholar]
- 14.Imarisio S, Carmichael J, Korolchuk V, Chen CW, Saiki S, Rose C, Krishna G, Davies JE, Ttofi E, Underwood BR, Rubinsztein DC. Huntington's disease: from pathology and genetics to potential therapies. Biochem. J. 2008;412:191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 16.Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 18.Khalili K, Del Valle L, Muralidharan V, Gault WJ, Darbinian N, Otte J, Meier E, Johnson EM, Daniel DC, Kinoshita Y, Amini S, Gordon J. Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol. Cell. Biol. 2003;23:6857–6875. doi: 10.1128/MCB.23.19.6857-6875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koob MD, Moseley ML, Schut LJ, Benzow KA, Bird TD, Day JW, Ranum LP. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nat. Genet. 1999;21:379–384. doi: 10.1038/7710. [DOI] [PubMed] [Google Scholar]
- 20.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladd AN, Stenberg MG, Swanson MS, Cooper TA. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev. Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- 22.Leroy O, Dhaenens CM, Schraen-Maschke S, Belarbi K, Delacourte A, Andreadis A, Sablonniere B, Buee L, Sergeant N, Caillet-Boudin ML. ETR-3 represses Tau exons 2/3 inclusion, a splicing event abnormally enhanced in myotonic dystrophy type I. J. Neurosci. Res. 2006;84:852–859. doi: 10.1002/jnr.20980. [DOI] [PubMed] [Google Scholar]
- 23.Leroy O, Wang J, Maurage CA, Parent M, Cooper T, Buee L, Sergeant N, Andreadis A, Caillet-Boudin ML. Brain-specific change in alternative splicing of Tau exon 6 in myotonic dystrophy type 1. Biochim. Bbiophys. Acta. 2006;1762:460–467. doi: 10.1016/j.bbadis.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin X, Ashizawa T. Recent progress in spinocerebellar ataxia type-10 (SCA10) Cerebellum. 2005;4:37–42. doi: 10.1080/14734220510007897. [DOI] [PubMed] [Google Scholar]
- 26.Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent postnatal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006 doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 27.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 28.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 29.Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 30.Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 31.Margolis JM, Schoser BG, Moseley ML, Day JW, Ranum LP. DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum. Mol. Genet. 2006;15:1808–1815. doi: 10.1093/hmg/ddl103. [DOI] [PubMed] [Google Scholar]
- 32.Margolis RL, Holmes SE, Rudnicki DD, O'Hearn E, Ross CA, Pletkikova O, Troncoso JC, editors. Huntinton's Disease-like 2. San Diego: Elsevier-Academic Pres; 2006. pp. 261–273. [Google Scholar]
- 33.Matsuura T, Yamagata T, Burgess DL, Rasmussen A, Grewal RP, Watase K, Khajavi M, McCall AE, Davis CF, Zu L, Achari M, Pulst SM, Alonso E, Noebels JL, Nelson DL, Zoghbi HY, Ashizawa T. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat. Genet. 2000;26:191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- 34.Maurage CA, Udd B, Ruchoux MM, Vermersch P, Kalimo H, Krahe R, Delacourte A, Sergeant N. Similar brain tau pathology in DM2/PROMM and DM1/Steinert disease. Neurology. 2005;65:1636–1638. doi: 10.1212/01.wnl.0000184585.93864.4e. [DOI] [PubMed] [Google Scholar]
- 35.Meola G, Sansone V. Cerebral involvement in myotonic dystrophies. Muscle Nerve. 2007;36:294–306. doi: 10.1002/mus.20800. [DOI] [PubMed] [Google Scholar]
- 36.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minnerop M, Luders E, Specht K, Ruhlmann J, Schneider-Gold C, Schroder R, Thompson PM, Toga AW, Klockgether T, Kornblum C. Grey and white matter loss along cerebral midline structures in myotonic dystrophy type 2. J. Neurol. 2008;255:1904–1909. doi: 10.1007/s00415-008-0997-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, Day JW, Ranum LP. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 39.Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 40.O'Rourke JR, Swanson MS. Mechanisms of RNA-mediated disease. The J. Biol. Chem. 2009;284:7419–7423. doi: 10.1074/jbc.R800025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orengo JP, Chambon P, Metzger D, Mosier DR, Snipes GJ, Cooper TA. Expanded CTG repeats within the DMPK 3' UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl. Acad. Sci. USA. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Ann. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 43.Osborne RJ, Lin X, Welle S, Sobczak K, O'Rourke JR, Swanson MS, Thornton CA. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum. Mol. Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 45.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Ann. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 46.Rudnicki DD, Holmes SE, Lin MW, Thornton CA, Ross CA, Margolis RL. Huntington's disease-like 2 is associated with CUG repeat-containing RNA foci. Ann. Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- 47.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 48.Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strack S, Zaucha JA, Ebner FF, Colbran RJ, Wadzinski BE. Brain protein phosphatase 2A: developmental regulation and distinct cellular and subcellular localization by B subunits. J. Comp. Neurol. 1998;392:515–527. [PubMed] [Google Scholar]
- 50.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vetting MW, Hegde SS, Fajardo JE, Fiser A, Roderick SL, Takiff HE, Blanchard JS. Pentapeptide repeat proteins. Biochemistry. 2006;45:1–10. doi: 10.1021/bi052130w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wakamiya M, Matsuura T, Liu Y, Schuster GC, Gao R, Xu W, Sarkar PS, Lin X, Ashizawa T. The role of ataxin 10 in the pathogenesis of spinocerebellar ataxia type 10. Neurology. 2006;67:607–613. doi: 10.1212/01.wnl.0000231140.26253.eb. [DOI] [PubMed] [Google Scholar]
- 53.Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J. Clin. Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA-dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J. Biol. Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zumwalt M, Ludwig A, Hagerman PJ, Dieckmann T. Secondary structure and dynamics of the r(CGG) repeat in the mRNA of the fragile X mental retardation 1 (FMR1) gene. RNA Biol. 2007;4:93–100. doi: 10.4161/rna.4.2.5039. [DOI] [PubMed] [Google Scholar]