Abstract
Ligands presented on biomaterials are a common method to facilitate and control the host response. In a gelatin and polyethylene glycol diacrylate (PEGdA) based semi-interpenetrating network (sIPN), the effects of extracellular matrix (ECM)-derived peptide amount on monocyte adhesion and subsequent protein and mRNA expression were examined. Peptide amount on the sIPN surface was controlled by varying the wt% ratio of the peptide-PEG grafted gelatin to PEGdA. We hypothesized that increasing bioactive peptide amount would modulate human blood derived monocyte adhesion, cytokine expression and gene regulation. Monocyte adhesion, release of gelatin degrading proteases matrix metalloprotease-2 (MMP-2), matrix metalloprotease-9 (MMP-9) and pro-inflammatory protein interleukin-1β (IL-1β), and mRNA expression of these proteins were evaluated. We found RGD-PEG grafted sIPNs with higher surface RGD concentrations showed increased adherent density. MMP-2 and IL-1β protein release was also influenced by the ligand concentration, as initial increase in protein concentration was observed at higher ligand concentrations. MMP-9 protein showed an initial increase that subsided then increased. A decreased IL-1β protein and mRNA expression was observed over time but MMP-2 mRNA was not detected at any time though MMP-2 protein concentrations showed an initial burst. Hence, monocyte behavior was modulated by surface ligand identity in tandem with ligand concentration.
Keywords: Human monocyte, receptor-ligand interactions, RGD, matrix metalloproteinase, interleukin-1beta
INTRODUCTION
Monocytes are one of the early cells to appear at the site of injury and actively respond to biomaterials including polymers, ceramics, and metals.[1-3] The highly complex monocytic response to biomaterials involves cellular attachment, recognition and secretion of a myriad of factors important in the wound healing process, such as matrix remodeling proteins and inflammatory cytokines. Monocytes communicate with the extracellular matrix (ECM) environment through various means, including through cell adhesion receptors called integrins. Integrin complexation with the ECM modulates cell adhesion, signaling, cytokine and gene expression and survival.[4] The cell adhesion motif found in many adhesive proteins, arginine-glycine-aspartic acid (RGD), has been utilized extensively to modify biomaterials to promote integrin-mediated cell attachment, focal adhesion contact formation, motility and protein expression.[5-10]
Previously, we have demonstrated the role of integrin mediated monocyte adhesion on materials grafted with ECM-derived peptides and subsequent cell response.[2,11,12] We showed that adherent monocyte density was increased by RGD tethered onto a flexible polyethylene glycol (PEG) linker on the gelatin based semi-interpenetrating network (sIPN), and that subsequent protein and genetic regulation was also influenced by sIPNs.[11,12] RGD-PEG grafted sIPNs also enhanced cellularity and organized ECM reconstruction over time in vivo in dermal wound healing models.[13] These data are consistent with literature describing RGD influence on cell-biomaterial interactions.[14,15] Cell-substratum interactions, specifically the effects of peptide density on cell adhesion and migration, have been investigated using fibroblasts, osteoblasts, and neurite cells.[6],[15-18] However, the effects of peptide density on monocyte adhesion and subsequent behavior has received little attention. As we have previously characterized integrin-mediated monocyte interaction with ECM-derived peptide-PEG presented on the sIPN surface, in this study, monocyte response to varied ECM-derived peptide amount presented on the sIPN surface was investigated.[11,12] The amount of cell adhesion peptides on the sIPN surface was controlled by varying the wt% ratio of ECM derived peptide-PEG grafted gelatin to polyethylene glycol diacrylate (PEGdA) in the total network. We hypothesized that the increasing amount of bioactive peptides would modulate monocyte adhesion, cytokine expression and gene regulation. In addition to altering peptide amount, the wt% of ligand-PEG modified gelatin or of PEGdA in the sIPN also affects structural properties such as crosslinking density, elasticity, topography and other physical properties of the sIPN. This may potentially affect subsequent cell behavior.[19]
In this study we specifically examine monocyte release of matrix metalloproteases type 2 and 9 (MMP-2 and MMP-9), which are also known as gelatinase A and B respectively, in the gelatin based sIPN environment. In addition to matrix remodeling, MMPs are also involved in cell signaling via cytokine regulation.[20] The interactions between MMP-2/-9 and other cytokines, including a major inflammatory cytokine interleukin-1β (IL-1β), have been described in literature.[21,22] IL-1β release in response to biomaterials has been a reliable marker of macrophage activation and allows prediction of potential in vivo immune reactions.[23] Inflammation is also influenced by the presence of MMPs during tissue remodeling and wound healing. In this study, monocyte expression of IL-1β and MMP-2/-9 and the relationship among these proteins in the context of ECM-derived peptide grafted sIPN were investigated.[24]
METHODS AND MATERIALS
Synthesis and characterization of ligand-PEG grafted gelatin-based sIPN
Synthesis and characterization of ligand-PEG grafted sIPNs have been previously described in detail.[2] Briefly, the terminal alcohol groups on PEG-diol (2kDa) were converted to ethyl acetate, then to carboxylic acid (bis-COOH-PEG) and finally to bis-N-hydroxysuccinimide (bis-NSu-PEG). PEG modification was followed using a reverse-phase column (Alltech, Inc.) on an high performance liquid chromatography (HPLC) system (Gilson, 10% to 100% acetonitrile, 0.7 ml/min) coupled with UV/Vis and evaporative light scattering (ELS) detectors. Peptide ligands obtained were: glycine-glycine-glycine (GGG; Bachem, King of Prussia, PA, 99% purity), RGD, and proline-histidine-serine-arginine-asparagine (PHSRN; University of Wisconsin Biotechnology center, 98% purity). Each peptide was grafted onto one of the two terminal NSu groups to form peptide-PEG-NSu by adding 1.2 eq. mol of peptide followed by drop wise addition of 1.2 eq. mol of N, N-diisopropyl ethylamine (DIPEA). The whole protein fibronectin (FN; Sigma-Aldrich) was not conjugated onto the bis-NSu -PEG as the molecular weight of FN was 225 times greater than the PEG used and would likely entrap the PEG molecules within the protein structure. Unreacted NSu of the peptide-PEG-NSu was grafted onto gelatin by adding one eq. mol of peptide-PEG-NSu to 1% gelatin in phosphate buffered saline (PBS) and by stirring at pH 8.0 for 1 h. Modified gelatin was purified using a pressurized ultrafiltration system with a 30 kDa membrane filter. Unreacted impurities were filtered with deionized water at room temperature under nitrogen at 60 psi. Synthesis of methoxy-PEG modified gelatin was conducted in a similar scheme but with monomethoxy PEG (MPEG) 2 kDa as the starting material rather than PEG diol 2 kDa and without peptide grafting. Methoxy-PEG and peptide-PEG grafted gelatin were verified by an ultrahydrogel column in a gel permeation chromatography (GPC) system (Waters, 80% 0.1 M NaNO3, 20% acetonitrile at 0.7 ml/min flow rate for 60 min) coupled with a refractive index (RI) detector. The degree of peptide-PEG grafted onto the lysyl residues of the gelatin backbone was characterized utilizing a well-established method based on trinitrobenzenesulfonic acid-based spectrophotometry.[25] Ligand-PEG-grafted gelatin was also characterized with 1H-NMR referring to peaks from Lin-Gibson et. al and Gattas-Asfura KM et. al, as well as with a reverse phase column on the HPLC.[26,27] Ligand-PEG grafted gelatin samples analyzed with 1H-NMR.
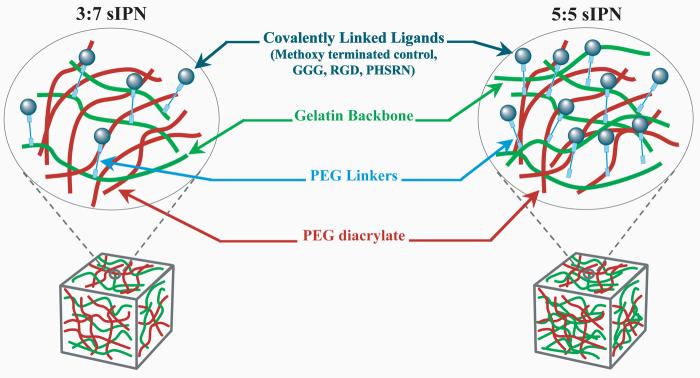
sIPNs were made by combining ligand-PEG-gelatin with PEGdA (600 Da, Sigma, St. Louis, MO, USA) and 0.1% 2,2-dimethoxy-2-phenyl-acetophenone initiator. The mixture of modified gelatin, PEGdA and photoinitiator were injected into Teflon® molds (7 mm dia × 0.75 mm thick) and crosslinked with UV light (maximal intensity, 365 nm, 21,700 μW/cm2, UVP, model B 100AP) for 5 min. sIPN discs were γ-irradiated (1 MRad) for sterility prior to cell culture. sIPN discs were made in three different formulations, based on the modified gelatin:PEGdA wt% ratio. 26:74, 35:65 and 41:59 wt% ratio sIPNs were made, noted as 3:7, 4:6, and 5:5 sIPN, respectively, in this paper. Between 3:7 and 4:6 sIPNs, there was a 9% increase in gelatin content from but between 4:6 and 5:5 sIPNs, there was a 6% increase in gelatin wt%. Beyond the 5:5 wt% ratio, increased gelatin content compromised the robustness of the sIPN to withstand handling. The schematics of the 3:7 and 5:5 sIPN are outlined in Figure 1. sIPNs were placed in 48-well tissue culture polystyrene (TCPS; BD Falcon) plates and incubated with RPMI 1640 at 37°C and 5% CO2 for 24 h prior to monocyte seeding.
Figure 1.
Schematic representation of the semi-interpenetrating network (sIPN) with varied ligand-PEG grafted gelatin:PEG wt%.
Accessible RGD concentration on sIPN surface
The RGD concentration on the surface of the sIPN discs was characterized via a previously reported indirect enzyme linked immunosorbent assay (ELISA).[28] sIPNs were equilibrated overnight in sterile PBS then incubated at room temperature for two hours in blocking solution containing 1% bovine serum albumin (BSA) in PBS. sIPNs were washed five times with wash buffer containing 1% Tween-20 in sterile PBS and incubated for 2 h at room temperature with RGD directed primary antibody (polyclonal rabbit anti-RGD domain antibody, EMD Biosciences), diluted 1:1000 in 1% BSA. Surfaces were then rinsed five times with wash buffer after incubation to remove non-adhered primary antibody. Secondary antibody (horseradish peroxidase-conjugated goat IgG fraction to rabbit whole IgG, MP Biomedicals) was diluted 1:20000 in 1% BSA then added to the sIPN surfaces for a two hour incubation. After incubation with secondary antibody, substrate solution (TMB-ELISA, Pierce) was added and then inactivated after 30 min with 2 M sulfuric acid in order to measure HRP concentration. Absorbance was measured using a 450 nm plate reader (BIO-TEK) and converted to surface RGD density from a second order polynomial standard curve (y = −0.0003x2 + 0.0068x + 0.0416, R2 = 0.9926). The curve was generated by adsorbing known amounts of RGD peptide, ranging from 0 to 5 μmole/ml, onto untreated polystyrene plates (Falcon).
Monocyte adhesion and protein measurement assay
Human peripheral blood monocytes were isolated from citrated whole blood of a healthy adult volunteer using a density-gradient, non-adhesion method previously described in detail.[29] The same donor was used throughout the experiment for adhesion, cytokine and gene expression assays to minimize donor-centered variability. To verify that the cytokines observed after seeding were not a result of the isolation procedure, cell lysates collected by sonication (5 pulses) as well as the media containing the freshly isolated monocytes prior to sonication were immediately saved then analyzed for MMP-2 and MMP-9 using ELISA. The data from these samples are noted as “0 h” (i.e. immediately upon isolation and prior to seeding on surface). Freshly isolated monocytes were seeded onto various ligand-PEG grafted sIPN at a concentration of 106 cells/ml in RPMI 1640 culture medium and 10% autologous serum at 37°C and 5% CO2.[2] At 2, 24, 96 and 168 h, supernatant was collected and analyzed with commercial ELISA kits (Raybiotech, Inc., Norcross, GA). Three 100 μl aliquots were obtained from each well in the experiment to test a variety of conditions, and these aliquots were used for the testing MMP-2/-9 and IL-1β concentrations with ELISA. The sensitivity of the kits ranged from 140-18,000 pg/ml for MMP-2, 8.2-6000 pg/ml for MMP-9 and 0.3-1000 pg/ml for IL-1β. ELISA was carried out according to manufacturer's instructions. After the supernatants were collected at 2, 24, 96 and 168 h, at each time point, all samples were washed twice with RPMI medium to remove nonadherent cells. Samples were resupplied with 10% autologous serum in RPMI and allowed to incubate further. At 2, 24, 96 and 168 h, sIPN samples were stained with Wright's stain. Adherent monocytes on all surfaces were imaged using a computer-assisted video analysis system (Metamorph v4.1) coupled to an inverted microscope (Nikon, Eclipse TE 300). The adherent cells were quantified and expressed as number of cells/mm2 surface area.
mRNA characterization with Reverse Transcription Polymerase Chain Reaction (RT-PCR)
All reagents employed in RT-PCR assays were purchased from Bio-Rad, USA, unless indicated otherwise. Adherent cells were lysed with TRIzol® reagent (InvitrogenTM, CA) for RT-PCR analysis. A modification of the Chomczynski and Sacchi method using TRIzol® reagent was carried out to isolate total RNA from adherent monocytes.[30] The quality of the purified RNA was tested by 1.5% agarose gel/ethidium-bromide staining with a 260/280 nm absorbance ratio. At the cDNA synthesis step, RNAse-free DNAse (Sigma, MO) was used to treat the RNA before being reverse transcribed with Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (InvitrogenTM, CA). Oligonucleotide primers for MMP-2/-9, IL-1β and internal standards β-actin, and glyceraldehydes-3-phosphate-dehydrogenase (GAPDH) were obtained using the DNA sequence database from NCBI (Bethesda, MD). The primer sequences, of which the reproducibility has been previously demonstrated, are described in Table I.[31,12] Primers3 Output computer program (Whitehead Institute for Biomedical Research, Cambridge, MA) which bases selection on G/C content, PCR product size, and melting temperature, was used to design primer sets. The specificity of the primer sequences was verified by the NCBI BLAST program. cDNA was amplified in a 25 μl PCR mixture containing 1x reaction buffer (Sigma, MO, USA), 0.24 μM of each primer, 0.2 mM of dNTP (InvitrogenTM, CA), and 0.75 U RedTaq DNA polymerase (Sigma, MO). The PCR reaction was started at 94°C for 3 min, after which followed 35 cycles for MMP-2, MMP-9, IL-1β, β-actin, and GAPDH. A denaturation step (at 94°C for 30 sec), an annealing step (at 56°C for 45 sec) and an extension step (at 72°C for 1 min), and a final elongation step (at 72°C for 7 min) took place at each cycle. The reaction was performed with iCycler thermal cycler. 15 μl of the amplified products were run under electrophoresis through a 1.5% agarose gel containing ethidium-bromide (0.5 μg/ml) and visualized under ultraviolet light. Contamination from genomic DNA was checked for with RT-PCR containing RNA but not M-MLV reverse transcriptase. The band densities in gels were quantified using the Scion imaging program (Frederick, MD). PCR analysis was carried out for sIPN samples at 2, 24, and 96 h for MMP-2/-9 and IL-1β. β-actin, a function of cell motility, and GAPDH, a function of cell metabolism, was observed for comparison as internal controls. Due to the unresolved issue over the consistency and the choice of housekeeping genes, MMP-2/-9, and IL-1β were normalized to β-actin as well as GAPDH.[32] mRNA levels in primary monocytes on different sIPNs were compared within each time point and within material formulation.
Table I.
Oligonucleotide primer sequences used for RT-PCR and expected PCR product sizes
| mRNA | Sense primer (5′ → 3′) | Antisense primer (5′ → 3′) | Product size (bp) |
|---|---|---|---|
| MMP-2 | AGGATCATTGGCTACACACC | AGCTGTCATAGGATGTGCCC | 535 |
| MMP-9 | CGCAGACATCGTCATCCAGT | GGATTGGCCTTGGAAGATGA | 406 |
| IL-1β | GAATCTCCGACCACCACTACA | CAACACGCAGGACAGGTACA | 416 |
| β-actin | AGGCATCCTCACCCTGAAGTA | AGCCTGGATAGCAACGTACA | 229 |
| GAPDH | CATCATCCCTGCCTCTACTG | GGTGGTCCAGGGGTCTTACT | 409 |
Statistical Analysis
All data, including surface RGD concentration, adherent cell density, cytokine concentration and transcriptional levels, were analyzed by two-way analysis of variance and Tukey post testing (SigmaStat, v2.03) and expressed as mean ± standard deviation (n=3). p-values of less than 0.001 were considered statistically significant for adherent cell density and protein concentrations and less than 0.05 for all other data. Transcriptional levels are expressed as arbitrary units (AU) as levels were normalized to either β-actin or GAPDH. Due to the semi-quantitative nature of RT-PCR, statistical comparisons were reserved within each RT-PCR runs, and thus were reserved within time points and within sIPN formulation groups.
RESULTS
Ligand-PEG grafted gelatin synthesis
HPLC elution times for PEG diol, bis-ethyl acetate-PEG, bis-COOH-PEG, bis-NSu-PEG were approximately 10.5, 12.2, 6.6, and 12.4 min, respectively. The percent of gelatin lysyl group modification for modified gelatin was 96% for methoxy-PEG-gelatin, 89% for GGG-PEG-gelatin, 78% for RGD-PEG-gelatin, and 92% for PHSRN-PEG-gelatin (n=3). 1H-NMR analysis showed PEG peaks approximately at 3.64 ppm for all ligand-PEG grafted gelatin samples. In addition to the characteristic PEG peaks, 1H-NMR spectra from modified gelatin samples exhibited gelatin peaks, which are characterized by a group of broad, asymmetrical peaks spanning from 0.885 to 4.803 ppm, a broad doublet peak at 7.250 ppm and a sharp peak at 8.411 ppm. Each ligand exhibited distinct sets of peaks in addition to the gelatin and PEG peaks. For GGG-PEG-gelatin samples, three singlets at 4.07, 4.12 and 4.17 ppm were observed. For RGD-PEG-gelatin, an overlapped doublet of triplet peaks at approximately 1.36 ppm, a broad group of peaks at 1.40 ppm, a peak at 2.79, 2.87, and 3.03 ppm, and two overlapping triplet peaks at approximately 3.38 ppm were displayed. PHSRN-PEG-gelatin samples yielded broad peaks composed of many overlapping small peaks at 1.7, 1.75, 1.92, 3.18, 4.19 and 4.2 ppm, in addition to two sharp peaks at 4.15 and 7.02 ppm.
RGD density on sIPNs
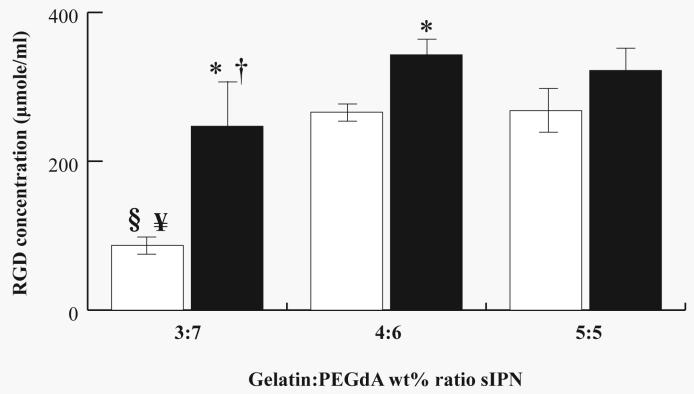
Accessible RGD concentration (μmole/ml) on the sIPN surface is shown in Figure 2. Unmodified sIPNs showed significantly (p<0.05) less RGD concentration compared to RGD-PEG grafted sIPNs for 3:7 and 4:6 but not for 5:5 sIPN. Unmodified 3:7 sIPN also exhibited significantly (p<0.05) less RGD on the surface compared to unmodified 4:6 and 5:5 sIPNs. Comparing across RGD-PEG grafted sIPN, 3:7 sIPN showed significantly (p<0.05) less RGD on the surface compared to 4:6 sIPN. Significant differences were not observed between RGD-PEG grafted 4:6 and 5:5 sIPN and this may be due to the less incremental increase in RGD-PEG grafted gelatin content wt% from 4:6 to 5:5 sIPN compared to the gelatin content increase from 3:7 to 4:6 sIPN.
Figure 2.
RGD concentration (μmole/ml) on unmodified (□) or RGD-PEG grafted gelatin (■) combined with varying wt % ratios of gelatin:PEGdA in the sIPN. Data expressed as mean ± s.d. *Significantly different (p<0.05) from respective unmodified sIPN. §Significantly different (p<0.05) from unmodified 4:6 sIPN. ¥Significantly different (p<0.05) from unmodified 5:5 sIPN. †Significantly different (p<0.05) from RGD-PEG grafted 4:6 sIPN.
Adherent monocyte density
Adherent cell density was dependent on ligand identity and concentration. RGD-PEG grafted 4:6 and 5:5 sIPNs exhibited significantly (p<0.001) greater adherent cell density as compared to all other ligand-PEG grafted respective sIPNs from 2 to 96 h (Table II). Adherent density of RGD-PEG grafted 3:7 sIPN was higher only compared to methoxy-PEG grafted sIPNs at 2 h. Comparing across RGD-PEG grafted sIPN formulations, significantly (p<0.001) lower adherent cell density was observed on 3:7 as compared to 4:6 and 5:5 sIPNs at all time points. Adherent cell density between RGD-PEG grafted 4:6 and 5:5 sIPN did not significantly differ over time and this may be contributed to the lack of significant differences in RGD concentration between these two formulations. Methoxy-PEG grafted sIPN did not show significant differences in adherent cell density across all formulations and time points. Comparing across GGG-PEG grafted sIPNs, 3:7 sIPNs showed significantly (p<0.001) less adherent cell density compared to both 4:6 and 5:5 sIPNs from 2 to 96 h, but at 168 h, only 5:5 sIPN had higher density than 3:7 sIPN. Adherent cell density on PHSRN-PEG grafted 3:7 sIPN was significantly (p<0.001) lower than PHSRN-PEG grafted 4:6 sIPNs at 2 and 24 h.
Table II.
Adherent monocyte density (cell/mm2) on sIPNs with varying ligand-PEG grafted gelatin:PEGdA wt% ratio.
| Culture Time: | |||||
|---|---|---|---|---|---|
| Ligand-PEG grafted gelatin:PEGdA wt% sIPN surface |
2 h | 24 h | 96 h | 168 h | |
| 3:7 sIPN | |||||
| Methoxy | 71 ± 14* | 73 ± 24 | 57 ± 26 | 33 ± 6 | |
| GGG | 155 ± 38B,C | 108 ± 14B,C | 46 ± 7B,C | 32 ± 8C | |
| RGD | 327 ± 76B,C | 166 ± 60B,C | 88 ± 8B,C | 66 ± 6B,C | |
| PHSRN | 172 ± 69B | 180 ± 33B | 105 ± 15 | 87 ± 9 | |
| 4:6 sIPN | |||||
| Methoxy | 145 ± 18* | 233 ± 14* | 106 ± 2* | 95 ± 12 | |
| GGG | 328 ± 69A* | 444 ± 16A* | 207 ± 52A* | 74 ± 40 | |
| RGD | 1321 ± 74A | 1071 ± 190A | 350 ± 42A | 157 ± 8A | |
| PHSRN | 442 ± 26A* | 399 ± 49A* | 154 ± 41* | 84 ± 14 | |
| 5:5 sIPN | |||||
| Methoxy | 113 ± 55* | 102 ± 51* | 67 ± 22* | 87 ± 8 | |
| GGG | 448 ± 189A* | 407 ± 195A* | 118 ± 14A* | 135 ± 31A | |
| RGD | 1192 ± 384A | 1133 ± 320A | 434 ± 77A | 173 ± 41A | |
| PHSRN | 314 ± 65* | 381 ± 91* | 169 ± 64* | 101 ± 35 | |
Significantly different from 3:7 sIPN (within each ligand and within each time point) p<0.001
Significantly different from 4:6 sIPN (within each ligand and within each time point) p<0.001
Significantly different from 5:5 sIPN (within each ligand and within each time point) p<0.001
Significantly different from RGD-PEG grafted sIPN (within ligand-grafted gelatin:PEGdA wt% ratio and within each time point) p<0.001
Protein concentration
MMP-2 concentration over time was influenced by ligand identity as well as by concentration, though the impact of ligand concentration was greater at later time points (Fig. 3). Both the cell lysate and the media yielded non-detectable MMP-2 concentrations (0 ± 0 pg/ml) immediately after isolation. From 0 to 2 h, a dramatic increase in MMP-2 secretion was observed for 4:6 and 5:5 but not for 3:7 sIPN. All except methoxy-PEG grafted 4:6 sIPNs showed significantly (p<0.001) higher MMP-2 concentrations than 5:5 sIPN, and methoxy-PEG grafted 5:5 sIPNs showed significantly (p<0.001) higher concentrations than RGD-, and PHSRN-PEG grafted 5:5 sIPN. From 24 to 168 h, MMP-2 concentrations for 3:7 sIPN gradually increased, but did not exceed 2000 pg/ml. At 24 and 96 h, all 4:6 sIPNs had significantly (p<0.001) higher concentrations compared to respective 3:7 and 5:5 sIPNs except versus GGG-PEG grafted 5:5 sIPN at 96 h. At 168 h, all 3:7 sIPNs showed significantly (p<0.001) higher concentrations than respective 5:5 sIPNs but lower than respective 4:6 sIPNs.
Figure 3.
MMP-2 concentrations (pg/ml) over time from human monocytes in the presence of sIPNs with various wt% ratios of ligand-PEG grafted gelatin:PEGdA: 3:7 sIPN ( ), 4:6 sIPN (
), 4:6 sIPN ( ), or 5:5 sIPN (
), or 5:5 sIPN ( ). Data are shown per ligand group and are expressed as mean ± s.d. Comparing within each time point and each ligand identity at p<0.001: i.) at 2 h, all ligand-PEG grafted 3:7 sIPNs showed a significantly lower concentration compared to that of 4:6 and 5:5 sIPNs and all except methoxy-PEG grafted 4:6 sIPNs showed significantly higher concentrations compared to 5:5 sIPNs while within 5:5 sIPNs, RGD-, PHSRN-PEG grafted sIPNs were significantly lower than methoxy-PEG grafted sIPNs; ii.) from 24 to 96 h, concentrations from all ligand-PEG grafted 4:6 sIPNs were significantly higher than all 3:7 sIPN and 5:5 sIPN samples except versus GGG-PEG grafted 5:5 sIPN at 96 h; iii.) at 168 h, all 4:6 sIPN samples showed significantly higher concentrations than all 3:7 and 5:5 sIPNs and all 3:7 sIPN samples were significantly higher than all 5:5 sIPN samples.
). Data are shown per ligand group and are expressed as mean ± s.d. Comparing within each time point and each ligand identity at p<0.001: i.) at 2 h, all ligand-PEG grafted 3:7 sIPNs showed a significantly lower concentration compared to that of 4:6 and 5:5 sIPNs and all except methoxy-PEG grafted 4:6 sIPNs showed significantly higher concentrations compared to 5:5 sIPNs while within 5:5 sIPNs, RGD-, PHSRN-PEG grafted sIPNs were significantly lower than methoxy-PEG grafted sIPNs; ii.) from 24 to 96 h, concentrations from all ligand-PEG grafted 4:6 sIPNs were significantly higher than all 3:7 sIPN and 5:5 sIPN samples except versus GGG-PEG grafted 5:5 sIPN at 96 h; iii.) at 168 h, all 4:6 sIPN samples showed significantly higher concentrations than all 3:7 and 5:5 sIPNs and all 3:7 sIPN samples were significantly higher than all 5:5 sIPN samples.
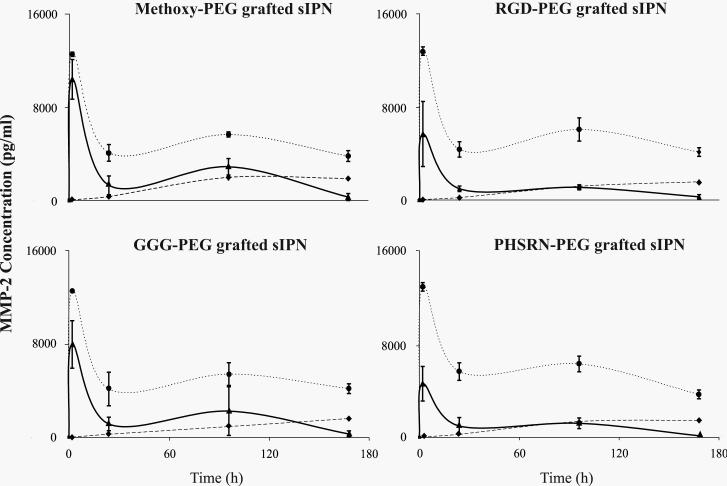
Similar to MMP-2 protein concentrations at earlier time periods, MMP-9 was non-detectable (0 ± 0 pg/ml) at 0 h, but a sharp increase in MMP-9 from 0 to 2 h was observed for all ligand type and material formulation (Figure 4). The increase in protein concentration from 0 to 2 h was followed by a drastic decrease from 2 to 24 h for all ligand-PEG grafted sIPN. At 2 h, methoxy- and RGD-PEG grafted 3:7 sIPN had significantly (p<0.001) higher concentrations than respective 4:6 sIPN. From 24 to 96 h, MMP-9 concentrations increased for all sIPNs, but to varying degrees depending on ligand type and concentration. For example, all methoxy-PEG grafted sIPN at 96 h were statistically similar while both GGG- and PHSRN-PEG grafted 4:6 sIPNs exhibited significantly (p<0.001) higher values compared to that of respective 3:7 and 5:5 sIPNs. RGD-PEG grafted 4:6 sIPN at 96 h had significantly (p<0.001) higher concentrations than that of 3:7 sIPN but significantly (p<0.001) lower than 5:5 sIPN. At 168 h, concentrations from methoxy-PEG grafted 4:6 sIPN were significantly higher than 3:7 but significantly lower than 5:5 sIPN. GGG-, PHSRN-PEG grafted 4:6 sIPN continued to show significantly (p<0.001) higher values than 3:7 and 5:5 sIPNs. RGD-PEG grafted 4:6 and 5:5 sIPNs displayed significantly (p<0.001) higher concentrations than 3:7 sIPN.
Figure 4.
MMP-9 concentrations (ng/ml) over time from human monocytes in the presence of sIPNs with various wt% ratios of ligand-PEG grafted gelatin:PEGdA: 3:7 sIPN ( ), 4:6 sIPN (
), 4:6 sIPN ( ), or 5:5 sIPN (
), or 5:5 sIPN ( ). Data are shown per ligand group and are expressed as mean ± s.d. Comparing within each time point and each ligand identity at p<0.001: i.) at 2 h, methoxy-, RGD-PEG grafted 3:7 sIPNs had significantly higher values than 4:6 sIPNs; ii.) at 96 h, both GGG- and PHSRN-PEG grafted 4:6 sIPNs exhibited significantly higher values compared to that of 3:7 and 5:5 sIPNs while for RGD-PEG grafted sIPN, 4:6 sIPNs had significantly higher values than that of 3:7 but lower than 5:5 sIPNs; iii.) at 168 h, GGG-, PHSRN-PEG grafted 4:6 sIPN had significantly higher values than 3:7 and 5:5 sIPNs while values for methoxy-PEG grafted 4:6 sIPN were significantly higher than those of 3:7 but significantly lower than those of 5:5 sIPNs, and RGD-PEG grafted 4:6 and 5:5 sIPN had significantly higher values than 3:7 sIPN.
). Data are shown per ligand group and are expressed as mean ± s.d. Comparing within each time point and each ligand identity at p<0.001: i.) at 2 h, methoxy-, RGD-PEG grafted 3:7 sIPNs had significantly higher values than 4:6 sIPNs; ii.) at 96 h, both GGG- and PHSRN-PEG grafted 4:6 sIPNs exhibited significantly higher values compared to that of 3:7 and 5:5 sIPNs while for RGD-PEG grafted sIPN, 4:6 sIPNs had significantly higher values than that of 3:7 but lower than 5:5 sIPNs; iii.) at 168 h, GGG-, PHSRN-PEG grafted 4:6 sIPN had significantly higher values than 3:7 and 5:5 sIPNs while values for methoxy-PEG grafted 4:6 sIPN were significantly higher than those of 3:7 but significantly lower than those of 5:5 sIPNs, and RGD-PEG grafted 4:6 and 5:5 sIPN had significantly higher values than 3:7 sIPN.
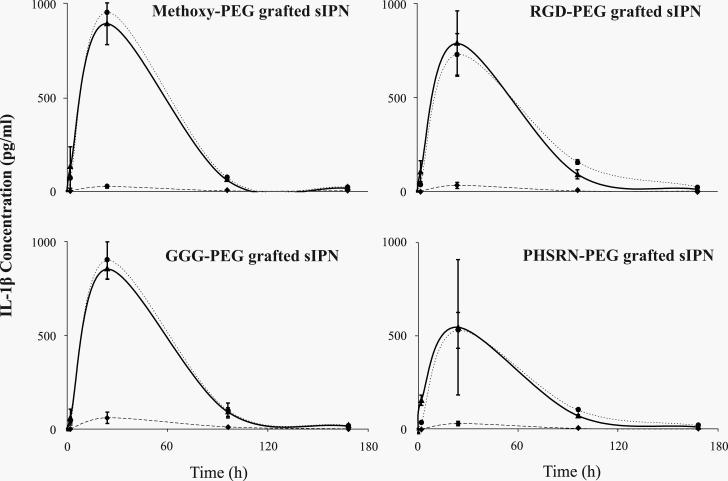
IL-1β protein concentrations were modulated by both ligand identity and concentration (Fig. 5). For 3:7 sIPN at 2 h and for 4:6 and 5:5 sIPNs, IL-1β was detected at concentrations below 200 pg/ml. At 2 h, PHSRN-PEG grafted 5:5 sIPNs had significantly (p<0.001) higher concentrations than PHSRN-PEG grafted 3:7 and 4:6 sIPNs. At 24 h, all 3:7 sIPN showed significantly (p<0.001) less IL-1β concentrations than respective 4:6 and 5:5 sIPN. Within 4:6 sIPNs at 24 h, RGD-, PHSRN-PEG grafted sIPN had significantly (p<0.001) lower concentrations than methoxy- and GGG-PEG grafted sIPN. At 24 and 96 h, no significant differences were observed between 4:6 and 5:5 sIPN. All 3:7 sIPNs continued to show significantly (p<0.001) less IL-1β concentrations compared to 4:6 and 5:5 sIPNs at 96 h. RGD-PEG grafted 4:6 sIPN exhibited significantly (p<0.001) higher concentrations than all other ligand-PEG grafted 4:6 sIPN at 96 h.
Figure 5.
IL-1β concentrations (pg/ml) over time from human monocytes in the presence of sIPNs with various wt% ratios of ligand-PEG grafted gelatin:PEGdA: 3:7 sIPN ( ), 4:6 sIPN (
), 4:6 sIPN ( ), or 5:5 sIPN (
), or 5:5 sIPN ( ). Data are shown per ligand group and are expressed as mean ± s.d. Comparing within each time point and each ligand identity at p<0.001: i.) at 2 h, PHSRN-PEG grafted 5:5 sIPNs had significantly higher concentrations than 3:7 and 4:6 sIPNs; ii.) at 24 h, all 4:6 and 5:5 sIPNs showed higher values than 3:7 sIPN, and within 4:6 sIPNs, RGD-, PHSRN-PEG grafted sIPNs had lower concentrations than methoxy- and GGG-PEG grafted sIPNs; iii.) at 96 h, all 4:6 and 5:5 sIPNs showed higher values than 3:7 sIPN, and within 4:6 sIPNs, only RGD-PEG grafted sIPNs exhibited higher concentrations than all other ligand-PEG grafted sIPN.
). Data are shown per ligand group and are expressed as mean ± s.d. Comparing within each time point and each ligand identity at p<0.001: i.) at 2 h, PHSRN-PEG grafted 5:5 sIPNs had significantly higher concentrations than 3:7 and 4:6 sIPNs; ii.) at 24 h, all 4:6 and 5:5 sIPNs showed higher values than 3:7 sIPN, and within 4:6 sIPNs, RGD-, PHSRN-PEG grafted sIPNs had lower concentrations than methoxy- and GGG-PEG grafted sIPNs; iii.) at 96 h, all 4:6 and 5:5 sIPNs showed higher values than 3:7 sIPN, and within 4:6 sIPNs, only RGD-PEG grafted sIPNs exhibited higher concentrations than all other ligand-PEG grafted sIPN.
mRNA expression
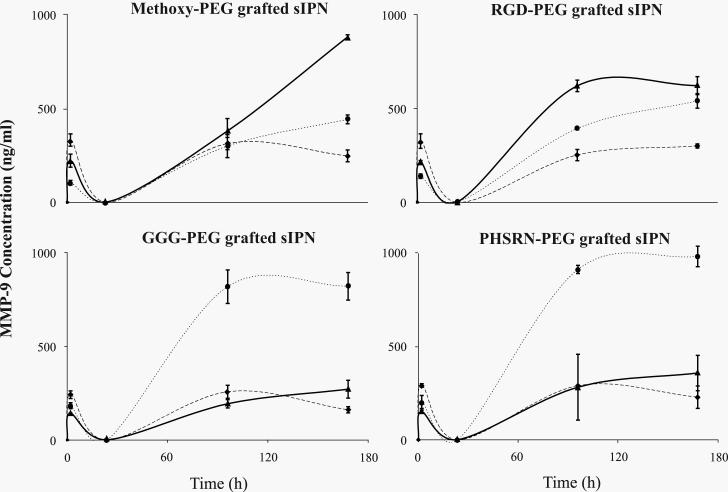
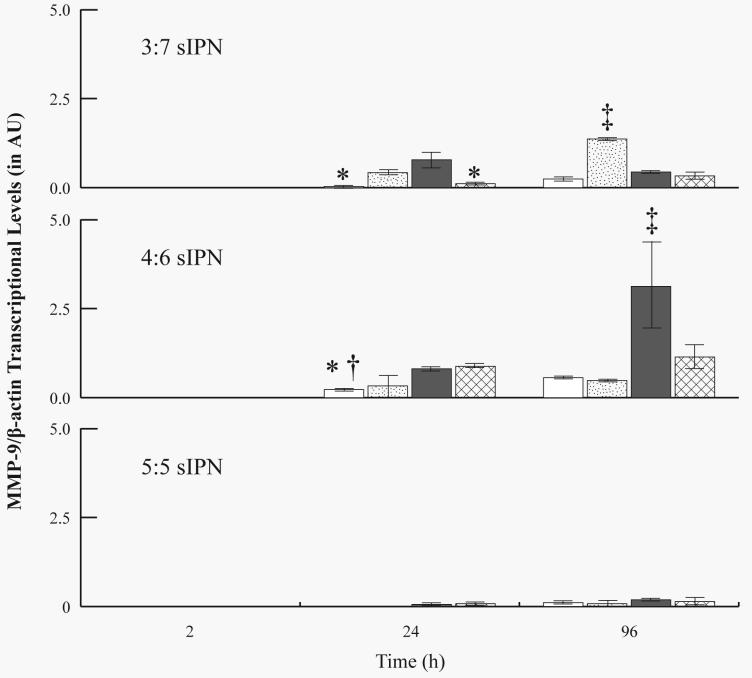
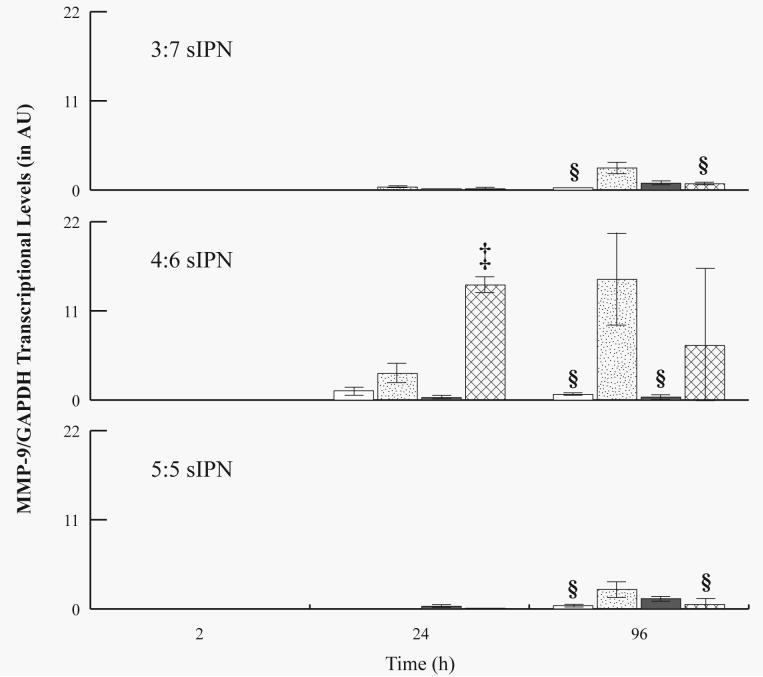
mRNA levels were investigated for all samples from 2 to 96 h, but not for 168 h, due to the low density of adherent cells from which mRNA was extracted. MMP-2 mRNA was not detectable (i.e. 0 ± 0 AU) for all sIPN at all time points. For all sIPN, MMP-9 mRNA levels were undetectable at 2 h, but generally increased over time (Figures 6 and 7). At 24 h, MMP-9/β-actin levels for RGD-PEG grafted 3:7 sIPN were significantly (p<0.05) higher than methoxy- and PHSRN-PEG grafted sIPN. RGD- and PHSRN-PEG grafted 4:6 sIPNs showed significantly (p<0.05) higher values than methoxy-PEG grafted sIPN at 24 h. At 96 h, GGG-PEG grafted 3:7 sIPN showed significantly (p<0.05) higher levels of MMP-9/β-actin than all other ligand-PEG grafted 3:7 sIPN. RGD-PEG grafted samples within 4:6 sIPN showed significantly higher transcriptional levels than all other ligand-PEG grafted 4:6 sIPNs at 96 h. When normalized to GAPDH, MMP-9 mRNA levels from PHSRN-PEG grafted 4:6 sIPN samples were significantly (p<0.05) higher than all other ligand-PEG grafted 4:6 sIPNs at 24 h (Fig. 7). At 96 h, MMP-9/GAPDH levels for GGG-PEG grafted 3:7 and 5:5 sIPN were significantly (p<0.05) higher than methoxy-, and PHSRN-PEG grafted sIPNs. GGG-PEG grafted 4:6 sIPNs were also significantly higher than methoxy-, and RGD-PEG grafted 4:6 sIPN at 96 h.
Figure 6.
RT-PCR analysis for MMP-9/ β-actin mRNA levels from adherent monocytes in the presence of 3:7, 4:6 and 5:5 wt% ratio of ligand-grafted gelatin:PEGdA sIPNs: methoxy-( ), GGG-(
), GGG-( ), RGD-(
), RGD-( ), and PHSRN-PEG grafted sIPN (
), and PHSRN-PEG grafted sIPN ( ). Data are expressed as mean ± s.d. *Significantly different (p<0.05) from RGD-PEG grafted sIPN (within sIPN group and time point). †Significantly different (p<0.05) from PHSRN-PEG grafted sIPN (within sIPN group and time point). ‡Significantly different (p<0.05) from all other ligand-PEG grafted sIPN (within sIPN group and time point).
). Data are expressed as mean ± s.d. *Significantly different (p<0.05) from RGD-PEG grafted sIPN (within sIPN group and time point). †Significantly different (p<0.05) from PHSRN-PEG grafted sIPN (within sIPN group and time point). ‡Significantly different (p<0.05) from all other ligand-PEG grafted sIPN (within sIPN group and time point).
Figure 7.
RT-PCR analysis for MMP-9/GAPDH mRNA levels from adherent monocytes in the presence of 3:7, 4:6 and 5:5 wt% ratio of ligand-PEG grafted gelatin:PEGdA sIPNs: methoxy-( ), GGG-(
), GGG-( ), RGD-(
), RGD-( ), and PHSRN-PEG grafted sIPN (
), and PHSRN-PEG grafted sIPN ( ). Data are expressed as mean ± s.d. §Significantly different (p<0.05) from GGG-PEG grafted sIPN (within sIPN group and time point). ‡Significantly different (p<0.05) from all other ligand-PEG grafted sIPN (within sIPN group and time point).
). Data are expressed as mean ± s.d. §Significantly different (p<0.05) from GGG-PEG grafted sIPN (within sIPN group and time point). ‡Significantly different (p<0.05) from all other ligand-PEG grafted sIPN (within sIPN group and time point).
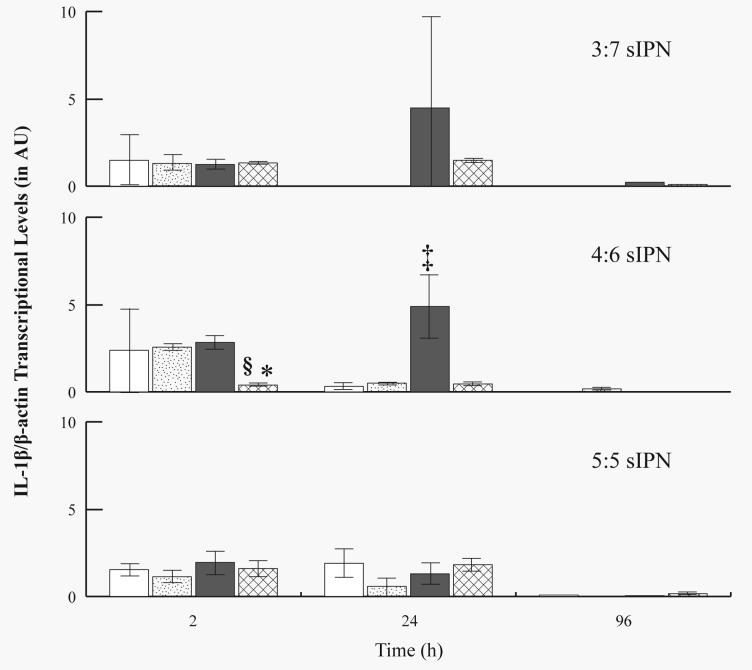
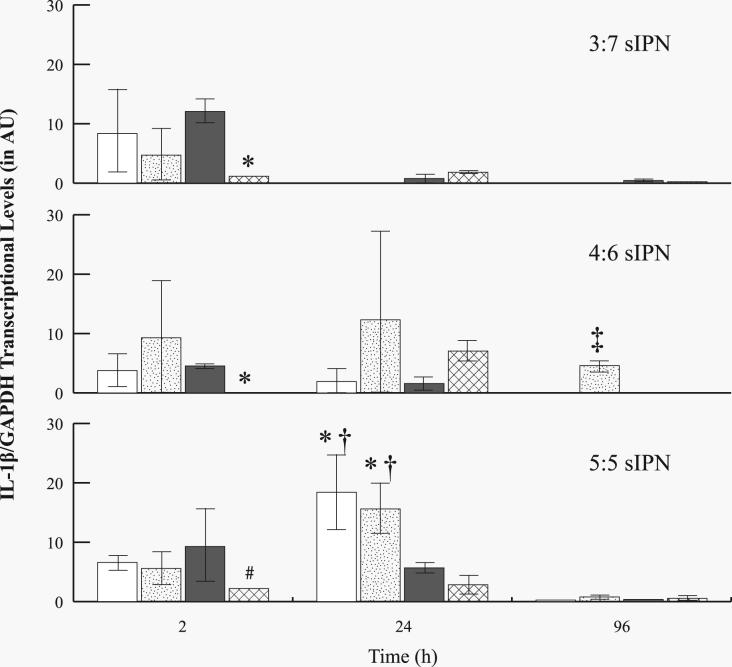
While MMP-9 mRNA levels increased over time in general, IL-1β mRNA levels generally decreased over time. IL-1β levels were also normalized to β-actin (Fig. 8) or to GAPDH (Fig. 9). At 2 h, PHSRN-PEG grafted 4:6 sIPNs showed significantly (p<0.05) lower levels as compared to GGG-, and RGD-PEG grafted 4:6 sIPNs. RGD-PEG grafted 4:6 sIPN showed significantly (p<0.05) higher IL-1β/β-actin levels compared to all other ligand-PEG grafted 4:6 sIPN at 24 h. Only a trace amount or no IL-1β/β-actin was detected by 96 h for all sIPN. Decrease in IL-1β mRNA over time was also observed when normalized to GAPDH (Fig. 9). At 2 h, both PHSRN-PEG grafted 3:7 and 4:6 sIPN samples showed significantly (p<0.05) lower IL-1β/GAPDH mRNA expression than the respective RGD-PEG grafted sIPN. PHSRN-PEG grafted 5:5 sIPNs also showed significantly (p<0.05) lower IL-1β/GAPDH levels compared to methoxy-PEG grafted sIPN at 2 h. At 24 h, IL-1β/GAPDH levels from methoxy- and GGG-PEG grafted 5:5 sIPN were significantly (p<0.05) higher than RGD-, and PHSRN-PEG grafted 5:5 sIPN. At 96 h, GGG-PEG grafted 4:6 sIPNs showed significantly (p<0.05) higher IL-1β/GAPDH levels compared to all other ligand-PEG grafted 4:6 sIPNs.
Figure 8.
RT-PCR analysis for IL-1β/β-actin mRNA levels from adherent monocytes in the presence of 3:7, 4:6 and 5:5 wt% ratio of ligand-PEG grafted gelatin:PEGdA sIPNs: methoxy-( ), GGG-(
), GGG-( ), RGD-(
), RGD-( ), and PHSRN-PEG grafted sIPN (
), and PHSRN-PEG grafted sIPN ( ). Data are expressed as mean ± s.d. §Significantly different (p<0.05) from GGG-PEG grafted sIPN (within sIPN group and time point). *Significantly different (p<0.05) from RGD-PEG grafted sIPN (within sIPN group and time point). ‡Significantly different (p<0.05) from all other ligand-PEG grafted sIPN (within sIPN group and time point).
). Data are expressed as mean ± s.d. §Significantly different (p<0.05) from GGG-PEG grafted sIPN (within sIPN group and time point). *Significantly different (p<0.05) from RGD-PEG grafted sIPN (within sIPN group and time point). ‡Significantly different (p<0.05) from all other ligand-PEG grafted sIPN (within sIPN group and time point).
Figure 9.
RT-PCR analysis for IL-1β/GAPDH mRNA levels from adherent monocytes in the presence of 3:7, 4:6 and 5:5 wt% ratios of ligand-PEG grafted gelatin:PEGdA sIPNs: methoxy-( ), GGG-(
), GGG-( ), RGD-(
), RGD-( ), and PHSRN-PEG grafted sIPN (
), and PHSRN-PEG grafted sIPN ( ). Data are expressed as mean ± s.d. #Significantly different (p<0.05) from methoxy-PEG grafted sIPN (within sIPN group and time point). *Significantly different (p<0.05) from RGD-PEG grafted sIPN (within sIPN group and time point). †Significantly different (p<0.05) from PHSRN-PEG grafted sIPN (within sIPN group and time point). †Significantly different (p<0.05) from all other ligand-PEG grafted sIPN (within sIPN group and time point).
). Data are expressed as mean ± s.d. #Significantly different (p<0.05) from methoxy-PEG grafted sIPN (within sIPN group and time point). *Significantly different (p<0.05) from RGD-PEG grafted sIPN (within sIPN group and time point). †Significantly different (p<0.05) from PHSRN-PEG grafted sIPN (within sIPN group and time point). †Significantly different (p<0.05) from all other ligand-PEG grafted sIPN (within sIPN group and time point).
DISCUSSION
This study has shown that human monocyte adhesion, MMP-2/-9 and IL-1β protein and MMP-9 and IL-1β mRNA expression were influenced by ligand identity and concentration. Increasing the wt% of RGD-PEG grafted gelatin in the sIPN increased accessible RGD concentration on the sIPN surface, but not beyond the 4:6 wt%. The lack of significant difference in accessible RGD between RGD-PEG grafted 4:6 and 5:5 sIPNs could be due to the less incremental wt% increase in RGD-PEG grafted gelatin content between 5:5 and 4:6 sIPNs compared to that between 3:7 and 4:6 sIPNs. There may also be a plateau after which RGD accessibility on the sIPN surface is limited and the ligand effects are diminished. Similar to this, no significant differences were observed in adherent cell density between 4:6 and 5:5 RGD-PEG grafted sIPNs. As shown in our previous work, initial monocyte adhesion was regulated by β1 and β3 containing integrin complexation onto RGD presented on the sIPN surface. However, other cell-material interaction involving various gelatin domains in the sIPN is likely to influence cell adhesion and subsequent behavior. For example, cells can adhere to gelatin via αVβ3-RGD complexation as well as α5β1-fibronectin bridge.[33,34] The mannose receptor expressed on the macrophage surface also specifically binds to gelatin.[35] Further, monocyte interactions with the sIPN are not limited to gelatin but also involve interactions with PEG. In another study, human monocytes are shown to adhere comparable to TCPS and respond to PEG-only hydrogels.[36]
Clear differences in MMP-2 and IL-1β protein release profiles over time were observed at initial time points from 3:7 sIPN as compared to those from 4:6 and 5:5 sIPN. Although this could be a result of the more significant incremental increase in gelatin wt% from 3:7 to 4:6 sIPNs compared to that between 4:6 and 5:5 sIPNs, a “threshold effect” of gelatin or PEG content at which MMP-2 and IL-1β release is suppressed may also exist. Though the kinetic pattern of MMP-2 protein release over time between 4:6 and 5:5 sIPN was similar, MMP-2 concentrations were generally higher for 4:6 compared to that of 5:5 sIPN. Even with the initial burst of MMP-2 protein release at 2 h for 4:6 and 5:5 sIPNs, MMP-2 mRNA was not observed from any sample at any time point. However, the disconnect between MMP-2 protein and mRNA expression has been reported by others.[20,38] The MMP-2 gene, containing few conserved cis-elements in the promoter region, has been speculated to transcribe MMP-2 protein, then to store the protein in the matrix until activation.[39, 40]
MMP-9 protein release was modulated by ligand identity, material formulation and time. Increasing MMP-9 protein concentration was consistent with generally increasing MMP-9/ β-actin and MMP-9/GAPDH over time. Ligand specific expression of MMP-9 protein and MMP-9/GAPDH mRNA was observed. For example, GGG-, and PHSRN-PEG grafted 4:6 sIPN showed a drastic increase in MMP-9 protein concentration from 24 to 96 h. This correlates with higher MMP-9/GAPDH mRNA levels at 96 h for GGG-, and PHSRN-PEG grafted 4:6 sIPNs, though the MMP-9/GAPDH level for PHSRN-PEG grafted 4:6 sIPN was not statistically significant.
Decreasing IL-1β protein concentrations were paralleled by decreasing IL-1β mRNA levels over time. At 24 h, RGD-PEG grafted 4:6 sIPNs showed higher IL-1β/β-actin mRNA levels compared to all other ligand-PEG grafted 4:6 sIPN. This was partly a result of low β-actin levels, perhaps due to specific receptor-ligand interactions and/or perhaps to the lack of cytoskeletal rearrangement on the sIPN surface. Ligand specific expression of IL-1β protein and mRNA expression was also observed. For example, relatively low IL-1β mRNA levels were observed for PHSRN-PEG grafted 4:6 sIPNs at 2 h and this correlated with significantly lower IL-1β protein concentrations for PHSRN-PEG grafted 4:6 sIPN as compared to methoxy-, and GGG-PEG grafted 4:6 sIPNs at 24 h. Only a trace amount or no IL-1β/β-actin and IL-1β/GAPDH mRNA were detected by 96 h, consistent with the decrease in IL-1β protein concentration at 96 h and beyond. The decrease in IL-1β protein levels was observed with an increase in MMP-9. The increase in MMP-9 protein from 24 to 96 h following the increase in IL-1β from 2 to 24 h correlates well with previous findings on IL-1β induction of MMP-9.[22] MMP-9 has also been shown to degrade IL-1β, and the decrease in IL-1β protein from 24 to 96 h could correspond with an increase in MMP-9 concentrations from 24 to 96 h.[21] Thus, in the context of sIPNs, monocyte expression of MMP-9 and IL-1β showed a potential autocrine regulation.
CONCLUSIONS
We have demonstrated that human monocyte adhesion, MMP-2/-9 and IL-1β protein and MMP-9 and IL-1β mRNA expression were influenced by ligand identity and concentration. These interrelationships are not always a positive correlation but a complex, time-dependent phenomenon.
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01HL77825, University of Wisconsin-Madison, and the NSF Graduate Research Fellowship Award to ASC.
List of Abbreviations
- Bis-COOH-PEG
bis-carboxylate-PEG
- Bis-NSu-PEG
bis-N-succinimidyl-acetate-PEG
- BSA
bovine serum albumin
- DIPEA
N, N-diisopropyl ethylamine
- ECM
extracellular matrix
- ELISA
enzyme linked immunosorbent assay
- ELS
evaporative light scattering
- FN
fibronectin
- GAPDH
glyceraldehydes-3-phosphate-dehydrogenase
- GGG
glycine-glycine-glycine
- GPC
gel permeation chromatography
- HPLC
high performance liquid chromatography
- IL-1β
interleukin-1beta
- M-MLV
moloney murine leukemia virus
- MMP-2
matrix metalloprotease-2
- MMP-9
matrix metalloprotease-9
- MPEG
polyethylene glycol monomethyl ether
- RT-PCR
reverse transcription polymerase chain reaction
- PBS
phosphate buffer saline
- PEG
polyethylene glycol (diol) 2kDa
- PEGdA
polyethylene glycol diacrylate
- PHSRN
proline-histidine-serine-arginine-asparagine
- RI
refractive index
- RGD
arginine-glycine-aspartic acid
- sIPN
semi-interpenetrating network
- TCPS
tissue culture polystyrene
References
- 1.Noda M, Wataha J, Lockwood P. Low-dose, long-term exposures of dental material components alter human monocyte metabolism. J Biomed Mater Res. 2002;62(2):237–243. doi: 10.1002/jbm.10065. [DOI] [PubMed] [Google Scholar]
- 2.Phillips JM, Kao WJ. Macrophage adhesion on gelatin-based interpenetrating networks grafted with PEGylated RGD. Tissue Engineering. 2005;11(5/6):964–973. doi: 10.1089/ten.2005.11.964. [DOI] [PubMed] [Google Scholar]
- 3.Xia Z, Triffitt JT. A review on macrophage responses to biomaterials. Biomed Mater. 2006;1(1):R1–R9. doi: 10.1088/1748-6041/1/1/R01. [DOI] [PubMed] [Google Scholar]
- 4.Kao WJ, Lee D, Schense JC, Hubbell JA. Fibronectin modulates macrophage adhesion and FBGC formation: the role of RGD, PHSRN, and PRRARV domains. J Biomed Mater Res. 2000;55(1):79–88. doi: 10.1002/1097-4636(200104)55:1<79::aid-jbm110>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 5.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113(10):1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 6.Massia SP, Hubbell JA. An RGD spacing of 440 nm is sufficient for integrin αvβ3-mediated fibroblast spreading and 140 nm for focal contact and stress fiber formation. J Cell Bio. 1991;114(5):1089–1100. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mann BK, Tsai AT, Scott-Burden T, West JL. Modification of surfaces with cell adhesion peptides alters extracellular matrix deposition. Biomaterials. 1999;20(23/24):2281–2286. doi: 10.1016/s0142-9612(99)00158-1. [DOI] [PubMed] [Google Scholar]
- 8.Mann BK, West JL. Cell adhesion peptides alter smooth muscle cell adhesion, proliferation, migration, and matrix protein synthesis on modified surfaces and in polymer scaffolds. J Biomed Mater Res. 2002;60(1):86–93. doi: 10.1002/jbm.10042. [DOI] [PubMed] [Google Scholar]
- 9.Cutler SM, Garcia AJ. Engineering cell adhesive surfaces that direct integrin α5β1 binding using a recombinant fragment of fibronectin. Biomaterials. 2003;24(10):1759–1770. doi: 10.1016/s0142-9612(02)00570-7. [DOI] [PubMed] [Google Scholar]
- 10.Liu JC, Heilshorn SC, Tirrell DA. Comparative cell response to artificial extracellular matrix proteins containing the RGD and CS5 cell-binding domains. Biomacromolecules. 2004;5(2):497–504. doi: 10.1021/bm034340z. [DOI] [PubMed] [Google Scholar]
- 11.Chung AS, Gao Q, Kao WJ. Macrophage matrix metalloproteinase-2/-9 gene and protein expression following adhesion to ECM-derived multifunctional matrices via integrin complexation. Biomaterials. 2006;28(2):285–298. doi: 10.1016/j.biomaterials.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 12.Chung AS, Gao Q, Kao WJ. Either integrin subunit beta1 of beta3 is involved in mediating monocyte adhesion, IL-1beta protein and mRNA expression in response to surface functionalized with fibronectin-derived peptides. J Biomater Sci Polym Ed. 2007;18(6):713–729. doi: 10.1163/156856207781034179. [DOI] [PubMed] [Google Scholar]
- 13.Waldeck HM, Chung AS, Kao WJ. Interpenetrating networks containing gelatin modified with PEGylated RGD and soluble KGF: synthesis: characterization, application in in vivo critical dermal wound. J Biomed Mater Res. 2007;82(4):861–871. doi: 10.1002/jbm.a.31054. [DOI] [PubMed] [Google Scholar]
- 14.Garcia AJ. Interfaces to control cell-biomaterial adhesive interactions. Adv Polym Sci. 2006;203:171–190. [Google Scholar]
- 15.Neff JA, Tresco PA, Caldwell KD. Surface modification for controlled studies of cell-ligand interactions. Biomaterials. 1999;20(23/24):2377–2393. doi: 10.1016/s0142-9612(99)00166-0. [DOI] [PubMed] [Google Scholar]
- 16.Shin H, Jo S, Mikos AG. Modulation of marrow stromal osteoblast adhesion on biomimetic oligo[poly(ethylene glycol) fumarate] hydrogels modified with Arg-Gly-Asp peptides and a poly(ethylene glycol) spacer. J Biomed Mater Res. 2002;61(2):169–179. doi: 10.1002/jbm.10193. [DOI] [PubMed] [Google Scholar]
- 17.Garcia AJ, Ducheyne P, Boettiger D. Cell adhesion strength increases linearly with adsorbed fibronectin surface density. Tissue Engineering. 1997;3(2):197–206. [Google Scholar]
- 18.Schense JC, Hubbell JA. Three-dimensional migration of neurites is mediated by adhesion site density and affinity. J Biol Chem. 2000;275(10):6813–6818. doi: 10.1074/jbc.275.10.6813. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Diaz GJ, Nelson D, Crone WC, Kao WJ. Mechanical and chemical analysis of gelatin-based hydrogel degradation. Macromol Chem Phys. 2003;204:1898–1908. [Google Scholar]
- 20.Parks WC. Matrix metalloproteinases in repair. Wound Repair Regen. 1999;7(6):423–432. doi: 10.1046/j.1524-475x.1999.00423.x. [DOI] [PubMed] [Google Scholar]
- 21.Ito A, Mukaiyama A, Itoh Y, Nagase H, Thogersen IB, Enghild JJ, Sasaguri Y, Mori Y. Degradation of interleukin 1β by matrix metalloproteinases. J Biol Chem. 1996;271(25):14657–14660. doi: 10.1074/jbc.271.25.14657. [DOI] [PubMed] [Google Scholar]
- 22.Rao VH, Singh RK, Delimont DC, Schaefer GB, Bridge JA, Neff JR, Sanger WG, Sappenfield JW, Buehler BA, Finnell RH. Interleukin-1β upregulates MMP-9 expression in stromal cells of human giant cell tumor of bone. J Interf Cytok Res. 1999;19(10):1207–1217. doi: 10.1089/107999099313154. [DOI] [PubMed] [Google Scholar]
- 23.Miller KM, Anderson JM. Human monocyte/macrophage activation and IL-1 generation by biomedical polymers. J Biomed Mater Res. 1988;22:713–731. doi: 10.1002/jbm.820220805. [DOI] [PubMed] [Google Scholar]
- 24.McCawley LJ, Matrisian LM. Matrix metalloproteinases: They're not just for matrix anymore. Curr Opin Cell Biol. 2001;13(5):531–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- 25.Einerson NJ, Stevens KR, Kao WJ. Synthesis and physicochemical analysis of gelatin-based hydrogels for drug carrier matrices. Biomaterials. 2002;24:509–523. doi: 10.1016/s0142-9612(02)00369-1. [DOI] [PubMed] [Google Scholar]
- 26.Lin-Gibson S, Bencherif S, Cooper JA, Wetzel SJ, Antonucci JM, Vogel BM, Horkay F, Washburn NR. Synthesis and characterization of PEG dimethacrylates and their hydrogels. Biomacromolecules. 2004;5(4):1280–1287. doi: 10.1021/bm0498777. [DOI] [PubMed] [Google Scholar]
- 27.Gattas-Asfura KM, Weisman E, Andreopoulos FM, Micic M, Muller B, Sirpal S, Pham SM, Leblanc RM. Nitrocinnamate-functionalized gelatin: synthesis and “smart” hydrogel formation via photo-cross-linking. Biomacromolecules. 2005;6(3):1503–1509. doi: 10.1021/bm049238w. [DOI] [PubMed] [Google Scholar]
- 28.Benoit DS, Anseth KS. The effect on osteoblast function of colocalized RGD and PHSRN epitopes on PEG surfaces. Biomaterials. 2005;26(25):5209–5220. doi: 10.1016/j.biomaterials.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 29.McNally AK, Anderson JM. Complement C3 participation in monocyte adhesion to different surfaces. P Natl Acad Sci USA. 1994;91(21):10119–10123. doi: 10.1073/pnas.91.21.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 31.Gao Q, Meijer MJW, Kubben FJGM, Sier CFM, Kruidenier L, van Duijn W, van den Berg M, van Hogezand RA, Lamers CBHW, Verspaget HW. Expression of matrix metalloproteinases-2 and -9 in intestinal tissue of patients with inflammatory bowel diseases. Digest Liver Dis. 2005;37(8):584–592. doi: 10.1016/j.dld.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 32.Bas A, Forsberg G, Hammarstroem S, Hammarstroem ML. Utility of the housekeeping genes 18S rRNA, β-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol. 2004;59(6):566–573. doi: 10.1111/j.0300-9475.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- 33.Davis GE. Affinity of integrins for damaged extracellular matrix: αvβ3 binds to denatured collagen type I through RGD sites. Biochem Bioph Res Co. 1992;182(3):1025–1031. doi: 10.1016/0006-291x(92)91834-d. [DOI] [PubMed] [Google Scholar]
- 34.Tuckwell DS, Ayad S, Grant ME, Takigawa M, Humphries MJ. Conformation dependence of integrin-type II collagen binding. Inability of collagen peptides to support α2β1 binding, and mediation of adhesion to denatured collagen by a novel α5β1-fibronectin bridge. J Cell Sci. 1994;107(4):993–1005. doi: 10.1242/jcs.107.4.993. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Pomares L, Wienke D, Stillion R, McKenzie EJ, Arnold JN, Harris J, McGreal E, Sim RB, Isacke CM, Gordon S. Carbohydrate-independent recognition of collagens by the macrophage mannose receptor. Eur J Immunol. 2006;36(5):1074–1082. doi: 10.1002/eji.200535685. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt DR, Kao WJ. Monocyte activation in response to PEG hydrogels grafted with RGD and PHSRN separated by interpositional spacers of various lengths. J Biomed Mater Res. doi: 10.1002/jbm.a.31270. e-published May 2007. [DOI] [PubMed] [Google Scholar]
- 37.Agren MS. Gelatinase activity during wound healing. Brit J Dermatol. 1994;131(5):634–640. doi: 10.1111/j.1365-2133.1994.tb04974.x. [DOI] [PubMed] [Google Scholar]
- 38.Van den Steen PE, Dubois B, Nelissen I, Rudd PM, Dwek RA, Opdenakker G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9) Crit Rev Biochem Mol. 2002;37(6):375–536. doi: 10.1080/10409230290771546. [DOI] [PubMed] [Google Scholar]
- 39.Werb Z. ECM and cell surface proteolysis: regulating cellular ecology. Cell. 1997;91(4):439–442. doi: 10.1016/s0092-8674(00)80429-8. [DOI] [PubMed] [Google Scholar]
- 40.Murphy G, Gavrilovic J. Proteolysis and cell migration: creating a path? Curr Opin Cell Biol. 1999;11(5):614–621. doi: 10.1016/s0955-0674(99)00022-8. [DOI] [PubMed] [Google Scholar]