Abstract
The incorporation of the DNA packaging connector complex during lambdoid phage assembly in vivo is strictly controlled - one and only one of the twelve identical icosahedral vertices is differentiated by the inclusion of a portal or connector dodecamer. Proposed control mechanisms include obligate nucleation from a connector containing complex, addition of the connector as the final step during assembly, and a connector mediated increase in the growth rate. The inability to recapitulate connector incorporation in vitro has made it difficult to obtain direct biochemical evidence in support of one model over another. Here we report the development an in vitro assembly system for the well characterized dsDNA phage Phi29 which results in the co-assembly of connector with capsid and scaffolding proteins to form procapsid-like particles (PLPs). Immuno-electron microscopy demonstrates the specific incorporation of connector vertex in PLPs. The connector protein increases both the yield and the rate of capsid assembly suggesting that the incorporation of the connector in Phi29 likely promotes nucleation of assembly.
Keywords: connector protein, dsDNA phage Phi29, in vitro capsid assembly, portal vertex incorporation, DNA packaging
Introduction
The genomes of icosahedral viruses are surrounded and protected by an outer shell of protein which is assembled from hundreds of copies of a single or small number of types of capsid protein subunits through the use of quasi-equivalent bonding interactions (Caspar, 1980; Caspar and Klug, 1962). The two primary mechanisms of genome encapsidation are concurrent assembly and packaging and packaging into a preformed shell termed a procapsid or prohead (Bazinet and King, 1985). In the latter case, typified by the dsDNA containing bacteriophages, ATP-dependent DNA translocation pumps DNA into the procapsid to liquid crystalline density. Recent crystallographic, electron microscopic, and single molecule studies have provided structural insights into the mechanism of DNA packaging (Cuervo et al., 2007; Hugel et al., 2007; Lander et al., 2006a; Lebedev et al., 2007; Poh et al., 2008; Smith et al., 2001; Sun et al., 2008; Sun et al., 2007; Xiang et al., 2006). The motor is composed of a dodecameric connector or portal protein (connector/portal vertex) which replaces the capsid subunits at one of the icosahedral vertices and forms a conduit for DNA translocation and egress and typically a two subunit terminase complex which recognizes the viral DNA, delivers it to the connector protein, and couples the hydrolysis of ATP to translocation. Once the genome is packaged conformational changes in the connector complex cause the dissociation of the terminase proteins and allow the attachment of the completion proteins necessary to form an infectious phage (Lander et al., 2006b; Lhuillier et al., 2009; Xiang et al., 2006).
To successfully package a full length genome incorporation of one and only one connector vertex into the procapsid is essential (Moore and Prevelige, 2002). In vivo, nearly every assembled procapsid has one and only one connector vertex and is able to package DNA and mature into an infectious phage (Casjens and King, 1974). This narrow distribution in which 95 % of particles have a single connector vertex cannot be explained by random statistical incorporation and implies the existence of a control mechanism. A number of possible control mechanisms have been proposed, including obligate coupling of connector incorporation to nucleation, the formation of an mRNA/protein nucleation complex (Bazinet, Villafane, and King, 1990), incorporation during lattice growth (Moore and Prevelige, 2002), and incorporation as the final assembly step (Cerritelli and Studier, 1996).
In the dsDNA phage procapsid assembly involves the capsid, connector, and scaffolding proteins (Fane and Prevelige, 2003). Mutational studies have indicated that scaffolding protein is involved either directly or indirectly in the incorporation of the connector vertex during procapsid assembly in a variety of phages (Bazinet and King, 1988; Earnshaw and King, 1978; Greene and King, 1996). However, with the exception of the Bacillus subtilis phage Phi29 a direct biochemical interaction between connector and scaffolding protein has not been reported (Lee and Guo, 1995).
The Phi29 procapsid is composed of 235 copies of capsid protein, ~180 copies of internal form-determining scaffolding protein, and 12 molecules of the connector protein arranged as a 43 nm×53 nm prolate procapsid particles with a triangulation number of T = 3 and an elongation factor of Q = 5 (Morais et al., 2005; Tao et al., 1998). The connector dodecamer occupies one pentameric vertex along the elongated axis and is complexed with 5 or 6 molecules of a small pRNA required for packaging linear dsDNA genome covalently linked to the terminal protein gp3. Packaging is powered by multiple copies of the ATPase gp16 (Grimes, Jardine, and Anderson, 2002; Guo, Erickson, and Anderson, 1987; Simpson et al., 2000; Tao et al., 1998; Zhang et al., 1998). We have previously developed an in vitro assembly system for Phi29 procapsids (Fu et al., 2007). In this manuscript we report the adaptation of that system to incorporate the connector protein and utilization of this system to gain insight into the mechanism of controlled connector incorporation.
Results
Connector Protein Assembled with Capsid Protein and Scaffolding Protein in vitro
An in vitro assembly system in which Phi29 capsid protein and scaffolding proteins assemble into prolate and T=3 isometric procapsid-like particles (PLPs) has previously been described (Fu et al., 2007). To investigate the role of the connector protein in capsid assembly the system was modified to include the connector protein. Recombinant connector protein was expressed and purified as previously described (Poliakov et al., 2007). Native mass spectrometry and sedimentation velocity experiments demonstrated that the purified connector protein was dodecameric (Poliakov et al., 2007).
The two-component assembly system utilizes polyethylene glycol to promote polymerization. In the three-component system the concentration of polyethylene glycol was lowered from 13% to 6% to achieve higher fidelity at the expense of some yield. Although connector protein requires high a concentration of NaCl (400 mM) to prevent it from aggregating, the interactions between scaffolding and capsid proteins are salt sensitive and particle formation drops by half at 80 mM NaCl when compared to 18 mM NaCl (Fu et al., 2007). As a tradeoff between connector solubility and assembly yield the NaCl concentration of the assembly reaction was maintained at 60–80 mM. The kinetics of assembly were followed by the time dependent increase in turbidity at 340 nm and the products were analyzed by centrifugation through a 10–40% sucrose gradient followed by SDS-PAGE.
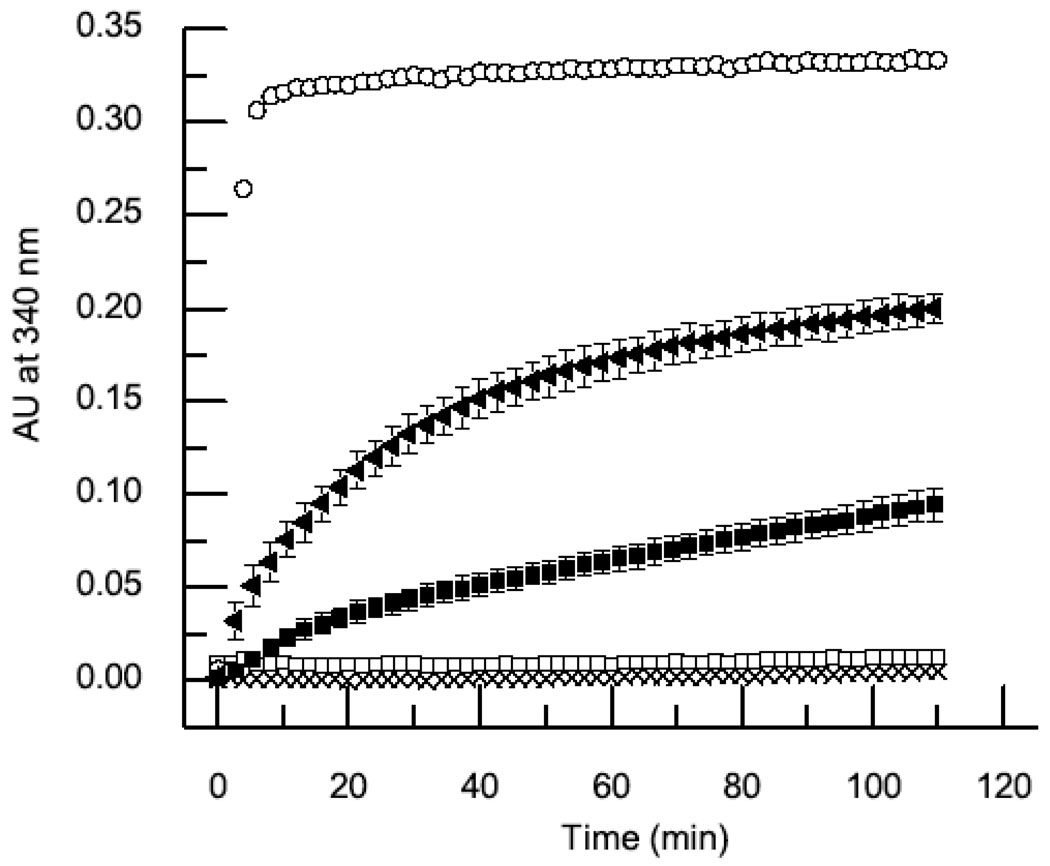
In accord with previous findings the capsid protein at 20µM (1 mg/ml) did not polymerize on its own as there was no apparent increase in turbidity over two hours of reaction (Fig 1) and the capsid protein remained at the top of the sucrose gradient, the expected position for a monomeric protein (Fig 2). However, capsid assembly was promoted by the addition of 40µM scaffolding protein as evidenced by a time dependent increase in turbidity (Fig 1) and co-sedimentation of the scaffolding and coat proteins to the middle of the sucrose gradient (Fig 2). When 4µM of connector protein was added to the mixture the assembly kinetics displayed a rate increase of approximately 3.5-fold and an approximately 2.5-fold increase in the final turbidity level compared to the reaction containing only capsid and scaffolding proteins (Fig 1). Analysis of the sucrose gradients demonstrated that the fraction of polymerized capsid protein increased from 47% to 74% in the presence of the connector. Sucrose gradient fractions 6 and 7 showed co-migration of the connector protein with the capsid and scaffolding proteins indicating association. The molar ratio of connector to capsid proteins was approximately 1:9 as determined by densitometry. This is higher than the 1:20 ratio expected for a single dodecameric connector incorporated in a 240 subunit capsid and suggests that connector may have been incorporated into aberrant or partially assembled shells. The fractions toward the bottom of the gradient had higher connector/capsid ratio but lower scaffolding content suggesting they were malformed. Indeed, unclosed or partially assembled particles were seen in these fractions by negative-stained EM (data not shown). Mutant scaffolding protein which could not bind the connector was not incorporated thereby providing evidence for specific incorporation.
Fig. 1. Connector Protein Increased Capsid Assembly Kinetics and Assembly Yield in Vitro.
The assembly kinetics of reactions containing 20 µM capsid protein (crosses), 20 µM capsid protein and 40 µM scaffolding protein (closed squares), 20 µM capsid protein, 40 µM scaffolding protein and 4 µM connector protein (closed triangles), 20 µM capsid protein and 4 µM connector protein (open squares), and 4 µM connector protein only (open circles) were followed by turbidity at OD 340 nm. The error bars display the standard deviation for five independent measurements.
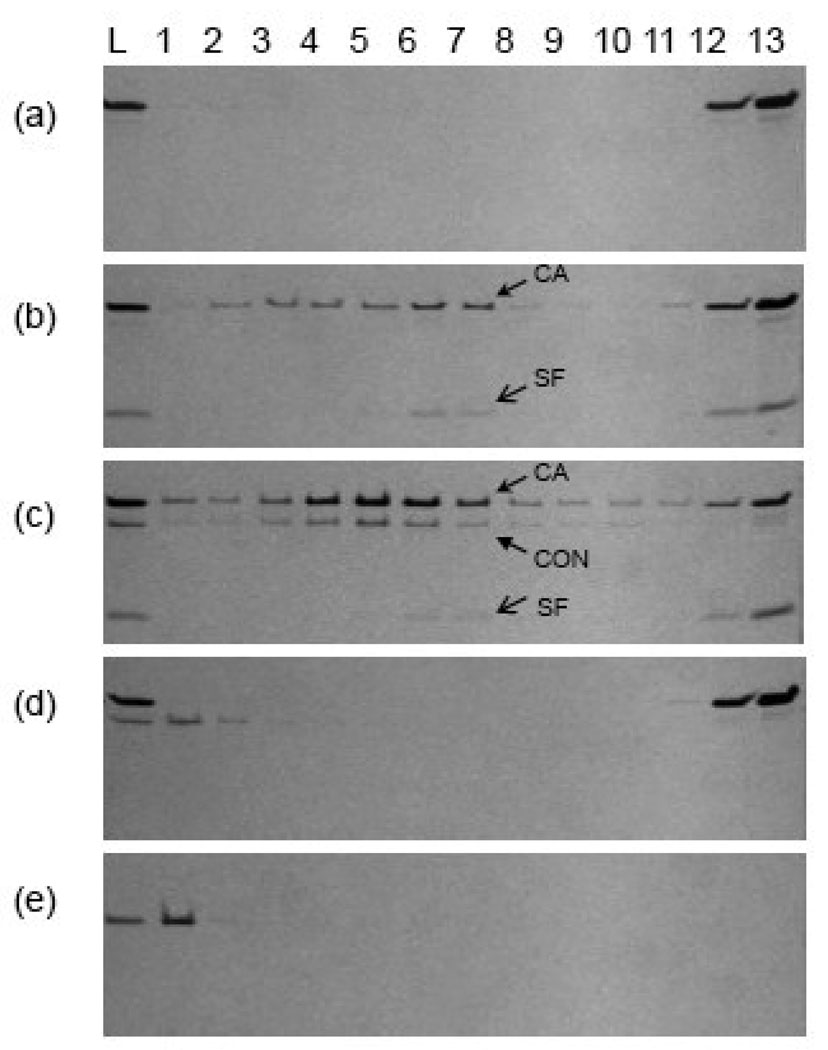
Fig. 2. Connector Protein Assembled with Capsid and Scaffolding Proteins in Vitro Assayed by Sucrose Gradient/SDS Page Analysis.
Assembly products of reactions containing (a) capsid protein, (b) capsid protein and scaffolding protein, (c) capsid protein, scaffolding protein and connector protein, (d) capsid protein and connector protein, and (e) connector protein only were analyzed by SDS-PAGE after sucrose gradient fractionation from bottom (fraction 1) to top (fraction 13). The samples loaded on to the gradients were shown in the lane L on the left. CA, capsid protein; SF, scaffolding protein; CON, connector protein.
The connector protein alone showed strong scattering and sedimented to the bottom of the gradient when diluted into assembly buffer consistent with aggregation. The aggregation of connector protein, proposed as a linear stacking of rings (Tsuprun, Anderson, and Egelman, 1994), is likely due to low NaCl concentration and the presence of the crowding agent PEG. Interestingly, mixing capsid protein with connector protein prevented aggregation as assayed by turbidity. However, when the mixture was analyzed by sucrose gradient centrifugation, the coat protein remained at the top of the gradient and the connector pelleted. These results suggest that there might be some interaction between capsid and connector proteins which prevents connector protein self-aggregation but that the interactions are weak and were disrupted during gradient centrifugation. It should be noted that the putative capsid/connector protein interaction alone was not sufficient to initiate or propagate assembly.
Connector Protein was Incorporated to Procapsid-Like Particles in Vitro
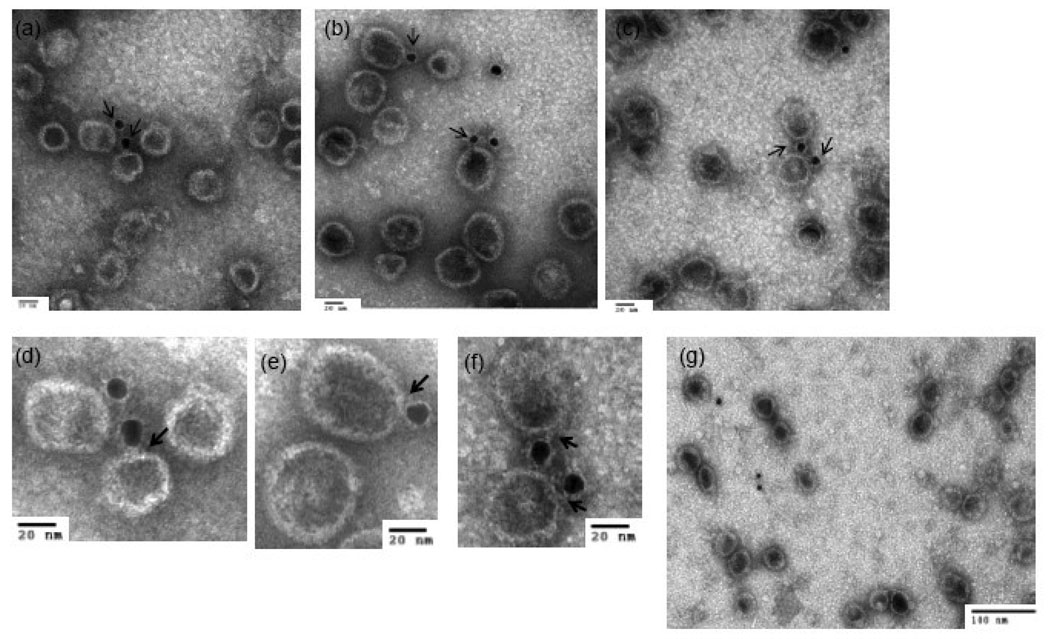
Immuno-electron microscopy was performed to directly demonstrate the specific incorporation of connector protein into PLPs. After absorption to grids a primary antibody against connector protein was used to tag the portal vertex in PLPs and visualization was accomplished using a secondary antibody conjugated to 10 nm gold particle followed by negative staining (Fig 3, and Table I).
Fig. 3. Connector Protein Incorporated to PLPs.
Electron micrographs of immuno-gold labeled of connector protein in (a) authentic procapsids, (b,c) procapsid-like particles assembled with added connector protein, and (d) procapsid-like particles assembled without added connector protein. Blown-up views of the particles in Fig a, b, c are shown in Fig e, f, g. Table I shows the distance distribution from the center of gold particles to the nearest PLPs in the samples assembled in the presence or absence of connector protein.
Table I.
Distance Distribution of Gold Particles
| Gold Particle Distance from PLP | With Connector | Without Connector |
|---|---|---|
| < 15 nm | 71 % | 2 % |
| 15 – 30 nm | 13% | 12 % |
| > 30 nm | 15% | 79 % |
| Number of Particles Counted | 45 | 86 |
In the case of procapsids purified in vivo (Fig 3a) one gold-particle appeared to be associated at one end of either the major or minor axis of the prolate particles. This difference in presentation likely arises from the propensity of the absorbed particles to adopt different orientations on the grid. In the case of the PLPs assembled in vivo in the presence of connector the micrographs showed a similar size and morphology for the PLPs as well as a similar gold-labeling pattern (Fig 3b, c). Only 10–15% labeling efficiency was achieved in the procapsid sample, perhaps partially due to inaccessibility of the connector vertex in particles absorbed on grid. Therefore, the efficiency of connector incorporation in vitro was hard to determine. However, the labeled PLPs had predominantly one gold-label and very rarely had two. The particles formed without connector protein were not immuno-gold labeled (Fig 3d).
Connector Protein Lowered Capsid Protein Concentration for Assembly
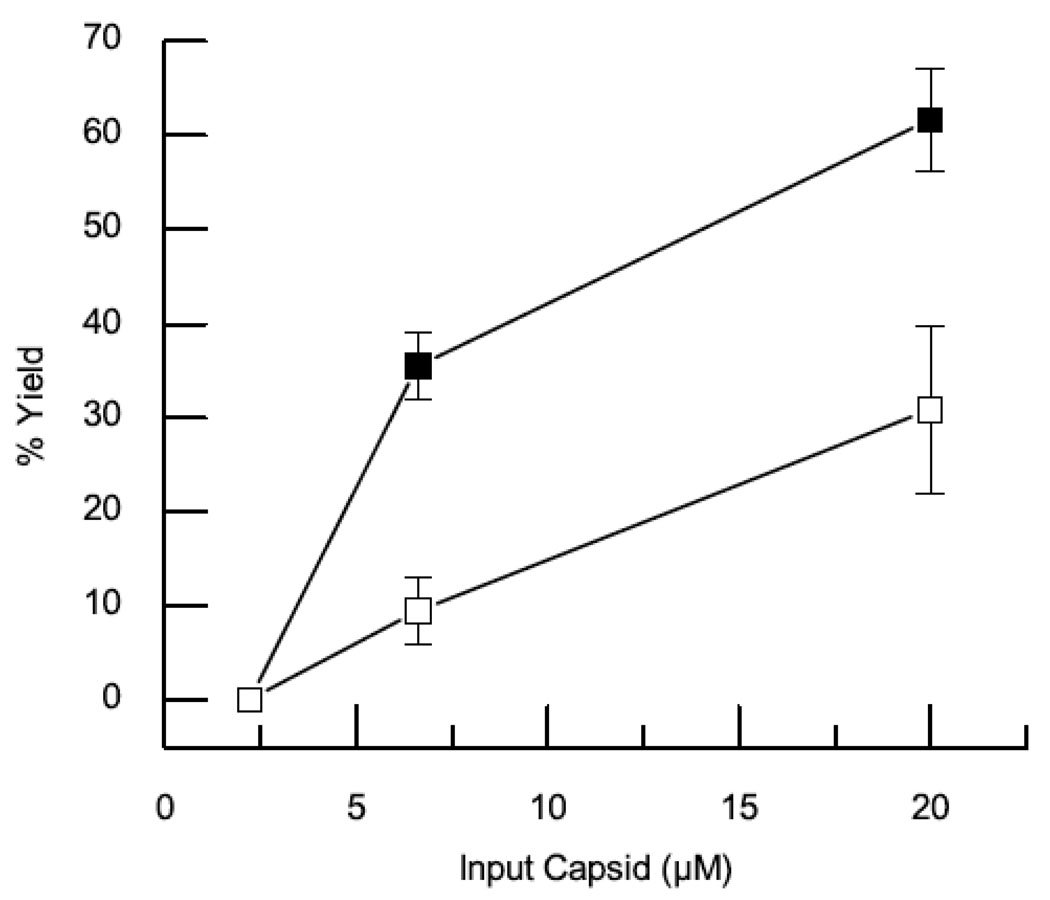
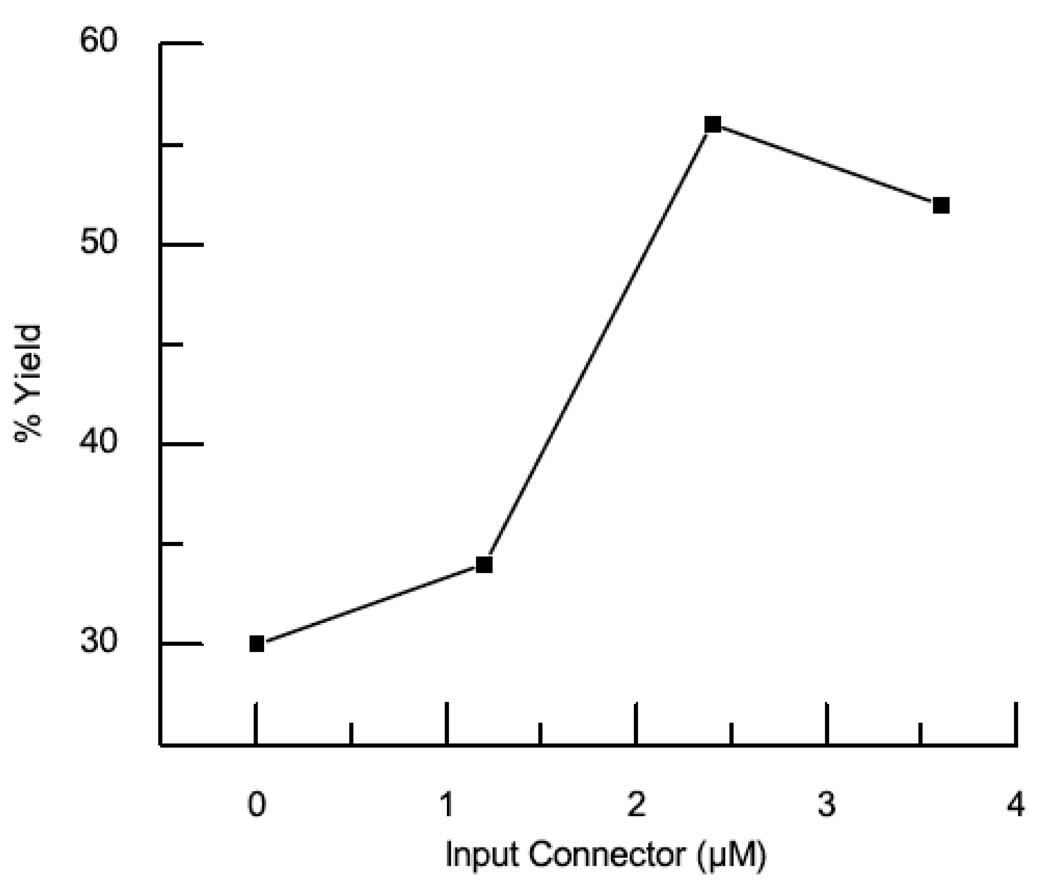
To determine whether the connector protein is involved in nucleating assembly, the effect of connector protein on the critical concentration of capsid protein required for assembly was investigated. Assembly reactions containing capsid protein concentrations ranging from 2.2–20 µM (0.1–1.0 mg/ml) were carried out in the presence or absence of 4 µM connector protein. The fraction of assembled capsid protein was determined by quantitative SDS-PAGE analysis following sucrose gradient separation and plotted as a function of input capsid protein (Fig 4a). The presence of connector protein increased the assembly yield at all capsid concentrations. At the lower capsid protein concentration (2.2 µM) the addition of connector resulted in an approximately five fold increase in yield whereas at higher concentration (20 µM) it resulted in an approximately two fold increase. A disproportionate effect of connector protein on yield as the capsid protein approaches the critical concentration is consistent with enhanced nucleation and lowered critical concentration. Complementary results were obtained when a fixed concentration of capsid protein (20 µM) was assembled in the presence of increasing concentrations of connector protein from 0–4 µM (Fig 4b). In this experiment the addition of connector protein resulted in an approximately doubling of the yield of assembled capsid.
Fig. 4. Connector Protein Lowered Capsid Concentration Required for Assembly.
(a) The fraction of assembled capsid protein formed with (closed squares) or without (open squares) connector protein was plotted as the function of input capsid protein 2.2 – 20 µM. The error bars display the standard deviation for five independent measurements. (b) The fraction of assembled capsid protein formed as the function of input connector protein (0–4 µM) at 20 µM capsid protein.
Discussion
Various mechanisms have been suggested to insure that one and only one portal complex is associated with a procapsid during assembly. The simplest scenario is that connector protein serves as a nucleus to initiate assembly thus insuring that every procapsid contains one and only one connector vertex. In phage T4, assembly was proposed to initiate at the connector vertex which is associated with the cell membrane through the protein gp40 (Black and Silverman, 1978; Traub and Maeder, 1984). In contrast, the rate of procapsid assembly for phages P22 and SPP1 is not affected by the presence or absence of connector protein (Bazinet and King, 1988; Droge et al., 2000; Droge and Tavares, 2000; Moore and Prevelige, 2002) and in Phi29 procapsid assembly can occur in the absence of connector (Guo et al., 1991). These results imply that incorporation of the connector is neither obligate nor the rate determining step in procapsid assembly. Moore et al. demonstrated that the P22 connector cannot be incorporated into pre-assembled procapsids ruling out incorporation as the final step (Moore and Prevelige, 2002) and connector incorporation as the final assembly step was ruled out for Phi29 based on its ability to determine whether prolate or isometric particles were produced (Guo et al., 1991). Moore et al. demonstrated that high level overexpression of P22 portal protein could result in multiple incorporation implying a kinetic rather than structural control mechanism (Moore and Prevelige, 2002).
The fact that it has proven difficult to incorporate the connector into procapsids assembled in vitro from purified components has made it difficult to dissect the mechanism of controlled incorporation. Prior to this report, the only documented case of in vitro incorporation was the scaffolding mediated incorporation of portal into herpes simplex virus (Singer et al., 2005). In the case of Phi29 we have been able to develop an in vitro assembly system in which the connector protein is incorporated. Purified recombinant dodecameric connector protein assembles with capsid and scaffolding proteins to form procapsid like particles. The incorporation of connector vertex is mediated by scaffolding protein, agreeing with the in vivo data that a temperature sensitive scaffolding protein can promote capsid assembly but fails to incorporate connector protein (Camacho et al., 1977; Guo et al., 1991; Hagen et al., 1976; Lee and Guo, 1995). Even though capsid and scaffolding proteins can assemble in the absence of connector (Fu et al., 2007; Guo et al., 1991), the presence of the connector protein affects the assembly kinetics and yield. The initial rate of assembly is increased ~3.5 fold by the presence of the connector with a resultant ~2 fold increase in the amount of assembled capsid. The data suggest that connector protein lowers the critical concentration of capsid protein required for assembly and thereby serves to nucleate assembly and ensure specific incorporation and proper positioning of one and only one connector vertex. As suggested by Guo (Guo et al., 1991) the likely nucleation complex is a ternary complex consisting of scaffolding, connector, and capsid protein. In the absence of connector nucleation can still occur, albeit less readily, through the interaction of scaffolding and capsid protein alone.
Materials and methods
Protein Preparations
Recombinant capsid protein was purified form Escherichia coli BL21 (DE3) pLysS as previously described (Fu et al., 2007). The capsid protein was purified from inclusion bodies and refolded at 0.5 mg/ml in 0.7 M Arginine at pH 8. The refolded proteins were dialyzed against 100 mM Tris, pH 8, 10 mM MgCl2 and 50 mM NaCl. Capsid proteins were bound and eluted from HiTrap SP HP column at around 150 mM NaCl in the same buffer. The purified capsid protein was dialyzed and stored in 100 mM Tris, pH 8, 10 mM MgCl2 and 150 mM NaCl.
Recombinant scaffolding proteins were expressed in Escherichia coli BL21 (DE3) pLysS cells harboring plasmid pARgp7 (Fu et al., 2007; Lee and Guo, 1995). The proteins were purified with HiTrap Q HP column using a NaCl gradient in 50 mM Tris-HCl pH 8, 1 mM EDTA. The scaffolding proteins eluted at 210 mM NaCl and were subsequently dialyzed against the same buffer at 50 mM NaCl. Further purification was carried out with Superdex G-75 size exclusion chromatography. The purified scaffolding proteins were stored in 50 mM Tris-HCl pH 8, 50 mM NaCl and 1 mM EDTA.
The recombinant connector protein was expressed in Escherichia coli harboring plasmid pPLc28D and purified as described (Poliakov et al., 2007). The cell pellets were resuspended in buffer containing 0.3 M NaCl, 50 mM Tris, pH 7.7. The protein was purified with HiTrap SP HP column, eluting at around 0.75 M NaCl. Following dialysis against 0.3 M NaCl and 50 mM Tris, pH 7.7, the second step of purification was carried out with HiTrap Q HP column where the protein was eluted at approximately 0.43 M NaCl. The pooled fractions were dialyzed against 0.4 M NaCl buffer for storage.
In Vitro Assembly Reactions
Typical assembly reactions were carried out at 20 µM (1 mg/ml) capsid protein, 40 µM scaffolding protein and 4 µM connector protein. 350 µl of 9% PEG-3350 in 100 mM Tris, pH 8, 10 mM MgCl2 was added to the cuvette and allowed temperature equilibrate to 13°C. 120 µl of capsid protein, 20 µl of scaffolding protein, and 35 µl of connector protein (or equivalent buffer in the reactions containing no connector protein) were mixed to a final buffer composition of 6% PEG-3350, 100 mM Tris, pH 8, 10 mM MgCl2 and 60 mM NaCl. The assembly kinetics were monitored by recording the turbidity at 340 nm at 40 second intervals for 2 hours using the Beckman DU640 spectrometer. The dead-time between mixing and the start of data acquisition was 40 seconds.
200 µl aliquots were overlaid on a 10–40% sucrose gradient. A 50 µl cushion of 60% CsCl in 50% sucrose was laid at the bottom. The samples were centrifuged at 4°C at 45,000 rpm for 45 min in a Beckman SW-55Ti rotor. The gradient was fractionated from the bottom and analyzed by SDS-PAGE. The intensity of the coomasssie blue stained protein bands was quantified using a Bio-Rad gel documentation system. The stoichiometry of the capsid to connector protein in PLPs was quantified by the relative protein intensity after subtracted background reading and normalized to the intensity reading obtained with known protein amount and ratio (lane L in Fig 2). The % yield displayed in Fig 4 was calculated as the ratio of the integrated capsid band intensity of gradient fraction 4–7 over all fractions.
Immuno-Electron Microscopy
In vitro assembled particles were purified by sucrose gradient centrifugation and dialyzed out of sucrose. The particles were absorbed to glow discharged carbon-coated Formvar layer supported nickel grids. The excess materials were removed with filter paper. The grids were washed with 1% ovalumin in PBS buffer, blocked with 10% ovalumin in PBS buffer for 1hr and incubated with primary antibody against connector protein for 1 hr. After washing, the grids were incubated with secondary anti-rabbit IgG conjugated to 10 nm conjugated gold particles for 45 min and washed out unbound antibody. The grids were stained with 2% uranyl acetate for 40 sec. The images were taken at 26, 000x and 52, 000x with Titan transmission electron microscope (FEI) operating with an accelerating voltage of 60 kV.
Acknowledgements
This work was supported by National Institutes of Health, GM47980. We thank Dr. Paul J. Jardine for Phi29 clones and Dr. José L. Carrascosa for antibody against connector protein.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bazinet C, King J. The DNA translocating vertex of dsDNA bacteriophage. Annu Rev Microbiol. 1985;39:109–129. doi: 10.1146/annurev.mi.39.100185.000545. [DOI] [PubMed] [Google Scholar]
- Bazinet C, King J. Initiation of P22 procapsid assembly in vivo. J Mol Biol. 1988;202(1):77–86. doi: 10.1016/0022-2836(88)90520-7. [DOI] [PubMed] [Google Scholar]
- Bazinet C, Villafane R, King J. Novel second-site suppression of a cold-sensitive defect in phage P22 procapsid assembly. J Mol Biol. 1990;216(3):701–716. doi: 10.1016/0022-2836(90)90393-Z. [DOI] [PubMed] [Google Scholar]
- Black LW, Silverman DJ. Model for DNA packaging into bacteriophage T4 heads. J Virol. 1978;28(2):643–655. doi: 10.1128/jvi.28.2.643-655.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho A, Jimenez F, De La Torre J, Carrascosa JL, Mellado RP, Vasquez C, Vinuela E, Salas M. Assembly of Bacillus subtilis phage phi29. 1. Mutants in the cistrons coding for the structural proteins. Eur J Biochem. 1977;73(1):39–55. doi: 10.1111/j.1432-1033.1977.tb11290.x. [DOI] [PubMed] [Google Scholar]
- Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J Supramol Struct. 1974;2(2–4):202–224. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- Caspar DL. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys J. 1980;32(1):103–138. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Cerritelli ME, Studier FW. Assembly of T7 capsids from independently expressed and purified head protein and scaffolding protein. J Mol Biol. 1996;258(2):286–298. doi: 10.1006/jmbi.1996.0250. [DOI] [PubMed] [Google Scholar]
- Cuervo A, Vaney MC, Antson AA, Tavares P, Oliveira L. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J Biol Chem. 2007;282(26):18907–18913. doi: 10.1074/jbc.M701808200. [DOI] [PubMed] [Google Scholar]
- Droge A, Santos MA, Stiege AC, Alonso JC, Lurz R, Trautner TA, Tavares P. Shape and DNA packaging activity of bacteriophage SPP1 procapsid: protein components and interactions during assembly. J Mol Biol. 2000;296(1):117–132. doi: 10.1006/jmbi.1999.3450. [DOI] [PubMed] [Google Scholar]
- Droge A, Tavares P. In vitro packaging of DNA of the Bacillus subtilis bacteriophage SPP1. J Mol Biol. 2000;296(1):103–115. doi: 10.1006/jmbi.1999.3449. [DOI] [PubMed] [Google Scholar]
- Earnshaw W, King J. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J Mol Biol. 1978;126(4):721–747. doi: 10.1016/0022-2836(78)90017-7. [DOI] [PubMed] [Google Scholar]
- Fane BA, Prevelige PE., Jr Mechanism of scaffolding-assisted viral assembly. Adv Protein Chem. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- Fu CY, Morais MC, Battisti AJ, Rossmann MG, Prevelige PE., Jr Molecular dissection of o29 scaffolding protein function in an in vitro assembly system. J Mol Biol. 2007;366(4):1161–1173. doi: 10.1016/j.jmb.2006.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene B, King J. Scaffolding mutants identifying domains required for P22 procapsid assembly and maturation. Virology. 1996;225(1):82–96. doi: 10.1006/viro.1996.0577. [DOI] [PubMed] [Google Scholar]
- Grimes S, Jardine PJ, Anderson D. Bacteriophage phi 29 DNA packaging. Adv Virus Res. 2002;58:255–294. doi: 10.1016/s0065-3527(02)58007-6. [DOI] [PubMed] [Google Scholar]
- Guo PX, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi 29 DNA. Science. 1987;236(4802):690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- Guo PX, Erickson S, Xu W, Olson N, Baker TS, Anderson D. Regulation of the phage phi 29 prohead shape and size by the portal vertex. Virology. 1991;183(1):366–373. doi: 10.1016/0042-6822(91)90149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen EW, Reilly BE, Tosi ME, Anderson DL. Analysis of gene function of bacteriophage phi 29 of Bacillus subtilis: identification of cistrons essential for viral assembly. J Virol. 1976;19(2):501–517. doi: 10.1128/jvi.19.2.501-517.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, Walter JM, Falk W, Anderson DL, Bustamante C. Experimental Test of Connector Rotation during DNA Packaging into Bacteriophage varphi29 Capsids. PLoS Biol. 2007;5(3):e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006a;312(5781):1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The Structure of an Infectious p22 Virion Shows the Signal for Headful DNA Packaging. Science. 2006b doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. Embo J. 2007;26(7):1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Guo P. Sequential interactions of structural proteins in phage phi 29 procapsid assembly. J Virol. 1995;69(8):5024–5032. doi: 10.1128/jvi.69.8.5024-5032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhuillier S, Gallopin M, Gilquin B, Brasiles S, Lancelot N, Letellier G, Gilles M, Dethan G, Orlova EV, Couprie J, Tavares P, Zinn-Justin S. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc Natl Acad Sci U S A. 2009;106(21):8507–8512. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SD, Prevelige PE., Jr Bacteriophage p22 portal vertex formation in vivo. J Mol Biol. 2002;315(5):975–994. doi: 10.1006/jmbi.2001.5275. [DOI] [PubMed] [Google Scholar]
- Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005;18(2):149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Poh SL, El Khadali F, Berrier C, Lurz R, Melki R, Tavares P. Oligomerization of the SPP1 Scaffolding Protein. J Mol Biol. 2008;378(3):551–564. doi: 10.1016/j.jmb.2008.02.028. [DOI] [PubMed] [Google Scholar]
- Poliakov A, van Duijn E, Lander G, Fu CY, Johnson JE, Prevelige PE, Jr, Heck AJ. Macromolecular mass spectrometry and electron microscopy as complementary tools for investigation of the heterogeneity of bacteriophage portal assemblies. J Struct Biol. 2007;157(2):371–383. doi: 10.1016/j.jsb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408(6813):745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer GP, Newcomb WW, Thomsen DR, Homa FL, Brown J. Identification of a region in the herpes simplex virus scaffolding protein required for interaction with the portal. J Virol. 2005;79(1):132–139. doi: 10.1128/JVI.79.1.132-139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413(6857):748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TL, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The Structure of the Phage T4 DNA Packaging Motor Suggests a Mechanism Dependent on Electrostatic Forces. Cell. 2008;135(7):1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol Cell. 2007;25(6):943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Tao Y, Olson NH, Xu W, Anderson DL, Rossmann MG, Baker TS. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95(3):431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub F, Maeder M. Formation of the prohead core of bacteriophage T4 in vivo. J Virol. 1984;49(3):892–901. doi: 10.1128/jvi.49.3.892-901.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuprun V, Anderson D, Egelman EH. The bacteriophage phi 29 head-tail connector shows 13-fold symmetry in both hexagonally packed arrays and as single particles. Biophys J. 1994;66(6):2139–2150. doi: 10.1016/S0006-3495(94)81009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG. Structural changes of bacteriophage phi29 upon DNA packaging and release. Embo J. 2006;25(21):5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lemieux S, Wu X, St-Arnaud D, McMurray CT, Major F, Anderson D. Function of hexameric RNA in packaging of bacteriophage phi 29 DNA in vitro. Mol Cell. 1998;2(1):141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]