Abstract
The interactions of beta2 glycoprotein I (B2GPI) with the receptors of the low-density lipoprotein receptor (LDLR) family are implicated in the clearance of negatively charged phospholipids and apoptotic cells and, in the presence of autoimmune anti-B2GPI antibodies, in cell activation, which might play a role in the pathology of antiphospholipid syndrome (APS). The ligand-binding domains of the lipoprotein receptors consist of multiple homologous LA modules connected by flexible linkers. In this study, we investigated at the atomic level the features of the LA modules required for binding to B2GPI. To compare the binding interface in B2GPI/LA complex to that observed in the high-resolution co-crystal structure of the receptor associated protein (RAP) with the LA modules 3 and 4 from the LDLR, we used the LA module 4 from the LDLR in our studies. Using solution NMR spectroscopy, we found that LA4 interacts with B2GPI and the binding site for B2GPI on the 15N-labeled LA4 is formed by the calcium coordinating residues of the LA module. We built a model for the complex between domain V of B2GPI (B2GPI-DV) and LA4 without introducing any experimentally derived constraints into the docking procedure. Our model, which is in the agreement with the NMR data, suggests that the binding interface of B2GPI for the lipoprotein receptors is centered at three lysine residues of B2GPI-DV, Lys 308, Lys 282 and Lys317.
Keywords: LDLR, lipoprotein receptors, B2GPI, beta2-glycoprotein I, PIPER, molecular docking, antiphospholipid syndrome, APS
INTRODUCTION
Beta2 glycoprotein I (B2GPI) is a soluble protein that circulates in plasma at a concentration of about 170 ug/ml 1. Interactions of B2GPI with the receptors of the low-density lipoprotein receptor (LDLR) family have been implicated in clearance of negatively charged lipids and apoptotic cells and, in the presence of autoimmune anti-B2GPI antibodies, in the pathology of antiphospholipid syndrome (APS) 2-8. B2GPI is the major antigen for the autoimmune antibodies in APS, an acquired autoimmune disorder associated with arterial and venous thromboembolic disease 9-12. The pathophysiological mechanisms underlying APS are not completely understood 11,12.
The lipoprotein receptors, which constitute a group of structurally homologous receptors, are expressed by a wide variety of cells and function both in endocytosis and signal transduction 13,14. The closely related members of the LDLR family include LDLR, ApoE receptor 2 (ApoER2), the very low-density lipoprotein receptor (VLDLR), the low-density lipoprotein receptor-related protein (LRP) and megalin. All lipoprotein receptors can bind B2GPI 15 and the complex formation results either in endocytosis or in signal transduction 2-8. Megalin, which is predominantly expressed in the kidneys and lungs, was shown to be an efficient clearance receptor for B2GPI 2. The lipoprotein receptors on macrophages are involved in B2GPI-dependent internalization of negatively charged lipids and apoptotic cells 4. B2GPI acquires its APS-related pathological properties only in the presence of anti-B2GPI autoimmune antibodies. In the presence of antibodies, B2GPI binds to platelets, endothelial cells and monocytes, inducing their pro-coagulant and pro-inflammatory state 16. Using chimeric dimers of B2GPI which mimic B2GPI/antibody complexes, it has been demonstrated that B2GPI complexes activate platelets through interactions with ApoER2, the member of the LDLR family on the surface of platelets 3,5-8.
The lipoprotein receptors interact with their ligands through the structurally homologous LA modules, which are arranged in sequence to form a ligand-binding domain 14,17. Although the primary sequences of different LA modules vary, all LA modules share common features. Each LA module requires a calcium ion for proper folding and to maintain the structural integrity of the module 18,19. In the presence of calcium, the LA modules, which are about 40 residues each, have similar fold determined by three disulfide bonds, six calcium coordinating residues and a conserved hydrophobic core 19-32.
Studies that employed deletion mutants of B2GPI localized the binding site for the lipoprotein receptors to domain V of B2GPI (B2GPI-DV) 8,15. Four N-terminal domains of B2GPI have similar folds with a characteristic antiparallel beta strand and two intradomain disulfide bonds 33,34. The fifth domain at the C-terminus of B2GPI belongs to a similar class, but has an inserted long loop and an additional disulfide bond. B2GPI-DV has an unusually large number of lysine residues, which constitute about 15 % of its primary sequence.
Like B2GPI, the receptor associated protein (RAP) interacts with all receptors of the LDLR family. Moreover, RAP interfered with the binding of B2GPI to the lipoprotein receptors 2,4,15. A high-resolution crystal structure of a complex between RAP and a pair of LA modules, LA3-4, from the LDLR revealed that electrostatic interactions play a major role in the binding of RAP by the LA3-4 modules 24. Since electrostatic interaction likely governs the binding of B2GPI by the lipoprotein receptors, we sought to determine whether the same residues that form the ligand-binding pocket on the LA modules in the crystal structure of the RAP/LA3-4 complex also interact with B2GPI. The comparison of the LA complexes with RAP and B2GPI will help to understand if the recognition mechanism observed in the RAP/LA3-4 structure is applicable to the B2GPI complexes with the lipoprotein receptors.
To directly compare the contact site on the LA module for B2GPI to that observed in the RAP/LA3-4 crystal structure, we used LA4 from the LDLR in our studies. We mapped the binding interface for B2GPI onto LA4 using solution NMR spectroscopy. In this study, we have demonstrated that the binding site for B2GPI on LA4 is formed by the calcium coordinating residues of LA4. The same residues form the binding pocket for RAP in the RAP/LA3-4 complex. Because it has been established that the domain V of B2GPI contains the binding site for the lipoprotein receptors 8,15, we built a model for the complex between B2GPI-DV and LA4 starting from the structures of B2GPI-DV and LA4 available in the PDB database. The docking model of the complex was calculated by the program PIPER without introducing any experimentally derived constraints into the docking protocol. Our model of the B2GPI-DV/LA4 complex is in agreement with the measured NMR data and suggests that the binding interface of B2GPI-DV for the lipoprotein receptors is centered at three lysine residues, Lys308, Lys282 and Lys317.
MATERIALS AND METHODS
Proteins
B2GPI was purchased from Hematologic Technologies, Inc. The purity of B2GPI was assessed by SDS-PAGE. The LA4 gene fragment of the LDLR encoding residues 126-165 was expressed in E.coli and purified from the inclusion bodies using a previously published protocol 18. For NMR spectroscopy, the LA4 samples uniformly labeled with 13C and/or 15N were produced by growth in M9 minimal media supplemented with 13C6-D-glucose and/or 15N-ammonium chloride, respectively.
NMR spectroscopy
All NMR experiments were conducted on a Varian Inova 500 MHz spectrometer equipped with four RF channels and a triple-resonance probe with pulsed-field gradients. Spectra were acquired at 298 K using BioPack (Varian Inc.) pulse sequences and processed with GIFA v4.3 software 35. Backbone 1H, 13C and 15N resonance assignments were carried out by analyzing HNCA, NHCOCA, HNCACB and HNCOCACB spectra. For spectral assignment, the NMR sample contained 0.9 mM 13C, 15N-labeled LA4 in 90% H2O/10%D2O, 10 mM bis-Tris buffer, pH 7.1, and 10 mM CaCl2. Chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt (DSS) used as an internal reference 36.
To prepare a 100 μM stock solution, B2GPI (Hematologic Technologies, Inc.) was dialyzed in 50 mM bis-Tris buffer, pH 7.0, 50 mM NaCl and 2 mM CaCl2, and concentrated. The concentrated B2GPI showed a single band about 54 kDa on SDS-PAGE under reducing conditions. The concentration was estimated using the extinction coefficient E280 =10.0 for a 1% solution.
In the NMR titration experiments, 65 μM of 15N-labeled LA4 in 90% H2O/10%D2O, 50 mM bis-Tris buffer pH 7.0, 50 mM NaCl and 2 mM CaCl2 were titrated with B2GPI. 15N-R2 relaxation was measured for LA4 resonances in the absence and presence of B2GPI. The NMR samples contained either 65 μM 15N-labeled LA4 or 100 μM 15N-labeled LA4 and 15 μM of unlabeled B2GPI. The samples were prepared in 90% H2O/10%D2O, 50 mM bis-Tris buffer pH 7.0, 50 mM NaCl and 2 mM CaCl2. Relaxation delays were 10, 30, 50, 90, 150, 210, 330 and 390 ms for the sample without B2GPI and 10, 30, 70, 90, 110 and 130 ms in the presence of B2GPI. To determine R2 relaxation rates, the experimentally measured resonance intensities were fitted to monoexponential decay using the xcrvfit program (developed by Boyko, R. and Sykes, B.D. at the University of Alberta), which can be found at: http://www.bionmr.ualberta.ca/bds/software/xcrvfit.
Molecular docking of the complex between domain V of B2GPI and the LA module
We generated the molecular models of the complex using the docking program PIPER which performes global sampling of the discrete 6D space of relative orientations of two rigid protein molecules 37. PIPER is freely available to academic researchers as stand-alone code or as a web server (http://cluspro.bu.edu). The docking algorithm was extensively validated at the Critical Assessment of Prediction of Interactions (CAPRI) competitions 38,39. The starting model of the fifth domain of B2GPI (residues from 245 to 326) was extracted from the crystal structure of the full length protein (PDB accession code 1c1z). The coordinates of LA4 (residues from 126 to 163) were taken from the X-ray structure of the RAP/LA3-4 complex (PDB accession code 2fcw). A calcium ion was explicitly included in the docking as part of the LA4 structure. The larger protein (B2GPI-DV) was centered at the coordinate origin, and rotations and translations of the smaller (LA4) were deterministically discretized. For the scoring function, we used a standard weighting scheme implemented in “Van der Waals plus electrostatics” option on the PIPER web server (http://cluspro.bu.edu). Initially, 70000 docking models were calculated and 1000 with the lowest score were retained. The calculated structures were clustered using a 9 Å cut-off pairwise RMSD for the backbone atoms in the interface of the complex. The central structure of the most populated cluster was minimized using the CHARMM program 40. Structures were vizualized using PyMOL 41. The intermolecular contact area was calculated with the AREAIMOL program which is part of the CCP4 program suite 42.
Molecular dynamics (MD) simulation was performed using CHARMM 40 in the presence of explicit water. The complex was surrounded by the 13134 TIP3 water molecules and the simulation was performed using cubic box with the periodic boundary conditions. The length of the MD simulation was 1 ns with a 2 fs integration step and each 100th step saved as a trajectory frame. The system was heated to 298 K during the first 20 ps of dynamics and equilibrated during the following 480 ps. The equilibration step was followed by 500 ps production run performed at a constant temperature of 298K and pressure of 1 atm. The last 500 ps (250000 steps or 2500 frames) of dynamics were analyzed and clustered.
RESULTS
Resonance assignments of the 1H-15N correlation spectrum of LA4
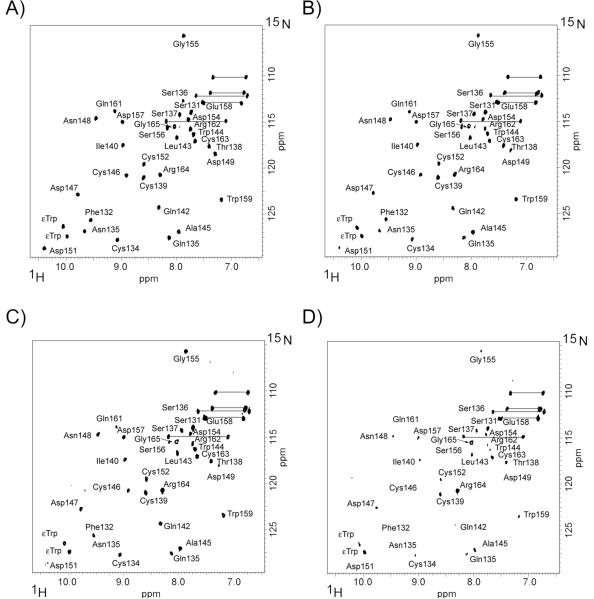
To map the binding site for B2GPI onto LA4, we first assigned the 1H-15N correlation spectrum of LA4. The backbone amides were sequentially assigned using the pairs of triple resonance experiments: (1H, 13C, 15N) 3D HNCA, HNCOCA and HNCACB, HNCOCACB. Thirty one out of forty backbone resonances are clearly visible on the spectra. The missing resonances include four proline residues Pro 129, 141, 150 and 160, Glu 153 and the stretch of residues at the N-terminus of the LA4, Thr 126, Cys 127, Gly 128 and Ala 130. Our previous studies of the relaxation properties of the modules LA5 and LA6 from the LDLR showed that the residues preceding the first cysteine of the LA modules are flexible and may be missing from the spectra 43. It is likely that the proline residue, Pro 129, at the N-terminus of LA4 undergoes conformational exchange between the trans and cis isomers. The exchange between multiple conformations results in broadening and disappearance from the spectra of the resonances for the neighboring residues. The assigned 1H-15N correlation spectrum of LA4 is presented on Figure 1. Two resonances corresponding to tryptophan sidechains are not assigned to the primary sequence of LA4 and labeled εTrp on the spectra.
FIGURE 1.
NMR titration of 15N-labeled LA4 with full-length B2GPI. The molar ratios of 15N-LA4 to B2GPI are 1:0 (A), 1:0.14 (B), 1:0.2 (C) and 1:0.25 (D). Residue numbers denote the backbone resonance assignments of LA4. The sidechain NH2 groups are shown by horizontal lines and the NH groups of tryptophan sidechains are labeled εTrp.
Titration of 15N-LA4 with B2GPI
Small aliquots of the stock solution of B2GPI were added to 15N-LA4 in increments such that the maximum dilution of 15N-LA4 corresponding to the last titration point was about 11 %. The spectra were acquired with the same number of scans, processed in the same fashion and scaled for plotting taking into account the dilution factor (Figure 1 A,B,C,D). The comparison of the 1H-15N correlation spectra of LA4 in the presence of increasing amounts of B2GPI shows that the majority of peaks corresponding to backbone resonances of LA4 gradually disappear from the spectra.
Interactions between a large protein and a smaller peptide are usually assessed by the decrease in the intensity of the NMR peaks of the observed smaller peptide 44. The molecular mass of unbound 15N-LA4 (~4400 Da) is much smaller than the molecular mass of the 15N-LA4/B2GPI complex (~60000 Da). The overall decrease of intensity of 15N-LA4 resonances clearly demonstrates that LA4 binds B2GPI. The rotational correlation time of 15N-LA4 in the complex increases significantly compared to that of the unbound LA4 causing the backbone resonances of a fraction of 15N-LA4 bound to B2GPI to broaden and disappear from the spectra. The titration studies were carried out until most of the resonances are no longer visible on the spectrum. Thus, the molar ratio of 15N-LA4 to B2GPI in Figure 1D does not reflect the stoichiometry of the complex.
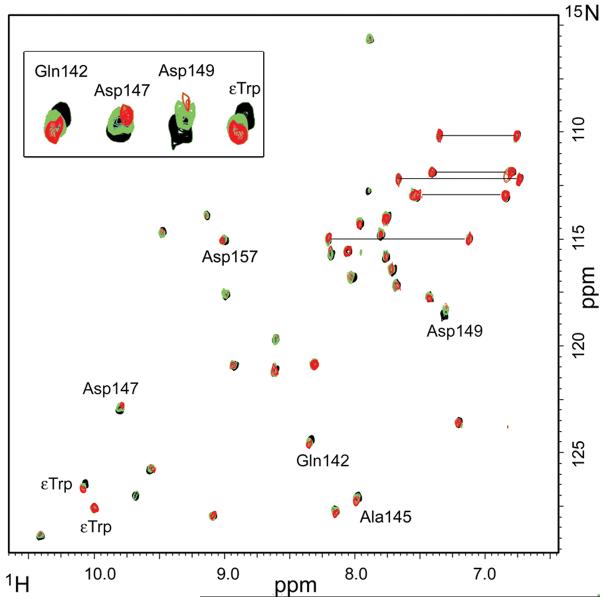
In addition to the overall decrease in intensity of the backbone resonances of 15N-LA4 upon titration with B2GPI, some resonances move gradually on the spectrum. The resonances corresponding to backbone amides of Gln142, Ala145, Asp147, Asp149, Asp157 and a sidechain of one of tryptophans of 15N-LA4 shifted more than a half peak-width in the presence of 0.2 molar equivalent of B2GPI (Figure 2). Previous studies have shown that the binding affinity of a chimeric dimer of B2GPI to the LDLR is about 341 ± 54 nM 15, suggesting that a monomeric B2GPI binds a single LA module from the ligand-binding domain of the LDLR with even lower affinity. A low affinity binding manifests itself as fast exchange between free and bound states on the NMR timescale. In the case of fast exchange, the titration spectrum has only one set of peaks for each observed nucleus. If the nucleus is affected by complex formation, its chemical shift moves on the spectrum proportionally to the fraction of bound molecules in solution. The observed shift of the backbone resonances on the titration spectra of LA4 suggests that the chemical environment of the backbone amides of Gln142, Ala145, Asp147, Asp149, Asp157, and one of tryptophan sidechains change when the complex is formed (Figure 2). The second cross-peak corresponding to the sidechain of a tryptophan residue does not move on the spectrum, suggesting that this residue is far from the contact interface. For comparison, in the crystal structure of the RAP/LA3-4 complex the sidechains of Asp147, Asp149, Asp151 and Trp144 form the binding site for RAP on LA4 24.
FIGURE 2.
Superimposed 15N-HSQC spectra of LA4. The molar ratios of 15N-LA4 to B2GPI are 1:0 (black); 1:0.1 (green); 1:0.2 (red). The residues that shifted more than a half peak width in the presence of 0.2 molar equivalent of B2GPI are labeled. The insert shows overlays of the shifted peaks enlarged 3 fold. The horizontal lines connect peaks corresponding to the sidechain NH2 groups.
The two peaks corresponding to residues at the C-terminus of LA4 immediately after the last cysteine, Arg164 and Gly165, change neither their chemical shifts nor intensity upon titration with B2GPI, strongly suggesting that the C-terminus of LA4 is distant from B2GPI and remains flexible in the B2GPI/LA4 complex.
15N-R2 relaxation data analysis
Comparison of the titration spectra shows that the intensity of the backbone resonances does not decrease uniformly upon titration. Instead, some peaks disappear faster from the spectra and are already beyond detection for the sample containing 1:0.25 molar ratio of 15N-LA4 to B2GPI, indicating that the R2 relaxation rates for these nuclei are higher than average (Figure 2 A,B,C,D). The values of transverse relaxation rate R2 are higher for residues that undergo chemical or conformational exchange. The residues that exhibit increased R2 relaxation rate only in the presence of B2GPI are likely in the vicinity of the binding interface in the B2GPI/LA4 complex.
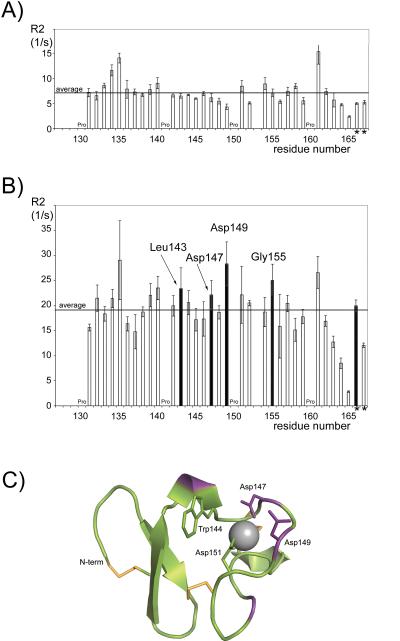
We compared the R2 relaxation rates for the backbone amides of 15N-LA4 in the absence and presence of B2GPI. The average value of R2 for the backbone amides of 15N-LA4 is 19.1 ± 1.7 (1/s) and 7.3 ± 2.6 (1/s) in the presence and absence of B2GPI, respectively (Figure 3 A, B). The average value of R2 is larger in the presence of B2GPI indicating that B2GPI interacts with LA4. The average value of R2 for the backbone amides of free 15N-LA4 is in agreement with previously published data for the LA5 and LA6 modules from the LDLR, which are similar in size and three dimensional structure to LA4 43.
FIGURE 3.
15N-R2 relaxation data. R2 relaxation rates for backbone amides of 15N-LA4 in the absence (A) and presence (B) of B2GPI, plotted as a function of the residue number. The last two bars on both panels correspond to the R2 values measured for the tryptophan sidechains (marked by asterisks). The residues affected the most by the presence of B2GPI are labeled and the corresponding bars are colored black. Residues identified by relaxation data analysis (colored purple) are mapped onto the structure of LA4 (C). The sidechains that form the binding pocket for RAP are labeled and shown in stick representation. The disulfide bonds are shown as orange sticks and the calcium ion is a gray sphere.
In the absence of B2GPI, the three residues of 15N-LA4, Cys134, Asp135 and Gln161, have R2 values that are more than 1.5* STD (standard deviation) above average (Figure 3 A). In the presence of B2GPI, the backbone amides of several other residues in addition to Cys134, Asp135 and Gln161 have values of the R2 relaxation rates more than 1.5*STD above average. These are Cys139, Ile140, Leu143, Asp147, Asp149, Asp151 and Gly155. Of these residues, Leu143, Asp147, Asp149 and Gly155 are affected the most by the presence of B2GPI since the measured values of R2 for these backbone amides are below average in the absence of B2GPI (Figure 3 A,B). Leu143, Asp147 and Asp149 are clustered on one side of the LA4 structure in the vicinity of the calcium-binding site (Figure 3 C). Gly155 is in the proximity to calcium-coordinating Asp157, which could explain an increased susceptibility of its R2 value to the presence of B2GPI. In the RAP/LA3-4 structure, Asp147 and Asp149 are the residues that form the lysine-binding pocket in the RAP/LA3-4 complex and Leu143 precedes Trp144 whose sidechain packs against the aliphatic portion of lysine from RAP in the RAP/LA3-4 structure (Figure 3 C).
We also measured the R2 relaxation rates for two tryptophan sidechains of LA4 (Figure 3 A, B). In the absence of B2GPI, the values of R2 for these sidechains are well below average, reflecting the general difference in the internal dynamics between the backbone and sidechains. In the presence of B2GPI, the measured R2 remains below average for one of these sidechains. The R2 value measured for the other sidechain approaches the average R2 of the backbone, suggesting that the internal dynamics of this sidechain is restricted in the presence of B2GPI (Figure 3 B, marked by asterisks). Interestingly, the cross-peak corresponding to this sidechain also moves on the spectra in the NMR titration experiments (Figure 2), strongly supporting our conclusion that this tryptophan sidechain is in the binding interface in the B2GPI/LA4 complex and the other tryptophan sidechain is far from the interface.
Model of the B2GPI/LA4 complex
B2GPI interacts with the lipoprotein receptors through its fifth domain 8,15. We modeled the complex between domain V from B2GPI and the LA4 module using the docking program PIPER and the standard “shape plus electrostatics” weighting scheme 37. Structures with the lowest scores were clustered using a 9 Å cut-off pairwise RMSD for the backbone atoms in the interface of the complex. The most populated cluster contained 297 structures out of the 1000 low energy conformations used for clustering. The second and third clusters contained only 158 and 123 structures, respectively, strongly suggesting that the first cluster represents the correct solution. In all three clusters the binding sites on LA4 are formed by the same residues. The first and the second clusters of structures have identical binding sites on both B2GPI-DV and LA4 but have the LA4 modules whose relative orientation differs by about 180°. Overall, of the structures comprising the three most populated clusters, 100 % have the same binding site on LA4 and 79 % have the same binding site on B2GPI-DV.
The central structure of the most populated cluster minimized with CHARMM is shown in Figure 4 A. The possible flexibility of the docking model was assessed by the molecular dynamics simulation in the presence of explicit water. The majority of the structures in the trajectory of the MD simulation are below 2.5 Å RMSD from the refined minimized structure calculated over the C-alpha atoms, indicating that the minimized model of the complex is stable (data not shown).
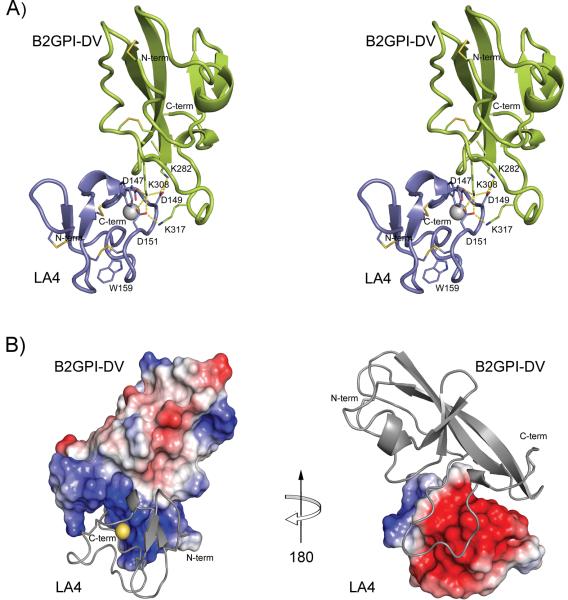
FIGURE 4.
Interface between B2GPI-DV and LA4 in the B2GPI-DV/LA4 complex. A) Overview of the structure shown in stereo. The backbone trace of B2GPI-DV is green, LA4 is blue. The disulfide bonds are shown as sticks and the calcium ion is a gray sphere. Hydrogen bonds between residues in the contact interface of the complex are shown by dashed yellow lines. B) Electrostatic complementarity. Surface representations of B2GPI-DV and LA4 are colored by electrostatic surface potential using a sliding scale from red (-85 kT) to blue (85 kT). The backbone ribbon trace is shown in gray. Bound calcium ion on LA4 is illustrated by yellow sphere. Vacuum electrostatics was calculated by PyMOL without accounting for the calcium ion.
Structural details of the B2GPI-DV/LA4 complex
The binding interface in the B2GPI-DV/LA4 complex is dominated by electrostatic interactions (Figure 4 B). In the complex, LA4 is docked into the cleft formed on B2GPI-DV by the C-terminal flexible loop and by the beta-hairpin. This region of B2GPI-DV contains surface-exposed lysines, Lys282, Lys284, Lys308 and Lys317, creating a positive surface potential on B2GPI-DV. The complementary charged surface on LA4 is composed predominantly by the sidechains of the calcium coordinating residues. The intermolecular contact area in the complex is 392 Å2, which is comparable to the contact areas between the helixes of RAP and each of the LA modules in the RAP/LA3-4 complex. The carboxylate oxygen atoms of Asp147, Asp149 and Asp151 encircle the ε-amino group Lys308 forming a tripartite salt bridge (Figure 4 A). Additional salt bridges stabilize the interactions between Lys308 and the acidic pocket on LA4. In particular, the ε-amino group of Lys282 forms hydrogen bonds with both carboxylate oxygens of the Asp149 sidechain, and Lys317 interacts with the sidechain of Asp151.
DISCUSSION
B2GPI interacts with the lipoprotein receptors that are expressed by many cells and tissues 15. These interactions support clearance of B2GPI and B2GPI-associated negatively charged particles and apoptotic cells and, in the presence of the autoimmune anti-B2GPI antibodies, promote cell activation 2-8. The binding of B2GPI to the lipoprotein receptors is inhibited by the receptor-associated protein RAP2,15.
Using solution NMR spectroscopy, we have investigated whether the same residues that form the binding pocket for RAP in the RAP/LA3-4 complex are in the contact interface with B2GPI. By monitoring changes in the chemical shifts and the relaxation properties of 15N-labeled LA4 in response to complex formation, we have shown that the same residues of LA4 interact with B2GPI and RAP. Specifically, the NMR titration studies of 15N-labeled LA4 with a full-length B2GPI revealed that the chemical shifts of the backbone amides of Gln142, Ala145, Asp147, Asp149 and Asp157, and the sidechain of one of two tryptophan residues of LA4 gradually move on the NMR spectra upon addition of B2GPI, suggesting that these residues are close to the binding interface in the complex. The chemical shifts observed in the current studies reflect only the beginning of titration. We could not follow the titration to saturation because of the line broadening. In our experiments, only a few resonances of 15NLA4 are shifted when titrated with B2GPI. Similarly, when the LA pair of modules from LRP was titrated with RAP domain 1 only a few residues of the 15N-labeled LA pair moved on the spectra 26. The measured values of the R2 relaxation rates of the backbone amides of Leu143, Asp147, Asp149 and the sidechain of the same tryptophan residue are affected the most by the presence of B2GPI. In the RAP/LA3-4 complex the sidechains of Asp147, Asp149, Asp151 and Trp144 from LA4 form the binding pocket for RAP 24. Our NMR data strongly suggests that LA4 binds B2GPI and RAP in a similar fashion.
Since the binding site for the lipoprotein receptors was localized to domain V of B2GPI 8,15, we used domain V to build a model of the complex between B2GPI-DV and LA4. The molecular docking program PIPER was used to calculate the structure of the complex without introducing any a priori information about the binding interface into the docking process. We selected a central structure of the most populated cluster and refined it with CHARMM. In the model, Lys308, Lys282 and Lys317 from B2GPI-DV form hydrogen bonds with the calcium coordinating residues of LA4. The docking model independently confirmed the binding interface of LA4 around the calcium coordinating residues, which is in agreement with our NMR measurements. The analysis of the NMR data predicted that Leu143, Ala145, Asp147, Asp149 and one of tryptophans of LA4 are in the close proximity to the binding interface (Figure 4 A). The model of the complex supports these observations. As expected, there is only one tryptophan of LA4, Trp144, in the binding interface. The second tryptophan of LA4, Trp159, is far from the contact site of the complex. The C-terminus of LA4 in the complex is distant from the interface in agreement with the NMR data that indicates that the C-terminus remains flexible in the complex and does not interact with B2GPI-DV.
In our docking model of the LA4/B2GPI-DV complex, LA4 interacts with B2GPI-DV by the sidechains. Therefore, the chemical shifts of the backbone resonances detected by the spectra are not expected to be large. A small contact interface and binding predominantly by the sidechains is seen in the available crystal structures of the LA complexes 24,29,31. Interactions of the LA modules with different ligands studied by NMR manifested in relatively small shifts of the observed backbone resonances of the LA modules. When the 15N-labeled LA module from LRP was titrated to saturation with a receptor-binding domain from alpha2-macroglobulin, none of the perturbations on the 15N-LA spectra were large 23.
The mode of interaction between B2GPI-DV and LA4 in the docking model of the B2GPI-DV/LA4 complex strongly resembles that observed in the crystal structure of RAP/LA3-4 24. In particular, Lys308 is inserted into the pocket created by the negatively charged sidechains of three calcium coordinating aspartates. These aspartates encircle Lys308 to form salt bridges with the ε-amino group of the lysine sidechain. The sidechain of Trp144 packs against the aliphatic portion of Lys308. The arrangement of sidechains around Lys308 and the resulting hydrogen bond geometry is similar to that observed for Lys256 of RAP entrapped by the calcium coordinating aspartates of LA4 in the RAP/LA3-4 complex. Two other lysines in the vicinity of Lys308, Lys282 and Lys317, augment the positive electrostatic potential created by Lys308. As in the RAP/LA3-4 structure, these lysines additionally stabilize the B2GPI-DV/LA4 complex, forming the salt bridges with carboxylate oxygens of Asp149 and Asp151.
According to our docking model, the binding interface of the B2GPI-DV/LA4 complex is close but not identical to the phospholipid-binding regions on B2GPI-DV 45,46. Therefore, mutations in the hydrophobic loop of B2GPI-DV will not affect the binding of B2GPI to the lipoprotein receptors. Indeed, a binding study utilizing a B2GPI mutant with Ser for Trp316 substitution in the hydrophobic phospholipids insertion loop, which is important for the binding of B2GPI to phospholipids, demonstrated that this mutation did not disrupt the binding between B2GPI and ApoER2 8. In our model, Trp316 of B2GPI-DV and the closest to it Pro150 of LA4 are 3.7 Å apart. Based on the model, the Trp316 to Ser mutation will increase the distance between this Ser and Pro150 of LA4 to about 7 Å. This mutation will not create any clashes or interfere with the existing contacts in the B2GPI-DV/LA4 complex.
It was previously shown that the cyclic peptide CKNKEKKC corresponding to a positively charged loop between Cys 281 and Cys 288 of B2GPI-DV interfered with the binding of dimeric B2GPI-DV and ApoER2 8. In the structure of the B2GPI-DV/LA4 complex, Lys282 and Lys317 of B2GPI-DV complement the interactions between Lys308 and the binding pocket on LA4. In the cyclic peptide, the relative orientation of the lysine residues is likely constrained in such a way that one of the lysines plugs into the calcium binding site of the LA modules similarly to Lys308 in the complex, precluding the interactions between ApoER2 and B2GPI. Other lysines contribute an additional positive potential. The scrambled cyclic peptide EKCKNKCK, which forms a tighter turn between the two cysteins, was less efficient in inhibiting these interactions, suggesting that the relative orientation of lysine sidechains is important for the binding of the LA modules.
Although the cyclic peptide can bind the LA module, our docking results indicate that it is very unlikely that the lysines from the loop between Cys281 and Cys288 of B2GPI-DV form the main contact site in the B2GPI-DV/LA complex. Out of the 1000 low energy structures clustered with 9 Å RMSD of the backbone atoms in the interface of the complex, only the third cluster with 123 structures in it (compared to 297 structures in the most populated cluster) represented a model with Lys287 of B2GPI-DV pointing into the binding pocket on the LA module.
In the complex, the contact interface is 392 Å2, which is comparable to the contact area of the individual LA modules observed in the crystal structures of the lipoprotein receptors and their ligands 24,29,31. The binding affinity in the protein complexes with a similar contact area is usually weak 23. We estimate that the affinity of a single LA4 for B2GPI is in the range from low mM to uM, since two LA modules bind ligands with low uM to nM affinity 47-49. It is typical for the lipoprotein receptors to use at least two LA modules to interact with their ligands with high affinity, which is facilitated by the modular organization of their ligand-binding domains 24,29,47-52. The B2GPI/antibody complexes and chimeric B2GPI dimers convert B2GPI into a multivalent ligand capable of binding several LA modules.
Recent structural studies revealed the mechanism used by the lipoprotein receptors in recognition of positively charged ligands 24,29,31. In each of these structures, the binding site on the structurally unrelated ligands is centered around a lysine residue that sticks into the binding pocket on the LA module created by conserved calcium coordinating acidic residues. Our data suggests that the general mode of recognition of basic ligands by the LA modules is directly applicable to B2GPI complexes with the lipoprotein receptors. Therefore, the LA modules capable of binding B2GPI could be predicted based on the presence of the required acidic and aromatic residues in their primary sequence. According to our results, the LDLR has the least number of the LA modules that can bind B2GPI and these modules, LA3 and LA4, reside side-by-side in the ligand-binding domain of the receptor. Since the avidity plays a major role in the binding of ligands by the lipoprotein receptors, the small number of the LA modules available for the interactions may explain why the LDLR has the weakest binding to B2GPI/antibody complexes 15. Distance between the two interacting LA modules may also be important for the efficient binding of B2GPI/antibody complexes.
In conclusion, we have shown that LA4 interacts with B2GPI in a similar fashion as observed in the high-resolution X-ray structure of the RAP/LA3-4 complex. The structure of the B2GPI-DV/LA4 complex reported in this paper is supported by three independent lines of evidence provided by NMR relaxation measurements, NMR titration studies and by molecular docking. Taken together, despite the experimental challenges associated with the large size of the full-length B2GPI, our results strongly support the presented structure of the complex. The model identifies three lysine residues of B2GPI-DV critical for binding the lipoprotein receptors. We are now working to confirm the model by side-directed mutagenesis of the identified Lys 282, Lys 308 and Lys 317.
GRANT SUPPORT
This work was supported in part by a Scientist Development Grant from the American Heart Association, a Junior Faculty Award from the American Society of Hematology and NIH GM061867 grant.
ABBREVIATIONS
- B2GPI
beta2-glycoproteinI
- B2GPI-DV
domain V of beta2-glycoproteinI
- APS
antiphospholipid syndrome
- LDLR
low-density lipoprotein receptor
- LA3
ligand-binding LA module 3 from the LDLR
- LA4
ligand-binding LA module 4 from the LDLR
- RAP
receptor associated protein
- RMSD
root mean square deviation
REFERENCES
- 1.Lin F, Murphy R, White B, Kelly J, Feighery C, Doyle R, Pittock S, Moroney J, Smith O, Livingstone W, Keenan C, Jackson J. Circulating levels of beta2-glycoprotein I in thrombotic disorders and in inflammation. Lupus. 2006;15(2):87–93. doi: 10.1191/0961203306lu2270oa. [DOI] [PubMed] [Google Scholar]
- 2.Moestrup SK, Schousboe I, Jacobsen C, Leheste JR, Christensen EI, Willnow TE. beta2-glycoprotein-I (apolipoprotein H) and beta2-glycoprotein-I-phospholipid complex harbor a recognition site for the endocytic receptor megalin. J Clin Invest. 1998;102(5):902–909. doi: 10.1172/JCI3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lutters BC, Derksen RH, Tekelenburg WL, Lenting PJ, Arnout J, de Groot PG. Dimers of beta 2-glycoprotein I increase platelet deposition to collagen via interaction with phospholipids and the apolipoprotein E receptor 2'. J Biol Chem. 2003;278(36):33831–33838. doi: 10.1074/jbc.M212655200. [DOI] [PubMed] [Google Scholar]
- 4.Maiti SN, Balasubramanian K, Ramoth JA, Schroit AJ. Beta-2-glycoprotein 1-dependent macrophage uptake of apoptotic cells. Binding to lipoprotein receptor-related protein receptor family members. J Biol Chem. 2008;283(7):3761–3766. doi: 10.1074/jbc.M704990200. [DOI] [PubMed] [Google Scholar]
- 5.Pennings MT, Derksen RH, Urbanus RT, Tekelenburg WL, Hemrika W, de Groot PG. Platelets express three different splice variants of ApoER2 that are all involved in signaling. Journal Thromb Haemost. 2007;5:1538–1544. doi: 10.1111/j.1538-7836.2007.02605.x. [DOI] [PubMed] [Google Scholar]
- 6.Pennings MT, Derksen RH, van Lummel M, Adelmeijer J, VanHoorelbeke K, Urbanus RT, Lisman T, de Groot PG. Platelet adhesion to dimeric beta-glycoprotein I under conditions of flow is mediated by at least two receptors: glycoprotein Ibalpha and apolipoprotein E receptor 2'. J Thromb Haemost. 2007;5(2):369–377. doi: 10.1111/j.1538-7836.2007.02310.x. [DOI] [PubMed] [Google Scholar]
- 7.Urbanus RT, Pennings MT, Derksen RH, de Groot PG. Platelet activation by dimeric beta(2)-glycoprotein I requires signaling via both glycoprotein Ibalpha and Apolipoprotein E Receptor 2'. J Thromb Haemost. 2008 doi: 10.1111/j.1538-7836.2008.03021.x. [DOI] [PubMed] [Google Scholar]
- 8.van Lummel M, Pennings MT, Derksen RH, Urbanus RT, Lutters BC, Kaldenhoven N, de Groot PG. The binding site in {beta}2-glycoprotein I for ApoER2' on platelets is located in domain V. J Biol Chem. 2005;280(44):36729–36736. doi: 10.1074/jbc.M504172200. [DOI] [PubMed] [Google Scholar]
- 9.Miyakis S, Lockshin MD, Atsumi T, Branch DW, Brey RL, Cervera R, Derksen RH, de Groot PG, Koike T, Meroni PL, Reber G, Shoenfeld Y, Tincani A, Vlachoyiannopoulos PG, Krilis SA. International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS) J Thromb Haemost. 2006;4(2):295–306. doi: 10.1111/j.1538-7836.2006.01753.x. [DOI] [PubMed] [Google Scholar]
- 10.Salmon JE, de Groot PG. Pathogenic role of antiphospholipid antibodies. Lupus. 2008;17(5):405–411. doi: 10.1177/0961203308090025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giannakopoulos B, Passam F, Rahgozar S, Krilis SA. Current concepts on the pathogenesis of the antiphospholipid syndrome. Blood. 2007;109(2):422–430. doi: 10.1182/blood-2006-04-001206. [DOI] [PubMed] [Google Scholar]
- 12.Urbanus RT, Derksen RH, de Groot PG. Current insight into diagnostics and pathophysiology of the antiphospolipid syndrome. Blood Rev. 2008;22(2):93–105. doi: 10.1016/j.blre.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 13.May P, Herz J, Bock HH. Molecular mechanisms of lipoprotein receptor signalling. Cell Mol Life Sci. 2005;62(19-20):2325–2338. doi: 10.1007/s00018-005-5231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeon H, Blacklow SC. Structure and physiologic function of the low-density lipoprotein receptor. Annu Rev Biochem. 2005;74:535–562. doi: 10.1146/annurev.biochem.74.082803.133354. [DOI] [PubMed] [Google Scholar]
- 15.Pennings MT, van Lummel M, Derksen RH, Urbanus RT, Romijn RA, Lenting PJ, de Groot PG. Interaction of beta2-glycoprotein I with members of the low density lipoprotein receptor family. J Thromb Haemost. 2006;4(8):1680–1690. doi: 10.1111/j.1538-7836.2006.02036.x. [DOI] [PubMed] [Google Scholar]
- 16.Meroni PL, Ronda N, De Angelis V, Grossi C, Raschi E, Borghi MO. Role of anti-beta2 glycoprotein I antibodies in antiphospholipid syndrome: in vitro and in vivo studies. Clin Rev Allergy Immunol. 2007;32(1):67–74. doi: 10.1007/BF02686083. [DOI] [PubMed] [Google Scholar]
- 17.Herz J, Bock HH. Lipoprotein receptors in the nervous system. Annu Rev Biochem. 2002;71:405–434. doi: 10.1146/annurev.biochem.71.110601.135342. [DOI] [PubMed] [Google Scholar]
- 18.Blacklow SC, Kim PS. Protein folding and calcium binding defects arising from familial hypercholesterolemia mutations of the LDL receptor. Nat Struct Biol. 1996;3(9):758–762. doi: 10.1038/nsb0996-758. [DOI] [PubMed] [Google Scholar]
- 19.Fass D, Blacklow S, Kim PS, Berger JM. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature. 1997;388(6643):691–693. doi: 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- 20.Beglova N, Jeon H, Fisher C, Blacklow SC. Cooperation between fixed and low pH-inducible interfaces controls lipoprotein release by the LDL receptor. Mol Cell. 2004;16(2):281–292. doi: 10.1016/j.molcel.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 21.Daly NL, Djordjevic JT, Kroon PA, Smith R. Three-dimensional structure of the second cysteine-rich repeat from the human low-density lipoprotein receptor. Biochemistry. 1995;34(44):14474–14481. doi: 10.1021/bi00044a025. [DOI] [PubMed] [Google Scholar]
- 22.Daly NL, Scanlon MJ, Djordjevic JT, Kroon PA, Smith R. Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proc Natl Acad Sci U S A. 1995;92(14):6334–6338. doi: 10.1073/pnas.92.14.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolmer K, Huang W, Gettins PG. NMR solution structure of complement-like repeat CR3 from the low density lipoprotein receptor-related protein. Evidence for specific binding to the receptor binding domain of human alpha(2)-macroglobulin. J Biol Chem. 2000;275(5):3264–3269. doi: 10.1074/jbc.275.5.3264. [DOI] [PubMed] [Google Scholar]
- 24.Fisher C, Beglova N, Blacklow SC. Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol Cell. 2006;22(2):277–283. doi: 10.1016/j.molcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Huang W, Dolmer K, Liao X, Gettins PG. NMR solution structure of the receptor binding domain of human alpha(2)-macroglobulin. J Biol Chem. 2000;275(2):1089–1094. doi: 10.1074/jbc.275.2.1089. [DOI] [PubMed] [Google Scholar]
- 26.Jensen GA, Andersen OM, Bonvin AM, Bjerrum-Bohr I, Etzerodt M, Thogersen HC, O'Shea C, Poulsen FM, Kragelund BB. Binding site structure of one LRP-RAP complex: implications for a common ligand-receptor binding motif. J Mol Biol. 2006;362(4):700–716. doi: 10.1016/j.jmb.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 27.North CL, Blacklow SC. Solution structure of the sixth LDL-A module of the LDL receptor. Biochemistry. 2000;39(10):2564–2571. doi: 10.1021/bi992087a. [DOI] [PubMed] [Google Scholar]
- 28.Querol-Audi J, Konecsni T, Pous J, Carugo O, Fita I, Verdaguer N, Blaas D. Minor group human rhinovirus-receptor interactions: geometry of multimodular attachment and basis of recognition. FEBS Lett. 2009;583(1):235–240. doi: 10.1016/j.febslet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 29.Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298(5602):2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 30.Simonovic M, Dolmer K, Huang W, Strickland DK, Volz K, Gettins PG. Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor. Biochemistry. 2001;40(50):15127–15134. doi: 10.1021/bi015688m. [DOI] [PubMed] [Google Scholar]
- 31.Verdaguer N, Fita I, Reithmayer M, Moser R, Blaas D. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat Struct Mol Biol. 2004;11(5):429–434. doi: 10.1038/nsmb753. [DOI] [PubMed] [Google Scholar]
- 32.Wolf CA, Dancea F, Shi M, Bade-Noskova V, Ruterjans H, Kerjaschki D, Lucke C. Solution structure of the twelfth cysteine-rich ligand-binding repeat in rat megalin. J Biomol NMR. 2007;37(4):321–328. doi: 10.1007/s10858-006-9129-3. [DOI] [PubMed] [Google Scholar]
- 33.Bouma B, de Groot PG, van den Elsen JM, Ravelli RB, Schouten A, Simmelink MJ, Derksen RH, Kroon J, Gros P. Adhesion mechanism of human beta(2)-glycoprotein I to phospholipids based on its crystal structure. Embo J. 1999;18(19):5166–5174. doi: 10.1093/emboj/18.19.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarzenbacher R, Zeth K, Diederichs K, Gries A, Kostner GM, Laggner P, Prassl R. Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome. Embo J. 1999;18(22):6228–6239. doi: 10.1093/emboj/18.22.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pons J-L, Malliavin TE, Delsuc MA, Gifa V. 4: A complete package for NMR data set processing. J Biomol NMR. 1996;8:445–452. doi: 10.1007/BF00228146. [DOI] [PubMed] [Google Scholar]
- 36.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6(2):135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 37.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65(2):392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 38.Comeau SR, Kozakov D, Brenke R, Shen Y, Beglov D, Vajda S. ClusPro: performance in CAPRI rounds 6-11 and the new server. Proteins. 2007;69(4):781–785. doi: 10.1002/prot.21795. [DOI] [PubMed] [Google Scholar]
- 39.Shen Y, Brenke R, Kozakov D, Comeau SR, Beglov D, Vajda S. Docking with PIPER and refinement with SDU in rounds 6-11 of CAPRI. Proteins. 2007;69(4):734–742. doi: 10.1002/prot.21754. [DOI] [PubMed] [Google Scholar]
- 40.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CRARMM- A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 41.DeLano WL. ThePyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 42.Collaborative Computational Project The CCP4 Suite:Programs for Protein Crystallography. 1994 doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.Beglova N, North CL, Blacklow SC. Backbone dynamics of a module pair from the ligand-binding domain of the LDL receptor. Biochemistry. 2001;40(9):2808–2815. doi: 10.1021/bi0027276. [DOI] [PubMed] [Google Scholar]
- 44.Shortridge MD, Hage DS, Harbison GS, Powers R. Estimating protein-ligand binding affinity using high-throughput screening by NMR. J Comb Chem. 2008;10(6):948–958. doi: 10.1021/cc800122m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehdi H, Naqvi A, Kamboh MI. A hydrophobic sequence at position 313-316 (Leu-Ala-Phe-Trp) in the fifth domain of apolipoprotein H (beta2-glycoprotein I) is crucial for cardiolipin binding. Eur J Biochem. 2000;267(6):1770–1776. doi: 10.1046/j.1432-1327.2000.01174.x. [DOI] [PubMed] [Google Scholar]
- 46.Sheng Y, Sali A, Herzog H, Lahnstein J, Krilis SA. Site-directed mutagenesis of recombinant human beta 2-glycoprotein I identifies a cluster of lysine residues that are critical for phospholipid binding and anti-cardiolipin antibody activity. J Immunol. 1996;157(8):3744–3751. [PubMed] [Google Scholar]
- 47.Andersen OM, Christensen PA, Christensen LL, Jacobsen C, Moestrup SK, Etzerodt M, Thogersen HC. Specific binding of alpha-macroglobulin to complement-type repeat CR4 of the low-density lipoprotein receptor-related protein. Biochemistry. 2000;39(35):10627–10633. doi: 10.1021/bi000498h. [DOI] [PubMed] [Google Scholar]
- 48.Andersen OM, Schwarz FP, Eisenstein E, Jacobsen C, Moestrup SK, Etzerodt M, Thogersen HC. Dominant thermodynamic role of the third independent receptor binding site in the receptor-associated protein RAP. Biochemistry. 2001;40(50):15408–15417. doi: 10.1021/bi0110692. [DOI] [PubMed] [Google Scholar]
- 49.Estrada K, Fisher C, Blacklow SC. Unfolding of the RAP-D3 helical bundle facilitates dissociation of RAP-receptor complexes. Biochemistry. 2008;47(6):1532–1539. doi: 10.1021/bi702076y. [DOI] [PubMed] [Google Scholar]
- 50.Andersen OM, Benhayon D, Curran T, Willnow TE. Differential binding of ligands to the apolipoprotein E receptor 2. Biochemistry. 2003;42(31):9355–9364. doi: 10.1021/bi034475p. [DOI] [PubMed] [Google Scholar]
- 51.Andersen OM, Christensen LL, Christensen PA, Sorensen ES, Jacobsen C, Moestrup SK, Etzerodt M, Thogersen HC. Identification of the minimal functional unit in the low density lipoprotein receptor-related protein for binding the receptor-associated protein (RAP). A conserved acidic residue in the complement-type repeats is important for recognition of RAP. J Biol Chem. 2000;275(28):21017–21024. doi: 10.1074/jbc.M000507200. [DOI] [PubMed] [Google Scholar]
- 52.Fisher C, Abdul-Aziz D, Blacklow SC. A two-module region of the low-density lipoprotein receptor sufficient for formation of complexes with apolipoprotein E ligands. Biochemistry. 2004;43(4):1037–1044. doi: 10.1021/bi035529y. [DOI] [PubMed] [Google Scholar]