Abstract
A central mechanistic paradigm of cysteine proteases is that the His – Cys catalytic diad forms an ion-pair NH(+)/S(−) already in the catalytically active free enzyme. Most molecular modeling studies of cysteine proteases refer to this paradigm as their starting point. Nevertheless, several recent kinetics and X-ray crystallography studies of viral and bacterial cysteine proteases depart from the ion-pair mechanism, suggesting general base catalysis. We challenge the postulate of the ion-pair formation in free papain. Applying our QM/SCRF(VS) molecular modeling approach, we analyzed all protonation states of the catalytic diad in free papain and its SMe derivative, comparing the predicted and experimental pKa data. We conclude that the His – Cys catalytic diad in free papain is fully protonated, NH(+)/SH. The experimental pKa=8.62 of His159 imidazole in free papain, obtained by NMR controlled titratin and originally interpreated as the NH(+)/S(−) ⇌ N/S(−) equilibrium, is now assigned to the NH(+)/SH ⇌ N/SH equilibrium.
Keywords: cysteine proteases, enzyme mechanism, solvation, pKa in proteins, molecular modeling, quantum mechanics
INTRODUCTION
Cysteine and histidine form the catalytic diad in the active site of all cysteine proteases. They are thought to be present as a thiolate-imidazolium ion pair (Cys25)-S−/(His159)-Im+H (papain numbering) in the free enzyme (FE).1–3 The thiolate anion in catalytically active free enzyme is prepared for nucleophilic attack at the carbonyl carbon of the scissile amide or ester bond. The His-Im+H functions as a general acid, facilitating the living group departure from the tetrahedral intermediate to produce an acyl enzyme intermediate. This widely accepted mechanistic paradigm, pioneered by Polgar and coworkers, was based on pH-dependence of the second-order rate constants of inactivation of papain and thiosubtilisin.4–6 The acidic pKa = 4 was assigned to the (Cys25)-SH/(His159)-Im+H ⇌ (Cys25)-S−/(His159)-Im+H equilibrium, and the basic pKa = 8.4 was assigned to the (Cys25)-S−/(His159)-Im+H ⇌ (Cys25)-S−/(His159)-Im equilibrium. This reversed order of pKa (Cys) < pKa (His) in the ion pair, compaired with 6.04 for histidine and 8.37 for cysteine in solution,7 was interpreted to reflect mutual electrostatic stabilization of the counter ions in the thiolate-imidazolium ion pair.1–3 Fluorescence spectroscopy8 and NMR9,10 titration experiments of free papain and its SSMe derivative, in which the Cys25 thiol was blocked by methylthiolation, further supported this interpretation. The pKa = 8.62 of free papain was assigned to histidine deprotonation from the ion-pair state. When the electrostatic influence of the thiolate anion was eliminated by the bound SMe, His pKa was decreased to 3.39.
Some kinetic experiments led to the speculation that ion-pair formation is not sufficient to generate catalytic competence in cysteine proteases and that other ionizable residues have to be deprotonated in order to affect substrate hydrolysis.11–16 Asp158 was considered as one such electrostatic modulating factor in papain.11–16 On the other hand, a study of Asp158 point mutations indicated that the charge of the Asp158 carboxylate plays a minor if at all role in the observed pKa shift in papain. The possible Asp158 influence was assigned to stabilization of the catalytic active conformation by forming local hydrogen bonds with the backbone amides of Ala136 and His159.17,18 A recent study suggested a key role for Trp177, influencing the hydrophobic shielding of the (Cys25)-S−/(His159)-Im+H ion pair.19 Recent interpretations of the catalytic mechanism of some viral and bacterial cysteine proteases based on kinetics20,21 and X-ray crystallography22 depart from the ion-pair mechanism.
The above experimental works provide macroscopic data derived from enzymatic kinetics and therefore they cannot directly identify the actual protonation state of the Cys-His catalytic diad in cysteine proteases. Moreover, currently there is no NMR or high resolution crystallographic 3D structure of free papain valuable for an unambiguous identification of protons position in the catalytic diad. Thus, theoretical analysis based on molecular modeling is the only way to address this issue.
A general theoretical principle regarding the role of electrostatic interactions in enzyme catalysis states that an enzyme active site can stabilize varied constellations of charges, including ion pairs and polar transition states, much better than bulk water does, by pre-oriented permanent dipoles either alone or in combination with several water molecules.23 These ideas were further developed24 and validated for various enzymes,25–28 including the proton transfer between Cys25 and His159 in papain.29–30 Applying molecular modeling methods calibrated on the experimental energy of the ion pair in solution using empirical valence bond model (EVB) for the reacting region and PDLD and other models for the protein, it was demonstrated that Cys25–His159 ion-pair is more stabilized in the enzyme active site than in water, i.e. the enzyme inverts the stabilization of the covalent and ionic states relative to their order in water solution.
The ion-pair paradigm dominates modern molecular QM/MM simulations of enzyme-substrate and enzyme-inhibitor interactions in the active site of cysteine protease.31–33 A recent QM/MM study of free cathepsin B concluded that the ion pair (Cys)-S−/(His)-Im+H is significantly more stable than its neutral counterpart (Cys)-SH/(His)-Im.34 A quantitative assessment of the various effects indicated that the enzyme environment stabilizes the ion pair by 40 kJ/mol in comparison with the ion pair in aqueous solution (QM region in COSMO environment). The stabilization was attributed to hydrogen bonding network involving the catalytic Cys, neighboring residues and solvent water molecules. It was observed that removal of even a single solvent water molecule destabilizes the ion pair and shifts the equilibrium to the neutral state (Cys)-SH/(His)-Im.
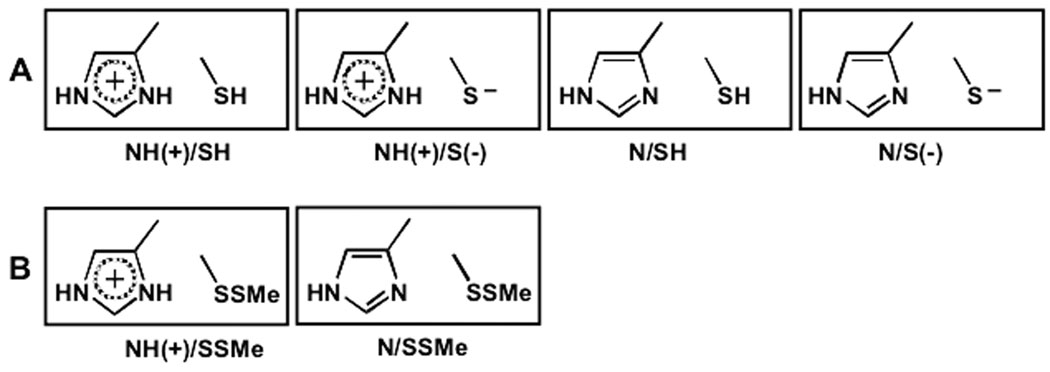
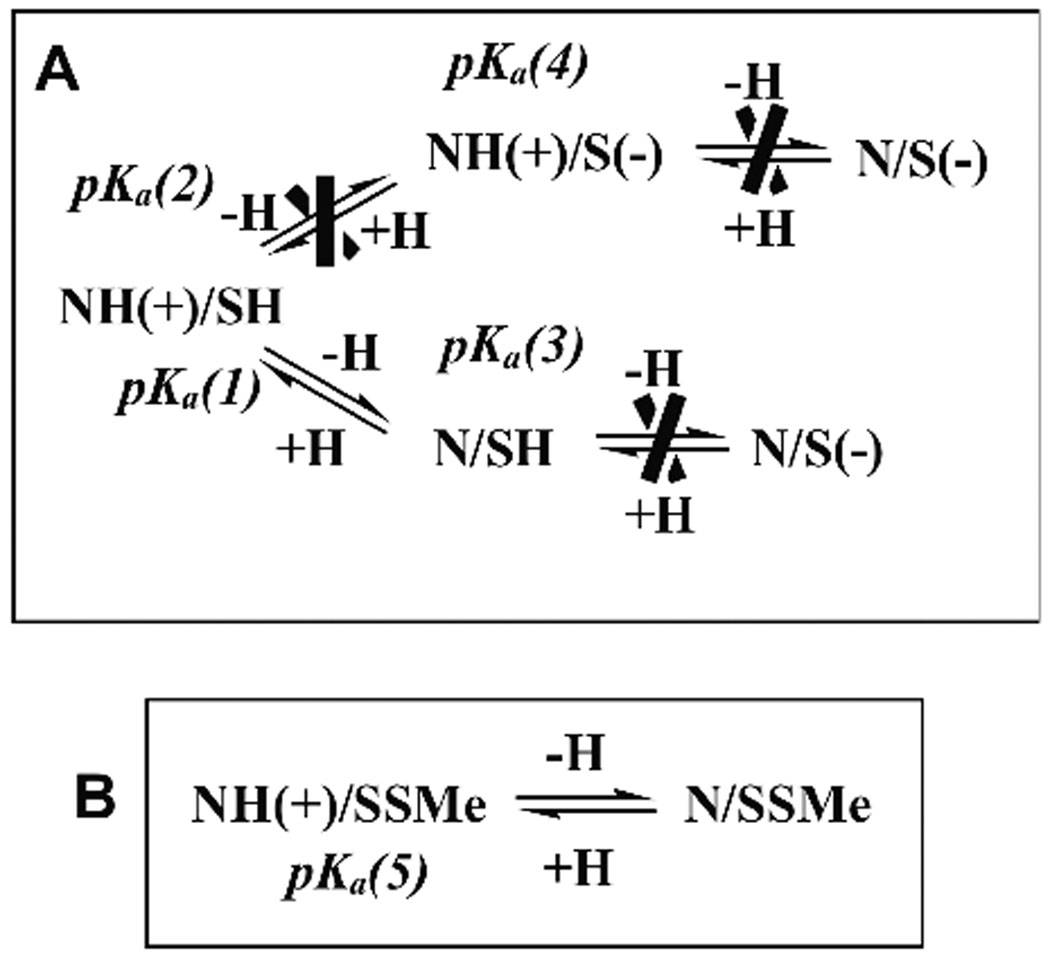
It is important to stress that the mechanistic discussions in all of the above experimental and theoretical studies circulates around Polgar’s hypothesis that the Cys–His catalytic diad in free cysteine proteases is more stable in its ion pair than in its neutral form. The idea was introduced in order to rationalize the observed pKa values of 4 and 8.4 of the bell shaped pH-dependence of the second-order rate constant of inactivation of papain.4–6 In the present study we shift the focus from the stability of the Cys-His ion-pair relative to the neutral form. Applying our quantum mechanical QM/SCRF(VS) molecular modeling approach35 we analyze all possible protonation states of the catalytic diad in free papain (FE) and its derivative (SSMe), in which the catalytic thiol is blocked by methylthiolation (Figure 1).9,10
Figure 1.
All possible protonation states of the Cys25—His159 catalytic diad in free papain (A), and its SMe derivative (B).
We have recently used our QM/SCRF(VS) approach for studying enzyme mechanisms,36,37 formulation of an enzyme mechanism based CADD method,38,39 and for the analysis of the role of active site polar interactions and bulk water solvation in modulating general acid / base catalysis in serine proteases.35,40 Combining the homogenious solvation Born model of spherical solute cavity with a model in which this cavity is located in the border between two different dielectric media, we have formulated analytical equation that describes the dependence of the pKa of catalytic residues on various environmental factors in the enzyme active site. These include the degree of exposure of the catalytic residue to the bulk solvent, the character of ionized groups in the enzyme active site, and the influence of the pre-oriented dipoles of the active site on the target catalytic residue.35 We have used this equation for the rationalization of the experimentally observed pKa shift of the catalytic histidine in serine proteases along the transformation from the free enzyme to the tetrahedral covalent complex, and the alterations of this His between acid and base catalytic functionality along the reaction coordinate.40 We have further demonstrated that ligand binding in the enzyme active site plays a triggering role in catalysis by screening of the catalytic His from the bulk water, leading to a large shift in its pKa35 in agreement with experimental observations.41,42
The ion-pair paradigm in cysteine proteases ignores a possible abrupt pKa shift in the catalytic His upon ligand binding; the Cys/His catalytic diad is suggested to have an ion-pair structure in both the free enzyme and the enzyme/ligand Michaelis-like non-covalent complex.1–6, 31–34 Thus, in the present study we challenge the NH(+)/S(−) ion pair paradigm.
METHODS
Construction of the modeling system
We have considered papain in two states: free enzyme (FE) and its derivative with the Cys25 thiol blocked by methylthiolation (SSMe). The geometries of FE and SSMe were derived from the 9pap43 PDB file (1.65 Å resolution). The initial X-ray structure was transformed into the two variants, SH group in free enzyme and SSMe derivative, by Maestro molecular editor.44
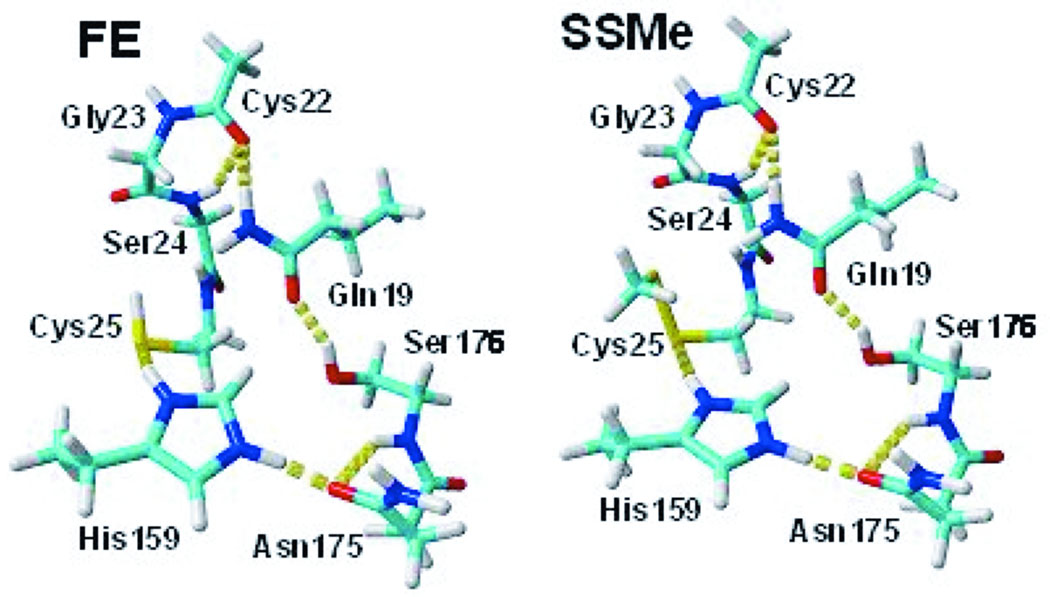
Preliminary geometry relaxation of the whole FE and SSMe protein molecules were conducted in vacuum with the backbone frozen by OPLS AA force field45 in the Macromodel 7 package.46 The applied cutoff was 7Å for vdW and 12Å for charge/charge interactions. All water exposed groups ionized at pH 7 (Asp, Glu, Lys and Arg) were neutralized to avoid an overestimation of ionic interactions in vacuum caused by the absence of screening water molecules and counterions. The Cys25 and His159 catalytic groups were examined in both protonated and deprotonated states. The position of the SSMe group was refined by conformational analysis on three torsional angles corresponding to rotations around Cα-Cβ, Cβ-S and S-S single bonds, applying 200 steps of random changes in torsional angles by MCMM algorithm47,48 implemented in the Macromodel package. The most stable structures of FE and SSMe proteins were used to extract molecular clusters corresponding to the enzyme active site – the quantum mechanical site (QM) in our modeling. In the QM/SCRF(VS) approach QM explicitly accounts for the preorganized local polar environment in the enzyme active site influencing recharging of the ionizable groups in the Cys/His catalytic diad. Thus, our model FE and SSMe molecular clusters include the Cys25 and His159 residues in their basic (B) and acidic (A) forms, as well as residues associated with them by a net of hydrogen bonds in the active site. Asp158, the closest ionic group to the Cys25-His159 catalytic diad, has negligible electrostatic influence on the ionization of His15917–19 (see Results and discussion) and thus was excluded from the QM site. The constructed QM molecular clusters FE and SSMe contain the Gln19, Cys22, Gly23, Ser24, Cys25, His159, Asp175, and Ser176 residues (Figure 2). Open valences of the broken covalent bonds at the “edges” of the modeling molecular clusters are capped with hydrogen atoms.
Figure 2.
The molecular clusters used for the modeling of the active site of free papain (FE), and its derivative with Cys25 covalently modified by methylthiolation (SSMe). The protonated form of His159 is illustrated. Color scheme: C - cyan, H - white, N - blue, O - red, S - yellow. H-bonds are marked as chains of yellow cylinders. Figures were prepared by the Yasara Dynamics software.
The QM/SCRF(VS) approach
In the QM/SCRF(VS) approach the water solvated enzyme is treated as a two layer system.35–37,40 The inner layer, the quantum mechanical region, is the enzyme active site (AS) molecular cluster, explicitly calculated by DFT. The outer layer, the classical region, includes the rest of the protein and the bulk water, and is accounted for on a macroscopic level as a uniform dielectric continuum – “virtual solvent” (VS). The molecular shaped solute cavity of the AS cluster is embedded in the virtual solvent, which is characterized by an effective dielectric constant, εeff. The empirical parameter εeff is calibrated by equalizing the theoretical pKa of the target ionizable group to the experimental value: pKatheor = pKaexp. The solute charge distribution, ρ(r), in the AS cluster (calculated in quantum mechanical Poisson-Bolzmann SCRF model implemented in the Jaguar software package49) polarizes the surrounding dielectric medium (virtual solvent), inducing surface point charges distributed over the solute-solvent interface.50,51 The induced surface charges give rise to an electrostatic field at the solute, called the reaction field, treated as a quantum mechanical perturbation of the solute Hamiltonian. SCRF uses an iterative self-consistent procedure until convergence in ρ(r) is achieved.
Geometries of the extracted QM molecular clusters FE and two SSMe were optimized at the B3LYP/6-31+G** level of DFT theory,52,53 implemented in the JAGUAR 5.1 package. B3LYP/6-31+G** can successfully reproduce geometries and energies of hydrogen bonds and proton positions on ionized groups in the enzyme active site.35–37,40 The geometries of the FE and the SSMe molecular clusters were optimized in vacuum. The Cα atoms were kept fixed during the QM optimization process in order to maintain biologically relevant molecular structures. Theoretical values, pKatheor, were calculated on the QM molecular clusters of the two models FE and SSMe by the QM/SCRF(VS) algorithm.35 A key feature of the QM/SCRF(VS) algorithm is its validation and calibration by experimental values of gas-phase deprotonation free energies, ΔGg, and pKa in water of small organic molecules before the estimation of pKatheor in the protein. Since we concentrate on the pKa’s of Cys25 and His159 of the catalytic diad in papain, we used training sets of five thiols and six nitrogen heteroaromatics with tabulated ΔGg 54,55 and pKa 56–59 values (Table I). Experimental pKa’s ranged from 7.0 to 11.5 in the former set, and from 0.7 to 9.7 in the latter. pKatheor of the training small molecules were calculated by the same formulas used for the FE and SSMe molecular clusters and at the same level of theory. We calculated pKatheor by the standard thermodynamic definition:
| (1) |
For small molecules, used for calibration:
| (2) |
The calculation of the deprotonation free energy of the catalytic residue in the enzyme active site is analogous to Eq. 2:
| (3) |
In Eq. 2 and Eq. 3, Δ represents differences between deprotonated (basic, B) and protonated (acidic, A) states: the gas-phase free energy of deprotonation, ΔGg = Gg(B) − Gg(A), and the difference of solvation free energy of the calibrating small molecules in water, ΔGws = Gws(B) − Gws(A). In Eq. 3 ΔGvs = Gvs(B) − Gvs(A) accounts for the difference in solvation free energy of the enzyme active site in a virtual solvent that according to the QM/SCRF(VS) algorithm simulates the target catalytic residue partially water exposed. The value of −262.5 kcal/mol was used for the free energy of proton hydration Ghyd(H+).60 The gas-phase deprotonation free energy of a small molecule, ΔGg in Eq. 2, or the catalytic residue in the enzyme active site, ΔGg(AS) in Eq. 3, is calculated explicitly by:
| (4) |
As above, Δ indicates difference between deprotonated and protonated species. The total electronic energy of the molecular cluster, zero point energy, enthalpy, and entropy (Etot, ZPE, H, and S, respectively) were calculated in harmonic approximation by standard procedures implemented in the Jaguar package. Eq. 4 also accounts for the gas-phase translational enthalpy and entropy of the released proton calculated at T = 298K and standard pressure in the gas phase: G(H+) = 5/2RT − TS(H+) = − 6.28 kcal/mol.61 The water solvation free energies of calibrating small molecules were calculated at ε = 80, and probe radius 1.4 Å by the self-consistent reaction field (SCRF) method50,51 implemented in the JAGUAR package.
Table I.
Experimental and theoretical gas-phase free energy of deprotonation of thiols and nitrogen heteroaromatics (in kcal/mol) and pKa in water
| Thiols | Gg expa | Gg theor | pKa expb | pKa theor |
|---|---|---|---|---|
| C2H5SH | 348.9 | 352.0 | 10.5 | 10.14 |
| C6H5SH | 333.8 | 336.0 | 7.78 | 6.58 |
| SH2 | 344.4 | 346.7 | 7.0 | 5.49 |
| t-C4H9-SH | 346.2 | 350.6 | 11.05 | 11.50 |
| n-C4H9-SH | 347.4 | 351.7 | 10.65 | 10.37 |
| nitrogen heteroaromatics |
Gg exp | Gg theor | pKa exp | pKa theor |
| 4-Me-imidazole | 220.0 | 221.2 | 7.45 | 7.49 |
| pyridine | 214.6 | 214.8 | 5.30 | 5.95 |
| pyrimidine | 204.4 | 204.3 | 1.30 | 1.04 |
| thiazole | 208.3 | 206.4 | 2.80 | 2.20 |
| 4-aminopyridine | 226.4 | 226.9 | 9.70 | 9.38 |
| pyrazine | 202.3 | 201.2 | 0.70 | 1.10 |
We used linear regression analysis on the training sets of the thiols and nitrogen heterocycles for the optimization of empirical corrections: pKatheor,corr = α pKatheor + β. The following values were obtained from linear regression analysis: α = 0.9940, β = 0.0427, SE = 0.52 pKa units, R2 = 0.983, F = 228.5 for nitrogen heteroaromatics, and α = 0.7085, β = 3.1506, SE = 0.19 pKa units, R2 = 0.991, F = 365.2 for the thiols.
MD simulations
All ionizable protein groups, except for Cys25 and His159, were protonated / deprotonated according to their tabulated pKa values and the accepted pH 7. Periodic boundary cell that is 20 Å larger than the protein along each axis was used for the MD simulation. The periodic box was filled with TIP3P water molecules. The free papain was put into a box of 54.5×71.2×62.3 Å with 7162 water molecules for NH(+)/S(−) and 54.5×71.4×62.4 Å with 7163 water molecules for NH(+)/SH, and the SSMe derivative into a box of 67.0×62.8×66.5 Å with 8397 water molecules. Counter ions were iteratively placed at the coordinates with the highest electrostatic potential until the cell was neutral. To remove bumps and correct the covalent geometry, the structure was energy-minimized with the AMBER 99 force field,62 using a 7.86 Å force cutoff and the Particle Mesh Ewald algorithm63,64 to treat longrange electrostatic interactions. After removal of conformational stress by a short steepest descent minimization, the procedure continued by simulated annealing (time step 2 fs, atom velocities scaled down by 0.9 every 10th step) until convergence was reached, i.e. the energy improved by less than 0.1% during 200 steps.
The molecular dynamics (MD) simulations were conducted with a total run time of 3.6 ns for NH(+)/S(−) and 1.6 ns for NH(+)/SH structures of free papain, and 3.5 ns for the NH(+)/SSMe derivative by the Yasara Dynamics software,65 using AMBER 99 force field.62 The MD equilibrated simulations were run at 298K and constant pressure. Multiple time step was used: 1.25 fs for intramolecular and 2*1.25 fs for intermolecular forces. The cutoff for van der Waals interactions was 7.86 Å. The electrostatic interactions were calculated without cutoff by particle mesh Ewald algorithm.63,64 Snapshots were collected every 3 ps. In the post processing analysis of the MD simulation trajectory we calculated the solvent accessible surface area (SASA) of the imidazolium cation of His159 in both free papain and its SMe derivative. The SASA calculation was done applying Gaussian approximation, and a probe radius of 1.4 Å.
1H NMR calculations
We calculated proton NMR chemical shifts, δ, in the FE and SSMe molecular clusters for the CHε proton of the His159 imidazole for all studied protonation states of the catalytic diad, applying a method for gas-phase calculations of the shielding constants, σ, implemented in the Jaguar software.66 Chemical shifts were calculated by standard formula, δi = σTMS − σi, relative to a calculated average proton shielding constant, 31.6443, of the TMS standard. The calculated proton chemical shift values of His159 in FE and SSMe were then corrected by a liner function: δicorr = aδi + b. The empirical coefficients a = 0.8102 and b = 1.4896 were previously calibrated by linear regression analysis used for the correlation of experimental vs. theoretical chemical shifts of all protons in pyridine and imidazole in both basic and acidic forms. All NMR calculations were done in the B3LYP/6-31+G** level of theory. Experimental values of imidazole and pyridine chemical shifts were determined in our lab at 25°C in water solution.
RESULTS AND DISCUSSIONS
MD simulations of free papain
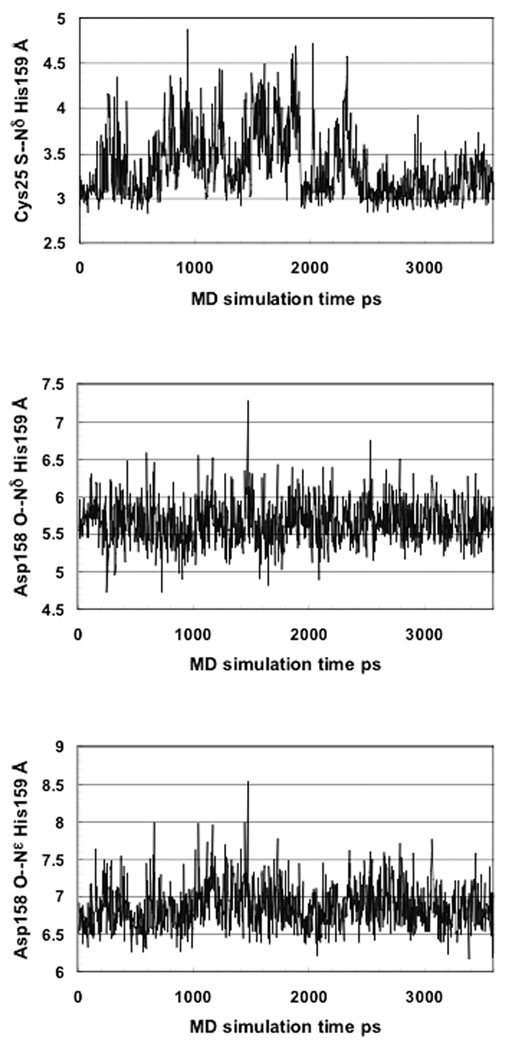
No experimentally-derived 3D structure of the catalytically active form of free papain is available. Therefore, we modeled the enzyme using molecular dynamics (MD). We carried out a 3.6 ns simulation with the suggested ion pair state.1–6 Figure 3A presents the time dependent Cys25 S(−)—NδH(+) His159 distance. The average value of 3.35Å over the whole trajectory and 3.13Å for the last 1 ns interval is too large for hydrogen bond formation. This is due to water exposure of the Cys25 thiolate and the His159 imidazolium, resulting in preferable formation of hydrogen bonds with solvent water molecules (Figure 4).
Figure 3.
Interatomic distances collected every 3ps on the MD simulation trajectory of free papain, assuming an NH(+)/S(−) ion pair state of the Cys25—His159 catalytic diad.
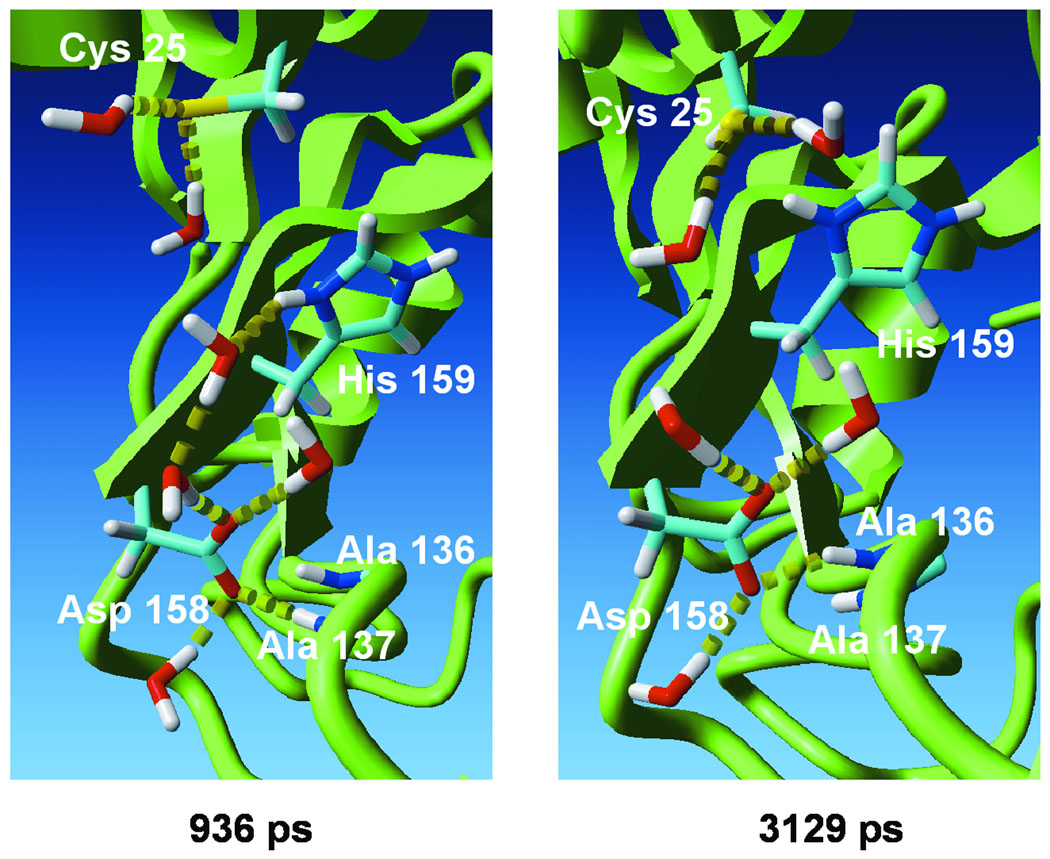
Figure 4.
MD snapshots collected at 936 ps and 3129 ps time points on the MD trajectory of the NH(+)/S(−) ion pair form of free papain. The Cys25 S—Nδ His159 distance is 4.86 Å at the first and 2.93 Å at the second time point. Residues irrelevant to the discussion and water molecules not forming hydrogen bonds with the considered residues are ommitted for clarity. H-bonds are marked as chains of yellow cylinders. Figures were prepared by the Yasara Dynamics software.
Asp158 is considered to be an electrostatic modulator of the pKa of the catalytic His159 (10–15). Figures 3B and 3C present fluctuations of the Asp158(−) O—Nδ His159(+) and Asp158(−) O—Nε His159(+) inter-atomic distances along the MD trajectory. The average values are 5.65 Å and 6.85 Å, respectively. The Asp158 carboxylate is highly exposed to the solvent, forming hydrogen bonds with water molecules and with the backbone amide of Ala136 or Ala137 (Figure 4). The average estimated Debye length (the distance at which the absolute value of the energy of an electrostatic interaction is ≤ RT) for solvent exposed charged groups in a protein at near physiological ionic strength (0.1M) is ~5Å.67 Thus, Asp158 is expected to play a negligible role in the electrostatic stabilization of the His159 imidazolium, in agreement with previous interpretations of kinetic experiments on wild-type and mutant papain.17–19
The protonation state of the catalytic diad in free papain
To study the protonation state of the catalytic diad in free papain, we applied our QM/SCRF(VS) approach.35 The current assignment of the pKa values of the catalytic Cys25 and His159 is based on comparison of two species, the free enzyme (FE) and the SMe derivative (SSMe), in which the catalytic thiol is blocked by methylthiolation.9,10 We examined all possible protonation states of the Cys—His catalytic diad in these two enzyme forms. FE has two centers of ionization – the His159 Nδ atom and the Cys25 thiol. His159 Nδ is the only protonation center in SSMe (Figure 1). The constructed QM molecular clusters are presented in Figure 2.
Table II contains pKatheor(ε) values calculated over a wide interval of ε values for all hypothetically possible protonation/deprotonation equilibria for FE and SSMe (Figure 5). It thus may be used to identify ε = εeff for which the pKatheor(εeff) = pKaexp condition is satisfied in both free papain and its SMe derivative. The values of pKatheor were calibrated on small organic molecules – thiols and amines - solvated in water according to the QM/SCRF(VS) algorithm (see Methods). Thus, in fact Table II contains pKatheor,corr, but we used the pKatheor designation for simplicity.
Table II.
pKatheor of FE and SSMe, solvated in VS at different dielectric constants εa
| ε | FE | SSMe | |||
|---|---|---|---|---|---|
|
pKa(1) NH(+)/SH ⇌ N/SH |
pKa(2) NH(+)/SH ⇌ NH(+)/S(−) |
pKa(3) N/SH ⇌ N/S(−) |
pKa(4) NH(+)/S(−)⇌ N/S(−) |
pKa(5) NH(+)/SSM⇌ N/SSMe |
|
| 1 | −13.9 | 0.0 | 41.9 | 44.8 | −13.9 |
| 2 | −2.5 | 6.5 | 31.1 | 32.0 | −2.9 |
| 3 | 1.4 | 8.6 | 26.9 | 27.1 | 0.9 |
| 4 | 3.5 | 9.6 | 24.5 | 24.4 | 2.8 |
| 6 | 5.4 | 10.6 | 22.1 | 21.5 | 4.5 |
| 8 | 6.4 | 11.1 | 20.8 | 20.1 | 5.6 |
| 10 | 7.1 | 11.3 | 20.0 | 19.2 | 6.1 |
| 20 | 8.5 | 12.0 | 18.5 | 17.5 | 7.2 |
| 30 | 8.9 | 12.2 | 18.2 | 17.2 | 7.6 |
| 40 | 9.2 | 12.3 | 17.8 | 16.8 | 7.7 |
Calculated at 25°C
Figure 5.
All possible variants of deprotonation/protonation of the Cys25—His159 catalytic diad in free papain (A) and its SMe derivative (B). The crossed equilibria represent computational variants that could not be fitted either to the experimental pKa or to the calculated relative stability.
We used Eq. 1–Eq. 4 in the QM/SCRF(VS) method for rigorous DFT calculations of the pKa values presented in Table I and Table II. Nevertheless, to rationally understand how changes of environmental factors in the enzyme active site influence the trend in pKa of catalytic residues, a more simplified model would be most helpful. We have recently demonstrated that a QM/SCRF(VS) model with molecular shaped solute cavity of the AS (QM molecular cluster) can be approximated by a spherical cavity of radius a embedded in the same VS with εeff.35 This new model is equivalent to the same spherical solute cavity located at the interface between different dielectric media: εeff = 1/(1–0.9875θ), where εeff grows hyperbolically with the increase of the normalized value of the surface fraction of the spherical solute cavity that faces the water, θ (0 ≤ θ ≤ 1). The SASA of a catalytic group in the enzyme active site, used for the pKa calibration in QM/SCRF(VS), changes proportionally with the values of θ and εeff. We have derived an approximated analytical expression for pKatheor:35
| (5) |
C = [ΔGg(AS) + Ghyd(H+)] / 2.303RT, where ΔGg(AS) is the free energy of deprotonation of the target catalytic residue calculated in the active site molecular cluster in the gas-phase approximation (see Eq. 1, Eq. 3, Eq. 4, and ref. 35), and Ghyd(H+) is the free energy of proton hydration. Q(A) and Q(B) are the total charges of the spherical solute cavity of radius a in Å, when the target ionizable functional group is in the acidic (A) or the basic (B) form, respectively. Eq. 5 rationalizes the dependence of pKa not only on the active site permanent dipoles (ΔGg(AS) in the second term), but also on the partial water solvation accounted for in the first term. The first term in Eq. 5 is the approximation of the rigorously calculated (by DFT) ΔGvs(AS) in Eq. 3. Being a combination of two functions, εeff (SASA) and (Q2(B) − Q2(A)), the first term of Eq. 5 accounts for the combined influence of two environmental factors on pKa: the degree of water exposure of the target catalytic residue, εeff (SASA); and the dependence of the free energy of solvation on the change of the total number of the ionized groups in the enzyme active site caused by the recharging of the target catalytic group during deprotonation. Eq. 5 enables prediction (used for the interpretations purpose only, but not for quantitative calculations) of the trend of pKa changes by knowing the value of Q2(B) − Q2(A) in the active site. If Q2(A) > Q2(B), pKa shifts in parallel to the change in εeff or θ, while if Q2(A) < Q2(B), a decrease in θ or εeff will lead to increase in pKa.35,40 Examining Table II and the corresponding protonation states (Figure 1) of the basic and acidic forms of the QM molecular clusters shows that the former trend applies to pKa(1), pKa(2), and pKa(5) (Q2(B) − Q2(A) = 0 − 1= −1), while the latter is represented by pKa(3) and pKa(4) (Q2(B) − Q2(A) = 1 − 0 = 1).
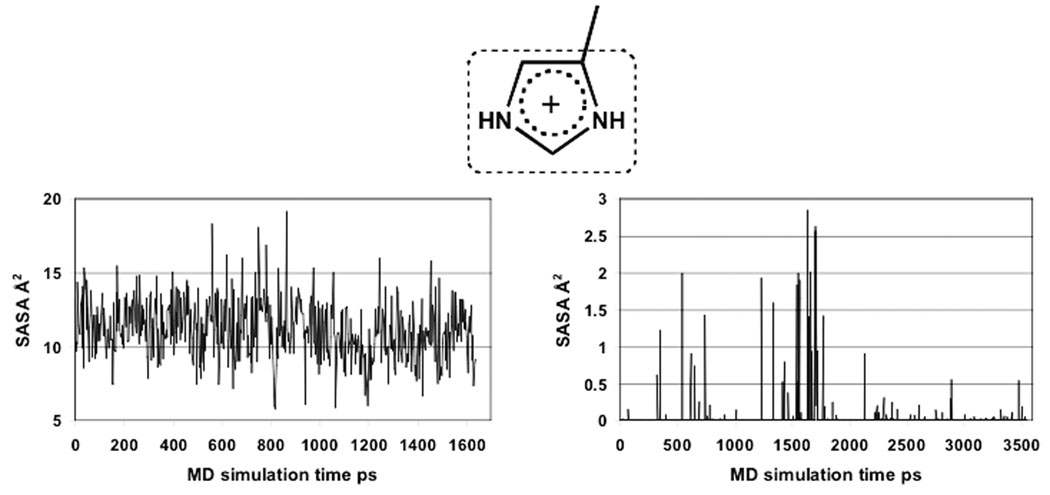
We used as a reference the experimental pKaexp = 9.2 at 25°C in FE (recalculated from pKa = 8.62 measured at 45°C by titration under 1H NMR analysis).10 Two alternative deprotonation variants of the free papain could potentially fit the experimental pKa in FE: pKa(1) corresponding to the equilibrium NH(+)/SH ⇌ N/SH, and pKa(2) corresponding to the equilibrium NH(+)/SH ⇌ N/S (Table II). These could be resolved by comparison of FE with its SMe derivative. The SMe group on Cys25 in the papain derivative screens the His159 imidazole from solvent. The time-averaged values of the solvent accessible surface area (SASA) collected on the MD trajectories are 11.13 Å2 in FE, and only 0.038 Å2 in the SMe derivative (Figure 6). The experimental pKa = 3.5 of His159 in SSMe at 25°C (recalculated from the measured value of 3.39 at 35°C9) was reproduced for the theoretical pKa(5) at εeff = 4.5. Thus, accepting the pKa(2) equilibrium, reached at εeff = 3.5, would indicate that the water exposure of His159 in free enzyme is smaller than in the SMe derivative, in contradiction to the above MD results. On the other hand, pKa(1) in FE fits the experimental pKa value at εeff ≥ 40, a value which is much larger than that identified for SSMe, in agreement with our theoretical findings that εeff is a measure of the SASA of the ionizable group used for the calibration.35
Figure 6.
Calculated SASA values of the His159 imidazolium along the MD simulation trajectory for free papain, and its SMe derivative.
The remaining pKa(3) and pKa(4) variants of FE are far beyond the range of the experimental pKa = 9.2 at all examined ε values (Table II). These high pKa values seem unreliable at first glance, if compared with the experimental values of the amino acids in aqueous solution (6.04 for histidine and 8.37 for cysteine)7 and of other small molecules (Table I). Nevertheless, these high values of pKa(3) and pKa(4) can be rationalized in the QM/SCRF(VS) approach considering energy contributions of the two cooperatively acting environmental effects – the local electrostatic environment in the enzyme active site (pre-oriented dipoles), ΔGg(AS), and the solvation of the target catalytic residue in a virtual solvent with calibrated εeff (partial water solvation), ΔGvs(AS) (Eq. 3).
Let us consider the N/SH ⇌ N/S(−) equilibrium. The calculated Cys pKa(3) = 41.9 at ε = 1 (Table II) is in fact smaller than what would be calculated without the effect of the active site environment. Indeed, the calculated gas-phase ΔGg(AS) = 343.1 kcal/mol corresponding to Cys pKa(3) is smaller than 348.9 kcal/mol, the analogous experimental gas-phase basicity of ethylmercaptane (Table I). The smaller value of ΔGg(AS) reflects the stabilization effect by neighboring polar groups of the thiolate anion of Cys in the enzyme active site. pKa(3) = 17.8 at ε = 40 is also not surprising. According to the MD simulations (Figure 4) the Cys thiolate in free papain is partially water solvated, forming several hydrogen bonds with water molecules. This observation correlates with QM/MM findings.34 Nevertheless, the partial water solvation of the catalytic Cys thiolate in free papain is considerably smaller than in the case of a surface cysteine residue or small molecules fully solvated in water. In addition there are no positively charged neighboring groups in the active site of free papain that could stabilize the Cys thiolate of the catalytic diad in the N/S(−) state.
The NH(+)/S(−)⇌N/S(−) equilibrium (corresponding to pKa(4), Table II) is commonly correlated with the experimental pKa = 8.62 at 45°C of free papain10 or pKa = 9.2 recalculated at 25° C in this work. This value was interpreted to reflect electrostatic stabilization in the Cys—His ion-pair in free cysteine proteases.4–6,9–10 pKa(4) is much larger at all considered ε values than the experimental value of 8.62 suggested for this interpretation. The calculated ΔGg(AS) = 330 kcal/mol contributing to the corresponding pKa(4) (Eq. 3) is much larger than the experimental gas-phase basicity of 220 kcal/mol of 4-Me-imidazole (Table I), confirming the expected stabilization of the imidazolium cation in the ion-pair. We would like to note in this context that we have previously calculated pKa = 11.5, and the corresponding ΔGg(AS) = 327.8 kcal/mol for the catalytic His195 in the modelling of enzyme-inhibitor covalent anionic tetrahedral complex in serine proteases, in very good agreement with the experimental value of pKa = 11.2 recalculated to 25°C.35 Thus the value of ΔGg(AS) = 330 kcal/mol in the present study is very reasonable.
Our results are only consistent with NH(+)/SH, the structure of the Cys25—His159 catalytic diad in free papain at neutral pH – the only state satisfying the pKatheor = pKaexp condition. This conclusion directly contradicts the NH(+)/S(−) ion pair paradigm. It should be noted that the acidic pKa value of papain was measured in kinetic experiments,4–6 but not in the NMR titration experiment.9,10 While the latter directly bears on the free enzyme structure, the former may be interpreted in different ways, relating to different species along the catalytic reaction. In the present study we considered only the structure of the free enzyme, while the observed acidic pKa issue will be addressed elsewhere.
It is interesting to note that while an ion-pair structure of the catalytic diad in free papain would render the enzyme fully active and thus amenable to non-specific reactivity, the suggested fully protonated structure may serve to protect the enzyme from undesired reactions. We suggest that papain-like cysteine proteases become active only upon substrate binding. The latter event reduces the water exposure of the catalytic diad in the Michaelis complex (MC) and thus triggers the enzymatic catalysis.35 Indeed, according to Eq. 5 the pKa change in the FE → MC transformation is characterized by the Q2(B) − Q2(A) = 0 − 1= −1 trend, if FE is initially in the NH(+)/SH protonation state as found in this work. Thus, two possible protonation states could be realized in the MC: N/SH or NH(+)/S(−). The latter is the ion pair, in which the thiolate is ready for the nucleophilic attack and the His imidazolium can activate the scissile amide by general acid catalysis. If N/SH is initially formed in the Michaelis complex, an alternative mechanism is feasible, in which the SH proton is transferred to the catalytic His concurrently with the nucleophilic attack of the S atom on the substrate. This mechanism is analogous to the corresponding catalytic mechanism of serine proteases.
1H NMR
We have also calculated chemical shifts of the CHε proton of His159 imidazole for all considered protonation states of the catalytic diad in both FE and SSMe molecular clusters (Table III). The theoretically calculated NMR chemical shifts nicely reproduce the experimentally observed chemical shifts and the difference between the δ values of the protonated and the deprotonated forms of His159 imidazole in free papain and its SMe derivative.9,10 Nevertheless, within computational error the calculated CHε proton chemical shifts cannot differentiate between the two examined equilibria: NH(+)/SH ⇌ N/SH and NH(+)/S(−) ⇌ N/S(−). This ambiguity demonstrates that the experimental NMR chemical shifts cannot unequivocally serve to support the paradigm of the presence of an NH(+)/S(−) ion pair in free papain.
Table III.
1H NMR chemical shifts (ppm) of CHε of the His159 imidazole
CONCLUSIONS
We have studied the protonation state of the His-Cys catalytic diad in free papain and its SSMe derivative. We demonstrated that the pKa values of catalytic residues are controlled by the cooperative effect of the active site polar local environment (pre-oriented dipoles) and the free energy of solvation of the partially water exposed target catalytic group in the active site. Based on our modeling simulations of all possible protonation states of the catalytic diad, calculated vs. experimental pKa’s, we suggest a novel structure to the catalytic Cys25-His159 diad in free papain at neutral pH, in which both residues are protonated, rather than in the ion pair paradigm. The experimental pKa 8.62 in free papain is now assigned to the equilibrium NH(+)/SH ⇌ N/SH. 1H NMR chemical shift calculations demonstrated that the experimental CHε proton chemical shifts cannot differentiate between the two equilibria NH(+)/SH ⇌ N/SH and NH(+)/S(−) ⇌ N/S(⇌), and therefore cannot support the ion pair paradigm. Thus, it is very difficult to provide definitive experimental data for the protonation state of the Cys-His catalytic diad in free cysteine proteases. One possible solution could be very high resolution X-ray crystallography of free papain, in which protons position could be identified. Such high resolution 3D structures of serine proteases with proton positioning were previously determined for serine proteases.68,69 Further application of the QM/SCRF(VS) approach to study the cysteine proteases mechanism is now underway.
Acknowledgments
Grant sponsor: National Institute of Health; Grant number: GM081329
References
- 1.Storer AC, Menard R. Catalytic mechanism in papain family of cysteine peptidases. Methods in Enzymology. 1994;244:486–500. doi: 10.1016/0076-6879(94)44035-2. [DOI] [PubMed] [Google Scholar]
- 2.Otto HH, Schirmeister T. Cysteine proteases and their inhibitors. Chem Rev. 1997;97:133–171. doi: 10.1021/cr950025u. [DOI] [PubMed] [Google Scholar]
- 3.Polgar L. Catalytic mechanisms of cysteine peptidases. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. 2nd ed. London: Elsevier; 2004. pp. 1072–1079. [Google Scholar]
- 4.Polgar L. On the mode of activation of the catalitically essential sulfhydryl group of papain. Eur J Biochem. 1973;33:104–109. doi: 10.1111/j.1432-1033.1973.tb02660.x. [DOI] [PubMed] [Google Scholar]
- 5.Polgar L. Spectrophotometric determination of mercaptide ion, an activated form of SH-group in thiol enzymes. FEBS Lett. 1974;38:187–190. [Google Scholar]
- 6.Polgar L. Mercaptide-imidazolium ion-pair: the reactive nucleophile in papain catalysis. FEBS Lett. 1974;47:15–18. doi: 10.1016/0014-5793(74)80415-1. [DOI] [PubMed] [Google Scholar]
- 7.Dawson RMC, Elliott DC, Elliott WH, Jones KM. Data for biochemical research. 3rd ed. Oxford Science Publications; 1986. pp. 1–31. [Google Scholar]
- 8.Johnson FA, Lewis SD, Shafer JA. Perturbations in the free energy and enthalpy of ionization of histidine-159 at the active site of papain as determined by fluorescence spectroscopy. Biochemistry. 1981;20:44–48. doi: 10.1021/bi00504a600. [DOI] [PubMed] [Google Scholar]
- 9.Johnson FA, Lewis SD, Shafer JA. Determination of low pK for histidine-159 in the S-methylthio derivative of papain by proton nuclear magnetic resonance spectroscopy. Biochemistry. 1981;20:44–48. doi: 10.1021/bi00504a008. [DOI] [PubMed] [Google Scholar]
- 10.Lewis SD, Johnson FA, Shafer JA. Effect of cysteine-25 on the ionization of histidine-159 in papain as determined by proton nuclear magnetic resonance spectroscopy. Evidence for His-159-Cys-25 ion pair and its possible role in catalysis. Biochemistry. 1981;20:48–51. doi: 10.1021/bi00504a009. [DOI] [PubMed] [Google Scholar]
- 11.Willenbrock F, Brocklehurst K. Natural structural variation in enzymes as a tool in the study of mechanism exemplified by a comparison of the catalytic-site structure and characteristics of cathepsin B and papain. pH-dependent kinetics of the reactions of cathepsin B from bovine spleen and from rat liver with a thiol-specific two-protonic-state probe (2,2'-dipyridyl disulphide) and with a specific synthetic substrate (N-alpha-benzyloxycarbonyl-L-arginyl-L-arginine 2-naphthylamide) Biochem J. 1984;222:805–814. doi: 10.1042/bj2220805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellor GW, Patel M, Thomas EW, Brocklehurst K. Clarification of the pH-dependent kinetic behaviour of papain by using reactivity probes and analysis of alkylation and catalysed acylation reactions in terms of multihydronic state models: implications for electrostatics calculations and interpretation of the consequences of site-specific mutations such as Asp-158-Asn and Asp-158-Glu. Biochem J. 1993;294:201–210. doi: 10.1042/bj2940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas MP, Topham CM, Kowlessur D, Mellor GW, Thomas EW, Whitford D, Brocklehurst K. Structure of chymopapain M the late-eluted chymopapain deduced by comparative modelling techniques and active-centre characteristics determined by pH-dependent kinetics of catalysis and reactions with time-dependent inhibitors: the Cys-25/His-159 ion-pair is insufficient for catalytic competence in both chymopapain M and papain. Biochem J. 1994;300:805–820. doi: 10.1042/bj3000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinitglang S, Watts AB, Patel M, Reid JD, Noble MA, Gul S, Bokth A, Naeem A, Patel H, Thomas EW, Sreedharan SK, Verma C, Brocklehurst K. A classical enzyme active center motif lacks catalytic competence until modulated electrostatically. Biochemistry. 1997;36:9968–9982. doi: 10.1021/bi9705974. [DOI] [PubMed] [Google Scholar]
- 15.Noble MA, Gul S, Verma CS, Brocklehurst K. Ionization characteristics and chemical influences of aspartic acid residue 158 of papain and caricain determined by structure-related kinetic and computational techniques: multiple electrostatic modulators of active-centre chemistry. Biochem J. 2000;351:723–733. [PMC free article] [PubMed] [Google Scholar]
- 16.Hussain S, Pinitglang S, Bailey TS, Reid JD, Noble MA, Resmini M, Thomas EW, Greaves RB, Verma CS, Brocklehurst K. Variation in the pH-dependent pre-steady-state and steady-state kinetic characteristics of cysteine-proteinase mechanism: evidence for electrostatic modulation of catalytic-site function by the neighbouring carboxylate anion. Biochem J. 2003;372:735–746. doi: 10.1042/BJ20030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menard R, Khouri HE, Plouffe C, Dupras R, Rippol D, Vernet T, Tessier DC, Lalberte F, Thomas DY, Storer AC. A protein engineering study of the role of aspartate 158 in the catalytic mechanism of papain. Biochemistry. 1990;29:6706–6713. doi: 10.1021/bi00480a021. [DOI] [PubMed] [Google Scholar]
- 18.Menard R, Khouri HE, Plouffe C, Laflamme P, Dupras R, Vernet T, Tessier DC, Thomas DY, Storer AC. Importance of hydrogen-bonding interactions involving the side chain of Asp158 in the Catalytic mechanism of papain. Biochemistry. 1991;30:5531–5538. doi: 10.1021/bi00236a028. [DOI] [PubMed] [Google Scholar]
- 19.Gul S, Hussain S, Thomas MP, Resmini M, Verma CS, Thomas EW, Brocklehurst K. Generation of nuclephilic character in the Cys25/His159 ion-pair of papin involves Trp177 but not Asp 158. Biochemistry. 2008;47:2025–2035. doi: 10.1021/bi702126p. [DOI] [PubMed] [Google Scholar]
- 20.Sarkany Z, Szeltner Z, Polgar L. Thiolate-imidazolium ion pair id not obligatory catalytic entity of cysteine peptidases: the active site of picornain 3C. Biochemistry. 2001;40:10601–10606. doi: 10.1021/bi010550p. [DOI] [PubMed] [Google Scholar]
- 21.Huang C, Wei P, Fan K, Liu Y, Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- 22.Eichinger A, Beisel HG, Jacob U, Huber R, Medrano FJ, Banbula A, Potempa J, Travis J, Bode W. Inhibition of Trypsin-Like Cysteine Proteinases (Gingipains) from Porphyromonas gingivalis by Tetracycline and Its Analogues. EMBO J. 1999;18:5453–5462. doi: 10.1093/emboj/18.20.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warshel A. Energetics of enzyme catalysis. Proc Natl Acad Sci USA. 1978;11:5250–5254. doi: 10.1073/pnas.75.11.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang J-K, Warshel A. Why ion pair reversal by protein engineering is unlikely to succeed. Nature. 1988;334:270–272. doi: 10.1038/334270a0. [DOI] [PubMed] [Google Scholar]
- 25.Warshel A. Calculations of enzymatic reactions: calculations of pKa, proton transfer reactions, and general acid catalysis reactions in enzymes. Biochemistry. 1981;20:3167–3177. doi: 10.1021/bi00514a028. [DOI] [PubMed] [Google Scholar]
- 26.Warshel A, Russel S. Theoretical correlation of structure and energetics in the catalytic reaction of trypsin. J Am Chem Soc. 1986;108:6569–6579. [Google Scholar]
- 27.Cutler RL, Davies AM, Creighton S, Warshel A, Moore JR, Smith M, Mauk AG. Role of arginine-38 in regulation of the cytochrome c oxidation-reduction equilibrium. Biochemistry. 1989;28:3188–3197. doi: 10.1021/bi00434a012. [DOI] [PubMed] [Google Scholar]
- 28.Schutz CN, Warshel A. The low barrier hydrogen bond (LBHB) proposal revisited: the case of the Asp … His pair in serine proteases. Proteins. 2004;55:711–723. doi: 10.1002/prot.20096. [DOI] [PubMed] [Google Scholar]
- 29.Warshel A. Computer modeling of chemical Reactions in enzymes and solutions. New York: John Wiley & Sons; 1991. pp. 140–146. [Google Scholar]
- 30.Lee FS, Chu ZT, Warshel A. Microscopic and semimicroscopic calculations of electrostatic energies in proteins by POLARIS and ENZYMIX programs. J Comput Chem. 1993;14:161–185. [Google Scholar]
- 31.Harrison MJ, Burton NA, Hillier IH. Catalytic Mechanism of the Enzyme Ppain: Predictions with a Hybrid Quantum Mechanical/Molecular Mechanical Potential. J Am Chem Soc. 1997;119:12285–12291. [Google Scholar]
- 32.Ma S, Devi-Kesavan LS, Gao J. Molecular dynamics simulations of the catalytic passway of a cysteine protease: a combined QM/MM study of human cathepsin K. J Am Chem Soc. 2007;129:13633–13645. doi: 10.1021/ja074222+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mladenovic M, Schirmeister T, Thiel S, Thiel W, Engels B. The importance of the active site histidine for the activity of epoxide- or aziridine-based inhibitors of cysteine proteases. ChemMedChem. 2007;2:120–128. doi: 10.1002/cmdc.200600159. [DOI] [PubMed] [Google Scholar]
- 34.Mladenovic M, Fink RF, Thiel W, Schirmeister T, Engels B. On the origin of stabilization of the zwitterionic resting state of cysteine proteases: a theoretical study. J Am Chem Soc. 2008;130:8696–8705. doi: 10.1021/ja711043x. [DOI] [PubMed] [Google Scholar]
- 35.Shokhen M, Khazanov N, Albeck A. Screening of the active site from water by the incoming ligand triggers catalysis and inhibition in serine proteases. Proteins. 2008;70:1578–1587. doi: 10.1002/prot.21727. [DOI] [PubMed] [Google Scholar]
- 36.Shokhen M, Albeck A. Is there a weak H-bond → LBHB transition upon tetrahedral complex formation in serine proteases? Proteins. 2004;54:468–477. doi: 10.1002/prot.10610. [DOI] [PubMed] [Google Scholar]
- 37.Shokhen M, Albeck A. Identification of Protons Position in Acid-Base Enzyme Catalyzed Reactions. The Hepatitis C Viral NS3 Protease. Proteins. 2004;55:245–250. doi: 10.1002/prot.20058. [DOI] [PubMed] [Google Scholar]
- 38.Ozeri R, Khazanov N, Perlman N, Shokhen M, Albeck A. Enzyme Isoselective Inhibitors: A Novel Tool for Binding Trend Analysis. ChemMedChem. 2006;1:631–638. doi: 10.1002/cmdc.200600029. [DOI] [PubMed] [Google Scholar]
- 39.Shokhen M, Khazanov N, Albeck A. Enzyme Isoselective Inhibitors: Application to Drug Design. ChemMedChem. 2006;1:639–643. doi: 10.1002/cmdc.200600030. [DOI] [PubMed] [Google Scholar]
- 40.Shokhen M, Khazanov N, Albeck A. The Cooperative Effect Between Active Site Ionized Groups and Water Desolvation Controls the Alteration of Acid/Base Catalysis in Serine Proteases. ChemBioChem. 2007;8:1416–1421. doi: 10.1002/cbic.200700241. [DOI] [PubMed] [Google Scholar]
- 41.Liang TC, Abeles RH. Complex of γ-chymotrypsin and N-acetyl-L-leucyl-L-phenylalanytr ifluoromethyl ketone: structural studies with NMR spectroscopy. Biochemistry. 1987;26:7603–7608. doi: 10.1021/bi00398a011. [DOI] [PubMed] [Google Scholar]
- 42.Cassidy CS, Lin J, Frey PA. A new concept for the mechanism of action of chymotrypsin: the role of the low-barrier hydrogen bond. Biochemistry. 1997;36:4576–4584. doi: 10.1021/bi962013o. [DOI] [PubMed] [Google Scholar]
- 43.Kamphuis IG, Kalk KH, Swarte MB, Drenth J. Structure of papain refined at 1.65 A resolution. J Mol Biol. 1984;179:233–256. doi: 10.1016/0022-2836(84)90467-4. [DOI] [PubMed] [Google Scholar]
- 44.Maestro 5. Portland, OR: Schrödinger, Inc; 1991–2004. [Google Scholar]
- 45.Kaminski GA, Friesner RA, Tirad-Rives J, Jorgensen WL. Evaluation of reparametrization of the OPLS AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 46.Macromodel 7. Portland, OR: Schrödinger, Inc; 1991–2004. [Google Scholar]
- 47.Chang G, Guida WC, Still WC. An internal-coordinate Monte Carlo method for searching conformational space. J Am Chem Soc. 1989;11:4379–4386. [Google Scholar]
- 48.Saunders M, Houk KN, Wu YD, Still WC, Lipton M, Chang G, Guida WC. Conformations of cycloheptadecane. A comparison of methods for conformational searching. J Am Chem Soc. 1990;112:1419–1427. [Google Scholar]
- 49.JAGUAR 5.1. Portland, OR: Schrödinger, Inc; 1991–2004. [Google Scholar]
- 50.Tannor DJ, Marten B, Murphy R, Friesner RA, Sitcoff D, Nicholls A, Honig B, Ringnalda M, Goddard WA., III Accurate first principles calculation of molecular charge distributions and solvation energies from ab initio quantum mechanics and continuum dielectric theory. J Am Chem Soc. 1994;116:11875–11882. [Google Scholar]
- 51.Marten B, Kim K, Cortis C, Friesner RA, Murphy RB, Ringnalda MN, Sitcoff D, Honig B. A new model for calculation of solvation free energies: correction of self-consistent reaction field continuum dielectric theory for short range hydrogen-bonding effects. J Phys Chem. 1996;100:11775–11788. [Google Scholar]
- 52.Friezner RA, Murphy RB, Beachy MD, Ringnalda MN, Pollard WT, Dunietz BD, Cao Y. Correlated ab initio electronic structure calculations for large molecules. J Phys Chem A. 1999;103:1913–1928. [Google Scholar]
- 53.Murphy RB, Cao Y, Beachy MD, Ringalda MN, Friesner RA. Efficient pseudospectral methods for density functional calculations. J Chem Phys. 2000;112:10131–10141. [Google Scholar]
- 54.Hunter EPL, Lias SG. Evaluated gas phase basicities and proton affinities of molecules: an update. J Phys Chem Ref Data. 1998;27:413–656. [Google Scholar]
- 55.Taft RW, Bordwell FG. Structural and solvent effects evaluated from acidities measured in dimethyl sulfoxide and in the gas phase. Acc Chem Res. 1988;21:463–469. [Google Scholar]
- 56.Jaguar 7.5 User Manual, The pKa prediction module. New York, NY: Schrodinger, LLC; 2008. http://yfaat.ch.huji.ac.il/jaguar-help/mank.html. [Google Scholar]
- 57.Lide DR. Editor-in-Chief CRC Handbook of chemistry and physiscs 88-th edition. New York: CRC Press; 2007–2008. [Google Scholar]
- 58.Danehly JP, Noel CJ. The relative nucleophilic character of several mercaptans toward ethylene oxide. J Am Chem Soc. 1960;82:2511–2515. [Google Scholar]
- 59.Kreevoy MM, Harper ET, Duvall RE, Wilgus HS, Ditsch LT. Inductive effects on the acid dissociation constant of mercaptans. J Am Chem Soc. 1960;82:4899–4902. [Google Scholar]
- 60.Topol IA, Tawa GJ, Burt SK, Rashin AA. Calculation of absolute and relative acidities of substituted imidazoles in aqueous solvent. J Phys Chem A. 1997;101:10075–10081. [Google Scholar]
- 61.Levin IN. Physical Chemistry. 3rd ed. New York: McGraw-Hill; 1988. Chapter 22. [Google Scholar]
- 62.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 63.Essman U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 64.Krieger E, Nielsen JE, Spronk CA, Vriend G. Fast empirical pKa prediction by Ewald summation. J Mol Graph Model. 2006;25:481–486. doi: 10.1016/j.jmgm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 65.Yasara Dynamics. ( www.yasara.org).
- 66.Cao Y, Beachy MD, Braden DA, Morril LA, Ringalda MN, Friezner RA. Nuclear-Magnetic-resonance shielding constants calculated by pseudospectral methods. J Chem Phys. 2005;122:224116–224126. doi: 10.1063/1.1924598. [DOI] [PubMed] [Google Scholar]
- 67.Lee KK, Fitch CA, Garcia-Moreno EB. Distance dependence and salt sensitivity of pairwise, coulombic interactions in a protein. Prot Sci. 2002;11:1004–1101. doi: 10.1110/ps.4700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuhn P, Knapp M, Soltis SM, Ganshaw G, Thoene M, Bott R. The 0.78 Å Structure of a Serine Protease: Bacillus lentus Subtilisin. Biochemistry. 1998;37:13446–13452. doi: 10.1021/bi9813983. [DOI] [PubMed] [Google Scholar]
- 69.Katona G, Wilmouth RC, Wright PA, Berglund GI, Hajdu J, Neutze R, Schofield CJ. X-ray structure of a serine protease acyl-enzyme complex at 0.95-Å resolution. J Biol Chem. 2002;277:21962–21970. doi: 10.1074/jbc.M200676200. [DOI] [PubMed] [Google Scholar]